Abstract
SNAREs provide energy and specificity to membrane fusion events. Fusogenic trans-SNARE complexes are assembled from Q-SNAREs embedded in one membrane and an R–SNARE embedded in the other. Regulation of membrane fusion events is crucial for intracellular trafficking. We identify the endosomal protein Varp as an R-SNARE-binding regulator of SNARE complex formation. Varp co-localises with and binds to VAMP7, an R-SNARE involved in both endocytic and secretory pathways. We present the structure of the second ankyrin repeat domain of mammalian Varp in complex with the cytosolic portion of VAMP7. The VAMP7 SNARE motif is trapped between Varp and the VAMP7 longin domain and hence Varp kinetically inhibits VAMP7’s ability to form SNARE complexes. This inhibition will be increased when Varp can also bind to other proteins present on the same membrane as the VAMP7 such as Rab32:GTP.
SNAREs are at the core of the vesicle:organelle and organelle:organelle membrane fusion machinery as a consequence of their ability to provide much of the mechanical energy and specificity to these events. They are small membrane proteins, which contain at least one 60 residue long SNARE motif. Four SNARE motifs come together to form a four helical bundle, the trans SNARE complex. One SNARE (R– or v–) is on one of the membranes to be fused and the three others (Q– or t– SNAREs) are on the other. The ‘zippering up’ of the SNARE motifs ‘pulls’ the two membranes into sufficiently close apposition to allow fusion to proceed (reviewed in [1, 2]). Only certain combinations of the 38 mammalian SNAREs are able to form fusogenic trans–SNARE complexes, and this plays a major role in conferring specificity to the fusion process [3, 4]. Both the fusogenic activity of SNAREs and their localisation to specific intracellular membranes must be carefully controlled if the vesicle transport system is to function correctly. Their subcellular distribution is determined by incorporation into or exclusion from the various types of route–specific transport vesicles and tubules within the cell.
In most SNAREs, the SNARE motif is preceded by either a short unstructured peptide of 10–30 residues or a folded domain of 100 to 150 residues [1]. The syntaxins and Vti1 (VPS10 Tail Interactor 1) SNAREs possess three–helical Habc domains, which in some cases bind back onto the SNARE’s own SNARE motif and so can inhibit their ability to participate in SNARE complex formation (reviewed in [5]). In the Qa SNAREs, regions of the Habc domain interact with proteins of the Munc18 family to regulate Qa SNARE incorporation into SNARE complexes with both inhibitory and activating roles having been proposed ([6-9]).
In mammalian cells, the major post–Golgi R–SNAREs are the VAMPs, of which only VAMP7 has a folded N–terminal region: a 120 residue mixed α/β longin domain, also present in the R–SNAREs Sec22b and Ykt6 [10]. VAMP7 is highly conserved across eukaryotes, is ubiquitously expressed and has been reported to be involved in a variety of membrane trafficking events. It is important for heterotypic fusion events between terminal endocytic compartments (i.e. late endosomes and lysosomes) by complexing with syntaxins 7 and 8 and vti1b [11], and between these organelles and other cellular membranes including autophagosomes [12, 13] and the cell’s limiting membrane (by complexing with syntaxin3 or syntaxin4 and SNAP23 [14]). The latter allows VAMP7 to play a role in ‘wound healing’ [14, 15], metastasis via secretion of the endo–lysosomal membrane type 1–matrix metalloproteinase protein [16] and in the expansion of the plasma membrane during mitosis [17]. VAMP7 also plays an important role in secretion in specialised cells (reviewed in [15]), in neurite outgrowth [18], in dendrite formation in melanocytes [19], and has been implicated in more generalised secretion to the plasma membrane from the trans–Golgi network [15]. Due to its involvement in such a diverse set of important membrane fusion events, it is logical that both the organelle localisation and fusogenic activity of VAMP7 should be tightly controlled.
In order to identify binding partners for VAMP7 that could regulate its localisation and its ability to form fusogenic SNARE complexes, we [20], and others [21-23] employed yeast 2–hybrid screening (Y2H) for both the entire cytosolic region and the isolated N–terminal longin domain of VAMP7. Screening with the longin domain identified two vesicle coat components, the heterotetrameric AP–3 adaptor complex δ subunit, and Hrb, a clathrin adaptor and ArfGAP, both of which direct the trafficking of VAMP7 along the endocytic pathway [20, 22-24]. Here we demonstrate that the widely expressed, multi–domain protein Varp (Vps9 and ankyrin repeat containing protein), which is an endosomal Rab21 GEF (Guanine nucleotide Exchange Factor) [25] and an effector for Rab32 and Rab38 [26, 27], is a binding partner for the full cytosolic portion of VAMP7, but not for the isolated longin domain nor the SNARE motif alone. We have solved the structure of the complex between the VAMP7 cytosolic domain and an ankyrin repeat containing portion of Varp. In the structure, the SNARE motif of VAMP7 binds back onto its longin domain and is trapped there by Varp. This results in a 65 times slower rate of SNARE complex formation for full length VAMP7 in the presence of Varp when compared to complex formation for those R–SNAREs not possessing a Varp–binding longin domain i.e a kinetic inhibitory effect. Potential roles of this inhibition in vivo are discussed in the light of Varp’s other functions.
Results
Varp binds only to the whole cytosolic domain of VAMP7
Our Y2H screens of cDNA libraries, using in parallel the VAMP7 longin domain (VAMP7LD) and the full VAMP7 cytosolic domain (VAMP7CD), gave completely non–overlapping results. Screening with the longin domain resulted in 37 identifications of δ–adaptin and 1 of Hrb as binding partners. Our screen with the full–length cytosolic domain of VAMP7 showed no interaction with Hrb or δ–adaptin but did demonstrate SNAP29 (1 identification) and Varp (11 identifications as binding partners, Supplementray figure 1). These data are in broad agreement with published studies[21-23]. We cloned the ORF of the full length human Varp gene from the IMAGE clone IMAGE:6067580 and confirmed the interaction with VAMP7 by Y2H (Supplementary figure 1) and GST pull downs, using both endogenous Varp from MNT–1 cells (Supplementary figure 1) and Varp expressed as a C–terminally His10 tagged form in E. coli (Figure 1). However, in both Y2H and GST pull down experiments only the full–length cytosolic portion of VAMP7, but neither the isolated longin domain nor the isolated SNARE motif (Figure 1) interacts with Varp. This is consistent with our subsequent structural work but not with published data [21].
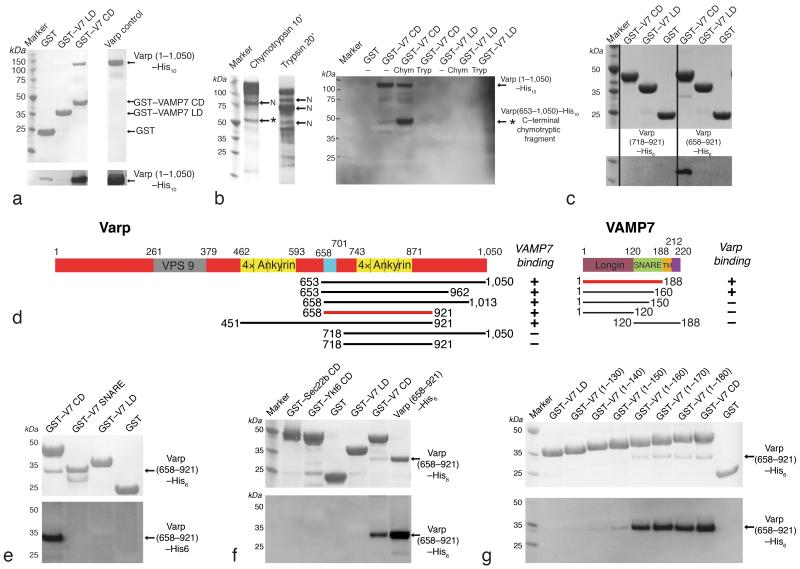
Figure 1. Mapping and confirming the binding site of Varp on VAMP7.
a,c,e,f,g Coomassie–stained (Top panel) and Western blot developed with HisProbe–HRP conjugate (bottom panel) of SDS PAGE of ‘GST pull down’ assays using the baits indicated.
a Full length heterologously expressed human Varp (lane ‘Varp control’) interacts with the GSTVAMP7 cytosolic domain (V7 CD) but not with GST nor the GSTVAMP7 longin domain (V7 LD)
b Coomassie–stained SDS–PAGE to show effects of digesting recombinant Varp His10 with chymotrypsin (10 minutes) and trypsin (20 minutes). Proteolytic products of Varp that have an N–terminus identical to that of the full–length protein are labelled with ‘N’. The C–terminal chymotryptic fragment of Varp (residues 653–1050) marked with * has an intact C–terminal His10–tag. These products were the input for pull–down assays using GST–VAMP7CD and GST–VAMP7LD as bait. Western blotting of the SDS–PAGE from these assays developed with HisProbe–HRP conjugate showed that the fragment * can bind to the VAMP7CD. Lanes are marked Chym or Tryp for chymotryptic or tryptic digestion products, “–” for undigested.
c His10-tagged residues 653–1050, 658–921 but not 718–921 of Varp can bind to the GST VAMP7CD while no Varp constructs bind to GSTVAMP7LD.
d Domain organisation of human Varp (left) and mouse VAMP7 (right) as indicated by pfam [30] and in [25]. The colour scheme used is replicated in all subsequent figures. A summary of the mapping of the interaction determined using the constructs indicated is shown underneath + indicates binding and − no binding.
e Varp(658–921)His6 is able to bind GSTVAMP7CD but not the GST–VAMP7 snare motif or GSTVAMP7LD.
f Varp(658–921)His6 cannot bind to the GST–tagged full cytosolic domains of the mammalian longin domain containing SNAREs Sec22b and Ykt6.
g Truncating GSTVAMP7 prior to residue 160 abolishes binding to Varp(658–921)His6.
