Abstract
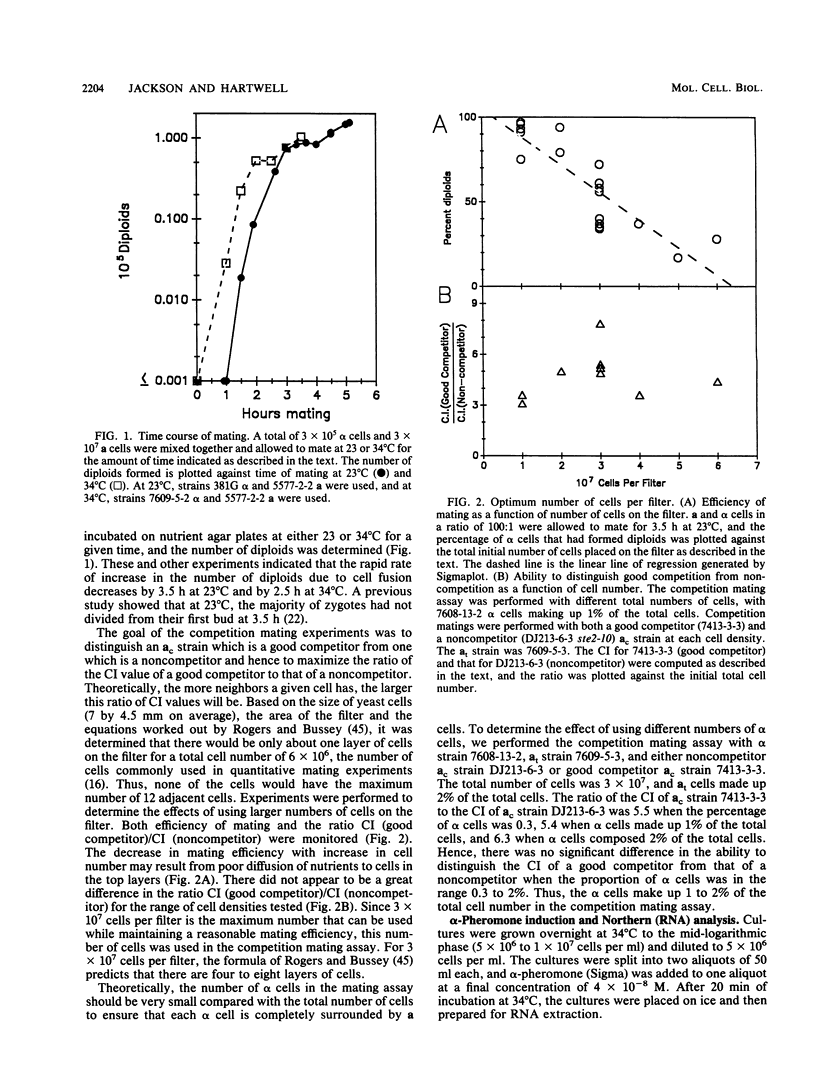
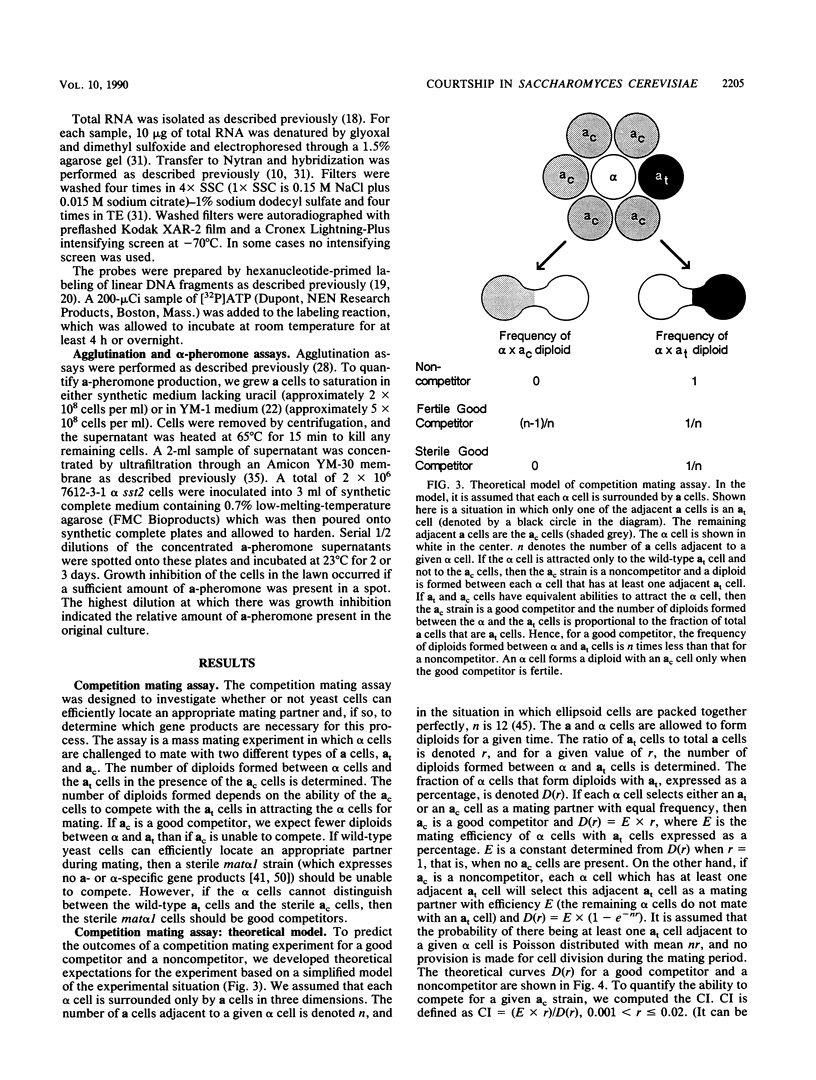
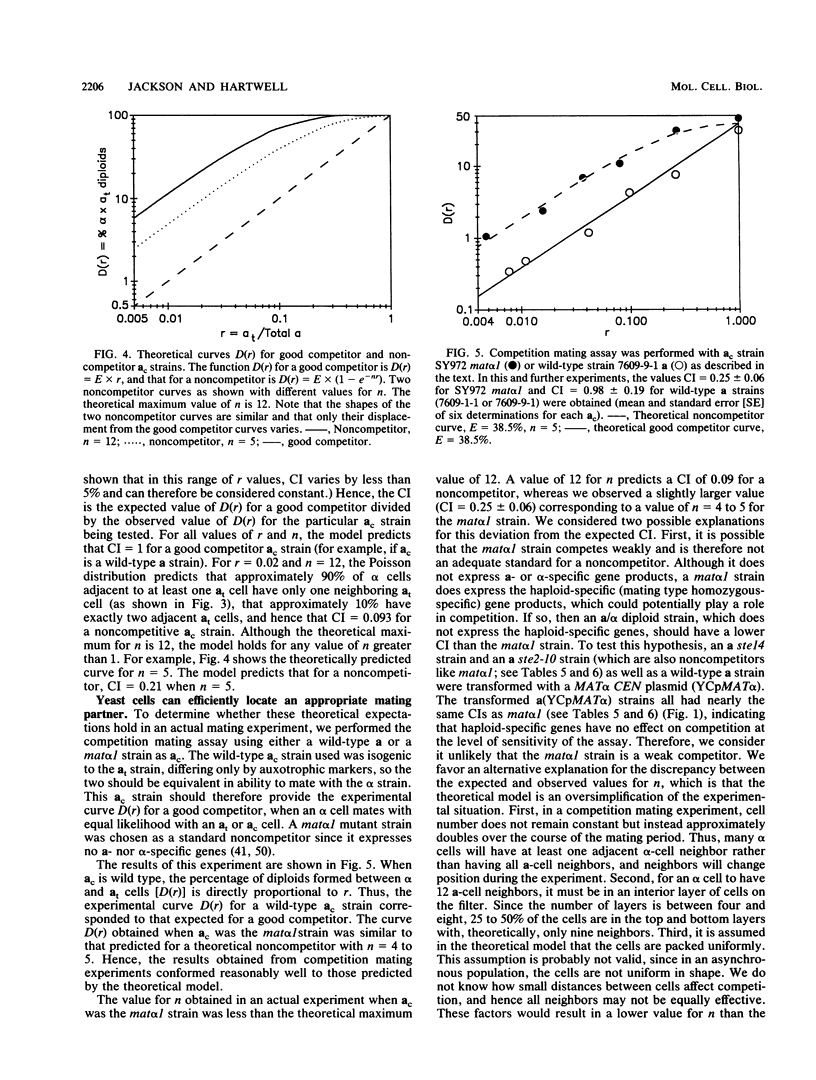
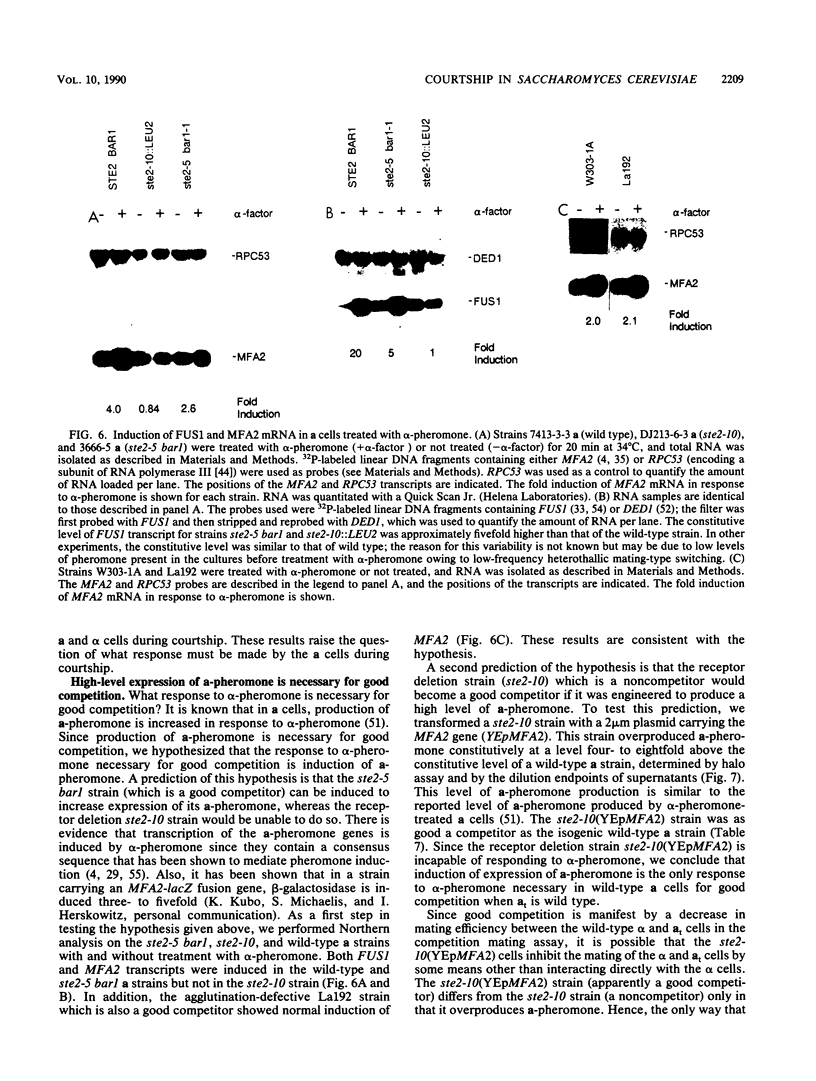
During conjugation in Saccharomyces cerevisiae, two cells of opposite mating type (MATa and MAT alpha) fuse to form a diploid zygote. Conjugation requires that each cell locate an appropriate mating partner. To investigate how yeast cells select a mating partner, we developed a competition mating assay in which wild-type MAT alpha cells have a choice of two MATa cell mating partners. We first demonstrated that sterile MAT alpha 1 cells (expressing no a- or alpha-specific gene products) do not compete with fertile MATa cells in the assay; hence, wild-type MATa and MAT alpha cells can efficiently locate an appropriate mating partner. Second, we showed that a MATa strain need not be fertile to compete with a fertile MATa strain in the assay. This result defines an early step in conjugation, which we term courtship. We showed that the ability to agglutinate is not necessary in MATa cells for courtship but that production of a-pheromone and response to alpha-pheromone are necessary. Thus, MATa cells must not only transmit but must also receive and then respond to information for effective courtship; hence, there is a "conversation" between the courting cells. We showed that the only alpha-pheromone-induced response necessary in MATa cells for courtship is production of a-pheromone. In all cases tested, a strain producing a higher level of a-pheromone was more proficient in courtship than one producing a lower level. We propose that during courtship, a MAT alpha cell selects the adjacent MATa cell producing the highest level of a-pheromone.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderegg R. J., Betz R., Carr S. A., Crabb J. W., Duntze W. Structure of Saccharomyces cerevisiae mating hormone a-factor. Identification of S-farnesyl cysteine as a structural component. J Biol Chem. 1988 Dec 5;263(34):18236–18240. [PubMed] [Google Scholar]
