Background: Progranulin is a secreted, anti-inflammatory glycoprotein, suggested to be a component of high density lipoproteins (HDL).
Results: Studies in cells and plasma revealed secreted progranulin exists as a homodimer, does not bind lipids, and is not detected on HDL.
Conclusion: Secreted progranulin exists as a homodimer and is not an HDL component.
Significance: These data provide insights into the molecular properties of secreted progranulin.
Keywords: HDL, Inflammation, Lipids, Neurodegenerative Diseases, Neuroinflammation, Progranulin
Abstract
Progranulin is a secreted glycoprotein, and the GRN gene is mutated in some cases of frontotemporal dementia. Progranulin has also been implicated in cell growth, wound healing, inflammation, and cancer. We investigated the molecular nature of secreted progranulin and provide evidence that progranulin exists as a homodimer. Although recombinant progranulin has a molecular mass of ∼85 kDa by SDS-PAGE, it elutes in fractions corresponding to ∼170–180 kDa by gel-filtration chromatography. Additionally, recombinant progranulin can be intermolecularly cross-linked, yielding a complex corresponding to a dimer (∼180 kDa), and progranulins containing different epitope tags physically interact. In plasma, progranulin similarly forms complexes of ∼180–190 kDa. Although progranulin partially co-fractionated with high density lipoproteins (HDL) by gel-filtration chromatography, we found no evidence that progranulin in mouse or human plasma is a component of HDL either by ultracentrifugation or by lipid binding assays. We conclude that circulating progranulin exists as a dimer and is not likely a component of HDL.
Introduction
Progranulin (also known as proepithelin, acrogranin, PC cell-derived growth factor, and glycoprotein 88) is implicated in a variety of processes, including embryogenesis, tumor proliferation, inflammation, wound healing, insulin resistance, and neurodegeneration (1, 2). At a cellular level, progranulin is a secreted growth factor expressed in epithelial cells, hematopoetic cells, adipocytes, immune cells, and certain types of neurons (3–7). Mutations in the GRN gene encoding progranulin are a leading cause of frontotemporal dementia (8, 9). Although research has uncovered much about the physiological functions of progranulin, much about the biochemical nature of progranulin remains to be explored.
Progranulin is an 88-kDa glycoprotein that comprises 7.5 tandem repeats of cysteine-rich granulin domains (10). Proteins with granulin domains are found in plants and throughout the animal kingdom including in insects, nematodes, fish, and mammals (11). Carp, granulins have a unique structure consisting of β-hairpin stacks (12, 13). However, the structures of mammalian granulins and full-length progranulin have not been determined. Proteases, including neutrophil-derived elastase and proteinase 3, cleave progranulin into granulin peptides in vitro (14, 15), and these granulins are thought to have a different function than full-length progranulin. For example, whereas the holoprotein is likely anti-inflammatory (16), some of the cleaved granulins are likely pro-inflammatory (14, 17). The significance of progranulin cleavage in vivo remains largely unknown.
Recent studies in mice have shed light on its role in diseases. Progranulin-deficient mice exhibit increased neuroinflammation and exhibit features of frontotemporal dementia (18–20). Additionally, progranulin-deficient mice are prone to inflammation and development of collagen-induced arthritis (16, 20). Moreover, progranulin-deficient mice are protected from high-fat diet-induced obesity and insulin resistance (6). The latter studies indicate a systemic role for progranulin. Indeed, progranulin has been detected in human blood (21–26) as well as in urine. Okura et al. (17) reported that circulating progranulin is a component of high density lipoproteins (HDL) and that this interaction prevents cleavage of progranulin into pro-inflammatory granulin peptides. These data suggested that the progranulin interaction with other proteins can affect its cleavage and bioactivity, which is relevant to understanding its function in the circulation.
In the present study, we characterized the molecular properties of secreted progranulin. First, we provide biochemical evidence that recombinant progranulin exists as a homodimer in conditioned cell culture media and in mouse and human plasma. Second, we utilize a variety of assays, including ultracentrifugation, gel-filtration chromatography, and lipid binding assays, to provide evidence that progranulin is not a component of HDL.
EXPERIMENTAL PROCEDURES
Materials
All materials were from Sigma unless indicated.
Plasma Samples
Mice were housed in a pathogen-free barrier-type facility with a 12-h light/12-h dark cycle and allowed food and water ad libitum. Grn−/− mice (18) were generated as described and Apoa1−/− mice (27) were obtained from Jackson Laboratories (Stock #002055). Mouse blood was collected from non-fasted, anesthetized mice by heart puncture in EDTA-coated tubes (Sarstedt). Human venous blood was collected from non-fasted, healthy donors. Plasma was obtained by centrifugation at 900 × g for 7 min at 4 °C.
Gel-filtration Chromatography
Samples (100 μl of plasma or 4 μg of purified protein) were separated by size-exclusion chromatography on a Superose 6 10/300 GL column (GE Healthcare) at a flow rate of 0.5 ml/min at 4 °C in a buffer containing 1× PBS, pH 7.4, 1 mm EDTA. Eluate was collected as 90 500-μl fractions. The column was calibrated by using protein standards (Sigma). For some experiments (Fig. 3, B and D), the fractions were pooled and concentrated using Amicon Ultra 0.5-ml columns (10-kDa cutoff). For lipoprotein analyses, the fractions corresponding to very low density lipoproteins, intermediate density lipoproteins, low density lipoproteins (LDL), and HDL were identified by measuring the cholesterol content of fractions 13–48 using the Cholesterol E kit (Wako). For the experiments in Fig. 3, fresh plasma was delipidated as described (28). Briefly, EDTA was added to plasma to obtain a final concentration of 0.1 mg/ml, and the sample was then mixed with two volumes of butanol/diisopropyl ether (40:60, v/v) for 1 h at room temperature. The mixture was centrifuged at 2000 rpm for 2 min, and the aqueous phase was collected with a syringe.