Mapping the VAMP7 interacting domain of Varp
We then used domain analysis and structure prediction [28, 29] in combination with N–terminal sequencing of limited tryptic and chymotryptic proteolytic digests of full length Varp to identify stable domains of Varp (Figure 1). The smallest of the designed stable domains that still bound strongly to VAMP7 consisted of residues 658–921 of Varp, which includes the second predicted set of four adjacent ankyrin repeats [30] and part of the linker between these and the first predicted set of ankyrin repeats. This domain also contains the smallest portion of Varp (641–707) common to all VAMP7–interacting clones identified in [21] and the smallest region (671–731) found in a single interacting clone in our own Y2H screen (Supplementary figure 1). Like the full length protein, residues 658–921 of Varp bound only to full length VAMP7 and not to the isolated SNARE motif nor to the longin domain and were also unable to bind the full length cytosolic portions of the other longin domain containing SNAREs Ykt6 and Sec22 (Figure 1 and supplementary figures 1 and 2).
Residues 658–921 of Varp showed robust binding to the VAMP7 full–length cytosolic domain (residues 1 to 188). We measured the dissociation constant and the stochiometry of the interaction by isothermal titration calorimetry (ITC) respectively as 2.3 ±0.6 μM and 1:1 (the latter confirmed by gel filtration: see Figure S4). ITC gave a similar KD for full–length Varp binding to VAMP7 (KD~3μM Supplementary figure 2), indicating that no additional VAMP7 binding sites are present. In addition, residues 1–650 of Varp showed no binding to VAMP7 (Supplementary figure 2) and we did not identify any Varp clones in our Y2H screening that did not contain residues 671–731 (Supplementary figure 1). Using C–terminal truncations of VAMP7, we also demonstrated that only residues 1–160 (i.e. containing the longin domain and the N–terminal portion of the SNARE motif) were necessary for binding of VAMP7 to Varp (Figure 1).
Varp and VAMP7 colocalise in the endocytic pathway
In the absence of an antibody that detects endogenous Varp in cells, we expressed Varp with an HA–tag appended at its C–terminus (Varp–HA) in NRK cells and showed by immunofluorescence microscopy good, although not complete, colocalisation with endogenous VAMP7 and also with Rab7, a marker of late endosomes (Figure 2). Varp–HA also partially co–localised with lgp110 (also known as LAMP2), a marker of terminal endocytic compartments but localises poorly with the TGN marker TGN38 (Supplementary figure 3). Faint Varp–HA staining could also be seen at the cell periphery. ImmunoEM of NRK cells expressing Varp–HA showed the presence of Varp on organelles with the characteristics of endosomes, vesicular tubular elements, lysosomes and the plasma membrane (Figure 2 and Supplementary figure 3). This localisation pattern is also consistent with sites at which VAMP7 has been localised previously and suggested to function [23, 31]. Varp’s localisation is consistent with it being an effector of the two highly similar Rabs, Rab32 and Rab38 (67% sequence identity [26, 27]), which together with Rab7 and Rab7L1, Rab29 and Rab23 are classified as the late endocytic group III rabs [32]. Rab32 is expressed at low levels in a wide variety of cell types [33-35] including NRK cells used in this study (Supplementary figure 3) and has been detected on late endocytic pathway organelles and along with Rab38 on melanosomes [26, 36-38]. Thus the recently reported physiological roles of the interaction of Varp with VAMP7, including the Rab32 and Rab38 dependent Tyrp1 trafficking in melanocytes [19], are consistent with this localisation of Varp. Finally, we found that a 75% depletion of Varp by siRNA treatment in NRK cells showed no readily detectable alteration in the trafficking of VAMP7, but only a subtle increase in the steady state colocalisation of VAMP7 with a terminal endocytic compartment marker (Supplementary figure 3), which is consistent with an increase in the amount of fusion between late and terminal endocytic compartments
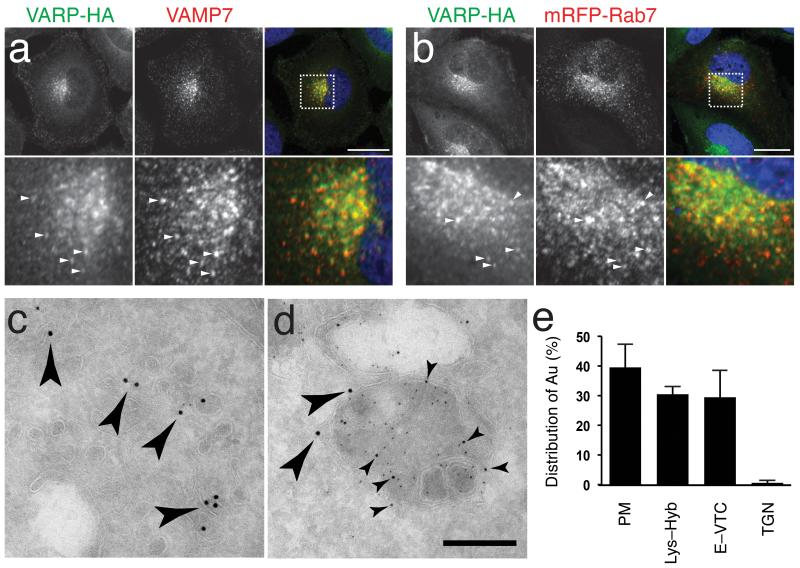
Figure 2. Varp–HA is localized to Vesicular–tubular elements of the endocytic pathway.
a,b Confocal immunofluoresence of NRK cells stably expressing Varp–HA stained for the HA tag and either endogenous VAMP7 a or with anti–mRFP following transfection with mRFP–Rab7 b. Pearson R correlation coefficient for Varp–HA and VAMP7 co–localisation, 0.67 and for Varp–HA and mRFP–Rab7, 0.51. Bottom panels of a and b are zoomed in images of the boxed regions, and arrowheads indicate representative examples of structures positive for both markers. Scale bars = 20μm
c,d Immunogold electron microscopy of NRK cells stably expressing Varp–HA. Dextran–Texas Red was endocytosed for 4h followed by a 20h chase to label lysosomes, and cells were then labeled with antibodies to Texas Red (5nm gold), lgp110 (10nm gold) or the HA tag (15nm gold). c Anti–HA immunoreactivity (large arrowheads show representative examples) associated with vesicular–tubular clusters (VTCs) and d dense core lysosomes co–immunolabelled with anti–Texas Red and anti–lgp110 (small arrowheads show representative examples) on ultrathin cryosections. Scale Bar = 200nm
e Quantitation of the labeling density of HA associated with membranous compartments: 62% of the immunogold labeling was associated with cytosol. The relative distribution of membrane associated gold label with the plasma membrane (PM), lysosomes, endolysosomes and hybrid organelles (Endo–Lys), endosomes and vesicular–tubular clusters (E–VTC) and the trans Golgi network (TGN) is shown. Bars represent the mean of 4 independent immunolabeling experiments ± SEM.
The structure of the Varp:VAMP7 complex
The robust interaction between the stable domain consisting of residues 658–921 of Varp and the VAMP7 cytosolic domain (KD ~2μM) allowed us to purify a complex of the two individually expressed proteins by gel filtration (Supplementary figure 4). We crystallised the complex between the two proteins and solved its structure by MIR using mercury derivatives (see Table 1). The structure of the VAMP7 longin domain was as expected from previous studies but Varp 658–921 actually forms a stack of six ankyrin repeats rather than the four predicted (Figure 3 and Supplementary figure 4) [30]. The first ankyrin repeat (coloured cyan in all figures), although separated from the next five repeats by a disordered region (residues 694–729) for which there is no electron density, contacts the second ankyrin repeat through an extensive and tightly packed hydrophobic interface (Figure 3). The VAMP7 binds ‘on top’ of the ankyrin repeat stack, with the binding being almost exclusively to the first repeat and represents a novel mode of ligand binding by ankyrin repeats (Figure 3 and Supplementary figure 5).