- Blinder D., Bouvier S., Jenness D. D. Constitutive mutants in the yeast pheromone response: ordered function of the gene products. Cell. 1989 Feb 10;56(3):479–486. doi: 10.1016/0092-8674(89)90250-x. [DOI] [PubMed] [Google Scholar]
- Blumer K. J., Reneke J. E., Thorner J. The STE2 gene product is the ligand-binding component of the alpha-factor receptor of Saccharomyces cerevisiae. J Biol Chem. 1988 Aug 5;263(22):10836–10842. [PubMed] [Google Scholar]
- Burkholder A. C., Hartwell L. H. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985 Dec 9;13(23):8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975 Oct;124(1):511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Melnick L. M., Blair L. C., Thorner J. Extracellular suppression allows mating by pheromone-deficient sterile mutants of Saccharomyces cerevisiae. J Bacteriol. 1983 Aug;155(2):903–906. doi: 10.1128/jb.155.2.903-906.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Otte C. A. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol Cell Biol. 1982 Jan;2(1):11–20. doi: 10.1128/mcb.2.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Otte C. A. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol Cell Biol. 1982 Jan;2(1):21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciejek E., Thorner J. Recovery of S. cerevisiae a cells from G1 arrest by alpha factor pheromone requires endopeptidase action. Cell. 1979 Nov;18(3):623–635. doi: 10.1016/0092-8674(79)90117-x. [DOI] [PubMed] [Google Scholar]
- Cross F. R. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F., Hartwell L. H., Jackson C., Konopka J. B. Conjugation in Saccharomyces cerevisiae. Annu Rev Cell Biol. 1988;4:429–457. doi: 10.1146/annurev.cb.04.110188.002241. [DOI] [PubMed] [Google Scholar]