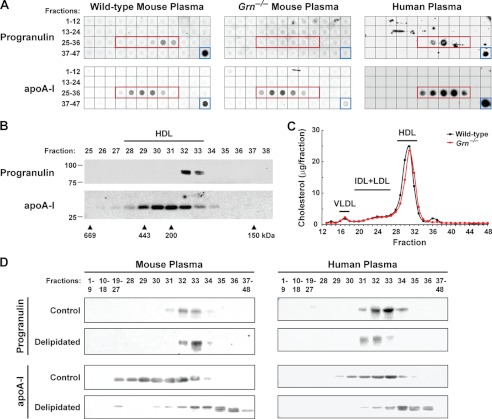
FIGURE 3.
Circulating progranulin is found in complexes of ∼180–190 kDa, partially co-elutes with HDL by gel-filtration, and is not associated with lipids. A, progranulin in mouse and human plasma is in a complex of ∼180–190 kDa and partially co-elutes with HDL. Dot blots of gel-filtration fractions 1–47 and plasma (blue box) from wild-type mice (left panel), Grn−/− mice (middle panel), and a human subject (right panel) are shown. The fractions corresponding to HDL are indicated by the red box. B, progranulin in mouse plasma partially co-elutes with HDL. Western blots of gel-filtration fractions 25–38 from wild-type mouse plasma are shown. These results are from a representative experiment that was performed four times with similar results. C, cholesterol profiles of plasma from wild-type (black line) and Grn−/− (red line) mice are shown. IDL, intermediate density lipoproteins. D, circulating progranulin is not associated with lipids. Delipidation of mouse (left panel) and human (right panel) plasma results in a shift in the gel-filtration elution profile of apoA-I but not in that of progranulin. Western blots of gel-filtration fractions 1–48 for control and delipidated mouse plasma. Similar results were obtained in two independent experiments.
Isolation of Lipoproteins by Ultracentrifugation
Plasma lipoproteins were isolated from mouse and human plasma by sequential ultracentrifugation in a TLA-100.2 rotor (Beckman Instruments) (29). Commercial preparations of HDL and LDL were obtained from Meridian Life Science and Fitzgerald Industries.
Expression Plasmids
The open reading frame of the human GRN gene was amplified using cDNA obtained from HEK293T human embryonic kidney cells and the Phusion DNA polymerase kit (New England Biolabs). The forward and reverse primers used in this reaction were CGTACGAATTCATGTGGACCCTGGTGAGCTGGGTGCTACGCGGCCGCCAGCAGCTGTCTCAAGGCTGG, respectively. The open reading frame of the human APOA1 gene was amplified using cDNA obtained from Huh7 human hepatocarcinoma cells. The forward and reverse primers used were CGTACGAATTCATGAAAGCTGCGGTGCTGACCT and GCTACGCGGCCGCCTGGGTGTTGAGCTTCTTAGTGTACTC, respectively. The PCR products were gel-purified, subjected to restriction digestion with EcoRI and NotI, and subcloned into the multiple cloning site of pcDNA3 upstream of three copies of the FLAG epitope tag. The resulting expression plasmids are denoted as pcDNA-human progranulin-3×FLAG and pcDNA-human apoA-I-3×FLAG. pcDNA-mouse progranulin-Myc/His encodes mouse progranulin with Myc and His epitope tags at the C terminus. The mouse progranulin cDNA was amplified from an IMAGE clone and subcloned into the multiple cloning site of pcDNA4 upstream of the Myc and His epitope tags. Plasmids encoding deletion mutants of mouse progranulin were generated by site-directed mutagenesis with the QuikChange Lightning kit (Stratagene) using pcDNA-mouse progranulin as the template. The forward and reverse primers used for these reactions were: GCTGGTAGCCGGAGATGGCTCCTGC and GCAGGAGCCATCTCCGGCTACCAGC for Δ31–70; ACGAGCCATCATCTAGTTTCACCCACGGGC and GCCCGTGGGTGAAACTAGATGATGGCTCGT for Δ71–190; TTCTGTCGTGTGCCCTGAAATGGGTATCCTCC and GGAGGATACCCATTTCAGGGCACACGACAGAA for Δ222–346; GAGTGATACACCTTGTGCTCGAGGTCACCC and GGGTGACCTCGAGCACAAGGTGTATCACTC for Δ377–602. The sequences of all plasmids were confirmed by DNA sequencing.
Cell Culture and Transfection
HEK293T cells were maintained in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected using Lipofectamine 2000 (Invitrogen).
Purification of Recombinant Progranulin
Recombinant FLAG-tagged human progranulin was purified from conditioned media of transfected HEK293T cells by using anti-FLAG M2 magnetic beads. Briefly, HEK293T cells were transfected with pcDNA-human progranulin-3×FLAG. After 3 days, conditioned medium was collected, cleared at 1500 × g at 4 °C for 10 min, concentrated using Amicon Ultra columns (10-kDa cutoff size), and incubated with anti-FLAG M2 magnetic beads (Sigma) at 4 °C on an end-over-end rotator for 4–6 h. The magnetic beads were washed 3 times with PBS for 5 min. The bound material was eluted from the magnetic beads by competition with 500 ng/μl of FLAG peptide (Sigma). The eluate was applied to an Amicon Ultra column (10-kDa cutoff) to remove the FLAG peptide (Sigma). The purity of the recombinant progranulin was >95%, as determined by Coomassie staining using the SimplyBlue kit (Invitrogen). Aliquots were stored at −20 °C.
