In a series of review articles on polyploid formation, Buggs & al. (2009, 2011) and Soltis & al. (2010) have repeatedly addressed the question of whether hybridization between divergent progenitors drives whole-genome duplication and criticized the conclusions of our paper, Paun & al. (2009: “Hybrid speciation in angiosperms […]”). As the third paper in less than two years has been published in the April issue of Taxon, we take the opportunity to respond and clarify further our conclusions on hybrid speciation. The consistent points of contention in the papers by Buggs, D.E. Soltis and P. S. Soltis referred to two main points: 1) oversimplification of the subject by excluding autopolyploids from our analyses, and 2) misinterpretation of results due to our incomplete analysis of data presented in Paun & al. (2009). We discuss these and other debated topics in the following paragraphs.
1) Sampling oversimplification
As the title clearly indicates, Paun & al. (2009) focused on “hybrid speciation in angiosperms”, and therefore analyzed only homoploid and polyploid hybrids (i.e., “hybrid status” was the first criterion for selecting the case studies, see Selection of taxa on page 509). However, we also discussed “the relevance of progenitor divergence as a determinant of ploidy in resulting hybrid species [only]” (page 513). In stark contrast, Buggs & al. (2009, 2011) and Soltis & al. (2010) commented on our results in the context of strict polyploid formation, not necessarily involving hybridization, and dismissed our study as an oversimplification for failing to include autopolyploids. The three papers therefore take our data and statements out of context. For example Buggs & al. stated “[Paun & al., 2009] support the hypothesis that parental divergence drives polyploidy” (p. 3337 in 2009 and p. 329 in 2011 paper), instead of the correct statement “in hybrid speciation […] parental divergence drives ploidy” (p. 507 of Paun & al., 2009). Moreover, in their figure 3A Buggs & al. (2011) indicated with a dashed line the approximate distribution of polyploids found by Chapman & Burke (2007) and Paun & al. (2009), whereas the original data cited comprised in fact only hybrid polyploids (i.e., allopolyploids). On the same page (p. 330) Buggs & al. (2011) stated that “Paun & al. (2009) argue that autopolyploids should not be included in these comparisons” (i.e., “regarding which conditions promote polyploidization”, p. 330), but in our discussions we clearly argued that “we regard inclusion of autopolyploids in analyses considering hybrid speciation as inappropriate” (p. 515 in Paun & al., 2009).
In this context, Buggs & al. (2009, 2011) and Soltis & al. (2010) overlooked the difference between hybrid and polyploid speciation. Indeed, the two processes overlap in allopolyploids, but major differences between the two types of speciation are obvious. Hybrid speciation (i.e., the focus of Paun & al., 2009) comprises evolutionary events when a new species is being established that combines divergent genomes in the same nucleus. In contrast, polyploid speciation includes events in which whole genome duplication triggers the formation of a new, evolutionarily successful species, including autopolyploid as well as allopolyploid formation. We stress here that we intentionally did not include autopolyploids in calculations regarding processes of hybrid speciation because they are not products of hybridization and do not combine divergent genomes in a single nucleus. This distinction is important; independently of the resulting ploidy, hybridization immediately triggers wide genomic re-patterning (e.g., Hegarty & al., 2006; Paun & al., 2007), together with transgressive segregation (Stelkens & Seehausen, 2009) and more dramatic novelties (e.g., Paun & al., 2006), whereas genome doubling alone has only rarely been reported to produce rapid and extensive genomic or phenotypic effects (see also Parisod & al., 2010). Moreover, unreduced gametes form at different rates in hybrids (resulting in allopolyploids) and non-hybrids (resulting in autopolyploids; Ramsey & Schemske, 1998), and the mechanisms responsible have most probably largely different cues. We acknowledge that there may be a continuum between auto- and allopolyploids with associated difficulties in drawing a clean line between the two (Stebbins, 1950; Wendel & Doyle, 2005; Buggs & al., 2011), although it is not yet clear if segmental allopolyploid species, the intermediates between allo- and autopolyploids, “exist, at least in the long-term” (p. 1388 in Soltis & al., 2010). In any case, under any definition, however broad, logic makes autopolyploids irrelevant for investigations regarding processes of hybrid speciation. Furthermore, although Paun & al. (2009) excluded autopolyploids from calculations, we discussed the issue appropriately (section Autopolyploids and allopolyploids, p. 515), and we never intended our general conclusions to be applied to the entire spectrum of polyploids or overall polyploid speciation. Simply stated, autopolyploids are irrelevant to hybrid speciation.
