Abstract
Mechanisms of protein recognition have been extensively studied for single-domain proteins1, but are less well characterized for dynamic multi-domain systems. Ubiquitin (Ub) chains represent a biologically important multi-domain system that requires recognition by structurally diverse Ub-interacting proteins (UbIPs)2,3. Ub chain conformations in isolation are often different from conformations observed in UbIP complexes, suggesting either great dynamic flexibility or extensive chain remodeling upon binding.
Using single-molecule FRET, we show here that Lys63-, Lys48- and Met1-linked diUb exist in several distinct conformational states in solution. Lys63- and Met1-linked diUb adopt extended ‘open’ and more compact ‘closed’ conformations, and Ub binding domains (UBDs) and deubiquitinases (DUBs) select pre-existing conformations. In contrast, Lys48-linked diUb adopts predominantly compact conformations. DUBs directly recognize existing conformations, but may also remodel Ub chains to hydrolyze the isopeptide bond. Disruption of the Lys48-diUb interface changes conformational dynamics and affects DUB activity. Hence, conformational equilibria in Ub chains provide an additional layer of regulation in the Ub system, and distinct conformations observed in differently linked polyUb may contribute to the specificity of UbIPs.
Ubiquitin (Ub) is involved in most aspects of cell biology, as it serves as a post-translational modification of lysine (Lys) residues, regulating many processes including protein degradation, cell signaling, trafficking and the DNA damage response2. Most of these functions are mediated by eight structurally and functionally distinct Ub chain types2, only two of which have been studied extensively. Lys48-linked Ub chains target proteins for proteasomal degradation4, while Lys63- and Met1-linked Ub chains have multiple non-degradative roles in cell signaling5,6. PolyUb signals are decoded by UbIPs including UBDs3 and DUBs7 that bind or hydrolyze Ub chains, respectively. Some UbIPs can distinguish between different linkage types, and trigger a specific downstream response to ubiquitination.
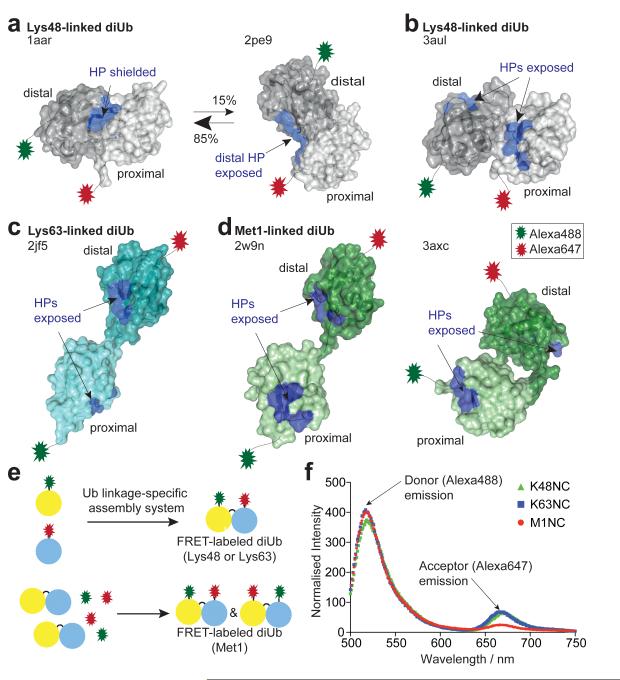
The distinct cellular roles of differently linked polyubiquitin have partly been explained by distinct chain conformations. Crystallographic, NMR and SAXS studies have proposed ‘compact’ conformations for Lys48-linked diUb8-10 (Fig. 1a, b). In the prevalent model for Lys48-linked diUb, Ub moieties interact via a hydrophobic patch (HP) on Ub8 (Fig. 1a), which is also the most common recognition site for UbIPs2. NMR residual dipolar coupling (RDC) measurements suggested that this conformation is in equilibrium with a second conformation that partly exposes the HP9. A recent crystal structure suggested a third compact conformation with exposed HPs on both moieties11 (Fig. 1b). In contrast, Lys63- and Met1-linked diUb are thought to adopt ‘open’ conformations with no interactions between Ub moieties, exposing the HPs10,12,13 (Fig. 1c, d), although a compact crystal structure of Met1-linked diUb has been reported recently14 (Fig. 1d).