- Dietzel C., Kurjan J. Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: a model for desensitization to pheromone. Mol Cell Biol. 1987 Dec;7(12):4169–4177. doi: 10.1128/mcb.7.12.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel C., Kurjan J. The yeast SCG1 gene: a G alpha-like protein implicated in the a- and alpha-factor response pathway. Cell. 1987 Sep 25;50(7):1001–1010. doi: 10.1016/0092-8674(87)90166-8. [DOI] [PubMed] [Google Scholar]
- Dutcher S. K., Hartwell L. H. Genes that act before conjugation to prepare the Saccharomyces cerevisiae nucleus for caryogamy. Cell. 1983 May;33(1):203–210. doi: 10.1016/0092-8674(83)90349-5. [DOI] [PubMed] [Google Scholar]
- Dutcher S. K., Hartwell L. H. The role of S. cerevisiae cell division cycle genes in nuclear fusion. Genetics. 1982 Feb;100(2):175–184. doi: 10.1093/genetics/100.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder R. T., Loh E. Y., Davis R. W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Field C., Schekman R. Localized secretion of acid phosphatase reflects the pattern of cell surface growth in Saccharomyces cerevisiae. J Cell Biol. 1980 Jul;86(1):123–128. doi: 10.1083/jcb.86.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980 Jun;85(3):811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Synchronization of haploid yeast cell cycles, a prelude to conjugation. Exp Cell Res. 1973 Jan;76(1):111–117. doi: 10.1016/0014-4827(73)90425-4. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness D. D., Burkholder A. C., Hartwell L. H. Binding of alpha-factor pheromone to Saccharomyces cerevisiae a cells: dissociation constant and number of binding sites. Mol Cell Biol. 1986 Jan;6(1):318–320. doi: 10.1128/mcb.6.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness D. D., Burkholder A. C., Hartwell L. H. Binding of alpha-factor pheromone to yeast a cells: chemical and genetic evidence for an alpha-factor receptor. Cell. 1983 Dec;35(2 Pt 1):521–529. doi: 10.1016/0092-8674(83)90186-1. [DOI] [PubMed] [Google Scholar]
