Abstract
Human serum albumin (HSA)-coated lipid nanoparticles (HSA-LNPs) loaded with phrGFP-targeted siRNA (HSA-LNPs-siRNA) were prepared and evaluated for gene downregulation effect in phrGFP-transfected breast cancer cells and the corresponding xenograft tumor model. HSA-LNPs-siRNA were successfully prepared with a particle size of 79.5±5.5 nm. In phrGFP-transfected MCF-7 cells, HSA-LNPs-siRNA significantly decreased cell fluorescence even in the presence of fetal bovine serum (FBS). Moreover, cell fluorescence and phrGFP mRNA expression were significantly downregulated by HSA-LNPs-siRNA in phrGFP-transfected MCF-7, MDA-MB-231, and SK-BR-3 cells in comparison with control or HSA-LNPs-siRNA (scrambled). In phrGFP-transfected MCF-7 xenograft tumor model, tumor fluorescence was significantly decreased after three IV administrations of HSA-LNPs-siRNA at a dose of 3 mg/kg in comparison with siRNA alone. HSA-LNPs-siRNA demonstrated a superior pharmacokinetic profile in comparison with siRNA at a dose of 1mg/kg. These results show that the novel nonviral carrier, HSA-LNPs, may be used for the delivery of siRNA to breast cancer cells.
Keywords: RNA interference (RNAi), Lipid nanoparticles, Human serum albumin (HSA), Breast cancer
Small interfering RNAs (siRNAs) have the potential for therapeutic application to numerous diseases, including cancer.1,2 However, despite quite efficient and reliable gene silencing in vitro, only limited siRNA delivery has been achieved because of rapid enzymatic degradation as well as poor cellular uptake of siRNA.3,4 Therefore, effective systems that can protect and transport siRNA to the cytoplasm of the target cells are needed to exploit the promising potential application offered by siRNA delivery. Recently, the use of viral vectors to deliver siRNAs has shown effective gene silencing in vitro and in vivo.5,6 However, safety concerns have been raised in clinical trials regarding their potential toxicity. The use of nonviral vectors is attractive for siRNA delivery. Lipid nanoparticles (NPs),7 chitosan NPs8 and polyethylenimine NPs9 have been evaluated for siRNA delivery in vitro and in vivo. Among these, lipid NPs are one of most promising vectors for the delivery of siRNA.10-12 However, their application in gene delivery is limited because of variable transfection rates13,14 and systemic toxicity when injected intravenously.15–17 Thus, improvements in delivery efficiency as well as decreased toxicity may be required for clinical application of lipid NPs' siRNA delivery.
Several kinds of cationic lipids have been synthesized and shown to be able to deliver genes into cells both in vitro and in vivo.18–20 Among those, 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA) are well characterized and widely used to deliver antisense oligonucleotides and siRNAs.21,22 In contrast, Dimethyldioctadecylammonium (DDAB) has not been extensively characterized. Recently, Marty et al examined the interaction of tRNA with cationic lipids and demonstrated that DDAB more stably binds with tRNA in comparison with DOTAP or 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE).23 Therefore, DDAB may facilitate more stable siRNA delivery to tumor cells. Furthermore, hydrophilic surfactants such as Tween 80 have been used to increase gene transfection efficiency.24 D-α-tocopheryl polyethylene glycol 1000 succinate (TPGS) is a hydrophilic surfactant that can be used as an emulsifier, drug solubilizer, absorption enhancer, and vehicle for lipid-based drug delivery.25-27 Nevertheless, the effect of TPGS on the delivery of siRNA has not been evaluated in vitro and in vivo.
On the other hand, coating cationic liposomes with albumin, the most abundant plasma protein, has shown enhancement of transfection efficiency of cationic liposome/DNA complexes as well as resistance to serum endonucleases.28-30 We have also demonstrated that the liposomes coated with heat-activated human serum albumin (HSA) can enhance the delivery of antisense oligonucleotides.31 In this study, HSA-coated lipid NPs with phrGFP siRNA (HSA-LNPs-siRNA) were prepared and evaluated for the effect of TPGS concentration and HSA coating on the transfection efficiency of siRNA. Furthermore, phrGFP down-regulation was investigated in phrGFP stably transfected breast cancer cells and a breast cancer xenograft tumor model. Pharmacokinetic properties of HSA-LNPs-siRNA were also evaluated.
Methods
Dimethyl dioctadecyl ammonium bromide (DDAB) and cholesterol were purchased from Avanti Polar Lipids, Inc. (Alabaster, Alabama). Lipofectamine was purchased from Invitrogen (Grand Island, New York). Polyethylenimine 2000, Agarose, and 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetra-zolium bromide (MTT) were purchased from Sigma (St. Louis, Missouri). D-α-tocopheryl-polyethylene glycol 1000 succinate (TPGS) was purchased from Eastman Chemical Company (Kingsport, Tennessee). phrGFP siRNA (sense: 5′-CAGGAGA-CAUGAGCUUCAAGGUGAA-3′,antisense: 5′-UUCACCUU-GAAGCUCAUGUCUCCUG-3′) and scrambled control siRNA (sense: 3′-UUCUCCGAACGUGUCACGUTT-5′, antisense: 3′-TTA AGA GGC UUG CAC AGU GCA-5′) were purchased from Invitrogen (Carlsbad, California). Other regents were all commercially available and were analytical grade.