Table 1. Data collection and refinement statistics.
| Native | EMTS1 | EMTS2 | PCMBS1 | PCMBS2 | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | C2221 | C2221 | C2221 | C2221 | C2221 |
| Cell dimensions | |||||
| a, b, c (Å) α=β=γ=90° |
69.6, 122.7, 158.6 |
69.1, 123.2, 158.6 |
69.7, 123.5, 158.7 |
69.5, 123.1, 158.8 |
69.7, 123.0, 158.6 |
| Resolution (Å) | 2.00 (2.05– 2.00) * |
2.45 (2.55– 2.45) |
2.09 (2.15– 2.09) |
2.29 (2.37– 2.29) |
2.30 (2.38– 2.30) |
| Resolution (Å) along a, b, c a |
2.69, 2.51, 2.00 |
3.37, 2.83, 2.45 |
2.86, 2.95, 2.09 |
3.07, 2.81, 2.29 |
3.04, 2.63, 2.30 |
| R merge | 0.103 (0.96) | 0.131 (1.50) | 0.117 (1.37) | 0.105 (1.09) | 0.104 (1.57) |
| Rmeas b | 0.134 (1.50) | 0.181 (1.88) | 0.147 (1.79) | 0.129 (1.39) | 0.135 (2.02) |
| CC1/2 c | 0.994 (0.59) | 0.994 (0.50) | 0.995 (0.42) | 0.996 (0.65) | 0.996 (0.77) |
| I / σI | 6.9 (0.9) | 7.9 (1.1) | 5.9 (0.7) | 8.4 (1.1) | 7.6 (1.1) |
| Completeness (%) | 99.7 (99.5) | 96.6 (98.4) | 99.5 (97.9) | 99.1 (97.2) | 99.4 (99.7) |
| Multiplicity | 4.0 (3.4) | 4.1 (4.1) | 4.4 (3.8) | 4.8 (4.3) | 4.0 (4.1) |
| Wilson <B> (Å2) | 33 | 55 | 38 | 47 | 44 |
| Anomalous completeness (%) |
– | 90.2 (93.2) | 96.1 (84.3) | 95.6 (91.3) | 96.4 (98.4) |
| CC1/2 on Δanom (inner bin) |
– | 0.28 (0.76) | 0.04 (0.31) | 0.05 (0.29) | 0.30 (0.82) |
| Refinement | |||||
| Resolution (Å) | 2.00 | ||||
| No. reflections | 43692 | ||||
| Rwork / Rfree | 0.213 / 0.251 | ||||
| No. atoms | 2924 | ||||
| Protein | 2715 | ||||
| Water | 209 | ||||
| B–factors (Å2) | |||||
| VAMP7, Varp | 45, 53 | ||||
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.018 | ||||
| Bond angles (°) | 1.8 |
Values in parentheses are for highest–resolution shell.
Resolution estimates based on half–dataset correlation coefficients CC1/2 > 0.5, calculated in cones along a*, b*, c* of semi–angle 20°
Rmeas = Σ(n/n–1)1/2 (Ihl – <Ih>)/Σ<Ih>
CC1/2 is correlation coefficient on <I> between random half–datasets
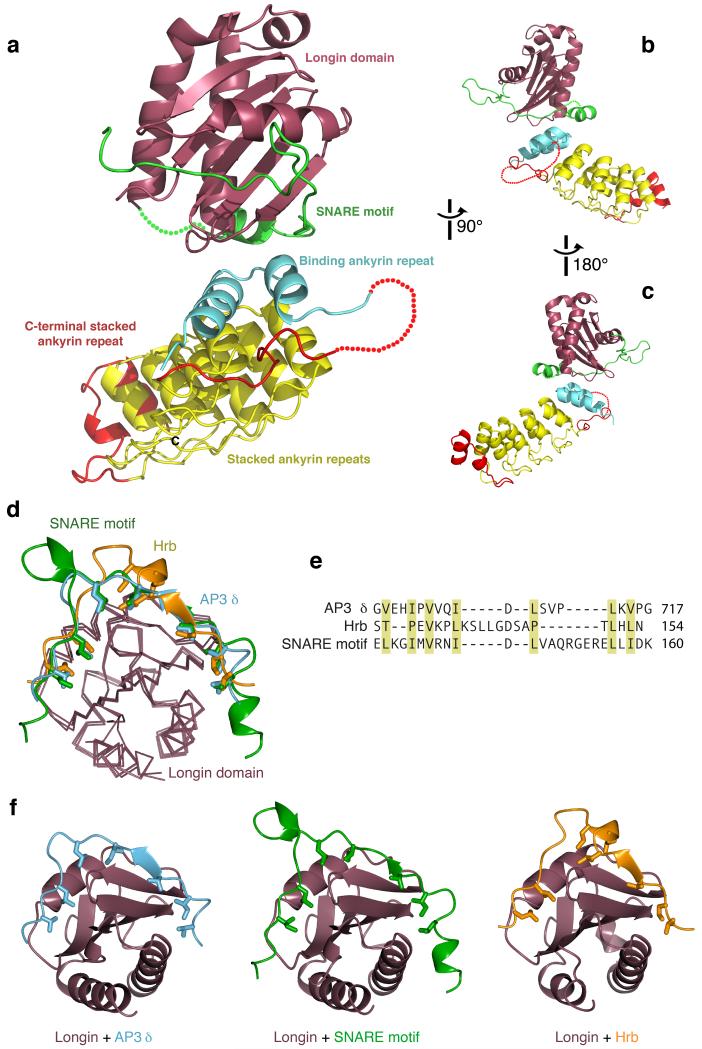
Figure 3. Molecular details of the interaction between Varp and VAMP7.
a–c Structure of the Varp–VAMP7 complex. VAMP7 (longin purple, SNARE motif in green) binds in a closed conformation to the top of the Ankyrin stack of Varp. The N–terminal Ankyrin repeat is in cyan, the four predicted Ankyrin repeats in yellow, and C–terminal Ankyrin repeat is in red.
d–f Comparisons with other VAMP7 complex structures. Superposition of the VAMP7 cytosolic domain with the VAMP7–Hrb [20] and VAMP7–δ–adaptin complex [24]. The VAMP7 SNARE motif (green), the Hrb peptide (gold) and the δ–adaptin peptide (cyan) all bind to the same groove of the VAMP7 longin domain (purple). Hydrophobic residues that slot into the hydrophobic groove of VAMP7 longin domain are shown.
e Structure–based alignment of the three VAMP7 longin domain binding sequences, showing that they share a pattern of hydrophobic side chains (yellow shading), but that the key residues are separated by very different sequences.
f Ribbon representations of the three complexes in e superimposed on the basis of the longin domains.
In the Varp–VAMP7 complex, the final 24 residues of the VAMP7 cytosolic region (164–188), which are dispensable for binding to Varp (Figure 1 and Supplementary figure 6) are not visible in the structure. Similarly, residues 121–128, which link the longin domain to the SNARE motif (Supplementary figure 6), also have no electron density. The complex structure shows that, when bound to Varp, VAMP7 adopts a closed conformation, whose existence has been previously suggested for the isolated protein free in solution [23, 39]. In this closed state, an ordered part of the SNARE motif (residues 129–163) binds back in an extended conformation in a groove stretching half way around the globular VAMP7 longin domain (Figure 3). The interaction between the longin domain and the SNARE motif is mediated largely by the hydrophobic side chains (Figures 3 and 4) burying ~2300 Å2 of accessible surface area (calculated with PISA [40]) explaining the stability of this interaction in solution. A subset of these longin domain interface residues are protected in NMR experiments confirming that the intramolecular ‘bound–back’ conformation of VAMP7 in the complex structure presented here is preformed in solution and is trapped by Varp [39]. Comparison with the VAMP7–Hrb structure [20] and the VAMP7–δ–adaptin structure [24] demonstrates a similar binding mode in the same groove on the VAMP7 longin domain for fragments of Hrb, δ–adaptin and the VAMP7 SNARE motif with the same longin domain residues being important to all three interactions. Thus, the interactions of the VAMP7 longin domain with Hrb or δ–adaptin and the SNARE motif are mutually exclusive (figure 3). There is however little homology between the three longin domain ligands: their key residues are differently spaced and so the mechanism of binding could not be predicted without detailed structural information. The major difference between the three interactions is that the interaction between the VAMP7 longin domain and its SNARE motif is intramolecular and the other two are intermolecular and hence the intramolecular interaction will predominate in solution. The SNARE motif residues I139 and I144 are key to its ‘binding back’ onto its own longin domain and this explains why the triple mutation of I139S, M140S and I144S disrupts the interaction between the VAMP7 SNARE motif and its longin domain, thus leaving its longin domain free to bind Hrb [20].
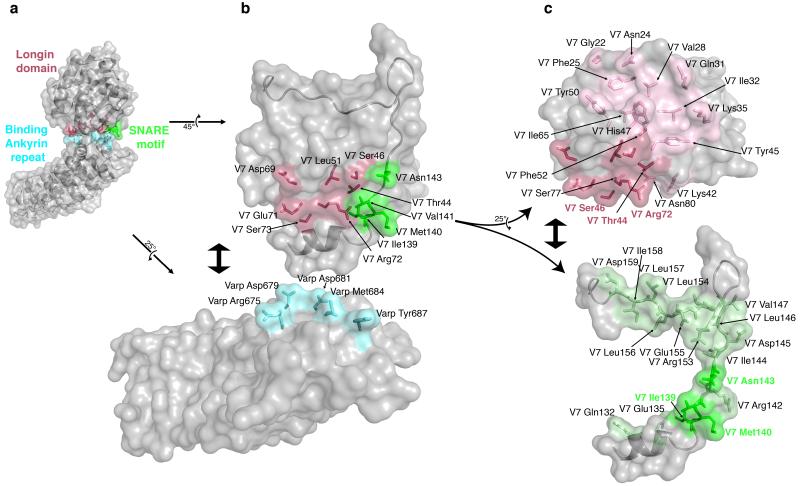
Figure 4. Analysis of the Varp–VAMP7 complex interface.
a Semi–transparent surface representation of the Varp–VAMP7 complex with the underlying backbone structure showing. Residues involved in the interface are shown in ball and stick representation and they and the parts of the protein surfaces they map to are highlighted in colour (VAMP7longin domain in purple, VAMP7SNARE motif in green and Varp in cyan)
b The ‘flipped open’ interaction interface of the Varp–VAMP7 complex. VAMP7 and Varp have been ‘rotated’ and ‘separated’ as shown by the arrows. Side chains involved in the interaction are shown and coloured as in a with only the VAMP7 SNARE motif backbone shown for clarity.
c VAMP7 SNARE motif and longin domain cytosolic domains ‘rotated’ and ‘separated’ as shown by the arrows. Residues involved in the binding to Varp are shown in darker purple (on longin domain) or green (on SNARE motif) and labelled accordingly. Residues involved in the interaction between the VAMP7 longin domain and the SNARE motif are shown in lighter purple or green and labelled in black.