- Konopka J. B., Jenness D. D., Hartwell L. H. The C-terminus of the S. cerevisiae alpha-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988 Aug 26;54(5):609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- Kronstad J. W., Holly J. A., MacKay V. L. A yeast operator overlaps an upstream activation site. Cell. 1987 Jul 31;50(3):369–377. doi: 10.1016/0092-8674(87)90491-0. [DOI] [PubMed] [Google Scholar]
- Kuchler K., Sterne R. E., Thorner J. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 1989 Dec 20;8(13):3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurjan J. Alpha-factor structural gene mutations in Saccharomyces cerevisiae: effects on alpha-factor production and mating. Mol Cell Biol. 1985 Apr;5(4):787–796. doi: 10.1128/mcb.5.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORTIMER R. K. Evidence of two types of x-ray-induced lethal damage in Saccharomyces cerevisiae. Radiat Res. 1955 Jun;2(4):361–368. [PubMed] [Google Scholar]
- Marsh L., Herskowitz I. STE2 protein of Saccharomyces kluyveri is a member of the rhodopsin/beta-adrenergic receptor family and is responsible for recognition of the peptide ligand alpha factor. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3855–3859. doi: 10.1073/pnas.85.11.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey G., Clay F. J., Kelsay K., Sprague G. F., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987 Aug;7(8):2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J. P., Varshavsky A. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature. 1989 Aug 3;340(6232):400–404. doi: 10.1038/340400a0. [DOI] [PubMed] [Google Scholar]
- Michaelis S., Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol Cell Biol. 1988 Mar;8(3):1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima I., Nakafuku M., Nakayama N., Brenner C., Miyajima A., Kaibuchi K., Arai K., Kaziro Y., Matsumoto K. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell. 1987 Sep 25;50(7):1011–1019. doi: 10.1016/0092-8674(87)90167-x. [DOI] [PubMed] [Google Scholar]
- Müller I. Parental age and the life-span of zygotes of Saccharomyces cerevisiae. Antonie Van Leeuwenhoek. 1985;51(1):1–10. doi: 10.1007/BF00444223. [DOI] [PubMed] [Google Scholar]