Preparation of HSA-LNPs-siRNA
HSA-LNPs-siRNA were prepared by ethanol injection method. Briefly, a lipid mixture of DDAB/Cholesterol/TPGS at varying molar ratios were dissolved in ethanol (EtOH) and then mixed with PBS solution at ratio of 1:9 (v/v) EtOH: PBS. Next, siRNA was complexed with polyethylenimine 2K in PBS solution (pH, 7.4) at a weight ratio of 1:0.3. The resulting siRNA solution was mixed with lipid NPs and incubated at room temperature (20°–25°C) for 15 minutes and then mixed with HSA solution at molar ratios to siRNA of 1:10, 2:10 and 4:10. The particle size was analyzed by dynamic light scattering (DLS) on a NICOMP 370 Submicron Particle Sizer (NICOMP, Santa Barbara, California). The zeta potential of the lipid NPs was determined by Zeta PALS (Brookhaven Instruments Corp., Worcestershire, New York). All measurements were carried out in triplicate.
Cell culture and establishment of phrGFP-transfected stable cell lines
MCF-7, MDA-MB-231 and SK-BR-3 cells were purchased form American Type Culture Collection (Rockville, Maryland). Cells were grown in phenol red free Dulbecco's Modified Eagle Medium (DMEM)/F12 supplemented with 5% fetal bovine serum (FBS), 100 ug/ml streptomycin at 37°C in a 5% CO2/95% air humidified atmosphere.
To establish phrGFP-transfected cells, breast cancer cells were transfected with phrGFP II-1 plasmid (Stratagene, La Jolla, California) in LipofectAMINE 2000 (Invitrogen) as described in the manufacturer's standard procedure. Stably transfected cells were selected with G418 and characterized by fluorescence microscopy.
Transfection studies
phrGFP transfected MCF-7, MDA-MB-231, and SK-BR-3 cells were used for transfection studies. Next, 2 × 105 cells were seeded in a 6-well plate and cultured overnight. Cells were transfected with 40 nM HSA-LNPs-siRNA, LNPs-siRNA, or HSA-LNPs-siRNA (scrambled) for 4 hours at 37°C in the presence or absence of FBS. After transfection, the medium was replaced with fresh growth medium and the cells were incubated for 48 hours at 37°C under 5% CO2 atmosphere. Then cells were collected and analyzed for phrGFP mRNA levels by real-time qRT–PCR and for cell fluorescence level by fluorescence spectrometry.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted using Trizol (Invitrogen) reagent. The reverse transcription step was carried out using the SuperScript™ III First-Strand Synthesis System from Invitrogen following the manufacturer's protocol. Two microliters of the resulting cDNA was added to 20 μl of the PCR reaction mixture containing 10 μL of SYBR green PCR Mastermix (Applied Biosystems, Foster City, California) and phrGFP or 36B4 (acidic ribosomal phosphoprotein) primer.
For phrGFP, forward primer (CAGGAGACAUGAGCUU-CAAGGUGAA) and reverse primer (UUCACCUUGAAGCU-CAUGUCUCCUG) were used. For 36B4, forward primer (AAACTGCTGCCTCATATCCG) and reverse primer (TTTCAGCAAGTGGGAAGGT) were used. PCR amplification was conducted as follows: 10 minutes at 95°C, followed by 40 cycles at 95°C for 15 seconds, 60°C for 60 seconds and finally at 72°C for 90 seconds. Fluorescence data generated were monitored and recorded by the Gene Amp 5700 sequence detection system (Applied Biosystems). All data were set up in triplicate and normalized to the 36B4 housekeeping gene.
Determine cell fluorescence
After transfection, cells were washed three times with cold PBS and lysed with protein lysis buffer. Fluorescence intensity of 100 μL cell lysates were determined by a microplate reader (λex: 485 nm, λem: 535 nm). The protein concentration of the samples was determined by using a protein assay kit (Micro BCA ™ protein assay kit, Pierce, Rockford, Illinois). Fluorescence intensity was normalized with the protein content.
In vitro cytotoxicity in phrGF- transfected breast cancer cells
In vitro cytotoxicity of HSA-LNPs-siRNA was determined by MTT method in phrGFP-transfected MCF-7, MDA-MB-231 and SK-BR-3 cells. Next, 4000 cells were seeded in 96-well plates and cultured overnight. Cells were incubated with 1:2 serial diluted scrambled siRNA alone or HSA-LNPs-siRNA (scrambled) for 4 hours at 37°C. After incubation, cells were washed twice with cold PBS and cultured for an additional 48 hours in fresh medium. For cell viability determination, 20 μL of 5 mg/ml MTT was added to each well and incubated for 4 hours at 37°C, and the formazan crystals were dissolved in 200 μL of DMSO. The absorbance was determined at 490 nm on a Dynatech MR-600 microplate reader.