Effect of mutating residues in the Varp–VAMP7 interface
The longin domain and the SNARE motif make approximately equal contributions to VAMP7 binding to Varp (see figures 3, 4 and 5). Since the Varp binding site on VAMP7 can only be formed when the extended SNARE motif passes through the groove on the longin domain, this explains why the complete cytosolic domain of VAMP7 is required for Varp binding and neither the longin domain nor the SNARE motif on their own are sufficient for the interaction (Figure 1). Thus our structural data are entirely consistent with our interaction data determined both by Y2H and ‘GST pull downs’.
Figure 5. Mechanistic details and quantitation of the Varp–VAMP7 interaction.
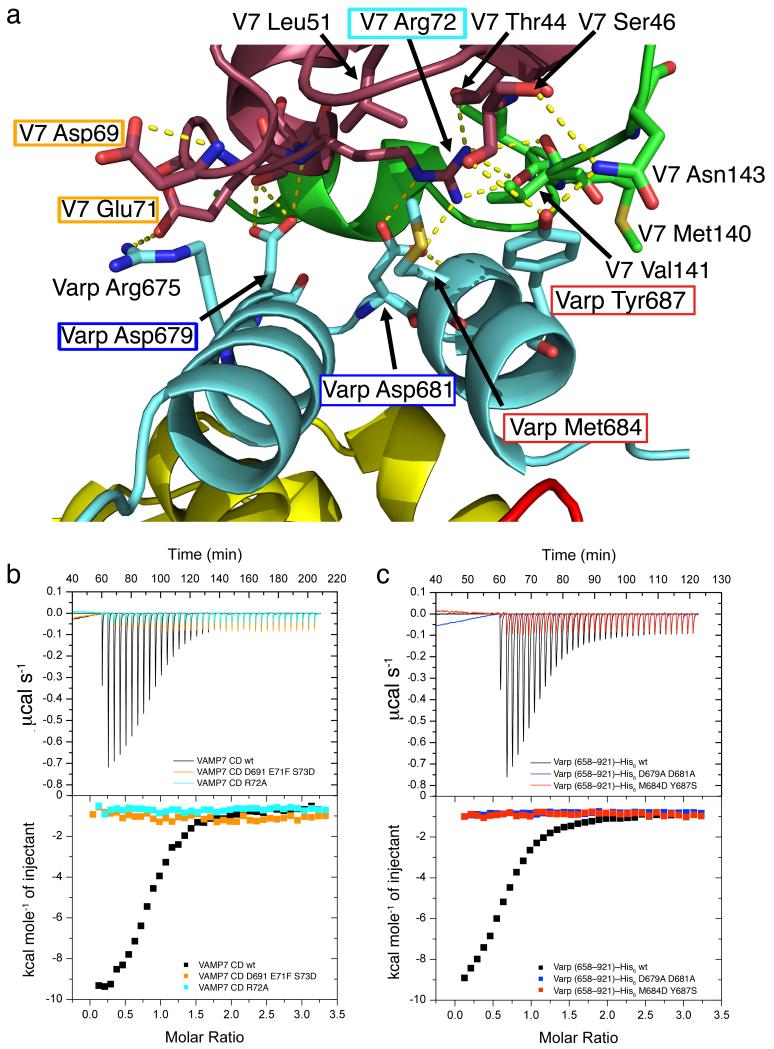
a The interaction interface between VAMP7 and Varp is composed of residues belonging to the first binding Ankyrin repeat of Varp and both the longin domain and the SNARE motif of VAMP7. Residues targeted by mutagenesis used to confirm the interface in solution are highlighted by a box of the same colour as the corresponding ITC trace in b and c.
b and c ITC analysis of VAMP7–Varp interaction. Mutations in the Varp:VAMP7 interface abolish the binding of Varp to VAMP7. The KD measured for the wild type proteins is 2.3 ±0.6 μM and all four mutations, which do not affect folding as judged by CD (data not shown) reduce binding below detectable levels (KD>300μM).
The interface buries a total of 1200Å2 of solvent accessible surface area and while not being inconsistent with the ~2μM KD for the interaction, is on the low side of what could be expected for this KD. This may be explained by the network of charged, hydrogen bonds, centred on Arg72 of the longin domain, which is 90% buried, with most of the hydrogen bonds consequently protected from solvent. (Figure 5 and Supplementary figure 6).
Regions of contact involve contributions from all three components of the complex (see Figure 5). We demonstrated using ITC experiments that mutations in residues critical to the interaction between all three components, M684D Y687S and D679A D681A in the case of Varp and R72A and D69A E71F S73D in VAMP7, abolished complex formation (Figure 5), but did not affect protein folding as judged by behaviour during purification and circular dichroism (data not shown). The effects of these mutations confirm that the complex seen in the crystal structure is the same as that in solution. Most of the residues in VAMP7 involved in Varp binding are not conserved in the other longin domain containing SNAREs Sec22 and Ykt6 (Supplementary figure 6), explaining why these SNAREs show no binding to Varp (Figure 1f)
Varp inhibits the ability of VAMP7 to form SNARE complexes
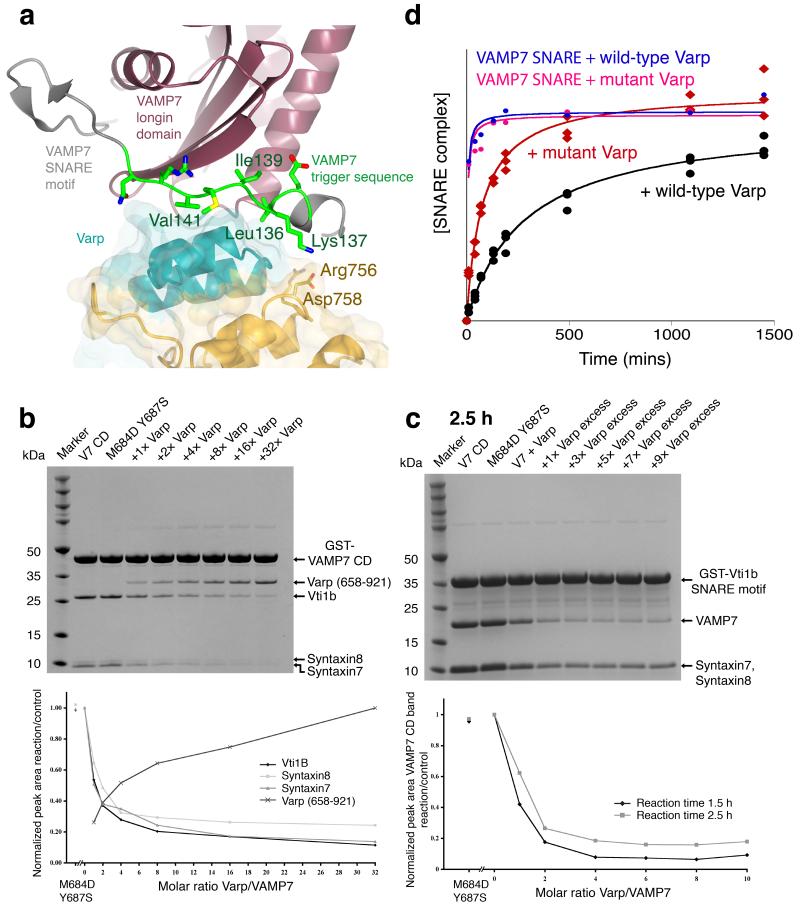
The intermolecular Hrb and AP3 δ–adaptin interactions with VAMP7 direct the trafficking of inactive cis–SNARE complexes containing VAMP7, suggesting that the intramolecular interaction of Varp with uncomplexed VAMP7 plays a different cellular role. The complex structure indicates that Varp traps a closed conformation of VAMP7 in which the SNARE motif would be unavailable for cognate trans SNARE complex formation. In VAMPs 2, 3 and 8 a trigger site sequence initiates SNARE complex formation [41]. The equivalent trigger sequence of VAMP7, residues 136 to 142 (LKGIMVR), is actually that which is sandwiched in the VAMP7:Varp interface with the critical isoleucine and methionine residues buried in the centre of the interface (Figure 6). To test the hypothesis that Varp binding inhibits the ability of VAMP7 to form SNARE complexes, we analysed the formation of the syntaxin7–syntaxin8–Vti1b–VAMP7 SDS stable SNARE complex (Supplementary figure 7) as a function of Varp(658–921)His6 concentration. As shown in Figure 6, the extent of SNARE complex formation is inversely correlated with the concentration of Varp(658–921)His6 present in the reaction whereas the VAMP7 non–binding mutant of Varp(658–921)His6 (M684D Y687S) did not inhibit SNARE complex formation. In a related set of experiments we analysed the time dependence of the SNARE complex formation on the full length cytosolic portion of GSTVAMP7 and on the GSTVAMP7–SNARE motif (ie missing the putative inhibitory longin domain), in the presence of wt and M684D Y687S mutant Varp(658–921)His6, and in the absence of Varp(658–921)His6 (Figure 6 and Supplementary figures 7 and 8). In assays with the full GSTVAMP7 cytosolic domain, wt Varp(658–921)His6 but not its (M684D Y687S) mutant reduced the rate of SNARE complex formation to about 30% of the rate with no Varp present (n.b. after 24 hours the level of SNARE complex attained in the presence of wt Varp is still only 80% of that achieved when the M684D,Y687S VAMP7 non–binding mutant was present). The SNARE complex formation rates were the highest when using the GSTVAMP7 SNARE motif (Figure 6d and Supplementary figure 8), thus confirming the proposed inhibitory role of the longin domain [23]. In this case the presence of wt or M684D,Y687S mutant Varp had no observable influence on the complex formation, again in line with our observations that the binding of Varp to VAMP7 occurs only to full length VAMP7 cytosolic domain. Thus, the presence of the longin domain on VAMP7 and addition of Varp together resulted in a rate of SNARE complex formation that was 60–70 times slower than that with the isolated VAMP7 SNARE motif (i.e. only 1.5% of the rate), demonstrating that in vitro at least, this two component system functions as an effective kinetic inhibitory system for VAMP7–mediated SNARE complex formation.
