Abstract
Let-7 is an important tumor-suppressive microRNA that acts as an on-off switch for cellular differentiation and regulates the expression of a set of human oncogenes. Binding of the human KSRP protein to Let-7 miRNA precursors positively regulates their processing to mature Let-7, thereby contributing to control cell proliferation, apoptosis and differentiation. Here we analyze the molecular basis for KSRP-pre-Let-7 selectivity and show how the third KH domain of the protein recognizes a G-rich sequence in the pre-let-7 terminal loop and dominates the interaction. The structure of the KH3-RNA complex explains the protein recognition of this non-canonical KH target sequence and we demonstrate that the specificity of this binding is crucial for the functional interaction between the protein and the miRNA precursor.
Keywords: KSRP, Let-7, miRNA biogenesis, KH domain, protein-RNA interactions
Introduction
miRNAs regulate the expression of more than half of all human genes by acting on the stability and translation of mRNA targets, and the cellular concentration of miRNAs is tightly controlled both at the transcriptional and post-transcriptional level (1; 2). The ~22 nucleotide-long miRNA is transcribed as part of a much larger RNA molecule, where the mature miRNA sequence base pairs with a quasi-complementary sequence to form a hairpin-like structure. The two-step processing of the initial transcript to mature miRNA can be selectively regulated by the interaction between effector proteins and the terminal loops of pre- and pri-miRNA hairpins (reviewed by Kim and co-workers, (1). The molecular information available on the processing complexes and the high sequence conservation observed for some but not all miRNA terminal loops support the proposed role of terminal loops as a platform for the binding of effector proteins and the regulation of miRNA biogenesis (3).
Originally discovered in C. elegans, where it has an important role in development, Let-7 was the first miRNA to be identified in humans. Let-7 provides an on-off switch for cell differentiation and has been shown to inhibit the expression of a set of important oncogenes in human. Here, Let-7 functions as a major tumor suppressor and its cellular concentration must be tightly regulated (4). At the post-transcriptional level, this regulation relies on proteins that bind to the terminal loop of the Let-7 precursors and either increase (KSRP) or decrease (Lin28 and hnRNPA1) the efficiency of Let-7 biogenesis (5; 6; 7; 8; 9). The key to establishing the selectivity of these post-transcriptional regulatory mechanisms is the recognition of the RNA terminal loop by different protein effectors.
Two recent studies have clarified the molecular basis of the recognition of the Let-7 terminal loop by the negative regulator Lin28, which interacts with the RNA using a CCHC double zinc finger domain and a Cold Shock RNA binding domain (10; 11). However, the recognition between Let-7 and the activator KSRP, which has a very different domain composition, is still to be elucidated (8; 9). KSRP is a multi-functional protein that interacts with its nucleic acid targets using four sequential KH domains (Figure 1) and has been shown to play a role in the decay, splicing and localisation of selected mRNAs (12). In miRNA biogenesis, KSRP interacts with a AGGGU sequence at the 5′ end of the Let-7 terminal loop (Figure 1c) and stimulates pri- and pre-Let-7 processing (9). How KSRP recognises this G-rich sequence is unclear, as the current structural understanding of KH-RNA recognition advocates a strict requirement for A or C nucleobases in the central positions of the target RNA sequences (13). Nor it is clear what role the different KH domains of KSRP play in the interaction.
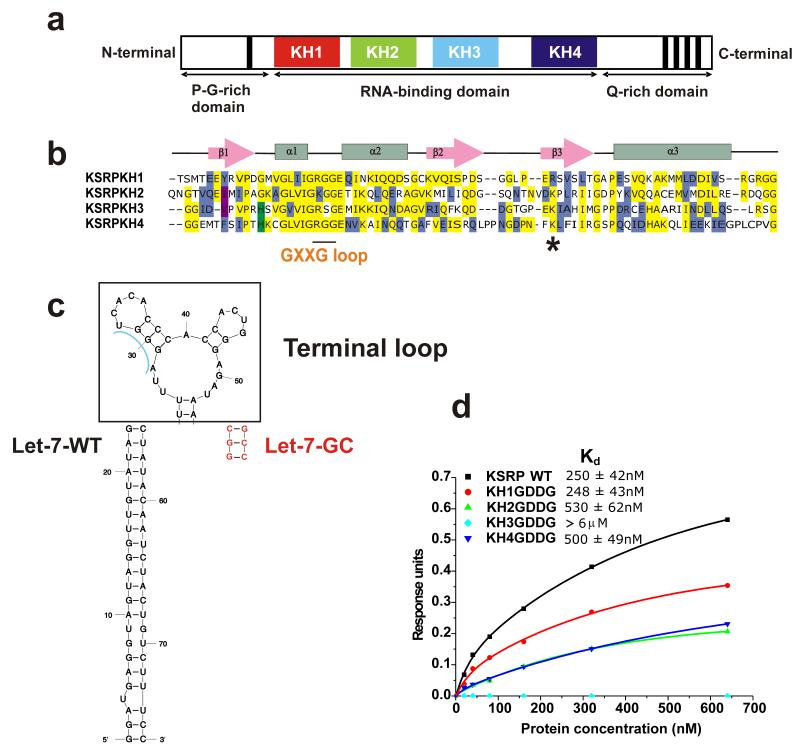
Figure 1.