In vivo imaging of phrGFP downregulation
The protocol for animal studies described in this article was approved by the Ohio State University's Institutional Animal Care and Use Committee. In vivo imaging of phrGFP downregulation was evaluated in a phrGFP-transfected MCF-7 xenograft tumor model. Female athymic mice (HSD:Athymic nude-FOXN1nd) were purchased from Harlan (Indianapolis, Indiana). Mice (18 g – 22 g) were subcutaneously inoculated on day 0 with 1×106 cells and then randomized to different treatment groups to avoid cage effects. Two days before cell inoculation, mice were implanted with estradiol pellets (0.36 mg/pellet, 60-day release). When a tumor reached a volume of 50 – 100 mm3, the mice (in groups of 3) were injected intravenously with HSA-LNPs-siRNA or free siRNA at a dose of 3 mg/kg every 3 days for 3 times via tail vein.
In vivo imaging of phrGFP downregulation in phrGFP-transfected MCF-7 xenografted tumor model was determined using the Xenogen IVIS 100 imaging System (Xenogen, Alameda, California). Mice were anesthetized with an initial dose of 2.5% isoflurane, followed by a maintenance dose of 1.5% isoflurane. Then, the mice were placed in an imaging chamber equipped with a CCD camera system (IVIS Imaging System equipped with a Series 100 intensified CCD camera; Xenogen, Alameda, California). The fluorescence intensity of the mice was measured by recording CCD photon counts. Total photon flux (photons/second) was acquired from fluorescence images using Living Image software 2.20 (Xenogen).
Pharmacokinetic studies
The pharmacokinetic profile of HSA-LNPs-siRNA was evaluated in female ICR mice (18 – 22 g, purchased from Harlan). Alexa Fluor-labeled siRNA was used in this study. Mice were randomized in two experimental groups and intravenously injected with HSA-LNPs-siRNA or siRNA at a dose of 1 mg/kg via tail vein. Mice were euthanized by CO2 inhalation and blood was obtained by cardiac puncture at various time points (0, 5, 15, 30, 60, 120, 240 minutes). Serum was isolated by centrifugation and stored at −20°C. Next, siRNA was extracted with 5% SDS and quantified by fluorescence spectrometry. The excitation and emission wavelengths were 550 and 555 nm, respectively. In all analyses, a calibration curve relating fluorescence intensity to the plasma siRNA concentration was established. Using WinNonlin software, pharmacokinetic parameters were determined, including area under the curve (AUC), mean residence time (MRT), total body clearance (CL), and plasma half-life for the distribution and elimination phases.
Statistical analysis
Data were represented as mean ± standard deviations (SD) and analyzed by 2-tailed Student's t-test using MiniTAB Program (Minitab Inc., State College, Pennsylvania). A P < 0.05 was considered statistically significant.
Results
Liposome characterization
LNPs-siRNA had an average diameter of 67.8±3.5 nm and zeta potential of +22.1 mV. A siRNA-to-lipid ratio of 1:15 was selected based on the gel retardation assay (data not shown). Addition of HSA to LNPs-siRNA changed the particle size and zeta potential. The particle size at HSA to siRNA molar ratios (w/w) of 1:10 and 2:10 was 70.3 and 79.5 nm, respectively. However, the particle size was significantly increased by increasing the HSA-to-siRNA ratio to 4:10 (121.6 nm). In contrast, the zeta potential was decreased to +19.4, +15.3, and + 9.8 mV at HSA-to-siRNA ratios of 1:10, 2:10, and 4:10, respectively.
Effect of TPGS and cationic lipids on the transfection efficiency of phrGFP siRNA
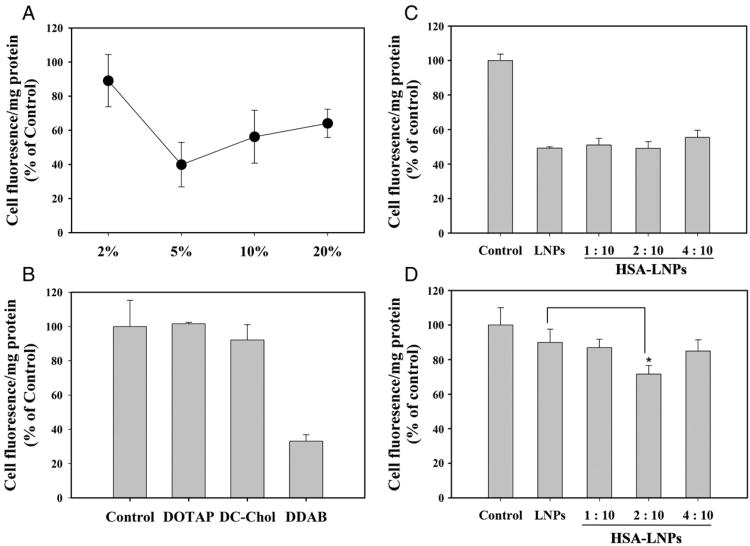
The effect of TPGS on phrGFP downregulation was investigated at different concentrations of TPGS in phrGFP-transfected MCF-7 cells. As shown in Figure 1, A, LNPs containing 5% TPGS significantly decreased phrGFP fluorescence in comparison with LNPs containing 2% TPGS. However, phrGFP fluorescence was slightly increased when increasing TPGS concentration from 5% to 30%. Therefore, 5% TPGS concentration was used in subsequent studies.