Figure 6. The effect of Varp on VAMP7 dependent SNARE complex formation.
a The trigger region of VAMP7 that initiates SNARE complex formation [41] (residues 136 LKGIMVR 142) is trapped at the centre of the Varp–VAMP7 interface. L136, I139 and V141 of the trigger region (green) are buried by interactions with the first ankyrin repeat of residues 658–921 of Varp.
b and c Coomassie blue staining of SDS PAGE gels (upper panels) and quantitation by densitometry (lower panels) of in vitro SDS–stable SNARE complex reconstitution on either GSTVAMP7 (b) or on GSTVti1b SNARE motif (c). The presence of Varp residues 658–921 at the molar ratios to VAMP7 shown reduces the level of SNARE complex formed whereas the presence of the M684D,Y687S mutant form of Varp does not inhibit SNARE complex formation.
d Quantitation by densitometry of Coomassie blue stained SDS PAGE gels (see Supplementary figure 8) of the time dependent effect of Varp (658–921) on SNARE complex formation on GSTVAMP7. The relative amount of SNARE complex is shown on the vertical axis (see online methods and Supplementary note). The presence of Varp (black) slows SNARE complex formation compared to the reaction in the presence of mutant Varp M684D Y687S (red). The essentially identical blue and pink curves show the equivalent time courses for the VAMP7 SNARE motif only with wt and M684D Y687S Varp present respectively.
Varp does not inhibit VAMP8’s ability to form SNARE complexes
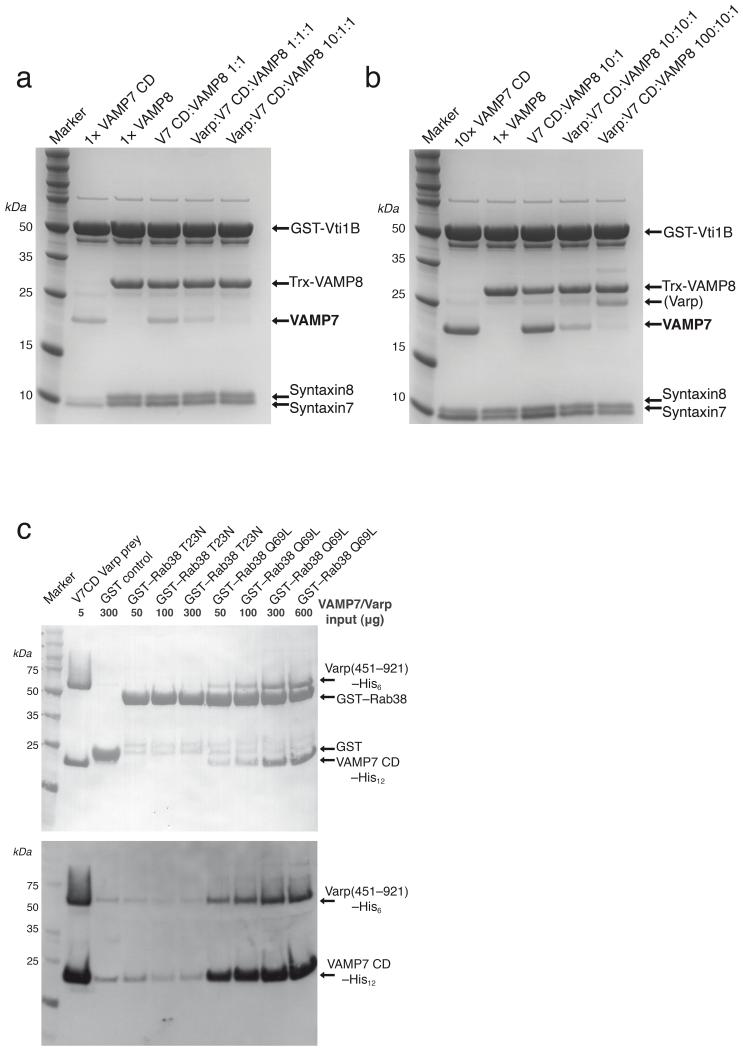
VAMP8, which is also found on compartments of the endocytic pathway [11, 42, 43] is closely related to VAMP7 (43 % sequence identity, 68 % similarity) in their SNARE motifs, can form complexes with the same Q–SNAREs (syntaxin7, syntaxin8 and Vti1b) [11, 42-44], but possesses no longin domain. We therefore investigated the rate of complex formation with syntaxin7 and syntaxin8 (SNARE motifs only) on GSTVti1b for VAMP7 and Trx–VAMP8 and tested whether they were sensitive to the presence of Varp(658–921)His6. VAMP8’s rate of SNARE complex formation was faster than that for VAMP7, with around 6 times more SNARE complex formed with VAMP8 than is formed with full length VAMP7 after 2.5 hours, and was a similar rate to that with the isolated VAMP7 SNARE motif (see Figures 6 and 7). Again as with VAMP7 SNARE motif only (Figure 6), the presence of Varp(658–921)His6 at either an equimolar or at a 10fold excess to the amount of VAMP caused no significant reduction in the amount of VAMP8 incorporated into SNARE complexes, but markedly reduced the amount of VAMP7 incorporated. In fact in order to achieve approximately equal amounts of VAMP7 and Trx–VAMP8 incorporation into SNARE complexes when they were both present in the same reaction and when Varp was absent, we needed to use 10 times as much VAMP7 as Trx–VAMP8. Under these conditions, the presence of Varp(658–921)His6 at an equimolar and at a 10fold excess of the amount of VAMP caused a 74% and a 95% reduction respectively in the amount of VAMP7 incorporated into SNARE complexes compared to the situation without Varp, whereas the amount of Trx–VAMP8 incorporated increased by 20% and 27% respectively.
Figure 7. Varp interacts with Rab38:GTP and VAMP7 but not VAMP8.
a Coomassie blue staining of SDS PAGE gels of SNARE complex reconstitutions with the SNARE motifs of syntaxin7 and syntaxin8 on an excess of GSTVti1b for 2.5 hours using equimolar amounts (1x) of VAMP7 CD to Trx–VAMP8 alone or together. The presence of Varp(658–921)His6 at an equimolar and at a 10fold excess of the amount of VAMP7 causes significant reductions in the amount of VAMP7 incorporated into SNARE complexes as compared to the situation without Varp(658–921)His6.
b Coomassie blue staining of SDS PAGE gels of SNARE complex reconstitutions as in a but using a 10–fold excess of VAMP7 CD over Trx–VAMP8. The presence of Varp(658–921)His6 again causes significant reductions in the amount of VAMP7 incorporated into SNARE complexes. ‘(Varp)’ indicates Varp(658–921)His6 that is non–specifically bound to the beads as a result of the very high amount used in the experiment.
a Coomassie blue (top) and Western blotting with HisProbe–HRP conjugate of SDS–PAGE of ‘GST pull downs’ with the amounts of GTP locked (Q69L) and the GDP locked (T23N) mutant versions of GST–Rab38 indicated used as bait. VAMP7CDHis12 is only detected in pull downs on GSTRab38GTP Q69L when Varp(451–921)His6, which contains both its sets of ankyrin repeats, is also included in the reaction.