Immunoblot Analyses
For Western blot analysis, 5× sample buffer (0.25 m Tris-HCl, pH 6.8, 10% (w/v) SDS, 0.05% (w/v) bromphenol blue, 50% glycerol, 12.5% β-mercaptoethanol) was added to each sample. After boiling, equivalent volumes of samples were subjected to SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). For dot blot analysis, equivalent volumes of gel-filtration fractions were transferred to nitrocellulose membranes using a dot blot apparatus (Schleicher & Schuell) in a buffer containing 48 mm Tris, 39 mm glycine, 1.3 mm SDS, and 20% (v/v) methanol. Membranes were incubated with the following antibodies: monoclonal anti-FLAG M2 (1:1000, Sigma), monoclonal anti-Myc (9B11 clone) (1:500, Cell Signaling), polyclonal anti-mouse progranulin (1:500, R&D Systems), polyclonal anti-mouse apoA-I2 (1:1000, gift from Dr. Karl Weisgraber), polyclonal anti-mouse apoB (1:5000, gift from Dr. Stephen Young), polyclonal anti-human progranulin linker 4 (1:500, see below), polyclonal anti-human progranulin linker 5 (1:1000, see below), polyclonal anti-human progranulin (1:100, Santa Cruz, N-19), polyclonal anti-human apoA-I IgG (1:500, Meridian Life Science), and polyclonal anti-human apoB (1:5000, Meridian Life Science). After incubations with primary antibodies and horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch) as indicated in the figure legends, bound antibodies were visualized with SuperSignal Chemiluminescent Substrate (Pierce) or SuperSignal West Femto Chemiluminescent Substrate (Pierce) using a Li-Cor ODYSSEY imager or x-ray film (Eastman Kodak Co., X-Omat).
Polyclonal anti-human progranulin antibodies were generated by immunizing rabbits with the peptides IVAGLEKMPARRGSLSHPR (amino acids 422–440, linker 4 region) and KEVVSAQPATFLARSPHVG (amino acids 497–515, linker 5 region) conjugated to keyhole limpet hemocyanin. The resulting sera were affinity-purified and referred to as anti-human progranulin linker 4 and anti-human progranulin linker 5, respectively.
Co-immunoprecipitation Assays
The conditioned medium of transfected HEK293T cells was collected and cleared at 1500 × g at 4 °C for 10 min. After adding a mixture of protease inhibitors (Roche Applied Science), the cleared conditioned medium was then subjected to immunoprecipitation with 50 μl of anti-FLAG M2 magnetic beads (Sigma) for 2–3 h at 4 °C. The bound material was washed 3 times with cold PBS and then eluted in 2× sample buffer at 100 °C for 3 min. Aliquots of the pellet or supernatant fractions of the immunoprecipitations were subjected to SDS-PAGE, and Western blot analysis was carried out as described above.
Cross-linking Assays
Cross-linking was carried out with disuccinimidyl suberate (Pierce), which was resuspended in DMSO at 100 mm. Disuccinimidyl suberate (or the DMSO control) was added to 2.5 μg of recombinant progranulin in PBS and incubated at room temperature for 20 min. The reaction was quenched by adding 1 m Tris-HCl, pH 8.0. Aliquots of the reaction were subjected SDS-PAGE and Western blot analysis as described above.
DMPC Clearance Assays
Interactions of purified proteins with DMPC was monitored as described with minor modifications (30). Dry DMPC was dispersed in Tris buffer (10 mm Tris, pH 7.4, 0.01% EDTA, 8.5% KBr, 0.01% NaN3) at a concentration of 0.5 mg/ml and vortexed to form multilamellar liposomes. Purified proteins were added to the DMPC liposomes with a mass ratio of 1:5 (w/w) in triplicate 96-μl reactions in a 96-well plate. The samples were maintained at 24.5 °C in a spectrophotometer, and the absorbance at 325 nm was measured at 2–5-min intervals for 1 h. Data are presented as the mean ± S.D. Comparisons between groups were made by one-way analysis of variance followed by the Tukey-Kramer post-hoc test.
RESULTS
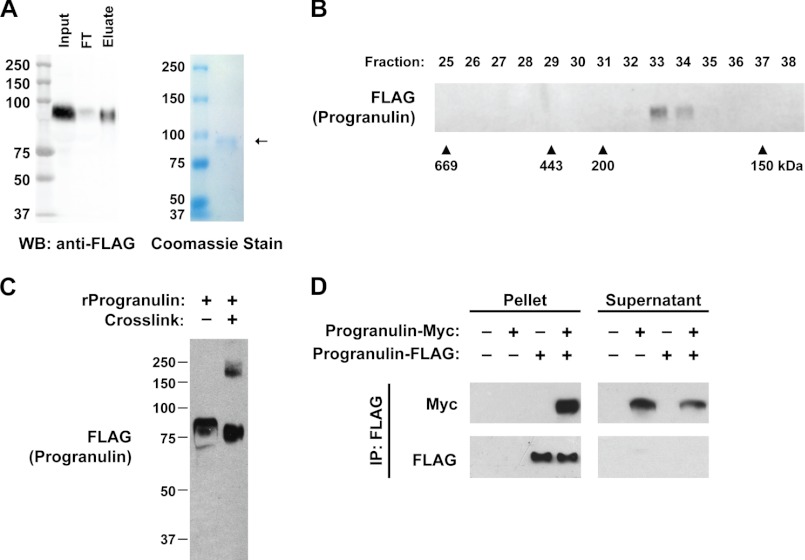
To determine the biochemical characteristics of secreted progranulin, we purified recombinant human progranulin, containing a 3×FLAG epitope tag at the C terminus from the culture supernatant of transfected HEK293T cells (Fig. 1A). Coomassie staining showed that the recombinant protein was >95% pure and migrated at ∼90 kDa by SDS-PAGE. When subjected to gel-filtration by size-exclusion chromatography, recombinant progranulin eluted from the column in fractions 33 and 34, corresponding to ∼170–180 kDa (Fig. 1B). This result suggested that secreted human progranulin is a dimer. We next performed in vitro cross-linking studies using disuccinimidyl suberate, a cross-linker that reacts with primary amines. Cross-linking recombinant progranulin resulted in a portion of progranulin shifting in size, from ∼85 to ∼180 kDa, as determined by SDS-PAGE (Fig. 1C), also consistent with secreted recombinant progranulin existing as a homodimer.
FIGURE 1.