Figure 1.
Effect of TPGS concentration (A), cationic lipid (B) and HSA coating (C, D) on phrGFP downregulation in phrGFP-transfected MCF-7 cells. Cells were treated with different LNPs-siRNA for 4 h at 37°C and then cultured for 48 h at 37°C. Fluorescence intensity of the cell lysates were determined by fluorescence spectrometry. The fluorescence intensity was normalized with the protein content.
The effect of cationic lipids on the transfection efficiency of phrGFP siRNA was also evaluated in phrGFP-transfected MCF-7 cells (Figure 1, B). LNPs containing DOTAP or DC-Chol did not decrease in cell fluorescence in comparison with the control. However, LNPs containing DDAB significantly decreased cell fluorescence in comparison with the control. Thus, DDAB was selected to evaluate phrGFP downregulation.
Effect of HSA on the phrGFP downregulation in phrGFP- transfected MCF-7 cells
Because of its negative charge, HSA may decrease the surface charge of LNPs and increase serum stability. Therefore, the effect of HSA coating on phrGFP downregulation was evaluated in the absence or presence of FBS. In the absence of FBS, LNPs-siRNA and HSA-LNPs-siRNA significantly decreased phrGFP florescence in phrGFP transfected MCF-7 cells. However, the addition of HSA did not change phrGFP fluorescence in the absence of FBS (Figure 1, C).
In the presence of FBS (5%), LNPs-siRNA did not significantly decrease phrGFP fluorescence in phrGFP-transfected MCF-7 cells. In contrast, HSA-LNPs-siRNA significantly decreased phrGFP fluorescence at a HSA-to-siRNA ratio of 2:10 (Figure 1, D). These results suggest that the decreased phrGFP fluorescence might be due to the increased serum stability stemming from HSA coating.
phrGFP downregulation in phrGFP-transfected breast cancer cells
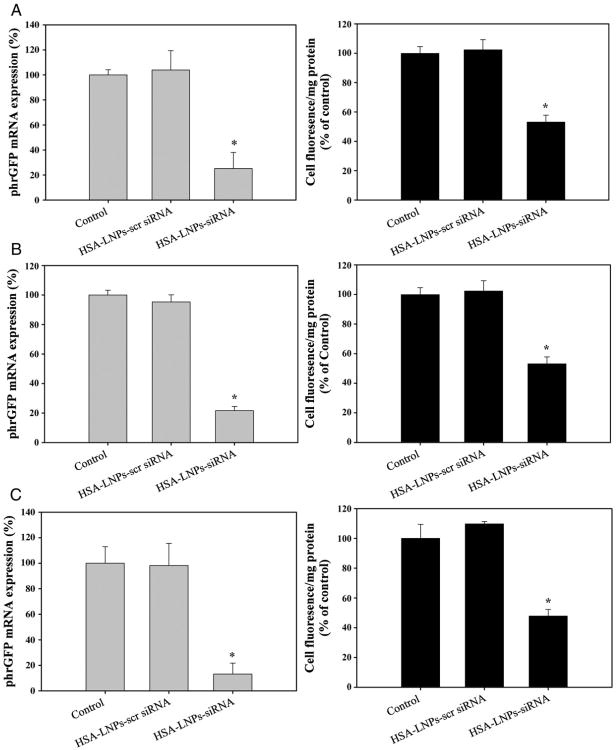
phrGFP down-regulation was also investigated in phrGFP-transfected MCF-7, MDA-MB-231, and SK-BR-3 cells. As shown in Figure 2, phrGFP mRNA levels and phrGFP fluorescence were significantly downregulated by treatment of HSA-LNPs-siRNA in comparison with the control in phrGFP-transfected MCF-7 cells. However, HSA-LNPs with scrambled siRNA did not change phrGFP mRNA levels and phrGFP fluorescence in comparison with control. Similar results were also observed in phrGFP-transfected MDA-MB-231 and SK-BR-3 cells. These results suggest that HSA-LNPs-siRNA could be used for the delivery of siRNA to breast cancer cells.
Figure 2.
In vitro phrGFP downregulation by treatment of HSA-LNPs-siRNA in phrGFP-transfected MCF-7 (A), MDA-MB-231 (B), and SK-BR-3 (C) cells. For the mRNA downregulation, total mRNA was extracted and the phrGFP mRNA expression was measured by qRT-PCR. For the phrGFP fluorescence, fluorescence intensity of the cell lysates was determined by fluorescence spectrometry and the results were normalized with the protein content.