- Nakayama N., Miyajima A., Arai K. Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 1985 Oct;4(10):2643–2648. doi: 10.1002/j.1460-2075.1985.tb03982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Shore D. Transcriptional regulation in the yeast life cycle. Science. 1987 Sep 4;237(4819):1162–1170. doi: 10.1126/science.3306917. [DOI] [PubMed] [Google Scholar]
- Osumi M., Shimoda C., Yanagishima N. Mating reaction in Saccharomyces cerevisiae. V. Changes in the fine structure during the mating reaction. Arch Mikrobiol. 1974 Apr 10;97(1):27–38. [PubMed] [Google Scholar]
- Powers S., Michaelis S., Broek D., Santa Anna S., Field J., Herskowitz I., Wigler M. RAM, a gene of yeast required for a functional modification of RAS proteins and for production of mating pheromone a-factor. Cell. 1986 Nov 7;47(3):413–422. doi: 10.1016/0092-8674(86)90598-2. [DOI] [PubMed] [Google Scholar]
- Riva M., Memet S., Micouin J. Y., Huet J., Treich I., Dassa J., Young R., Buhler J. M., Sentenac A., Fromageot P. Isolation of structural genes for yeast RNA polymerases by immunological screening. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1554–1558. doi: 10.1073/pnas.83.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D., Bussey H. Fidelity of conjugation in Saccharomyces cerevisiae. Mol Gen Genet. 1978 Jun 14;162(2):173–182. doi: 10.1007/BF00267874. [DOI] [PubMed] [Google Scholar]
- Schekman R., Brawley V. Localized deposition of chitin on the yeast cell surface in response to mating pheromone. Proc Natl Acad Sci U S A. 1979 Feb;76(2):645–649. doi: 10.1073/pnas.76.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena E. P., Radin D. N., Fogel S. Synchronous mating in yeast. Proc Natl Acad Sci U S A. 1973 May;70(5):1373–1377. doi: 10.1073/pnas.70.5.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano P. G., Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984 Jul;37(3):969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- Strathern J., Hicks J., Herskowitz I. Control of cell type in yeast by the mating type locus. The alpha 1-alpha 2 hypothesis. J Mol Biol. 1981 Apr 15;147(3):357–372. doi: 10.1016/0022-2836(81)90488-5. [DOI] [PubMed] [Google Scholar]