KSRP, pre-Let-7a and their interaction (a) Domain organisation of KSRP. (b) Sequence alignment of the four KH domains of KSRP. Conserved amino acids are shadowed in yellow (identity) or other colours (similarity). The short continuous line below the sequence alignment highlights the position of the conserved GxxG loop, while the asterisk marks the position of the K368R mutant. (c) Comparison of sequence and secondary structure of wild type pre-let-7a miRNA (Let-7-SL) and a mutant with a shorter GC-rich stem (Let-7-GC) which binds with the same affinity as the wild type and has been used in BLI studies. The terminal loop is in bold, the wild type pre-Let-7a in black and the mutations used for the short stable Let-7-GC are in red. (d) The contribution of different KH domains to KSRP-Let-7 binding is measured using BioLayer Interferometry. A streptavidin-coated sensor is derivatized with biotinylated Let-7-GC and exposed to different concentrations of KSRP (wild type and mutants). Response Units (RU) are plotted against protein concentration and Kds are calculated for the different mutants. KH3 binding is too weak for data fitting and only a lower limit for the Kd could be estimated.
Here, we set out to explain the molecular bases of KSRP recognition of the Let-7 miRNA precursor. We clarify that the different KH domains of KSRP play very different roles in the interaction. We then analyse the interaction between the key KH3 domain and its pre-miRNA target sequence and explain how KSRP can use a conserved KH scaffold to recognise a non-canonical G-rich sequence. Our data highlight that even moderate changes in this recognition have very important effects on the ability of KSRP to promote Let-7 biogenesis.
Results
KSRP KH3 dominates the KSRP–pre-Let-7 RNA interaction
To clarify the role of the four KH domains of KSRP in pre-Let-7a recognition we have used biophysical binding assays and a set of KSRP point mutants. We had previously proposed that KSRP recognizes its various RNA targets by combinatorial and diverse use of its KH domains (14). More recently, we have established that by systematically mutating the conserved GxxG loop in each of the KH domains of KSRP to GDDG we can eliminate RNA binding by a single domain in the context of the full-length protein without altering the structural framework of recognition (15). This allows an accurate evaluation of the importance of the different domains in the interaction. Here, we test the binding of the four GDDG KSRP mutants to the Let-7 terminal loop using BioLayer Interferometry (BLI). To reduce non-specific RNA absorption to the surface of the sensor used in the BLI measurements we have substituted the wild type pre-Let-7a stem with a shorter stable GC-rich stem (Let-7-GC, Figure 1c), after having confirmed by CD that this change does not affect KSRP binding to the terminal loop (Supplementary Figure 1). BLI data show that wild-type KSRP and the KH1 mutant have equivalent affinities for the Let-7-GC (Kd ~250 nM), suggesting that KH1 does not contribute to binding, while the KH2 and KH4 bind with an affinity equal to half of the one of the wild-type protein (Figure 1d, Supplementary Figure 1). In marked contrast we did not detect any interaction between the KH3 mutant and the RNA even at the highest tested concentration (600 nM), indicating a Kd >6000 nM. Hence, our biophysical binding assays show that KH3 dominates the KSRP–pre-Let-7 interaction in the context of the full-length protein, although both KH2 and KH4 provide a small contribution to protein binding affinity. The modest contribution of the KH2 and KH4 domains to binding is consistent with the difference in affinity we measure between the KSRP–Let-7-GC and the KH3-AGGGU interactions. Further, KH3 binds with the same affinity to AGGGU and Let-7-GC RNAs and binding to the two RNAs result in analogous chemical shift changes in the KH3 15N-1H NMR spectrum (Supplementary Figure 2), indicating that no appreciable interaction is taking place between KH3 and nucleotides outside the AGGGU sequence. Finally, the comparative analysis of methyl resonances in the 13C-1H spectrum of KSRP, alone and in complex with the AGGGU RNA, shows that only resonances of KH3 are affected by AGGGU binding and confirms that KH2 and KH4 do not make appreciable interactions with the short KH3 target sequence (Supplementary Figure 2).
KH3 recognition of the target pre-Let-7 sequence
The different contributions of the four KH domain of KSRP to the interaction with pre-Let-7 RNA clarify that, in order to identify the key determinants of KSRP–Let-7 recognition and to understand regulation of Let-7 biogenesis we need to dissect the KH3-RNA interaction. KH domains can interact with five or more nucleotides but only four nucleobases make contact with the hydrophobic groove of the protein (13). Of these, the two central bases (positions 2 and 3) are recognised with higher selectivity and are either an A or a C in all the published structures (13; 16; 17). However, we have previously shown that the isolated KSRP KH3 domain prefers Gs in these central positions, which is consistent with the identified AGGGU target sequence on pre-Let-7a (9; 14), and we have now confirmed that the contacts KH3 makes with the AGGGU RNA recapitulate the interaction with the larger pre-Let-7 terminal loop. To explain how KSRP KH3 can achieve the recognition of non-canonical G nucleobases within a canonical KH structural framework we have determined the structure of the KH3-AGGGU complex (Table 1, Figure 2, Supplementary Figure 3), that we describe below with a focus on the two central and more specific positions.