Recombinant progranulin is a homodimer. A, purification of secreted recombinant human progranulin from conditioned media of transfected HEK293T cells is shown. Anti-FLAG Western blot (WB, left) and Coomassie-stained gel (right) of purified progranulin are shown. FT, flow-through. B, recombinant human progranulin migrates at ∼170–180 kDa. Western blot of gel-filtration fractions. Similar results were obtained in two independent experiments. C, cross-linking partially shifts the size of recombinant progranulin from ∼85 to ∼180 kDa. Shown is a Western blot of recombinant human progranulin subjected to cross-linking with disuccinimidyl suberate. D, expressed progranulin-FLAG physically interacts with progranulin-Myc. Co-immunoprecipitation (IP) of conditioned medium from transfected HEK293T cells with anti-FLAG magnetic beads is shown. These results are from a representative experiment that was performed three times with similar results.
To further test this model, we carried out immunoprecipitation assays using the conditioned media of HEK293T cells co-transfected with human and murine progranulin, tagged at the C termini with FLAG and Myc epitopes, respectively (Fig. 1D). Immunoprecipitation with anti-FLAG M2 magnetic beads pulled down nearly all of the progranulin-FLAG (pellets) and a large proportion of progranulin-Myc. Although this assay does not exclude the possibility of indirect interactions, this result demonstrates that progranulin interacts intermolecularly in the conditioned media of cultured cells. Taken together, the results of Fig. 1 indicate that recombinant progranulin exists as a homodimer.
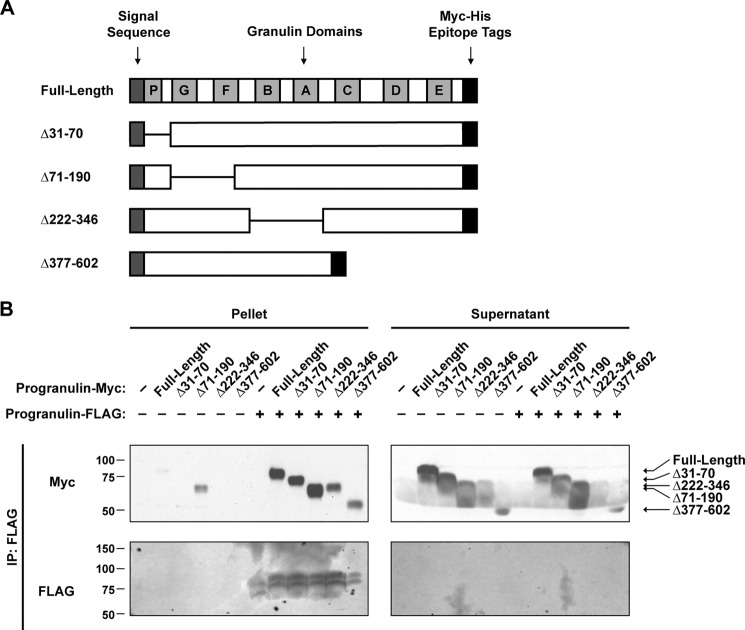
To identify the region(s) in progranulin that is required for dimerization, we generated a series of deletion mutants (Fig. 2A). We co-transfected FLAG-tagged full-length progranulin and Myc-tagged versions of these deletion mutants into HEK293T cells and performed immunoprecipitation assays. Immunoprecipitation of the full-length progranulin-FLAG pulled down full-length progranulin-Myc, Δ31–70 progranulin-Myc, Δ71–190 progranulin-Myc, Δ222–346 progranulin-Myc, or Δ377–602 progranulin-Myc (Fig. 2B). Thus, despite testing deletion mutants, which span ∼90% of the mature protein, we were unable to identify any regions required for the progranulin-progranulin interaction. Although it is possible that the region(s) required for dimerization was not covered by the deletion mutants tested, these results suggest multiple regions are sufficient for mediating the interaction.
FIGURE 2.
Mapping of the dimerization domain of progranulin. A, a schematic representation of deletion mutants of mouse progranulin is shown. B, mapping of dimerization domain of progranulin in co-immunoprecipitation (IP) assays is shown. Conditioned media from transfected HEK293T cells was subjected to immunoprecipitation with anti-FLAG magnetic beads. Similar results were obtained in three independent experiments.
To determine whether progranulin exists as a dimer in vivo, we analyzed plasma from wild-type and progranulin-deficient (Grn−/−) mice by size-exclusion chromatography. As revealed in the dot blots of Fig. 3A, top panel, and the western blots of Fig. 3B, top panel, almost all of the progranulin immunoreactivity was found in fractions 32 and 33, which corresponds to ∼180–190 kDa and is consistent with the molecular mass of a dimer. Progranulin was not detected in plasma from Grn−/− mice. In human plasma, progranulin was similarly found in fractions 31–33 (Fig. 3A, top panel). We did not detect progranulin immunoreactivity in fractions 39–40, where progranulin monomers would be expected to elute.
Progranulin was previously reported to be part of human HDL, as determined by Western blot analysis of HDL isolated by ultracentrifugation (17). To test this, we probed the dot blot and Western blot membranes with anti-apolipoprotein (apo) A-I antibodies, which detect the major HDL protein. The immunoblots in Fig. 3, A and B, revealed that the progranulin-containing fractions partially overlapped with the apoA-I-containing fractions of mouse and human plasma. We also measured the cholesterol content of the gel-filtration fractions to identify the lipoprotein-containing fractions. Indeed, progranulin partially co-eluted with both mouse HDL-cholesterol (fractions 28–33) (Fig. 3C) and human HDL-cholesterol (fractions 30–34) (data not shown). Of note, the plasma cholesterol profile of Grn−/− mice was not different than that of wild-type mice, indicating that progranulin deficiency does not appreciably affect plasma lipoprotein levels.