- Strazdis J. R., MacKay V. L. Induction of yeast mating pheromone a-factor by alpha cells. Nature. 1983 Oct 6;305(5934):543–545. doi: 10.1038/305543a0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Nasmyth K. A., Hall B. D., Astell C., Smith M. In vitro mutation analysis of the mating-type locus in yeast. Cell. 1981 Nov;27(1 Pt 2):25–35. doi: 10.1016/0092-8674(81)90357-3. [DOI] [PubMed] [Google Scholar]
- Trueheart J., Boeke J. D., Fink G. R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987 Jul;7(7):2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arsdell S. W., Stetler G. L., Thorner J. The yeast repeated element sigma contains a hormone-inducible promoter. Mol Cell Biol. 1987 Feb;7(2):749–759. doi: 10.1128/mcb.7.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzele M., Klis F., Tanner W. Purification and characterization of the inducible a agglutinin of Saccharomyces cerevisiae. EMBO J. 1988 May;7(5):1483–1488. doi: 10.1002/j.1460-2075.1988.tb02966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Hougan L., Dignard D., Thomas D. Y., Bell L., Saari G. C., Grant F. J., O'Hara P., MacKay V. L. The STE4 and STE18 genes of yeast encode potential beta and gamma subunits of the mating factor receptor-coupled G protein. Cell. 1989 Feb 10;56(3):467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- Wilson K. L., Herskowitz I. Negative regulation of STE6 gene expression by the alpha 2 product of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2420–2427. doi: 10.1128/mcb.4.11.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. L., Herskowitz I. STE16, a new gene required for pheromone production by a cells of Saccharomyces cerevisiae. Genetics. 1987 Mar;115(3):441–449. doi: 10.1093/genetics/115.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]