Varp binds simultaneously to VAMP7 and Rab23 or Rab38
It has been demonstrated that residues 378–717 of Varp, which are predicted to contain another set of ankyrin repeats, interact with the GTP bound form of the closely related endosomal Rabs Rab32 and Rab38 [27]. Since avidity effects through a single protein molecule binding multiple ligands on the same membrane can greatly increase apparent affinity of a protein for a membrane [45, 46], we investigated whether Varp could bind to both VAMP7 and Rab38:GTP or Rab32:GTP at the same time. As shown in Figure7c, VAMP7 CD was only detected in pull downs on GSTRab38GTP Q69L in the presence of Varp(451–921)His6, which contains both the VAMP7 binding site and the Rab binding site. Identical results were obtained using Rab32 instead of Rab38 and neither Rab was able to bind directly to VAMP7 (data not shown). These data therefore establish that Varp can bind simultaneously to both VAMP7 and to Rab32:GTP or Rab38:GTP.
Discussion
If VAMP7–mediated fusion events of various organelles with endolysosomes (reviewed in [47, 48]) took place at the wrong time and/or place there would be serious consequences for the cell with inappropriate luminal content digestion occurring. It therefore seems logical that VAMP7–mediated fusion events should be tightly and accurately controlled in both a spatial and temporal manner and that inappropriate fusogenic behaviour of VAMP7 must be effectively inhibited. This regulation must, however, occur in the context of the low nM KD, i.e. quasi–irreversible formation, of SNARE complexes [49].
In this study we have shown how the folded longin domain of VAMP7 binds back onto the unstructured ‘trigger sequence’ of its own SNARE motif, so inhibiting the SNARE motif from participating in SNARE complex formation, and how this intramolecular interaction can be stabilised by the subsequent binding of Varp. The binding site for Varp on VAMP7 is only created when the longin domain and the SNARE motif bind to each other, which explains why the binding to Varp can only be detected with the full length cytosolic domain of VAMP7. The result of this two component inhibitory system (longin domain and Varp) is that the rate of SNARE complex formation is 60–70 times slower than that of a VAMP7 form lacking its longin domain (Figure 6 and [23]). However, under in vitro experimental conditions in which only VAMP7, its cognate Q–SNAREs and Varp are present in solution, Varp cannot prevent the eventual incorporation of the VAMP7 present into SNARE complexes, i.e. thermodynamically, Varp cannot in the long term compete with the SNARE complex and exerts only a kinetic inhibitory effect. This means that even when working in concert, the ~2μM KD Varp–VAMP7 interaction plus the VAMP7 SNARE motif–longin domain interaction must be weaker than the low nM KD of SNARE complex formation [49]. The effective KD for the intramolecular VAMP7 interaction is hard to determine but an upper limit to the effective KD in intact VAMP7 can be estimated from the 10μM KD for the Hrb–longin domain interaction that ϕαιλδ to compete with the intramolecular binding between the VAMP7 SNARE motif and its longin domain [20].
In vivo the capacity of Varp to inhibit VAMP7’s participation in SNARE complex formation and consequently inhibit VAMP7–dependent fusion events can be enhanced over its ability to do so in vitro by an increase in its effective local concentration. This may be achieved through coincidence detection by the simultaneous binding of Varp to VAMP7 and to other factors, that are present on the same membrane as VAMP7 or on a membrane to which the VAMP7 containing organelle or vesicle has been tightly tethered, such as GTP–bound Rab32 and Rab38 (Figure 7). Like Varp, Rab32 is present on late endocytic compartments as well as on early and late stage autophagosomes and both Rab32 and VAMP7 have been shown to be involved in autophagy [13, 36] [37]. That Rab32 is involved in membrane fusion of endocytic compartments with endolysosomes is indicated by the observation that increased expression of the Rab32 GAP RUTBC1 leads to an increase in mannose–6–phosphate receptor turnover in terminal endocytic compartments instead of the mannose–6–phosphate being recycled back from endosomes to the TGN [33, 50]. This can be explained by the GAP reducing the amount of Rab32:GTP thus causing a reduction in the binding of Rab32 to Varp. This would in turn lead to a reduction in the Varp concentration on late endocytic compartment membranes and so reduce Varp’s inhibition of VAMP7’s fusogenic ability, which is involved in the fusion of endosomes and autophagosomes with lysosomes[11-13]. Rab32, its close homologue Rab38 and Varp are all enriched in lysosome related organelle (LRO)–containing cell types such as platelets [51] and especially melanocytes where they show good colocalisation with VAMP7 [38]. This likely reflects the importance of preventing the undesirable fusion of melanosomes with lysosomes, which would lead to degradation of the melanosomes’ luminal contents and indeed, the Varp–Rab32 and Varp–Rab38, and Varp–VAMP7 interactions have been shown to be important for the correct trafficking of melanosomal enzymes [26]. Although the interactions of Varp with Rab38:GTP or Rab32:GTP have KDs of 2–5μM (Figure 7 and data not shown), it has so far proved technically impossible to measure the degree to which Rab38 or Rab32 immobilised on a membrane enhances the binding of Varp to VAMP7 embedded in the same membrane. Measurements of similar low μM KD avidity–driven interaction systems with liposome–based SPR [45, 46] suggest that the increase in binding from co–recognising Rab32:GTP or Rab38:GTP and VAMP7 present on the same membrane would be between 10–50 fold. A logical extension of an increased inhibitory effect on SNARE complex formation involving VAMP7 by Varp caused by simultaneous Rab32 and Rab38 binding is that in vivo it could be subsequently negated by GTP hydrolysis on Rab32 and Rab38 as indicated by [33], thereby allowing spatial and/or temporal regulation of Varp’s inhibitory role on VAMP7. Indeed such weakening would have the effect of releasing free, fusogenically–competent VAMP7, which could drive membrane fusion events. That Varp can therefore ultimately play a role in driving VAMP7–mediated membrane fusion events at the correct time and place is supported by the recent demonstration that the VAMP7–Varp interaction is important for dendrite formation in melanocytes [19]. Finally, given the large size of Varp and the presence of multiple protein–protein interaction domains within it (Figure 1), other binding partners for Varp may exist that could function analogously to Rab32 and Rab38 in providing additional factors to recruit Varp to other VAMP7–containing membranes. In vivo, both VAMP7 and VAMP8 are found throughout the endocytic pathway [11, 42, 43]. VAMP7 has been proposed to be involved in fusion events later down the pathway to the degradative endolysosome [11, 31, 47] whereas VAMP8 is believed to be involved in the fusion of early endosomes and endosomal carriers with later endocytic compartments in order to deliver cargo and for endosome remodelling [11, 42, 43]. VAMP8, in that it has no longin domain and has a high rate of SNARE complex formation that is insensitive to the presence of Varp, resembles a version of VAMP7 from which the longin domain has been deleted. Therefore, the kinetic inhibition by Varp of VAMP7’s (but not VAMP8’s) participation in SNARE complex formation on endosomes would favour VAMP8-driven maturation of endosomes, where they acquire luminal contents. A combination of factors including depletion of the pool of free VAMP8 and reducing the inhibitory effect of Varp through removing it from an endosome membrane, would switch the endosome to a state in which fusion with dense core lysosomes and endolysomes would predominate and thus the endosomes’ luminal contents would be degraded (reviewed in [47, 48]). Reduction in the levels of Varp on an endosomal membrane could be brought about by hydrolysis of GTP on Rab32 or Rab38 or by binding Varp to a non–endosome localised protein.
In conclusion, the structural and biochemical studies described here demonstrate that in vitro Varp is a kinetic inhibitor of SNARE complex formations uniquely involving VAMP7. We predict that in vivo Varp should sequentially inhibit but then at the right time effectively promote fusion of late endocytic organelles, where Varp is predominantly found, with target membranes. The switching from inhibiting to potentiating VAMP7 dependent fusion will depend on the complement of other proteins that can bind directly or indirectly to Varp and/or of SNAREs that can compete with VAMP7 for inclusion into SNARE complexes on an endosome’s surface. Varp could also function as a ‘chaperone’, in the Victorian sense of the word, for VAMP7 in that it could inhibit ‘improper’ interactions that VAMP7 could indulge in i.e. off–path VAMP7–mediated fusion events. That this is important for the cell is suggested by the observation that during its trafficking down the endocytic pathway from the plasma membrane via first Hrb and then AP3, VAMP7 is kept in an inactive state as part of cis–SNARE complexes. At the physiological level, the list of cellular processes in which Varp functions is very likely not complete since published studies have focussed on specialised cell types. Nonetheless, in all those processes that have been characterised, Varp’s ability to bind VAMP7 is essential. The work presented here suggests that is because of Varp’s ability to regulate VAMP7’s participation in membrane fusion events.
Online Methods
DNA constructs and antibodies
For a comprehensive description of the DNA constructs and antibodies used in this study see Supplementary note.
Protein Expression and Purification
Expression of all described constructs was performed in the E. coli strain Rosetta 2 (DE3) pLysS (Novagen) using 2xTY media supplemented with the suitable combination of antibiotics. Cells were initially grown at 37 °C until an OD600nm of 0.6 – 1.0 was reached. The temperature was then lowered to 16°C and the culture cooled down for at least an hour before induction of protein expressions with 500 μM IPTG. After induction cells were grown for 20 – 24h at 16°C. Cells were lysed at 4 °C using either 6 × 30 sec bursts of sonication or a continuous–flow French Press and the insoluble material removed by centrifugation. Supernatants were filtered through 0.45 μm cut–off syringe filters. His tagged only proteins (e.g. Varp –10xHis) were purified on Ni–NTA agarose (Qiagen) using an Imidazole step gradient for elution. The protein was further purified by anion exchange chromatography on a Sepharose Q column followed by a Superdex 200 gel filtration (column equilibrated in 500 mM NaCl, 20 mM Tris pH 8.5, 1 mM EDTA, 10 mM DTT).