Table 1. NMR and refinement statistic for the KSRPKH3-AGGGU RNAcomplex.
| Protein | RNA | |
|---|---|---|
| NMR distance and dihedral constraints | ||
| Distance constraints | ||
| Total NOE | 2366 | 47 |
| Unambiguous | 2296 | |
| Ambiguous | 70 | |
| Intra residues | 895 | 33 |
| Inter-residue | 1471 | 14 |
| Sequential (|i-j|=1) | 486 | 14 |
| Nonsequential (|i-j|>1) | 985 | |
| Hydrogen-bonds protein | 23 | |
| Protein-RNA intermolecular | 67 | |
| Dihedral angles RNA | ||
| Sugar pucker | 5 | |
| Backbone | 10 | |
| Structure statistics | ||
| Violations | ||
| Number of distance constraints > 0.3 Å | 1 | |
| Max. distance constraint violation (Å) | 0.367 | |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.002±0.000 | |
| Bond angles (°) | 0.363±0.003 | |
| Improper (°) | 0.249±0.006 | |
| Average pairwise r.m.s. deviation (Å) | ||
| Protein | ||
| Backbone (residues 11-46, 55-82) |
0.48±0.10 | |
| Heavy (residues 11-46, 55-82) |
1.26±0.18 | |
| RNA | ||
| Heavy | 0.74±0.17 | |
| Complex | ||
| Heavy (residues 11-46, 55-82, RNA) |
1.21±0.17 |
Statistics from the final 20 water-minimised structures
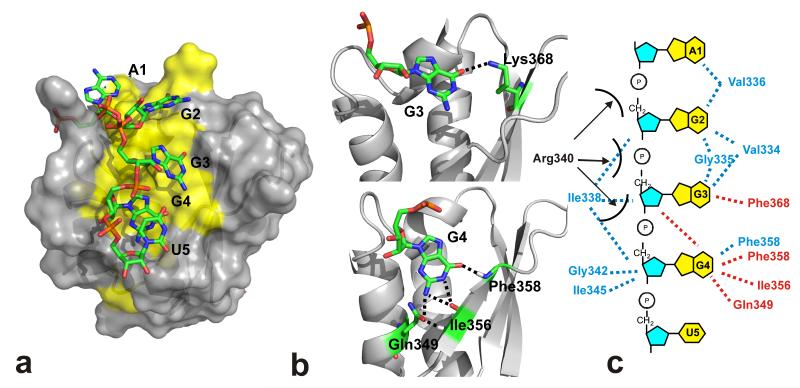
Figure 2.
Structure of the KH3-RNA complex. (a) The protein surface of the lowest energy conformer is in grey (hydrophobic amino acids in yellow) while the RNA heavy atom representation is in colour code. The RNA nucleotides are labelled. (b) Two enlargements of the structure highlight the hydrogen bonds we observe between the RNA G3 in position 2 (top) and G4 in position 3 (bottom) and the protein. (c) Protein-RNA contacts observed in the KH3–AGGGU structure. Hydrogen bonds are in red, hydrophobic interactions in blue. The position of the solvent-exposed side chain of the GxxG arginine residue (Arg340) is not well-defined and an interaction with an RNA phosphate group is possible.
Our NMR structure shows that AGGGU RNA (referred to below as nucleobases A1, G2, G3, G4 and U5, in positions 0, 1, 2, 3 and 4 of the specifically recognised sequence) interacts with the nucleic acid recognition groove of KSRP KH3. While the A1 nucleobase makes limited contacts with the protein (Figure 2 and Supplementary Figure 3) and its position is not well defined in our solution structure, the position and the relative arrangement of the four RNA nucleobases in the hydrophobic groove (G2, G3, G4 and U5) is similar to the one reported for the equivalent nucleobases of other KH-RNA complexes (Figure 3) (16; 17). However, the hydrophobic groove of KH3 is longer than the grooves of the Nova-1 and hnRNPK KH3 domains and broadens in a wide pocket that allows the protein to accommodate a G (G2 and G3) in positions 1 and 2 (Figures 2 and 3 and Supplementary Figure 3). G2 rests on a broad hydrophobic platform comprising the Val334, Gly335 and Val336 amino acids (Figure 2, Supplementary Figure 3) and its Watson-Crick edge does not engage in H-bonding in our structure, which is consistent with the lack specificity we have previously observed by Scaffold Independent Analysis (SIA) (14). Several of the hydrophobic amino acids contacting G2 also make contact with G3, but G3-protein contacts include a hydrogen bond between the Watson-Crick edge of the base and a protein side chain (Figure 2). KH3 shows a limited specificity in position 2 and can tolerate mutation of G3 to A and U but not C (Supplementary Figure 4 and Supplementary Table 1) most likely because of a steric clash between the C amino group and a methyl group of Val334. In contrast to G3, G4 forms four intermolecular hydrogen bonds, three of which are with the protein backbone. This creates a recognition pattern which is very specific but also different from the canonical pattern observed in the known KH-RNA structures (Figures 2 and 3, Supplementary Figure 4). Indeed, rather than binding the backbone amide and carboxy group of the same amino acid this RNA nucleobase recognises the carboxy and amide groups of two separate amino acids, Ile356 and Phe358 (Figures 2 and 3). NMR binding assays confirm our structural observations, indicating that G4 recognition is highly specific and any mutation of the nucleobase in this position causes a drop in affinity of between one and two orders of magnitude (Supplementary Figure 4, Supplementary Table 1). Finally, the U5 nucleobase stack onto G4 (Figure 2, Supplementary Figure 3), a common arrangement in KH-NA complexes, and it is possible that further contacts take place with the surrounding solvent exposed protein side chains. Globally, the KH3-AGGGU structure explains how the canonical KH-RNA recognition can be adapted to select a G-rich sequence.