2) Incomplete data analyses
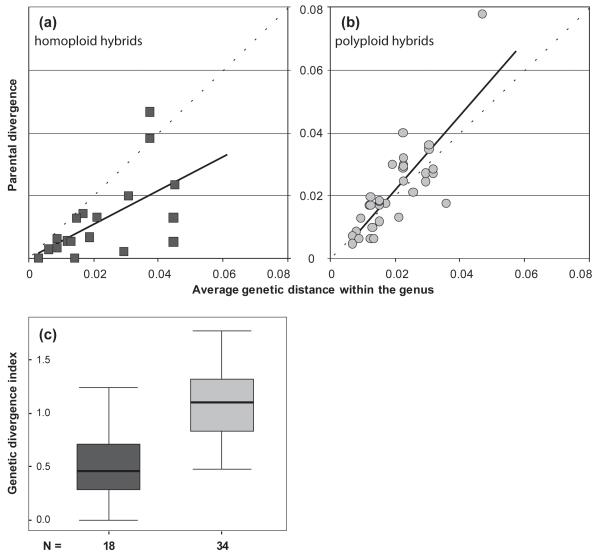
To support their conclusions on formation of polyploids, Buggs & al. (2009) carried out an additional statistical test using the data of Paun & al. (2009) with the average divergence within each genus as a null hypothesis for the expected divergence between parents of allopolyploids versus homoploid hybrids. They employed a two-tailed paired t-test and compared genetic distances between parental pairs and the average genetic distance between all pairs of genera in Paun & al. (2009); they reported a significant result for homoploid hybrids (P < 0.01, see also Fig. 1A), whereas no significance for allopolyploids (P > 0.1, see also Fig. 1B) (i.e. parental divergence for allopolyploids could not be distinguished from the average genetic distance within these genera). They interpreted these results to indicate that “homoploid hybrid formation occurs at low parental divergence, but polyploid formation fits a model of random hybridization” (p. 3337). These tests and interpretations are reviewed and restated in Soltis & al. (2010) and Buggs & al. (2011).
Fig. 1.
a)-b) Relationship of the parental genetic divergence and average genetic distance within the respective genera for homoploid hybrid species, and, respectively, polyploid hybrid species, based on the data from Paun & al. 2009. The correlation in b) is suggested by Buggs & al. 2009, 2011 and Soltis & al. 2010 to represent a random process. c) The distribution of genetic divergence index (the ratio between parental divergence and the average genetic divergence in the respective genus) of parental pairs for homoploid (dark grey) and polyploid hybrid species (light grey), modified from Paun & al. 2009. The two groups have an asymmetric dispersion range, supporting the view that parents producing allopolyploids are more divergent than parents of homoploid hybrid species (Mann-Witney U test, P < 0.0001).
A closer look at the original data of Paun & al. 2009 proves that this interpretation of Buggs & al. is flawed; the result of the t-test for allopolyploids indicate a correlation between parental divergence and the average divergence in the respective genera (Fig. 1B), which is not the same as random. Indeed, there is no significant difference between parental divergence for allopolyploids and the average genetic distance for the respective genera because these two are statistically significantly correlated (Pearson’s r = 0.81, P < 0.01, Figure 1B). This result was in fact easily observed in Figure 1 of Paun & al. (2009; redrawn here as Figure 1C), which shows that the parental divergence index (i.e., the ratio between parental divergence and the average divergence for the genus) for allopolyploids has a median value slightly higher than 1 and a normal distribution. This non-random, significant correlation indicates that polyploid hybrid speciation is more likely when parental divergence is similar to the average of divergence in the respective genus, whereas homoploid hybrid speciation is more frequent when parental divergence is much lower than the average divergence in the genus (Figure 1C). We therefore maintain our previous conclusions on hybrid speciation (Paun & al., 2009).
In addition to the two major points considered above, Buggs & al. (2009) criticized Paun & al. (2009) for underestimating the importance of natural selection in hybrid speciation. In order to investigate only “speciation” (i. e., production of evolutionarily successful entities), the 50 data points from Paun & al. (2009) included in Selection of taxa (p. 509) only “fertile, successful hybrids that have a long species history”, in other words only hybrid species that are “natural and stable, with proven evolutionary success” (point 4). We therefore excluded obvious “neopolyploids and unnamed suspected hybrids”, together with triploids and other hybrids types that have not (yet) become established. We even stressed: “Long-term success in meiosis is key to operation of the mechanism that governs ploidal shifts, which means that only taxa that appear to be valid species in their own right should be included in the calculations” (p. 509 in Paun & al., 2009). Moreover in the Discussion section of Paun & al. (2009) we have commented on the role of selection in hybrid speciation.
Nonetheless, in their abstract and discussions (p. 3338), Buggs & al. (2009) criticized previous studies (including Chapman & Burke, 2007, Buggs & al, 2008 and Paun & al., 2009) because they did not consider sufficiently “the subsequent selection on the newly formed hybrid or polyploid”. In fact, the only study to which this criticism would apply is the one conducted by the authors themselves (Buggs & al., 2008), which considered in their analysis any naturally occurring hybrid/polyploid individual (even triploids) reported in the literature, thereby investigating simple (stochastic) formation and not speciation. By investigating only successful and stable hybrid/polyploid species, the other two studies (Chapman & Burke, 2007; Paun & al., 2009) accounted for the effects of selection.