In vitro cytotoxicity of HSA-LNPs-siRNA in phrGFP-transfected breast cancer cells
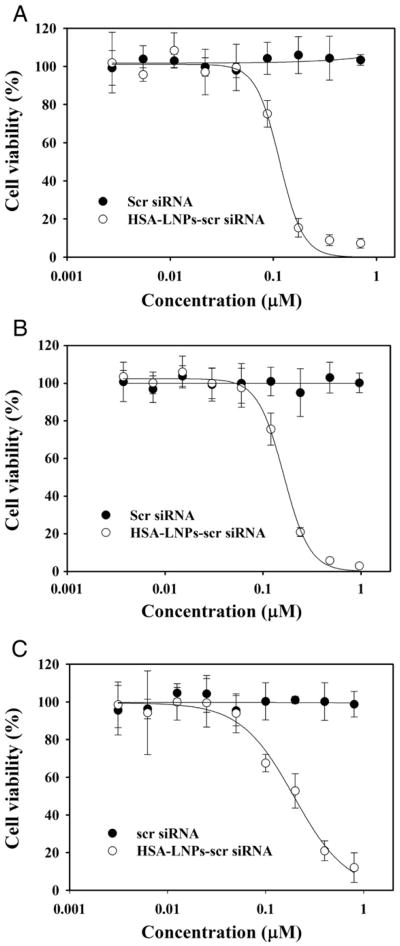
Cell toxicity was determined by MTT assay after treatment of HSA-LNPs-siRNA (scrambled). As shown in Figure 3, scrambled siRNA alone did not exhibit cytotoxicity at tested concentrations (1.5 to 1000 nM). In contrast, HSA-LNPs-siRNA (scrmbled) in phrGFP-transfected MCF-7, MDA-MB-231, and SK-BR-3 cells showed cytotoxicity with IC50 of 134.8± 11.8, 164.6±15.7, and 171.9±20.7 nM, respectively.
Figure 3.
Cytotoxicity of HSA-LNPs with scrambled siRNA to phrGFP-transfected MCF-7 (A), MDA-MB-231 (B), and SK-BR-3 (C) cells. Cytotoxicity was determined using MTT assay as described in Methods. Data represent the mean±SD (n=4).
In vivo imaging of phrGFP downregulation in phrGFP-transfected MCF-7 xenograft tumor model
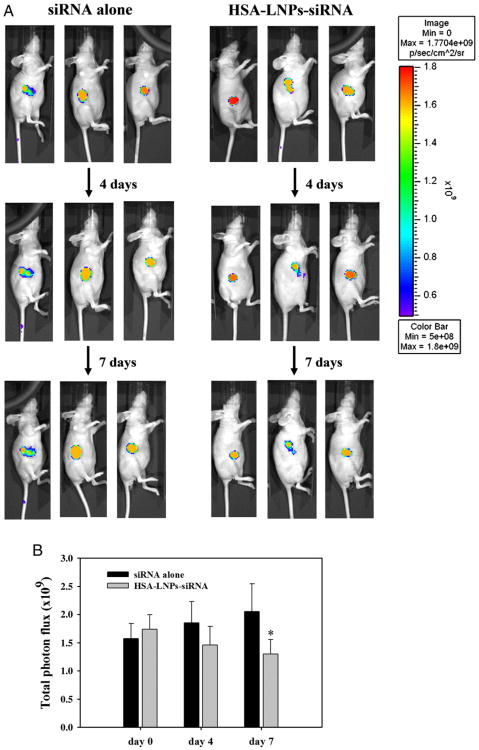
In vivo imaging of phrGFP downregulation in the phrGFP-transfected MCF-7 xenograft tumor model was evaluated following IV administration of HSA-LNPs-siRNA. All animals tolerated the treatments well with no observable signs of toxicity and no change in body weight over the course of the study (data not shown). As shown in Figure 4, A, phrGFP fluorescence did not decrease, but gradually increased by treatment of siRNA alone. In contrast, phrGFP fluorescence decreased with treatment of HSA-LNPs-siRNA. The total photon flux (photons/second) was significantly decreased with treatment of HSA-LNPs-siRNA in comparison with siRNA alone after 7 days' treatment (P < 0.05, Figure 4, B). However, the total photon flux was not decreased with treatment of HSA-LNPs with scrambled siRNA (data not shown). These results suggest that HSA-LNPs-siRNA could be used for in vivo gene silencing.
Figure 4.
In-vivo imaging of phrGFP downregulation in phrGFP-transfected MCF-7 xenograft tumors. (A) In vivo imaging of MCF-7/phrGFP tumors was determined using the Xenogen IVIS 100 imaging System. Tumor xenograft mice were intravenously injected with HSA-LNPs-siRNA on 1, 4, and 7 days, and the fluorescence imaging was determined on days 0, 5, and 8. (B) The total photon flux (photons/second) by treatment of siRNA alone and HSA-LNPs-siRNA. The fluorescence intensity of the mice was measured by recording CCD photon counts. Total photon flux (photons/second) was acquired from fluorescence images using Living Image software 2.20.