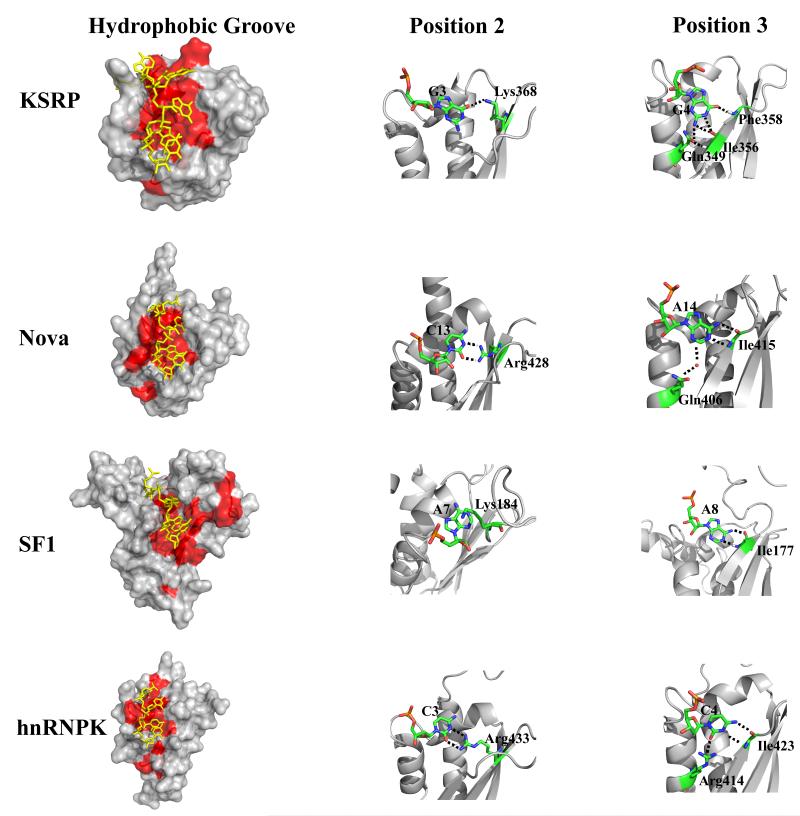
Figure 3.
Comparison of the KSRP KH3-AGGGU complex with three representative KH-nucleic acid (NA) complexes (i.e. the Nova KH3, SF1 and hnRNPK KH3-NA complexes). Left – Surface representations of the three proteins (grey) with hydrophobic residues (L, I, V, A, F) in red. The nucleic acids are in yellow. KSRP has a broader hydrophobic groove that accommodates a G in positions 1 and 2 (G2 and G3 of the AGGGU). Middle – The specific recognition of a C in position 2 by Nova1 KH3 and hnRNPK KH3 is mediated by a set of H-bonds to the Watson-Crick edge of the base. Here we display the two H-bonds formed between the guanidinium group of an arginine in the protein β3 strand and the O2 and N3 groups of the base. The equivalent amino acid in KSRP KH3 is a lysine (Lys368) and only one H-bond is observed, with the O3 of the G. The different degree of specificity of the interactions is consistent with our published SIA data as well as with the binding data reported in Supplementary Table 1, that indicate that the protein tolerates a U, an A and a G in position 2. Right – In all the complexes the nucleobase in position 3 is recognised specifically by a network of intermolecular H-bonds but in KSRP KH3 the pattern of H-bonds is different from that observed for other KH-NA complexes – where the amide and carboxy group of the same amino acid are interacting with the nucleobase.
KH3-AGGGU recognition and Let-7 processing
Next, we assessed the functional significance and contribution of KH3-AGGGU recognition to the regulation of Let-7 biogenesis. In order to evaluate how sensitive the system is to changes in the KH3–RNA interaction we tested the processing activity of two KSRP mutants, one with a fully compromised RNA binding capability of KH3 (the KH3 GDDG mutant) and the other where the KH3 affinity for the target RNA is reduced to a third. The second mutant was designed based on the novel structural information provided by the KH3-AGGGU complex. The structure shows that Lys368, which is solvent exposed in the free protein and whose mutation does not change KH3 structure and stability (Supplementary Figure 5), forms a hydrogen bond with G3 O6. In most other canonical KH-RNA complexes an Arginine residue is present in this position and is associated to the specific recognition of a C (Figure 3, Supplementary Figure 5). We mutated Lys368 to Arginine and, using NMR and ITC, compared the binding of wild type and the stable K368R mutant to the AGGGU RNA (and to the three nucleobase permutations of G3, Supplementary Table 1, Supplementary Figure 5). The Lysine-to-Arginine mutation results in a 3-to-4 decrease in affinity for the target, a result that we have validated also in the context of the pre-Let-7 terminal loop (Supplementary Figure 5).