Figure 1. Ub chain conformations and ensemble FRET measurements.
(a-d) Distal (dark) and proximal (light) Ub moieties are shown in surface representation with hydrophobic patch (HP) residues (Leu8, Ile44, His68, Val70) in blue. N- and C-terminal Alexa dye attachment points are indicated. (a) Lys48-linked diUb derived from crystallographic and NMR analysis. (pdb-id 1aar8, 2pe99). (b) Alternative ‘compact’ Lys48-linked diUb (3aul11). (c) ‘Open’ Lys63-linked diUb (2jf513). (d) Met1-linked diUb from crystallography (2w9n13, 3axc14). (e) Schematic representation of FRET-labeled diUb assembly. See Supplementary Fig. 1 and Methods. (f) Uncorrected ensemble FRET measurements for diUb used in this study (see Supplementary Methods).
Importantly, the first crystal structures of Ub and diUb bound to UbIPs (reviewed in 2,3,7) suggest that in some cases known diUb structures are incompatible with binding to UbIPs. In particular, DUBs must interact with the isopeptide linkage between Ub moieties, which is not accessible in known models of Lys48-linked polyUb (Fig. 1a, b). This suggests that compact Ub chain types might undergo remodeling (‘opening’) in order to be hydrolyzed by DUBs. Whether such chain opening is induced by DUB binding, or whether Lys48-linked diUb pre-exists in open conformations is unclear.
To understand the principles governing Ub chain recognition, we here generated Lys48-, Lys63- and Met1-linked diUb (termed K48NC, K63NC, M1NC, respectively) each containing a Förster resonance energy transfer (FRET) compatible dye pair (Alexa488/Alexa647, R0=5.6 nm). Linkage-specific assembly reactions and/or selective purification resulted in pure dual-labeled diUb (see Fig. 1e, Supplementary Fig. 1, Methods). Mass-spectrometric and enzymatic analysis, interchanging dye positions, and ensemble measurements of fluorescence lifetime and dye anisotropy confirmed sample quality and dye stability (Supplementary Figures 1, 2, Supplementary Methods). Dye photophysics were not significantly altered in labeled diUb (see Supplementary Fig. 2, Supplementary Methods). All samples showed a FRET signal in ensemble measurements (Fig. 1f).
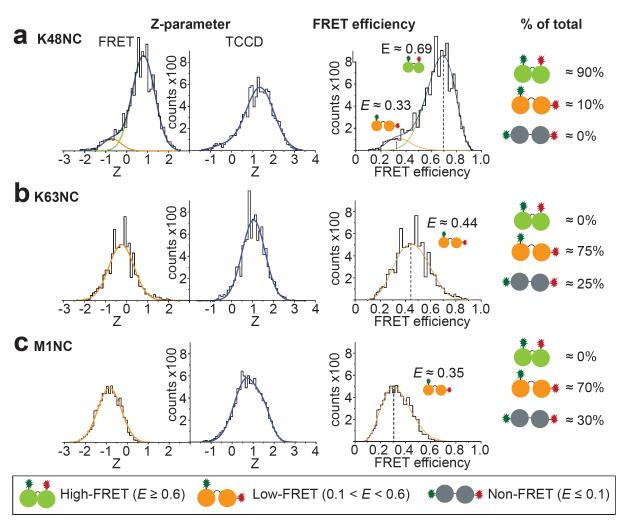
Distinct Ub chain conformations that underwent FRET were detected at the single molecule level by excitation of the donor fluorophore, with a single laser, and monitoring emission of both donor and acceptor fluorophores. The resulting FRET histograms were fitted to Gaussian functions representing distinct populations of diUb conformations (Fig. 2, see Methods). To estimate the proportion of molecules in conformations that gave no detectable FRET (termed ‘non-FRET’), two color coincidence detection (TCCD, see Methods)15 was used on the same sample. In TCCD, two lasers are used to directly excite the two fluorophores in the diUb independently, allowing direct quantification of the number of molecules with both donor and acceptor fluorophores. Combined use of TCCD and FRET measurements thus enabled estimation of the proportion of molecules that were in non-FRET conformations (see Supplementary Material).
