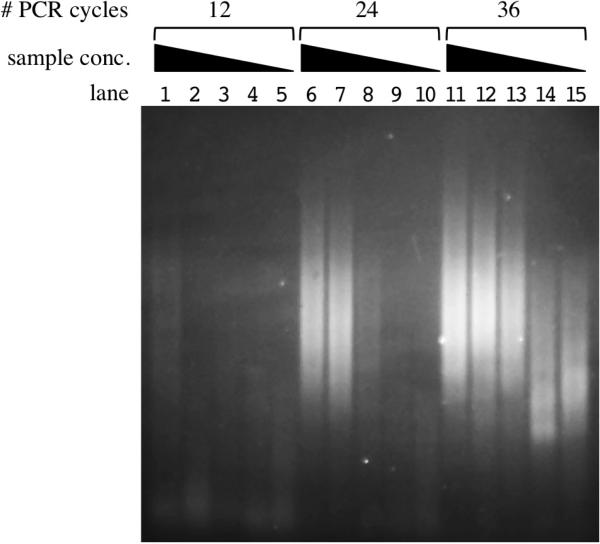
Figure 2. Sensitivity of HTML-PCR using Vibrio cholerae genomic DNA.
HTML-PCR products generated using a range of starting sample amounts as visualized by 2% agarose gel electrophoresis. The following amounts of fragmented V. cholerae E7946 gDNA were used as input for the homopolymer tail addition step of HTML-PCR: 100 ng for lanes 1, 6 and 11; 10 ng for lanes 2, 7 and 12; 1 ng for lanes 3, 8 and 13; 0.1 ng for lanes 4, 9 and 14; and 0.01 ng for lanes 5, 10 and 15. Lanes 1, 6, 7, 8, 11, 12 and 13 show products of a successful reaction, namely DNA smears ranging from approximately 140–600 base pairs that represent V. cholerae library fragments (see text). The lower molecular weight DNA smears in lanes 14 and 15 represent a mixture of V. cholerae library fragments plus artifactual products (see text).