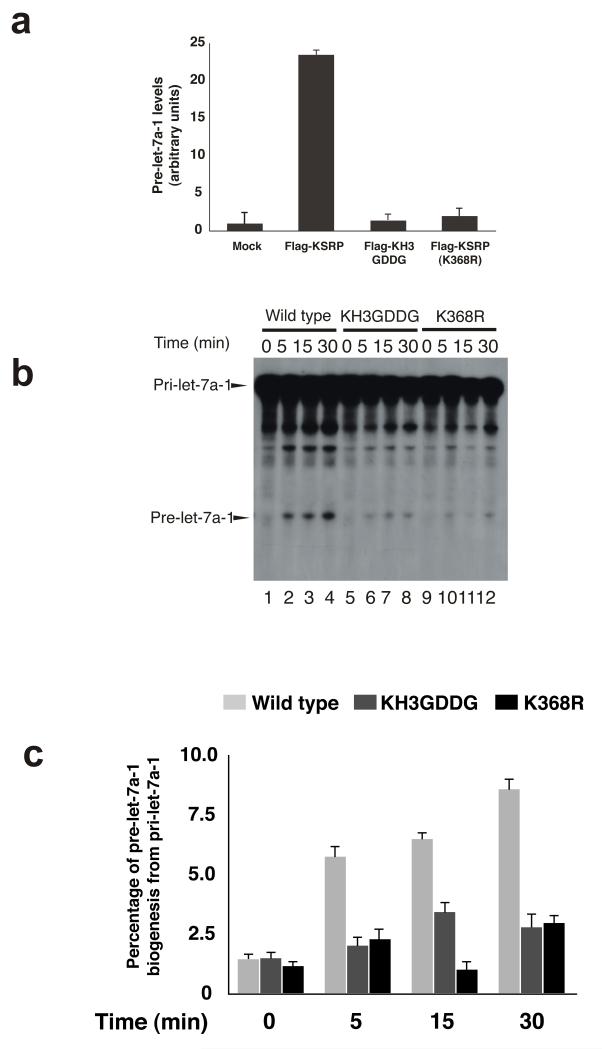
We then explored in cell extract whether KSRP-pre-Let-7 association is affected by the mutation. We used Ribonucleoprotein immunoprecipitation (RIP) assays to assess the association of KSRP, wild-type, KH3 GDDG and K368R mutatants, to pre-Let-7a (Figure 4a). The results indicate that K368R mutant association to pre-Let-7a-1 is strongly affected by the mutation. Finally we tested the efficiency of pri-Let-7a processing by wild-type KSRP and by the KH3 GDDG and KH3 K368R mutants in HEK-293 cells and found that while pri-Let-7a was efficiently processed by cell extracts from cells expressing wild-type KRSP, its processing was greatly reduced by both KH3 GDDG and KH3 K368R mutations (Figure 4b and c, Supplementary Figure 5). These results suggest that a relatively small decrease in affinity of the KH K368R mutant for the RNA has a strong effect on KSRP activity, and demonstrate that KH3 recognition of the G-rich target sequence is key to the role of KSRP in regulating Let-7 biogenesis.
Figure 4.
KH3 interaction with an AGGGU RNA in the Let-7 precursor hairpin domain is essential for KSRP function in Let-7 biogenesis. (a) Immunoprecipitation assay of pre-Let-7a-1 using extracts from HEK-293 cells transfected with either pTAG2B-KSRP, pTAG2B-KH3GDDG or pTAG2BKSRPK368R. The values shown are averages (± standard error mean) of three independent experiments. (b) In vitro pri-Let-7a-1 processing assays performed using total extracts from HEK-293 cells transiently transfected with either pTAG2B-KSRP (lanes 1-4), pTAG2B-KH3GDDG (lanes 5-8), pTAG2B-KSRPK368R (lanes 9-12). Internally 32P-labeled RNA substrate was added and used to monitor pri-Let-7a-1 processing. (c) The intensity of the bands corresponding to pri-let-7a-1 and pre-let-7a-1 were quantitated using ImageJ software (http://rsb.info.nih.gov/ij/index.html) and expressed as percentage of pre-let-7a-1 generated from pri-let-7a-1 at each time point. To avoid signal saturation, the quantitation has been performed on underexposed autoradiograms.
Discussion
The interaction of protein regulators with the terminal loop of the Let-7 miRNA precursors determines the concentration of the mature Let-7 miRNA, but our understanding of this interaction is still incomplete. Here we define the contribution of the individual KH domains of the protein KSRP, a positive regulator of Let-7 biogenesis, to its interaction with the Let-7 miRNA precursor. We show that, on the contrary to what we observe in KSRP-TNFα ARE recognition (15), one of the domains of the protein (KH3) plays a dominant role in the interaction. The different use of KH domains in KSRP recognition of the two TNFα ARE and pre-Let-7 RNAs emphasises the ability of the protein to target a broad range of RNAs and to act at different post-transcriptional regulatory steps. We determine the structure of the functionally well-characterised KH3-AGGGU complex, which encompasses the key recognition elements of the KSRP–pre-Let-7 interaction and we show that even a moderate impairment in the ability of KH3 to recognize a specific G-rich sequence has a strong effect on KSRP ability to promote pri-Let-7 processing. This assay both provides a functional validation of our molecular data and highlights the exquisite sensitivity of recognition in this multi-component system.