Pharmacokinetic properties of HSA-LNPs-siRNA
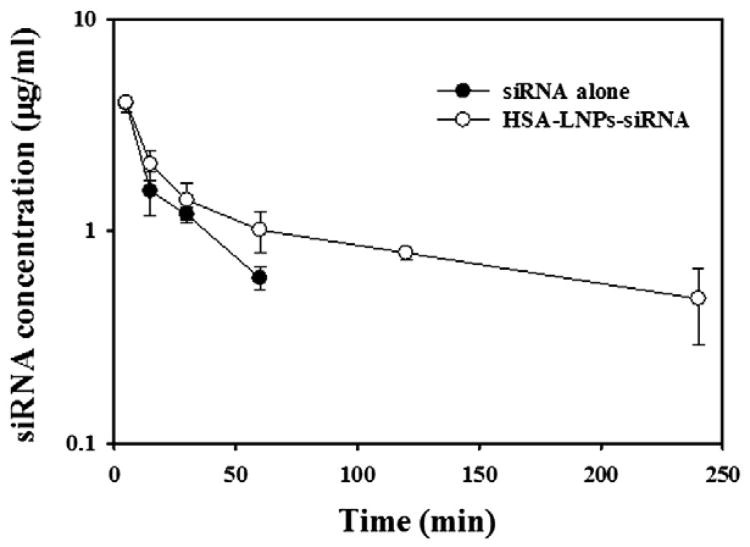
To assess pharmacokinetic properties of HSA-LNPs-siRNA, mice were intravenously administrated with HSA-LNPs-siRNA (Alexa Fluor labeled siRNA) at a dose of 1 mg/kg. The serum concentrations of siRNA after IV injection of HSA-LNPs-siRNA or siRNA alone are shown in Figure 5, and pharmacokinetic parameters are summarized in Table 1. HSA-LNPs-siRNA showed a biphasic pattern with a rapid distribution phase (t1/2α=5.8±0.5 minutes) and a slow terminal elimination phase (t1/2β=150.8±21.7 minutes). The area under the concentration-time curve (AUC) of siRNA by treatment of HSA-LNPs-siRNA was significantly increased in comparison with free siRNA (P < 0.05). The mean residence time (MRT) of HSA-LNPs-siRNA was increased by 11.6-fold in comparison with free siRNA.
Figure 5.
Temporal serum profile of Alexa Fluor-labeled phrGFP siRNA following IV administration of siRNA alone or HSA-LNPs-siRNA at a dose of 1mg/kg. Each result represents the mean±SD (n=3)
Table 1. Pharmacokinetic parameters of phrGFP siRNA following IV administration of HSA-LNPs-siRNA to ICR mice at a dose of 1 mg/kg.
| Group | siRNA alone | HSA-LNPs-siRNA |
|---|---|---|
| AUC (μg·minr/ml) | 84.6±18.7 | 341.5±29.5 |
| t½(α) (min) | 11.5±4.0 | 5.8±0.5 |
| t½(β) (min) | - | 150.8±25.7 |
| CL (ml/min/kg) | 11.8±2.6 | 2.9±0.2 |
| MRT (0-24min) | 16.6±5.8 | 192.8±28.5 |
Data represents the mean±SD (n=3).
Discussion
LNPs are one of most promising vectors for the delivery of plasmid DNA, oligonucleotides, and siRNA. However, their application in gene delivery is limited because of variations in transfection efficiency and systemic toxicity when injected intraveneously.15–17 Therefore, improvements in delivery efficacy as well as decreases in toxicity are required for clinical application of LNPs for delivery of siRNA. Recently, Bilati et al demonstrated smaller-sized NPs showed higher uptake than the larger ones, but a combination of surface charge and polymer hydrophilicity are necessary for favorable to drug delivery.32 In this study, we prepared novel HSA-coated LNPs with cationic lipid, hydrophilic surfactant, and cholesterol for increased siRNA delivery.
To maximize transfection efficiency a hydrophilic surfactant, TPGS, was added to the LNPs. Addition of TPGS to LNPs significantly downregulates phrGFP expression in phrGFP-transfected MCF-7 cells. Moreover, the maximum phrGFP downregulation was observed by addition of 5% TPGS (Figure 1, A). Furthermore, the effect of commonly used cationic lipids on the transfection of phrGFP siRNA was evaluated in phrGFP-transfected MCF-7 cells. LNPs containing DDAB showing a strong phrGFP downregulation in phrGFP-transfected MCF-7 cells in comparison with LNPs containing other cationic lipids such as DOTAP (Figure 1, B), might be due to the increased binding with siRNA. Finally, HSA coating significantly downregulates phrGFP expression in the presence of 5% FBS, probably due to the increased serum stability (Figure 1, C, 1, D).