Figure 2. Single-molecule FRET of K48NC (a), K63NC (b) and M1NC (c).
Left: FRET and TCCD signals in histogram counts against the Z-parameter -(ln[IA/γID]) for fitting to Gaussian functions, normalized to equal area in TCCD. Gaussian functions (see Supplementary Methods) are shown in green (high-FRET), orange (low-FRET) and blue (cumulative fit). Middle: Histograms of FRET species plotted against the FRET efficiency. The curves derived from the Z-parameter Gaussian fits are shown (see Supplementary Methods). Right: Estimate of the relative abundance of each population. Non-FRET refers to dual-labeled molecules (detected by TCCD) without FRET emission.
For K48NC, two distinct FRET populations could be resolved (Fig. 2a); a high-FRET population (FRET efficiency E≈0.69) representing ~90% of all molecules, and a low-FRET population (E≈0.41) accounting for the remaining ~10%. We were unable to detect any non-FRET populations in these experiments, however, we cannot rule out the existence of a small population of ’hidden’16 non-FRET K48NC species which would be beyond the detection limit (see Supplementary Methods). ‘Hidden’ protein conformations may modulate enzyme activity but are not easily detected16.
Similar to the results for K48NC, multiple conformations were also observed for K63NC and M1NC with ~70-75% low-FRET and ~25-30% non-FRET populations (Fig. 2b,c). Models of extended Lys63- or Met1-linked diUb (Fig. 1c,d) are compatible with non-FRET populations, while the compact crystal structure of Met1-linked diUb14 (Fig. 1d) may represent a low-FRET M1NC species. The prevalence of compact Lys63-linked diUb conformations in FRET is surprising, and has not been observed by other methods10,12,13. This may be due to multiple compact and semi-compact conformations that collectively result in the observed low-FRET populations.
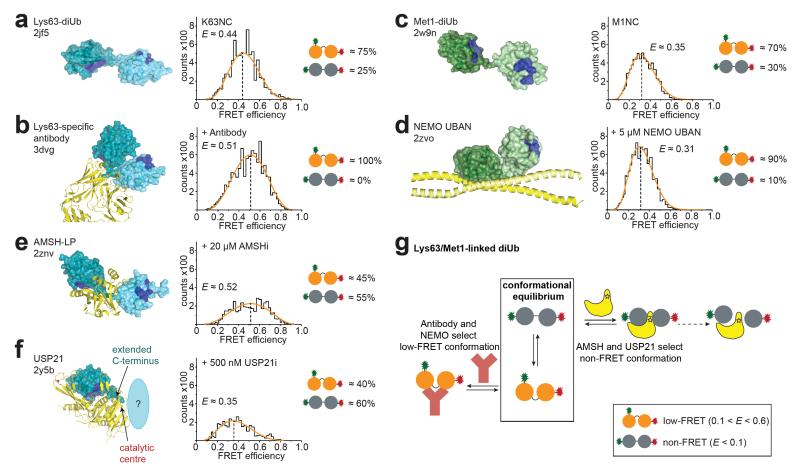
To test whether the observed diUb conformations are relevant for UbIP interaction, single-molecule measurements were performed using pM concentrations of K63NC, K48NC or M1NC mixed with UbIPs at concentrations exceeding the KD of the interaction (Supplementary Fig. 7). With our methodology we can follow relative changes in the populations upon addition of UbIPs, none of which affected dye photophysics (Supplementary Fig. 8). Several UbIPs interacted with pre-existing compact conformations of diUb. A Lys63-linkage-specific antibody increased the FRET population relative to the non-FRET population of K63NC, in agreement with structural work17 (Fig. 3a,b). Similarly, the Ub binding UBAN domain of NEMO18 enriched the low-FRET population of M1NC, consistent with UBAN binding to a compact conformation of linear diUb18 (Fig. 3c,d).
Figure 3. UbIP binding to Lys63- and Met1-linked chains.