In this work, we dissect the structural contacts between KSRP KH3 and its G-rich target sequence. The structure of several KH domains in complex with nucleic acids have been solved, and the analysis of the KH-RNA (and KH-DNA) contacts advocates a requirement for A or C nucleobases in the central positions of the bound nucleic acid (13). However, the KH3-AGGGU structure shows that relatively minor changes in the geometry and the hydrophobicity of the nucleic acid recognition groove of a KH domain facilitates the binding and recognition of G nucleobases. Particularly striking is how the previously described use of a Watson-Crick-like recognition of the base in position 3 is here maintained by shifting the contacts made by the RNA base along the protein β-strand. This finding provides a more general understanding of KH-RNA recognition and facilitates the design of KH domains with different sequence specificities.
The regulation of Let-7 biogenesis involves several proteins, including hnRNPA1, Lin28 and KSRP, and is a prototype for the post-transcriptional regulatory mechanisms that selectively control miRNA concentration. We have previously shown that high levels of Lin28 can counteract KSRP-mediated up-regulation of Let-7 biogenesis in P19 cells (9). However in the recently reported structure of the Lin28-Let7 complex (11), the AGGGU sequence target of KH3 does not directly interact with Lin28. We expect that future work will define the molecular interplay between these factors at the molecular level, providing further insight into how Let-7 biogenesis is regulated.
ONLINE METHODS
RNA and Protein samples
The different KSRP constructs were cloned into a modified pETM-30 vector (EMBL Heidelberg, protein expression facility) and expressed as His-GST fusion proteins. Unlabeled and uniformly 15N, 13C-labeled samples of KSRP KH3 domain were obtained as previously described (18). Briefly, a His-GST fusion protein comprising amino acids 423 to 525 of human KSRP protein (NM_003685) was expressed in E. coli and purified by Ni-IMAC. The tag was removed by overnight TEV protease digestion at 4°C followed by a second Ni-IMAC step and by sixe exclusion chromatography (Superdex 75 16/60 column, Pharmacia). The KH1-KH4 KSRP construct (amino acids G68-Q525) which is use in our biophysical assays and is referred in the paper as ‘KSRP’ was expressed and purified as above, except that a MonoQ 5/50GL anion exchange column (GE Healthcare) followed by a 1 ml HiTrap Heparin column (GE Healthcare) was used. The final proteins were stored in 10 mM Tris-HCl pH 7.4, 50 mM NaCl (or LiCl), 2 mM Tris(2-carboxyethyl)phosphine (TCEP) 0.05% (w/v) NaN3. The concentration of the protein samples was determined from their 280 nm absorbance and their molecular weight and purity were confirmed by electrospray mass spectrometry. KSRP K368R and GxxG-to-GDDG constructs were prepared using the Quikchange Site-Directed Mutagenesis Kit (Stratagene) according to manufacturer’s instructions as described in Hollingworth et al. (15) and were expressed and purified as the wild type protein. Let-7-SL RNA was synthesized by run off transcription and purified using denaturing acrylamide gels, as previously described (19). All other non-biotinylated or biotinylated oligonucleotides were purchased from Dharmacon and IDT (Coralvill, IA).
NMR spectroscopy
NMR experiments were recorded at 25°C, 15°C and 5°C on Bruker Avance and Varian Inova spectrometers operating at 700, 600 and 800 MHz 1H frequency. Protein and RNA samples were in 10mM Tris at pH 7.4, 50mM NaCl (or LiCl) buffer. Backbone and side-chain resonance assignments were obtained using 2D 1H-15N HSQC, 2D 1H-13C HSQC, 3D HNCA, 3D CBCACONH, 3D HCCH-TOCSY, 3D 15N NOESY-HSQC experiments, as well as, 3D 13C NOESY-HSQC experiments optimized for either aliphatic or aromatic resonances. Assignment of the resonances of the free and KH3-bound AGGGU RNA were obtained using 2D 1H-1H TOCSY and 2D 1H-1H NOESY experiments. NOESY experiments were recorded with mixing times between 100ms and 250ms, TOCSY experiments with mixing times of 30, 50, 60, 70 and 100 ms. Intramolecular restraints were derived from 15N- and 13C-NOESYHSQC spectra with a mixing time of 100 ms at 25 °C. Intermolecular NOEs were obtained from the analysis of decoupled and non-decoupled 2D 1H-1H NOESY (mixing times: 50, 100, 200, and 250 ms), 3D 15N NOESY-HSQC (mixing time 100 ms), 3D 13C NOESY-HSQC (mixing times 100 and 120 ms) and 3D-filtered 13C NOESY on samples of unlabeled RNA and 15N-13C labeled protein (mixing times 100 and 150 ms) at 25 °C. The temperature dependence of backbone amide chemical shift was calculated as the ratio between chemical shift and temperatures in 15N-1H correlated spectra recorded at 10, 20, 24, 30, 35, 40 and 45 °C. NMR spectra were processed using the NMRpipe suite of programs (20) and analysed using the Sparky (21) and XEASY (22) programs.