The effect of HSA-LNPs-siRNA on the phrGFP downregulation was confirmed in phrGFP-transfected breast cancer cells (Figure 2). In MCF-7, MDA-MB-231, and SK-BR-3 cells, HSA-LNPs-siRNA showed significant phrGFP downregulation at 40-nM siRNA concentrations in coparison with control and HSA-LNPs-siRNA (scrambled). Moreover, decreased phrGFP fluorescence was also observed in phrGFP-transfected MCF-7 xenograft tumors from mice treated with HSA-LNPs-siRNA in which phrGFP fluorescence was reduced by approximately 37% (Figure 4). In addition, pharmacokinetic data showed that the terminal half-life of HSA-LNPs-siRNA was 150.8 min (Figure 5).
The toxicity of HSA-LNPs-siRNA was also investigated in this study. MTT assay data showed that the IC50 of HSA-LNPs-siRNA in phrGFP-transfected MCF-7, MDA-MB-231 and SK-BR-3 cells was 134.8±11.8, 164.6±15.7, and 171.9±20.7 nM, respectively. These results indicate that HSA-LNPs-siRNA are toxic to breast cancer cells. However, the IC50 values were much higher than effective siRNA concentrations (40 nM). Even at higher siRNA concentration (100 nM), cell viability of HSA-LNPs-siRNA in phrGFP-transfected MCF-7, MDA-MB-231, and SK-BR-3 cells was 70.9, 85.1 and 74.1 %, respectively. Moreover, all animals tolerated the treatments well with no observable signs of toxicity and no change in body weight over the course of the study. These results suggest that HSA-LNPs-siRNA have low toxicity to breast cancer cells.
In summary, HSA-LNPs-siRNA significantly downregulate phrGFP expression and fluoresence in phrGFP-transfected breast cancer cells and phrGFP-transfected MCF-7 xenograft tumors. Moreover, no significant cytotoxicity was observed both in vitro and in vivo at tested concentrations. Together, these findings demonstrate the potential of HSA-LNPs for safe and effective siRNA delivery.
Acknowledgments
This work was supported in part by DOD grant W81XWH-08-0610 to Robert J. Lee and grant W81XWH-08-1-0521 to Robert W. Brueggemeier.
References
- 1.Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest. 2007;117:3623–32. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–53. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirchhoff F. Silencing HIV-1 in vivo. Cell. 2008;34:566–8. doi: 10.1016/j.cell.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010;277:4814–27. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- 5.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–10. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 6.Barton GM, Medzhitov R. Retroviral delivery of small interfering RNA into primary cells. Proc Natl Acad Sci U S A. 2002;99:14943–5. doi: 10.1073/pnas.242594499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao W, Davide JP, Cai M, Zhang GJ, South VJ, Matter A, et al. Noninvasive imaging of lipid nanoparticle-mediated systemic delivery of small-interfering RNA to the liver. Mol Ther. 2010;18:1657–66. doi: 10.1038/mt.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Howard KA, Dong M, Andersen MØ, Rahbek UL, Johnsen MG, et al. The influence of polymeric properties on chitosan/siRNA nanoparticle formulation and gene silencing. Biomaterials. 2007;28:1280–8. doi: 10.1016/j.biomaterials.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Werth S, Urban-Klein B, Dai L, Höbel S, Grzelinski M, Bakowsky U, et al. A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. J Control Release. 2006;112:257–70. doi: 10.1016/j.jconrel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Simões S, Filipe A, Faneca H, Mano M, Penacho N, Düzgünes N, et al. Lipid nanoparticles for gene delivery. Expert Opin Drug Deliv. 2005;2:237–54. doi: 10.1517/17425247.2.2.237. [DOI] [PubMed] [Google Scholar]
- 11.Tseng YC, Huang L. Self-assembled lipid nanomedicines for siRNA tumor targeting. J Biomed Nanotechnol. 2009;5:351–63. doi: 10.1166/jbn.2009.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musacchio T, Torchilin VP. Recent developments in lipid-based pharmaceutical nanocarriers. Front Biosci. 2011;16:1388–412. doi: 10.2741/3795. [DOI] [PubMed] [Google Scholar]
- 13.Uchida E, Mizuguchi H, Ishii-Watabe A, Hayakawa T. Comparison of the efficiency and safety of non-viral vector mediated gene transfer into a wide range of human cells. Biol Pharm Bull. 2002;25:891–7. doi: 10.1248/bpb.25.891. [DOI] [PubMed] [Google Scholar]