Our results indicate that circulating progranulin elutes in the observed fractions because it exists as a dimer, but whether a portion of progranulin physically associates with HDL is unclear. Therefore, we performed experiments to determine whether circulating progranulin in the HDL-containing fractions is lipid-associated. We delipidated mouse and human plasma with a mixture of butanol and diisopropyl ether (28). This procedure removes lipids but does not denature proteins, and was previously used as one criterion to determine if candidate proteins are bona fide HDL proteins (31). For lipid-free proteins, the gel-filtration elution profile is not affected by delipidation. On the other hand, the delipidation procedure causes a shift in the elution profiles of lipid-associated proteins. Delipidation effectively removed all cholesterol from mouse and human plasma (data not shown). As expected, delipidation of plasma caused a shift of the major HDL protein, apoA-I, to smaller molecular mass (higher numbered) fractions (Fig. 3D, lower panels). However, delipidation of mouse and human plasma did not cause a substantial shift in the elution profiles of progranulin (upper panels). These results indicate that progranulin is likely not lipid-associated and, therefore, not likely a component of HDL.
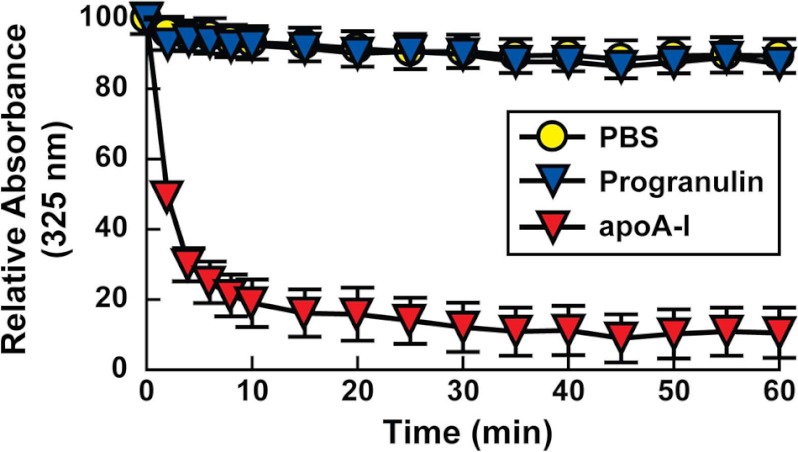
We also tested in vitro whether progranulin associates with lipids using phospholipid clearance assays. In these assays, purified proteins are added to DMPC multilamellar vesicles. Lipid-associated proteins can bind and solubilize the phospholipid vesicles, converting the vesicles into discoidal-shaped particles. The kinetics of this conversion is reflected in the clearance of the turbid solution of vesicles and is monitored by measuring the absorbance at 325 nm. Unlike purified apoA-I, which cleared the DMPC vesicles (p < 0.001, compared with PBS control), recombinant progranulin did not clear the DMPC vesicles (p > 0.05, compared with PBS control) (Fig. 4), suggesting that it does not directly associate with phospholipids.
FIGURE 4.
Recombinant progranulin does not reorganize DMPC vesicles. A phospholipid clearance assay using recombinant human progranulin and purified human apoA-I is shown. ApoA-I cleared the DMPC vesicles (p < 0.001 compared with PBS), but progranulin did not (p > 0.05, compared with PBS). Data are presented as the mean ± S.D. Comparisons between groups were made by one-way analysis of variance followed by the Tukey-Kramer post-hoc test. Similar results were obtained in three independent experiments.
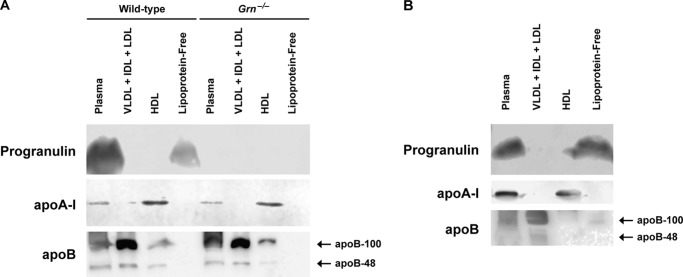
To independently confirm that circulating progranulin is not a component of HDL, we isolated lipoproteins by ultracentrifugation. We found that both murine (Fig. 5A) and human (Fig. 5B) progranulin were found exclusively in the lipoprotein-free fraction. As expected, apoB was enriched in the fractions containing very low density lipoproteins (VLDL), intermediate density lipoproteins, and LDL, and apoA-I was enriched in the HDL fractions. Additionally, in Western blots using three different progranulin antibodies, we did not detect progranulin in two commercially obtained preparations of HDL isolated by ultracentrifugation (data not shown). Together, our results indicate that circulating progranulin is not physically associated with HDL.
FIGURE 5.
Circulating progranulin is not a component of murine or human lipoproteins isolated by ultracentrifugation. Western blots of plasma and lipoprotein fractions isolated from mouse (A) and human (B) plasma are shown. These results are from representative experiments that were performed three times (A) and two times (B) with similar results. IDL, intermediate density lipoproteins.