Moreover, in an confusing, interchangeable use of “formation” and “speciation”, Soltis & al. (2010) discussed the relevance of the results of Paun & al. (2009) in the section on “what factors promote/facilitate polyploidization” (i.e., mere formation of a polyploid and not necessarily speciation) and concluded based on our data that “polyploid formation fits a model of random hybridization, whereas homoploid hybrid formation tends to occur successfully at lower parental divergence” (p. 1391). Later (2011), Buggs & al. restated these conclusions (abstract and p. 328) in their review on “the formation of polyploids”, disregarding once again the original selection of data to include putatively successful speciation events, rather than simple, stochastic hybrid formation.
Conclusions
The criticisms of Buggs & al. (2009, 2011) and Soltis & al. (2010) are unjustified and their conclusions have to be refuted. We maintain here our previous conclusions, fully supported by our results (Paun & al., 2009): parental divergence does influence the ploidy of offspring in hybrid speciation in angiosperms. Formation of polyploids is, most likely, a random process, but this has nothing to do with the hypothesis we were interested in evaluating; we purposefully tried to eliminate hybrids untested by selection from our data set. We also emphasize here again that our conclusions do not apply to the frequency of successful autopolyploids in nature. We here reexamined our results in the light of the criticisms by Buggs & al. (2009, 2011) and Soltis & al. (2010) and restate our previous hypothesis that polyploid frequency plotted against parental divergence will have a bimodal distribution, with a putative adaptive valley between auto- and allopolyploid species (in possibly unfit polyploids that will exhibit more or less equal combinations of bivalents and quadrivalents in meiosis; Paun & al.,2009). Indeed, such segmental polyploid types have not (yet) been documented (Soltis & al., 2010). At the same time, we criticize here figure 3C of Buggs & al. (2011), which incorrectly illustrated our prediction, as no data were available on the comparative frequency of homoploid hybrids, allopolyploids and autopolyploids (to justify the reference points on the Y axis from Fig. 3C in Buggs & al., 2011).
Acknowledgements
O.P. is funded by the Austrian Science Fund (FWF grant P2226021) and a Marie Curie Reintegration grant (PolyAdaptation, PERG-GA-2010-268462).
References
- Buggs RJA, Soltis PS, Mavrodiev EV, Symonds VV, Soltis DE. Does phylogenetic distance between parental genomes govern the success of polyploids? Castanea. 2008;73:74–93. [Google Scholar]
- Buggs RJA, Soltis PS, Soltis DE. Does hybridization between divergent progenitors drive whole-genome duplication? Molec. Ecol. 2009;18:3334–3339. doi: 10.1111/j.1365-294X.2009.04285.x. [DOI] [PubMed] [Google Scholar]
- Buggs RJA, Soltis PS, Soltis DE. Biosystematic relationships and the formation of polyploids. Taxon. 2011;60:324–332. [Google Scholar]
- Chapman MA, Burke JM. Genetic divergence and hybrid speciation. Evolution. 2007;61:1773–1780. doi: 10.1111/j.1558-5646.2007.00134.x. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, Barker GL, Wilson ID, Abbott RJ, Edwards KJ, Hiscock SJ. Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr. Biol. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- Parisod C, Holderegger R, Brochmann C. Evolutionary consequences of autopolyploidy. New Phytol. 2010;186:5–17. doi: 10.1111/j.1469-8137.2009.03142.x. [DOI] [PubMed] [Google Scholar]
- Paun O, Fay MF, Soltis DE, Chase MW. Genetic and epigenetic alterations after hybridization and genome doubling. Taxon. 2007;56:649–656. [PMC free article] [PubMed] [Google Scholar]
- Paun O, Forest F, Fay MF, Chase MW. Hybrid speciation in angiosperms: parental divergence drives ploidy. New Phytol. 2009;182:507–518. doi: 10.1111/j.1469-8137.2009.02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O, Stuessy TF, Hörandl E. The role of hybridization, polyploidization and glaciation in the origin and evolution of the apomictic Ranunculus cassubicus complex. New Phytol. 2006;171:223–236. doi: 10.1111/j.1469-8137.2006.01738.x. [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Ann. Rev. Ecol. Syst. 1998;29:467–501. [Google Scholar]
- Soltis DE, Buggs RJA, Doyle JJ, Soltis PS. What we still don’t know about polyploidy. Taxon. 2010;59:1387–1403. [Google Scholar]
- Stebbins GL. Variation and Evolution in Plants. Columbia Univ. Press; New York: 1950. [Google Scholar]
- Stelkens R, Seehausen O. Genetic distance between species predicts novel trait expression in their hybrids. Evolution. 2009;63:884–897. doi: 10.1111/j.1558-5646.2008.00599.x. [DOI] [PubMed] [Google Scholar]
- Wendel J, Doyle J. Polyploidy and evolution in plants. In: Henry RJ, editor. Plant diversity and evolution. CABI; Wallingford: 2005. pp. 97–117. [Google Scholar]