Structure calculations
The structure of the KH3-AGGGU complex was calculated using a semi-automated ARIA 1.2-based protocol (23). Experimental distance restraints were obtained from the integration of NOE peaks in 3D and 2D NOESY spectra using the XEASY program (22). All intra-protein NOE cross-peaks were calibrated automatically and assigned iteratively within ARIA, while the peaks arising from RNA proton resonances were calibrated manually in a semi-quantitative fashion (24). Protein angle restraints were derived from chemical shifts analysis using the program TALOS. α, ζ and δ RNA angle restraints were derived from 31P-1H correlation spectra and 1H-1H TOCSY spectra as described by Varani and coworkers (24). Structures were initially calculated without the use of H-bond restraints; only if a proton was hydrogen bonded in at least 50% of this initial set of structures the corresponding H-bond restraint was added in the final set of calculations. Inter-molecular H-bond constraints were validated by a downfield shift of the proton resonance upon RNA binding (25) and a low temperature dependence of the chemical shifts (26, 27). ARIA 1.2 was used to calculate 100 conformers of the complex (iterations 0-7). The 40 conformers with the lowest restraint energies were refined in a shell of explicit water. The 20 conformers with the lowest restraint energies, restraint violations and rms deviations from the ideal covalent geometry were taken as representative of the converged structures and selected for structural analysis. The structures were analysed using the programs Molmol (28), InsightII (Accelerys) and PyMOL (http://www.pymol.org). A Ramachandran analysis of the structure shows 83.8%, 14.9%, 0.8% and 0.8% of the protein residues in the most favoured, additional, generously and disallowed regions, respectively.
ITC binding assays
ITC experiments were recorded in 10mM Tris, pH 7.4, 50mM NaCl (or LiCl for the AGGGU RNA) by titration of a concentrated RNA solution into a cell containing KSRP KH3. For the AGGGU RNA, which can form G-tetrads at high concentrations, a concentrated solution of protein was added to the RNA. Protein concentration was optimized according to the expected binding affinities of the different RNAs, as derived from NMR experiments. Dissociation constants were determined by fitting the measured heat of reaction at 25°C using the Origin analysis package and a 1:1 binding model. The heat of dilution of the different RNAs was evaluated in control experiments where RNAs solutions were injected into buffer.
NMR binding assays
1H-15N correlation experiments (typically sofast 15N HMQC spectra) were recorded at the different steps of a titration of 15N-labeled samples of KH3 with the different RNA oligonucleotides. The titrations were performed in 10mM Tris at pH 7.4, 50mM NaCl, 2mM TCEP at protein concentration between 50 and 100 μM. To obtain Kd values the chemical shift changes (Δδave=[(ΔNH)2+(Δ15N/10)2]1/2) of 5-10 peaks in the fast regime of exchange were measured and plotted against increasing RNA:protein ratios. A non-linear two-parameter least-square fit procedure was applied to each dataset using the program Origin and a single site binding model (29). Kd values are reported as mean ± 2 standard deviations.
CD binding assays
CD spectra were recorded on a Jasco J-715 spectropolarimeter equipped with a Peltier system for temperature control. The titrations were performed in 10mM Tris at pH 7.4, 150mM NaCl, 2mM TCEP by adding increasing amount of protein to RNA samples. RNA concentration varied between 1.5μM and 3μM. Experiments were recorded at 5°C to maximize signal-to-noise and the CD signal at 260nm was fitted to the protein:RNA ratio using non linear regression and in house software as described in (30). Thermal denaturation of the KH3 and KH K368R mutant was monitored using the ellipticity at 220nm as a function of temperature between 5 and 90°C at a rate of 1°C per min. The data were fitted to a two state-model and in-house software as reported in (18).
BLI binding assays
BLI experiment were recorded in 10mM Tris at pH 7.4, 150mM NaCl, 2mM TCEP, 0.5mg/ml BSA, 0.005% TWEEN 20 at 25°C on a Octet Red instrument (ForteBio, Inc). Briefly, the biotinylated RNAs were immobilized on Streptavidin coated sensors and exposed to different concentrations of the wt and mutant four-domain proteins. Dissociation constants were derived by fitting the increase in BLI signal as a function of protein concentration (Figure 1 and Supplementary Figure 1) using non-linear regression and in-house software as described in (31).
Pri-miRNA in vitro processing assays
Pri-Let-7a-1 processing assays were performed essentially as reported in (32). Briefly, total cell extracts from HEK-293 cells were prepared in 50mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% Triton X-100, 1X Complete (Roche, Mannheim, Germany), 10% glycerol and incubated (typically 40 μg per 25 μl reaction at 37° C for the indicated times) with in-vitro synthesized and uniformly labeled pri-Let-7a-1 (5 fmol) in processing buffer containing 100 mM potassium acetate, 2 mM magnesium acetate, 10mM Tris-Cl (pH 7.6), 2 mM DTT, 10 mM creatine phosphate, 1 μg creatine phosphokinase, 1 mM ATP, 0.4 mM GTP, 0.1 mM spermine, 2 units Stop RNase Inhibitor (5 PRIME, Hamburg, Germany). For some experiments, 4 μg of HEK-293 total cell extracts were preincubated (1h at 16 C under continuous agitation in a Thermomixer [Eppendorf, Hamburg, Germany]) with immunocomplexes derived from anti-Flag immunoprecipitation of 250 μg aliquots of total extracts of HEK-293 cells transiently transfected (Lipofectamine Plus, Invitrogen, Carlsbad, CA) with either empty pTAG2B vector (Stratagene, Santa Clara, CA), or pTAG2B-KSRP, pTAG2B-KH3GDDG, pTAG2B-KSRPK368R. Immediately after preincubations pri-Let-7a-1 processing assays were performed.