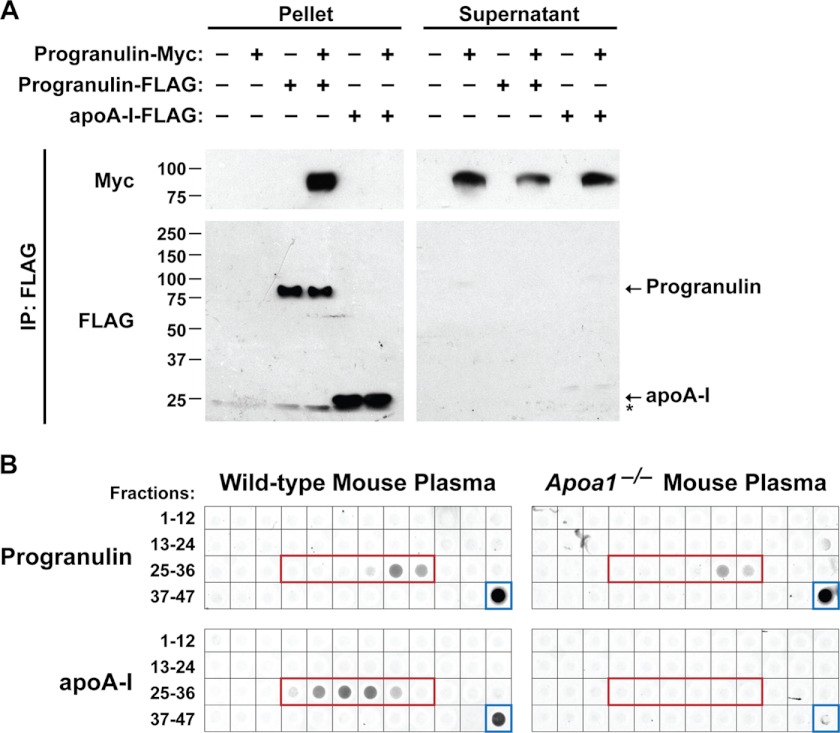
Progranulin was reported to directly bind to apoA-I (17), a major HDL protein. To test this, we performed co-immunoprecipitation assays with the conditioned media of transfected HEK293T cells by using anti-FLAG magnetic beads. As expected, immunoprecipitation of progranulin-FLAG pulled down progranulin-Myc (Fig. 6A). In contrast, immunoprecipitation of apoA-I-FLAG did not pull down progranulin-Myc (lane 6), indicating these proteins do not interact in this assay. We next examined progranulin in plasma isolated from apoA-I-deficient (Apoa1−/−) mice. When we subjected the plasma to gel-filtration and subsequently performed dot blot analysis, we found no difference in the elution profile of progranulin with wild-type and Apoa1−/− plasma (Fig. 6B). These findings indicate that, independently of apoA-I, circulating progranulin exists in a complex of ∼180–190 kDa. Taken together, these results suggest that progranulin does not physically interact with apoA-I in cultured cells and mouse plasma.
FIGURE 6.
Progranulin and apoA-I do not physically interact. A, progranulin and apoA-I do not physically interact in co-immunoprecipitation (IP) assays. Conditioned medium from transfected HEK293T cells was subjected to immunoprecipitation with anti-FLAG magnetic beads. The asterisk indicates a cross-reactive band. These results are from a representative experiment that was performed six times with similar results. B, gel-filtration elution profile of progranulin is not affected by loss of apoA-I. Dot blots of gel-filtration fractions 1–47 and plasma (blue box) from wild-type (left) and Apoa1−/− (right) mice are shown. The fractions corresponding to HDL are indicated by the red box. Similar results were obtained in two independent experiments.
DISCUSSION
The current study provides evidence that the majority of secreted and circulating progranulin exists as a homodimer. Recombinant human progranulin that was purified from the conditioned media of transfected HEK293T cells existed in a complex with a molecular mass of ∼170–180 kDa, which is consistent with that of a dimer. Additionally, cross-linking of recombinant progranulin yielded a higher molecular mass band that corresponds to a dimer (∼180 kDa) by SDS-PAGE. Moreover, in co-immunoprecipitation assays, progranulin molecules with different epitope tags were found to physically interact. In vivo, circulating progranulin in mouse and human plasma similarly exist in complexes that are ∼180–190 kDa.
Our findings agree with a previous observation that progranulin is potentially a dimer (17). In that study, progranulin was found to migrate at ∼80 kDa under reducing SDS-PAGE and at ∼130 kDa under non-reducing conditions. However, because the ∼130-kDa band was observed with protein mixtures and not with purified progranulin, it was not clear whether higher molecular mass progranulin was due to homo- or heterodimers. Our studies with purified recombinant progranulin demonstrate that progranulin is indeed a homodimer. It remains uncertain what regions of progranulin are required for dimerization. To investigate this, we generated a series of deletion mutants that span ∼90% of the mature protein, but we were unable to identify a region required for the progranulin-progranulin interaction in co-immunoprecipitation assays (Fig. 2), suggesting that multiple regions are sufficient. Furthermore, the addition of a strong reducing agent (1 mm DTT) to the recombinant progranulin and the buffer did not affect the progranulin gel-filtration elution profile (data not shown), suggesting that dimerization is not mediated by disulfide bonds.
The functional importance of progranulin dimerization remains unknown. Progranulin has been reported to bind to receptors at the cell surface including sortilin (32) and tumor necrosis factor receptors (16). It is conceivable that dimerization may affect the ability of secreted progranulin to bind to these receptors. Moreover, it is possible that dimerization may modulate progranulin proteolysis by affecting its binding to secretory leukocyte peptidase inhibitor, a protein that complexes with progranulin and thereby prevents its cleavage into granulin peptides by proteases (14).
Progranulin was reported to be a component of human HDL (17). Progranulin anti-inflammatory activities (16, 18, 20) raised the possibility that progranulin at least partially confers the known anti-inflammatory properties to HDL (33). However, our results indicate that progranulin is not a component of mouse and human HDL. Despite partially co-fractionating with HDLs by gel-filtration chromatography, we determined that progranulin in mouse and human plasma is not a component of HDL by several independent methods. First, the delipidation of plasma did not result in a shift in the gel-filtration elution profile of progranulin (Fig. 3D). In contrast, delipidation causes a clear shift in the elution profile of the major HDL protein apoA-I. Second, progranulin was unable to reorganize DMPC in phospholipid clearance assays, suggesting that it does not interact with phospholipids in vitro (Fig. 4). Third, we could not detect progranulin in mouse or human HDL isolated by ultracentrifugation (Fig. 5). Rather, progranulin is found in the lipoprotein-free fraction.
Progranulin was also reported to interact with apoA-I in co-immunoprecipitation assays (17). In our studies we were unable to detect any physical interaction between progranulin and apoA-I. In co-immunoprecipitation assays, immunoprecipitation of apoA-I-FLAG did not pull down progranulin-Myc (Fig. 6A). Moreover, the gel-filtration elution profile of progranulin was not different between wild-type and Apoa1−/− plasma (Fig. 6B), indicating that the molecular mass of mouse progranulin in plasma is not influenced by loss of apoA-I.