For the reconstitution of the Varp/VAMP7 complex VARP (658–921)–6xHis and VAMP7 CD were purified separately and the complex reconstituted in a final gelfiltration step. GST– Varp (658–921)–6xHis was purified on GSH–Sepharose and the protein released form the GST tag by proteolytic cleavage with human rhinovirus 3C protease. The Varp (658–921)–6xHis was further purified on Ni–NTA agarose and Superdex 75 gel filtration. The protocol used for the purification of VAMP7 CD (1–188) is similar to that described for VARP (658–921)–6xHis except that the protein was not purified via a Ni–NTA column but immediately run over the size exclusion column after cleaving of the GST–tag with Prescission protease. VAMP7 CD (1–188) was mixed in a molar excess of 1.25:1 with VARP (658–921)–6xHis (total volume of reaction 2 ml, total amount of protein ~ 20 mg). The reaction was incubated for 1 h at 4 °C on a roller incubator to allow the complex to form. The sample was then applied on a Hi Load Superdex 75 16/60 prep grade gel filtration column (column equilibrated in 200 mM NaCl, 20 mM HEPES pH 7.4, 5 mM TCEP). The presence of complex of a 1:1 complex was assessed by SDS–PAGE gel electrophoresis (Supplementary figure 4).
SNARE proteins were purified in a similar fashion as outlined above depending on their respective affinity tag. All SNARE proteins were gel filtered into SNARE complex reconstitution buffer (500 mM NaCl, 20 mM HEPES pH 7.4, 10 mM DTT, 1 mM EDTA) after the affinity chromatography step. Rab38 constructs were purified in a similar way as described for the VAMP7 cytosolic domain with the difference that all buffers contained the matching nucleotide (GDP for T23N for example) and MgCl2 at a concentration of at least 10 μM and 0.5 mM respectively. GST tagged bait proteins for pull down experiments were purified as described above with the exception that the GST–tag was not removed but the proteins eluted off the GSH–Sepharose resin using a suitable buffer containing 20 mM reduced GSH and then run over a Superdex 75 gelfiltration column (equilibrated in 500 mM NaCl, 20 mM HEPES pH 7.4, 1 mM DTT).
Limited proteolysis of Varp (1–1050)–His10
Varp (1–1050)–His10 was digested with Chymotrypsin (mass ratio of 5000:1) at 23 °C for the indicated time spans. Samples were blotted on PVDF membrane and N–terminally sequenced. For a full description, see Supplementary note.
ITC and pull downs
GSH–sepharose pull downs were conducted according to commonly established protocols e.g. [20]. Briefly, 50 μg of the GST–tagged bait was incubated with 300 μg of the untagged prey–protein and 60 μl of a 50 % (v/v) GSH–Sepharose–bead suspension (GE Healthcare) for 1 h at 4 °C. The beads were washed 4 to 5 times with 1 ml of pull–down buffer. The proteins were eluted off the beads using 30 mM reduced GSH in the pull–down buffer. Binding was assessed by SDS PAGE followed by coomassie staining or western blotting with HisProbe–HRP conjugate and quantification using ImageJ (http://rsbweb.nih.gov/ij/). For ITC measurements all proteins were gel filtered into the appropriate ITC buffer prior to the measurements. All measurements were performed with a VP–ITC isothermal titration calorimeter (MicroCal) at 4 °C. To determine the KD of a binding reaction the mean of four independent ITC measurements that showed clear signs of saturation of the binding by the end of the run were used and the SD calculated. For a full description, see Supplementary note.
Crystallisation and structure determination
The best crystals of the Varp–VAMP7 complex grew at a concentration of 10 mg/ml complex from wells containing 5 to 7.5 % (v/v) Isopropanol and 0.1 M Imidazole pH 6.5 −7.8 in hanging and sitting drops at 4 °C. Crystals were cryoprotected in mother liquor containing additionally 25 % (v/v) ethylene glycol. Mercury derivatives were prepared by soaking crystals in mother liquor containing additionally 0.2 – 0.5 mM ethylmercurythiosalicylate (EMTS) or p–chloromercuribenzenesulfonate (PCMBS) for 5 to 30 minutes, followed by cryoprotection. Datasets were collected at beamline ID23 of the ESRF light source at 1.0Å wavelength. Data were indexed and integrated using Mosflm [52], scaled with Scala [53]. Heavy–atom sites were located with ShelxD [54] and phases calculated using autoSHARP [55]. Building and refinement were carried out with programs from the CCP4 suite, Coot and Refmac [56-58]. The final refined R–factor (Rfree) was 0.213 (0.251), with 1 Ramachandran outlier (Varp Asp743, which is on the edge of the acceptable region) and a Molprobity clashscore 10.5 (79th percentile) [59].
Pictures of the refined structure were prepared with PyMOL [HTTP://WWW.PYMOL.ORG/] and CCP4mg [60].
SNARE complex inhibition assay
GST tagged SNARE proteins where incubated with their SNARE partners in a molar ratio of 1:4:4:4 (:4 in the case of experiments in which VAMP8 was also present). Varp was added in the concentrations described. One hour before the end of the experiments the reactions were supplemented with 1/3 volume of 50 % (v/v) glutathione Sepharose beads if not mentioned otherwise. After washing the beads 5 to 6 times with reconstitution buffer containing 1 % (w/v) NP40, bound proteins were eluted using wash buffer supplemented with 30 mM glutathione. Samples were analyzed on 4–12 % Nu–PAGE gradient gels (Invitrogen). The SDS stability of the complexes was demonstrated (see Supplementary figure 7). For a full description, see Supplementary note.
Immunofluorescent confocal microscopy examining colocalization of Varp–HA and endogenous markers
NRK cells stably expressing Varp–HA were created using the pLXIN retroviral system (Clontech). For immunofluorescence, stable cells seeded on glass coverslips were fixed with 3.7% formaldehyde in PBS for 20 minutes. Cells were then permeabilized by incubation with 0.1% Triton X–100 in TBS–T for 20 minutes and incubated with blocking solution (5% non–fat dry milk powder in TBS–T) for 1 hour. Cells were incubated with primary antibody followed by incubation with fluorescently conjugated secondary antibodies and images captured using a confocal microscope.
siRNA mediated gene knock–down in NRK cells
Endogenous genes (Varp and Rab32) were knocked–down in wild–type NRK cells using siRNA oligos at 100 nM transfected with DharmaFECT 2 (Dharmacon, Thermo) reagent according to the manufacturer’s instructions. Levels of knock–down were compared to cells transfected with a non–targeting control siRNA oligo at 100 nM.
Electron microscopy
For immunogold labeling of cryosections, NRK cells stably expressing Varp–HA were incubated with Dextran conjugated to Texas Red for 4h followed by a 20h chase as previously described [61]. Cells were fixed with 0.1% Glutaraldehyde / 4% Paraformaldehyde in 0.1M Na Cacodylate pH7.2 at 20°C, scraped, pelleted, embedded in 10% gelatin and infused with 1.7 M sucrose/15% poly–vinyl pyrolidone prior to being frozen on aluminium stubs in liquid nitrogen. Ultra–thin frozen sections were cut using an ultramicrotome equipped with an FCS cryochamber attachment (Leica, Milton Keynes, UK) and collected from the knife–edge with 2% methyl cellulose and 2.3M sucrose in a ratio of 50:50 [62]. The sections were immunolabelled using rabbit anti–Texas Red, rabbit anti–lgp110/LAMP2 and mouse monoclonal anti–HA, and detected using protein A conjugated to 5nm colloidal gold (Department of Cell Biology, University of Utrecht, NL), protein A conjugated to 10nm colloidal gold and rabbit anti–mouse immunoglobulin followed by protein A conjugated to 15nm colloidal gold, respectively, using a sequential immunolabelling protocol. The immunolabelled sections were contrasted, dried and observed in a Philips CM100 transmission electron microscope at 80 kV.
Quantitation of the degree of immunolabelling was performed by counting gold particles overlying defined cellular components by scanning sections sequentially and recording images with a CCD camera (MegaView III). The labelling was repeated 4 times and the distribution of 200 gold particles for each labelling experiment was determined.
Supplementary Material
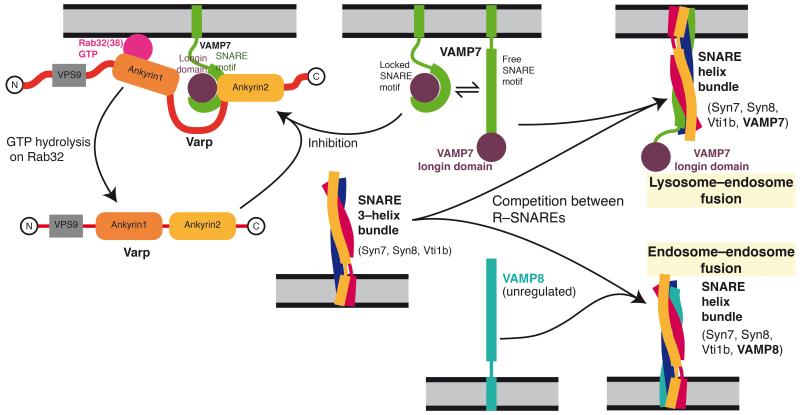
Figure8. Model of Varp function on an endosomal membrane.