Ribonucleoprotein complexes immunoprecipitation (RIP) assays
RIP assays were performed as previously described in (32) with the following modifications. Briefly, cell lysates were immunoprecipitated with magnetic beads-Protein G-coupled anti-Flag (M2, Sigma, St. Louis, MO, USA) mouse monoclonal antibody at 4° C overnight. Pellets were washed four times with 50mM Tris-HCl [pH 8.0], 150mM NaCl, 0.5% Triton X-100. Total RNA was prepared from immunocomplexes using miRNeasy mini kit (Qiagen, Milano, Italy), retro-transcribed and amplified by qPCR utilizing miScript Precursor Assays for pre-Let-7a-1 (Qiagen, Milano, Italy).
Supplementary Material
Acknowledgements
All the NMR experiments were recorded at the MRC Biomedical NMR Centre at the MRC NIMR and we thank T. Frenkiel and A. Oregioni for assistance. We thank S. Howell for mass spectrometry analysis of protein samples. The work of A.R. has been funded by the MRC grant U117574558 and the Wellcome Trust Grant WT082088MA. The work of P.B. and R.G. has been supported by grants from AICR (# 10-0527) and AIRC (I.G. # 10090). We thank B. Faust for help in the expression and purification of the KH3 K368R mutant.
Footnotes
Accession codes
Atomic coordinates for the NMR ensemble have been deposited in the Protein Data Bank with accession code 4B8T and the corresponding chemical shift assignments have been deposited in the Biological Magnetic Resonance Data Bank with accession code 18702
Author Contributions
Cloning and RNA synthesis were performed by D.H. Protein purification was performed by G.N. and D.H. Recording and analysis of the NMR spectra were performed by G.N., M.G. and G.K. Structure calculations and recording of the ITC, CD and BLI data were performed by G.N. Analysis of the CD and ITC data was performed by S.M and G.N. RIP and pri-miRNA processing assays were performed by L.B and R.G. The manuscript was written by A.R. and G.N.
References
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Michlewski G, Guil S, Semple CA, Cáceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:262–263. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Büssing I, Slack FJ, Grosshans H. Let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michlewski G, Cáceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loughlin FE, et al. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat. Struct. Mol. Biol. 2011;19:84–89. doi: 10.1038/nsmb.2202. [DOI] [PubMed] [Google Scholar]
- 11.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gherzi R, Chen CY, Trabucchi M, Ramos A, Briata P. The role of KSRP in mRNA decay and microRNA precursor maturation. Wiley Interdiscip. Rev. RNA. 2010;1:230–239. doi: 10.1002/wrna.2. [DOI] [PubMed] [Google Scholar]
- 13.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 14.García-Mayoral MF, Díaz-Moreno I, Hollingworth D, Ramos A. The sequence selectivity of KSRP explains its flexibility in the recognition of the RNA targets. Nucleic Acids Res. 2008;36:5290–5296. doi: 10.1093/nar/gkn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollingworth D, et al. KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks368. doi:10.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis HA, et al. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 17.Backe PH, Messias AC, Ravelli RB, Sattler M, Cusack S. X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure. 2005;13:1055–1067. doi: 10.1016/j.str.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 18.García-Mayoral MF, et al. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure. 2007;15:485–98. doi: 10.1016/j.str.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Nikonowicz EP, et al. Preparation of 13C and 15N labelled RNAs for heteronuclear multidimensional NMR studies. Nucleic Acids Res. 1992;20:4507–1113. doi: 10.1093/nar/20.17.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 22.Bartels C, Xia T, Billeter M, Guntert P, Wuthrich K. The program XEASY for computer supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 23.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 24.Varani G, Fareed Aboul-ela., Allain Frederic-H-T. NMR investigation of RNA structure. Progress in Nuclear Magnetic Resonance Spectroscopy. 1996;29:51–127. [Google Scholar]
- 25.Liu Z, et al. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 26.Baxter NJ, Williamson MP. Temperature dependence of 1H chemical shifts in proteins. J. Biomol. NMR. 1997;9:359–69. doi: 10.1023/a:1018334207887. [DOI] [PubMed] [Google Scholar]
- 27.Cierpicki T, Zhukov I, Byrd RA, Otlewski J. Hydrogen bonds in human ubiquitin reflected in temperature coefficients of amide protons. J. Magn. Reson. 2002;157:178–80. doi: 10.1006/jmre.2002.2597. [DOI] [PubMed] [Google Scholar]
- 28.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 29.García-Mayoral MF, Díaz-Moreno I, Hollingworth D, Ramos A. The sequence selectivity of KSRP explains its flexibility in the recognition of the RNA targets. Nucleic Acids Res. 2008;36:5290–6. doi: 10.1093/nar/gkn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin SR, Masino L, Bayley PM. Enhancement by Mg2+ of domain specificity in Ca2+-dependent interactions of calmodulin with target sequences. Protein Sci. 2000;9:2477–88. doi: 10.1110/ps.9.12.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cukier CD, et al. Molecular basis of FIR-mediated c-myc transcriptional control. Nat. Struct. Mol. Biol. 2010;17:1058–64. doi: 10.1038/nsmb.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Briata P, et al. PI3K/AKT signaling determines a switch between distinct KSRP functions favoring skeletal miogenesis. Cell Death Diff. 2012;19:478–87. doi: 10.1038/cdd.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.