It is unclear why our results contrast those of a previous report claiming progranulin is a component of HDL and binds to apoA-I (17). Consistent with our findings, proteomic studies have failed to detect progranulin in human HDL isolated by ultracentrifugation (34) and by gel-filtration followed by a phospholipid binding resin (31).
In summary, our data provide insights into the biochemical properties of secreted progranulin. Such insights provide a foundation for further elucidating the biochemistry, receptor interactions and signaling, and proteolytic cleavage of progranulin. For example, do truncated forms of progranulin act as dominant negatives? How does dimerization relate to cleavage of progranulin by proteases? Identification of a dimerization domain and/or dimerization-defective mutants will allow for studies on the effects of dimerization on progranulin activity. Future studies on secreted progranulin should be directed toward understanding it in the context of a circulating dimer rather than in the context of binding lipoproteins.
Acknowledgments
We thank Karl Weisgraber and Stephen Young for antibodies, the UCSF Memory and Aging Center for blood samples, Robert Mahley, Yadong Huang, Jay Heinecke, and Daniel Rader for advice, Laura Mitic and members of the Farese laboratory for helpful discussions, Anna Lisa Lucido, and Gary Howard for editorial assistance, John Carroll for assistance with graphics, and Walter Brecht, Grisell Diaz-Ramirez, and Michael Ward for technical assistance. The Gladstone Institutes received support from National Center for Research Resources Grant RR18928.
This work was supported, in whole or in part, by National Institutes of Health Grants P50 AG023501 (to R. V. F.), GMO100909 (to R. V. F.), R01 AG029547 (to G. Y.), R01 HL63762 (to J. H.), R01 GM097194 (to T. C. W.), and F32 HL116197 (to A. D. N.). This work was also supported by Consortium for Frontotemporal Dementia Research (to R. V. F., G. Y., and J. H.), the Alzheimer's Disease Research Center (to R. V. F.), the American Diabetes Association (to R. V. F.), the Alzheimer's Association (to G. Y.), the Welch Foundation (to G. Y.), the American Health Assistance Foundation (to J. H.), the Murchison Foundation (to J. H.), the Ted Nash Longlife Foundation (to J. H.), and the Gladstone Institutes.
- apo
- apolipoprotein
- DMPC
- 1,2-dimyristoyl-rac-glycero-3-phosphocholine.
REFERENCES
- 1. He Z., Ong C. H., Halper J., Bateman A. (2003) Progranulin is a mediator of the wound response. Nat. Med. 9, 225–229 [DOI] [PubMed] [Google Scholar]
- 2. Cenik B., Sephton C. F., Cenik B. K., Herz J., Yu G. (2012) Progranulin. A proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J. Biol. Chem. 287, 32298–32306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ryan C. L., Baranowski D. C., Chitramuthu B. P., Malik S., Li Z., Cao M., Minotti S., Durham H. D., Kay D. G., Shaw C. A., Bennett H. P., Bateman A. (2009) Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neuroscience 10, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. He Z., Bateman A. (1999) Progranulin gene expression regulates epithelial cell growth and promotes tumor growth in vivo. Cancer Res. 59, 3222–3229 [PubMed] [Google Scholar]
- 5. Ong C. H., He Z., Kriazhev L., Shan X., Palfree R. G., Bateman A. (2006) Regulation of progranulin expression in myeloid cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 291, R1602–E1612 [DOI] [PubMed] [Google Scholar]
- 6. Matsubara T., Mita A., Minami K., Hosooka T., Kitazawa S., Takahashi K., Tamori Y., Yokoi N., Watanabe M., Matsuo E., Nishimura O., Seino S. (2012) PGRN is a key adipokine mediating high fat diet-induced insulin resistance and obesity through IL-6 in adipose tissue. Cell Metab. 15, 38–50 [DOI] [PubMed] [Google Scholar]
- 7. Petkau T. L., Neal S. J., Orban P. C., MacDonald J. L., Hill A. M., Lu G., Feldman H. H., Mackenzie I. R., Leavitt B. R. (2010) Progranulin expression in the developing and adult murine brain. J. Comp. Neurol. 518, 3931–3947 [DOI] [PubMed] [Google Scholar]
- 8. Baker M., Mackenzie I. R., Pickering-Brown S. M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A. D., Rollinson S., Cannon A., Dwosh E., Neary D., Melquist S., Richardson A., Dickson D., Berger Z., Eriksen J., Robinson T., Zehr C., Dickey C. A., Crook R., McGowan E., Mann D., Boeve B., Feldman H., Hutton M. (2006) Mutations in progranulin cause τ-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919 [DOI] [PubMed] [Google Scholar]
- 9. Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J.-J., van Duijn C., Peeters K., Sciot R., Santens P., De Pooter T., Mattheijssens M., Van den Broeck M., Cuijt I., Vennekens K., De Deyn P. P., Kumar-Singh S., Van Broeckhoven C. (2006) Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442, 920–924 [DOI] [PubMed] [Google Scholar]
- 10. Bhandari V., Bateman A. (1992) Structure and chromosomal location of the human granulin gene. Biochem. Biophys. Res. Commun. 188, 57–63 [DOI] [PubMed] [Google Scholar]
- 11. Bateman A., Bennett H. P. (1998) Granulins. The structure and function of an emerging family of growth factors. J. Endocrinol. 158, 145–151 [DOI] [PubMed] [Google Scholar]
- 12. Tolkatchev D., Ng A., Vranken W., Ni F. (2000) Design and solution structure of a well-folded stack of two β-hairpins based on the amino-terminal fragment of human granulin A. Biochemistry 39, 2878–2886 [DOI] [PubMed] [Google Scholar]
- 13. Vranken W. F., Chen Z. G., Xu P., James S., Bennett H. P., Ni F. (1999) A 30-residue fragment of the carp granulin-1 protein folds into a stack of two β-hairpins similar to that found in the native protein. J. Pept. Res. 53, 590–597 [DOI] [PubMed] [Google Scholar]