- 14.Loisel S, Floch V, Le Gall C, Ferec C. Factors influencing the efficiency of lipoplexes mediated gene transfer in lung after intravenous administration. J Liposome Res. 2001;11:127–38. doi: 10.1081/LPR-100108457. [DOI] [PubMed] [Google Scholar]
- 15.Eliyahu H, Servel N, Domb AJ, Barenholz Y. Lipoplex induced hemagglutination: potential involvement in intravenous gene delivery. Gene Ther. 2002;9:850–8. doi: 10.1038/sj.gt.3301705. [DOI] [PubMed] [Google Scholar]
- 16.Kedmi R, Ben-Arie N, Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials. 2010;31:6867–75. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 17.Tousignant JD, Gates AL, Ingram LK, Johnson CL, Nietupski JB, Cheng SH, et al. Comprehensive analysis of the acute toxicities induced by systemic administration of cationic lipid: plasmid DNA complexes in mice. Hum Gene Ther. 2000;11:2493–513. doi: 10.1089/10430340050207984. [DOI] [PubMed] [Google Scholar]
- 18.Masotti A, Mossa G, Cametti C, Ortaggi G, Bianco A, Grosso ND, et al. Comparison of different commercially available cationic liposome-DNA lipoplexes: parameters influencing toxicity and transfection efficiency. Colloids Surf B Biointerfaces. 2009;68:136–44. doi: 10.1016/j.colsurfb.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–6. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Xu Y, Wang B, Qiao W, Liu D, Li Z. Cationic compounds used in lipoplexes and polyplexes for gene delivery. J Control Release. 2004;100:165–80. doi: 10.1016/j.jconrel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Simberg D, Weisman S, Talmon Y, Barenholz Y. DOTAP (and other cationic lipids): chemistry, biophysics, and transfection. Crit Rev Ther Drug Carrier Syst. 2004;21:257–317. doi: 10.1615/critrevtherdrugcarriersyst.v21.i4.10. [DOI] [PubMed] [Google Scholar]
- 22.Ozpolat B, Sood AK, Lopez-Berestein G. Nanomedicine based approaches for the delivery of siRNA in cancer. J Intern Med. 2010;267:44–53. doi: 10.1111/j.1365-2796.2009.02191.x. [DOI] [PubMed] [Google Scholar]
- 23.Marty R, N'soukpoé-Kossi CN, Charbonneau DM, Kreplak L, Tajmir-Riahi HA. Structural characterization of cationic lipid-tRNA complexes. Nucleic Acids Res. 2009;37:5197–207. doi: 10.1093/nar/gkp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding W, Izumisawa T, Hattori Y, Qi X, Kitamoto D, Maitani Y. Non-ionic surfactant modified cationic liposomes mediated gene transfection in vitro and in the mouse lung. Biol Pharm Bull. 2009;32:311–5. doi: 10.1248/bpb.32.311. [DOI] [PubMed] [Google Scholar]
- 25.Varma MV, Panchagnula R. Enhanced oral paclitaxel absorption with vitamin E-TPGS: effect on solubility and permeability in vitro, in situ and in vivo. Eur J Pharm Sci. 2005;25:445–53. doi: 10.1016/j.ejps.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Feng SS. Nanoparticles of poly(lactide)/vitamin E TPGS copolymer for cancer chemotherapy: synthesis, formulation, characterization and in vitro drug release. Biomaterials. 2006;27:262–70. doi: 10.1016/j.biomaterials.2005.05.104. [DOI] [PubMed] [Google Scholar]
- 27.Mu L, Feng SS. PLGA/TPGS nanoparticles for controlled release of paclitaxel: effects of the emulsifier and drug loading ratio. Pharm Res. 2003;20:1864–72. doi: 10.1023/b:pham.0000003387.15428.42. [DOI] [PubMed] [Google Scholar]
- 28.Simoes S, Slepushkin V, Pires P, Gaspar R, Pedroso de Lima MC, Duzgunes N. Human serum albumin enhances DNA transfection by lipoplexes and confers resistance to inhibition by serum. Biochim Biophys Acta. 2000;1463:459–69. doi: 10.1016/s0005-2736(99)00238-2. [DOI] [PubMed] [Google Scholar]
- 29.Faneca H, Simões S, Pedroso de Lima MC. Association of albumin or protamine to lipoplexes: enhancement of transfection and resistance to serum. J Gene Med. 2004;6:681–92. doi: 10.1002/jgm.550. [DOI] [PubMed] [Google Scholar]
- 30.Sahin S, Selek H, Ponchel G, Ercan MT, Sargon M, Hincal AA, et al. Preparation, characterization and in vivo distribution of terbutaline sulfate loaded albumin microspheres. J Control Release. 2002;82:345–58. doi: 10.1016/s0168-3659(02)00141-4. [DOI] [PubMed] [Google Scholar]
- 31.Weecharangsan W, Yu B, Zheng Y, Liu S, Pang JX, Lee LJ, et al. Efficient delivery of antisense oligodeoxyribonucleotide g3139 by human serum albumin-coated liposomes. Mol Pharm. 2009;6:1848–55. doi: 10.1021/mp900150g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acharyaa S, Sahoo SK. PLGA nanoparticles containing various anticancer agents and tumor delivery by EPR effect. Adv Drug Deliv Rev. 2011;63:170–83. doi: 10.1016/j.addr.2010.10.008. [DOI] [PubMed] [Google Scholar]