Through its simultaneous and direct binding to Rab32:GTP or Rab38:GTP (pink) and to VAMP7 (purple and green), Varp will trap VAMP7 in an inactive form so allowing VAMP8 (turquoise) to outcompete VAMP7 for SNARE complex formation with syntaxin7, syntaxin8 and Vti1b. Hence Varp could act to inhibit the amount of endosome:lysosome fusion and promote endosome:endosome fusion. The effect would be reversed when the GTP bound Rab was converted to its GDP bound form, which cannot bind Varp : the interaction between Varp and VAMP7 would become effectively weaker, and the resulting free, uninhibited VAMP7 could then be used in the formation of SNARE complexes that will cause endosome:lysosome fusion.
Acknowledgements
We thank beamline staff at IO3 (SLS Diamond plc) H. Kent and R. Suckling (MRC LMB) and L. Jackson and A. Peden (CIMR, Cambridge) for assistance and useful discussions. MNT–1 cells were a kind gift of D. Cutler (LMCB, University College, London). DJO is funded by a Wellcome Trust PRF, JPL by an MRC programme grant (G0900113), IBS and PRE by MRC grant U105178845, and GGH by a postdoctoral fellowship from the Canadian Institutes of Health Research. The Cambridge Institute for Medical Research is supported by a Wellcome Trust Strategic Award (079895).
Footnotes
Coordinates and structure factors have been deposited in the Protein Data Bank with accession code 4B93.
References
- 1.Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744(3):493–517. [PubMed] [Google Scholar]
- 2.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7(9):631–43. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 3.McNew JA, et al. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407(6801):153–9. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 4.Paumet F, Rahimian V, Rothman JE. The specificity of SNARE-dependent fusion is encoded in the SNARE motif. Proc Natl Acad Sci U S A. 2004;101(10):3376–80. doi: 10.1073/pnas.0400271101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacDonald C, Munson M, Bryant NJ. Autoinhibition of SNARE complex assembly by a conformational switch represents a conserved feature of syntaxins. Biochem Soc Trans. 2010;38(Pt 1):209–12. doi: 10.1042/BST0380209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkhardt P, et al. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27(7):923–33. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu SH, et al. Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation. Proc Natl Acad Sci U S A. 2011;108(3):1040–5. doi: 10.1073/pnas.0914906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404(6776):355–62. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 9.Shen J, et al. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128(1):183–95. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Filippini F, et al. Longins: a new evolutionary conserved VAMP family sharing a novel SNARE domain. Trends Biochem Sci. 2001;26(7):407–9. doi: 10.1016/s0968-0004(01)01861-8. [DOI] [PubMed] [Google Scholar]
- 11.Pryor PR, et al. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5(6):590–5. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fader CM, et al. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793(12):1901–16. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Moreau K, et al. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146(2):303–17. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao SK, et al. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J Biol Chem. 2004;279(19):20471–9. doi: 10.1074/jbc.M400798200. [DOI] [PubMed] [Google Scholar]
- 15.Chaineau M, Danglot L, Galli T. Multiple roles of the vesicular-SNARE TI-VAMP in post-Golgi and endosomal trafficking. FEBS Lett. 2009;583(23):3817–26. doi: 10.1016/j.febslet.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Steffen A, et al. MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr Biol. 2008;18(12):926–31. doi: 10.1016/j.cub.2008.05.044. [DOI] [PubMed] [Google Scholar]
- 17.Boucrot E, Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proc Natl Acad Sci U S A. 2007;104(19):7939–44. doi: 10.1073/pnas.0702511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Arca S, et al. Role of tetanus neurotoxin insensitive vesicle-associated membrane protein (TI-VAMP) in vesicular transport mediating neurite outgrowth. J Cell Biol. 2000;149(4):889–900. doi: 10.1083/jcb.149.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohbayashi N, et al. The Rab21-GEF activity of Varp, but not its Rab32/38 effector function, is required for dendrite formation in melanocytes. Mol Biol Cell. 2012;23(4):669–78. doi: 10.1091/mbc.E11-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pryor PR, et al. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell. 2008;134(5):817–27. doi: 10.1016/j.cell.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgo A, et al. Role of Varp, a Rab21 exchange factor and TI-VAMP/VAMP7 partner, in neurite growth. EMBO Rep. 2009;10(10):1117–24. doi: 10.1038/embor.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaineau M, et al. Role of HRB in clathrin-dependent endocytosis. J Biol Chem. 2008;283(49):34365–73. doi: 10.1074/jbc.M804587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Arca S, et al. A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc Natl Acad Sci U S A. 2003;100(15):9011–6. doi: 10.1073/pnas.1431910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent HM, et al. Structural basis of the intracellular sorting of the SNARE VAMP7 by the AP3 adaptor complex. Dev Cell. 2012;22(5):979–88. doi: 10.1016/j.devcel.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, et al. Varp is a Rab21 guanine nucleotide exchange factor and regulates endosome dynamics. J Cell Sci. 2006;119(Pt 6):1053–62. doi: 10.1242/jcs.02810. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, et al. Varp is a novel Rab32/38-binding protein that regulates Tyrp1 trafficking in melanocytes. Mol Biol Cell. 2009;20(12):2900–8. doi: 10.1091/mbc.E08-12-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, et al. Varp interacts with Rab38 and functions as its potential effector. Biochem Biophys Res Commun. 2008;372(1):162–7. doi: 10.1016/j.bbrc.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Deleage G, Blanchet C, Geourjon C. Protein structure prediction. Implications for the biologist. Biochimie. 1997;79(11):681–6. doi: 10.1016/s0300-9084(97)83524-9. [DOI] [PubMed] [Google Scholar]
- 29.Linding R, et al. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31(13):3701–8. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36(Database issue):D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Advani RJ, et al. VAMP-7 mediates vesicular transport from endosomes to lysosomes. J Cell Biol. 1999;146(4):765–76. doi: 10.1083/jcb.146.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klopper TH, et al. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol. 2012;10:71. doi: 10.1186/1741-7007-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nottingham RM, et al. RUTBC1 protein, a Rab9A effector that activates GTP hydrolysis by Rab32 and Rab33B proteins. J Biol Chem. 2011;286(38):33213–22. doi: 10.1074/jbc.M111.261115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101(16):6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirota Y, Tanaka Y. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions. Cell Mol Life Sci. 2009;66(17):2913–32. doi: 10.1007/s00018-009-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, Liu Z, Huang X. Rab32 is important for autophagy and lipid storage in Drosophila. PLoS One. 2012;7(2):e32086. doi: 10.1371/journal.pone.0032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wasmeier C, et al. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J Cell Biol. 2006;175(2):271–81. doi: 10.1083/jcb.200606050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vivona S, et al. The longin SNARE VAMP7/TI-VAMP adopts a closed conformation. J Biol Chem. 2010;285(23):17965–73. doi: 10.1074/jbc.M110.120972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Wiederhold K, et al. A coiled coil trigger site is essential for rapid binding of synaptobrevin to the SNARE acceptor complex. J Biol Chem. 2010;285(28):21549–59. doi: 10.1074/jbc.M110.105148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antonin W, et al. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19(23):6453–64. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antonin W, et al. The R-SNARE endobrevin/VAMP-8 mediates homotypic fusion of early endosomes and late endosomes. Mol Biol Cell. 2000;11(10):3289–98. doi: 10.1091/mbc.11.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antonin W, et al. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat Struct Biol. 2002;9(2):107–11. doi: 10.1038/nsb746. [DOI] [PubMed] [Google Scholar]
- 45.Honing S, et al. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol Cell. 2005;18(5):519–31. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 46.Miller SE, et al. The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell. 2011;147(5):1118–31. doi: 10.1016/j.cell.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30(17):3481–500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8(8):622–32. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 49.Wiederhold K, Fasshauer D. Is assembly of the SNARE complex enough to fuel membrane fusion? J Biol Chem. 2009;284(19):13143–52. doi: 10.1074/jbc.M900703200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirst J, Futter CE, Hopkins CR. The kinetics of mannose 6-phosphate receptor trafficking in the endocytic pathway in HEp-2 cells: the receptor enters and rapidly leaves multivesicular endosomes without accumulating in a prelysosomal compartment. Mol Biol Cell. 1998;9(4):809–16. doi: 10.1091/mbc.9.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao X, et al. Molecular cloning, bacterial expression and properties of Rab31 and Rab32. Eur J Biochem. 2002;269(1):259–71. doi: 10.1046/j.0014-2956.2001.02645.x. [DOI] [PubMed] [Google Scholar]
- 52.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 53.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 54.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):479–85. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vonrhein C, et al. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–30. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McNicholas S, et al. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):386–94. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bright NA, Gratian MJ, Luzio JP. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr Biol. 2005;15(4):360–5. doi: 10.1016/j.cub.2005.01.049. [DOI] [PubMed] [Google Scholar]
- 62.Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol. 1996;106(1):41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.