- 14. Zhu J., Nathan C., Jin W., Sim D., Ashcroft G. S., Wahl S. M., Lacomis L., Erdjument-Bromage H., Tempst P., Wright C. D., Ding A. (2002) Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell 111, 867–878 [DOI] [PubMed] [Google Scholar]
- 15. Kessenbrock K., Fröhlich L., Sixt M., Lämmermann T., Pfister H., Bateman A., Belaaouaj A., Ring J., Ollert M., Fässler R., Jenne D. E. (2008) Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J. Clin. Invest. 118, 2438–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang W., Lu Y., Tian Q.-Y., Zhang Y., Guo F.-J., Liu G.-Y., Syed N. M., Lai Y., Lin E. A., Kong L., Su J., Yin F., Ding A.-H., Zanin-Zhorov A., Dustin M. L., Tao J., Craft J., Yin Z., Feng J. Q., Abramson S. B., Yu X.-P., Liu C.-J. (2011) The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science 332, 478–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okura H., Yamashita S., Ohama T., Saga A., Yamamoto-Kakuta A., Hamada Y., Sougawa N., Ohyama R., Sawa Y., Matsuyama A. (2010) HDL/apolipoprotein A-I binds to macrophage-derived progranulin and suppresses its conversion into proinflammatory granulins. J. Atheroscler. Thromb. 17, 568–577 [DOI] [PubMed] [Google Scholar]
- 18. Martens L. H., Zhang J., Barmada S. J., Zhou P., Kamiya S., Sun B., Min S. W., Gan L., Finkbeiner S., Huang E. J., Farese R. V., Jr. (2012) Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J. Clin. Invest. 122, 3955–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahmed Z., Sheng H., Xu Y.-F., Lin W.-L., Innes A. E., Gass J., Yu X., Wuertzer C. A., Hou H., Chiba S., Yamanouchi K., Leissring M., Petrucelli L., Nishihara M., Hutton M. L., McGowan E., Dickson D. W., Lewis J. (2010) Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am. J. Pathol. 177, 311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yin F., Banerjee R., Thomas B., Zhou P., Qian L., Jia T., Ma X., Ma Y., Iadecola C., Beal M. F., Nathan C., Ding A. (2010) Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J. Exp. Med. 207, 117–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coppola G., Karydas A., Rademakers R., Wang Q., Baker M., Hutton M., Miller B. L., Geschwind D. H. (2008) Gene expression study on peripheral blood identifies progranulin mutations. Ann. Neurol. 64, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghidoni R., Benussi L., Glionna M., Franzoni M. (2008) Low plasma progranulin levels predict progranulin mutations in frontotemporal lobar degeneration. Neurology 71, 1235–1239 [DOI] [PubMed] [Google Scholar]
- 23. Finch N., Baker M., Crook R., Swanson K., Kuntz K., Surtees R., Bisceglio G., Rovelet-Lecrux A., Boeve B., Petersen R. C., Dickson D. W., Younkin S. G., Deramecourt V., Crook J., Graff-Radford N. R., Rademakers R. (2009) Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain 132, 583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Youn B.-S., Bang S.-I., Klöting N., Park J. W., Lee N., Oh J.-E., Pi K.-B., Lee T. H., Ruschke K., Fasshauer M., Stumvoll M., Blüher M. (2009) Serum progranulin concentrations may be associated with macrophage infiltration into omental adipose tissue. Diabetes 58, 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carecchio M., Fenoglio C., De Riz M., Guidi I., Comi C., Cortini F., Venturelli E., Restelli I., Cantoni C., Bresolin N., Monaco F., Scarpini E., Galimberti D. (2009) Progranulin plasma levels as potential biomarker for the identification of GRN deletion carriers. A case with atypical onset as clinical amnestic mild cognitive impairment converted to Alzheimer's disease. J. Neurol. Sci. 287, 291–293 [DOI] [PubMed] [Google Scholar]
- 26. Hsiung G. Y., Fok A., Feldman H. H., Rademakers R., Mackenzie I. R. (2011) rs5848 polymorphism and serum progranulin level. J. Neurol. Sci. 300, 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williamson R., Lee D., Hagaman J., Maeda N. (1992) Marked reduction of high density lipoprotein cholesterol in mice genetically modified to lack apolipoprotein A-I. Proc. Natl. Acad. Sci. U.S.A. 89, 7134–7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cham B. E., Knowles B. R. (1976) A solvent system for delipidation of plasma or serum without protein precipitation. J. Lipid Res. 17, 176–181 [PubMed] [Google Scholar]
- 29. Havel R. J., Eder H. A., Bragdon J. H. (1955) The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34, 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pownall H. J., Massey J. B., Kusserow S. K., Gotto A. M., Jr. (1978) Kinetics of lipid-protein interactions. Interaction of apolipoprotein A-I from human plasma high density lipoproteins with phosphatidylcholines. Biochemistry 17, 1183–1188 [DOI] [PubMed] [Google Scholar]
- 31. Gordon S. M., Deng J., Lu L. J., Davidson W. S. (2010) Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J. Proteome Res. 9, 5239–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu F., Padukkavidana T., Vægter C. B., Brady O. A., Zheng Y., Mackenzie I. R., Feldman H. H., Nykjaer A., Strittmatter S. M. (2010) Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron 68, 654–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cockerill G. W., Rye K. A., Gamble J. R., Vadas M. A., Barter P. J. (1995) High-density lipoproteins inhibit cytokine-induced expression of endothelial cell adhesion molecules. Arterioscler. Thromb. Vasc. Biol. 15, 1987–1994 [DOI] [PubMed] [Google Scholar]
- 34. Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., Chea H., Knopp R. H., Brunzell J., Geary R., Chait A., Zhao X.-Q., Elkon K., Marcovina S., Ridker P., Oram J. F., Heinecke J. W. (2007) Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]