(a-f) Structural models (colored as in Fig. 1a with yellow UbIP), FRET efficiency histograms and population estimates are shown. All experiments are normalized to equivalent TCCD areas (see Supplementary Fig. 12 for Z-parameter plots). (a) Lys63-linked diUb (pdb-id 2jf513). (b) Lys63-linkage specific antibody (3dvg17). (c) Met1-linked diUb (2w9n13). (d) NEMO UBAN (2zvo18, only one diUb shown). (e) AMSH-LP (2znv19), and K63NC single-molecule FRET with AMSHi. (f) USP21 (2y5b21, proximal Ub indicated). (g) Model for Lys63- and Met1-linked diUb interaction with binding partners.
Access to the isopeptide bond is essential for DUB activity. AMSH-like protein (AMSH-LP), a Lys63-specific JAMM family DUB, binds an open Lys63-linked diUb conformation19 (Fig. 3e). Indeed, inactivated AMSH20 (AMSHi) depleted the FRET and increased the non-FRET population of K63NC in single-molecule measurements (Fig. 3e). Structures of Ub specific protease (USP) DUBs with diUb bound across the active site are unavailable, but monoUb complexes show that the C-terminus of a distal Ub is extended7 (Fig. 3f), suggesting that USPs also bind open conformations. Indeed, inactivated USP2121 (USP21i) enriched non-FRET K63NC conformations in our single-molecule measurements (Fig. 3f). Therefore, while the Lys63 linkage-specific antibody or the UBAN domain of NEMO selects existing compact conformations of K63NC and M1NC respectively, DUBs select pre-existing open conformations of K63NC (Fig. 3g).
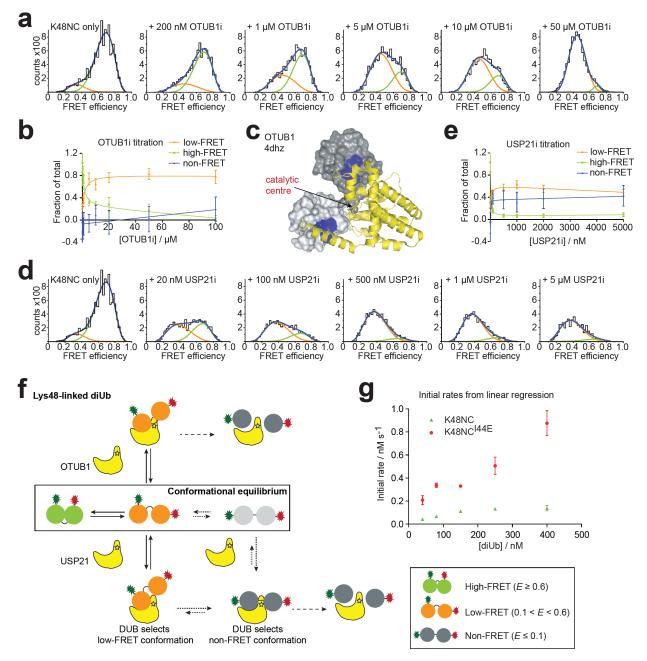
K48NC is in equilibrium between predominantly compact conformations (Fig. 2a). Inactivated OTUB1 (OTUB1i), a Lys48-specific DUB, enriched the low-FRET and depleted the high-FRET population of K48NC in single-molecule measurements (Fig. 4a,b, Supplementary Fig. 9). This is consistent withrecent crystal structures of OTUB1 in complex with E2 and two Ub molecules22,23 that revealed a relatively compact conformation of the two Ub moieties when bound to OTUB1 (Fig. 4c).
Figure 4. DUB interaction with Lys48-linked chains.
(a) FRET efficiency histograms of K48NC with increasing OTUB1i concentration. Estimated FRET efficiencies: Ehigh-FRET≈0.69, Elow-FRET≈0.45, Supplementary Fig. 13, Methods). (b) Relative changes of K48NC populations from a. Errors represent standard deviation in curve fitting. (c) Structure of OTUB1 (yellow) bound to two Ub molecules (grey) (derived from 4dhz22,23). (d) FRET efficiency histograms of K48NC with increasing USP21i concentrations as in a (Ehigh-FRET≈0.71, Elow-FRET≈0.37). (e) Relative changes of K48NC populations derived from d, shown as in b. (f) Model for interaction of DUBs with Lys48-linked chains. (g) Initial rate analysis of USP21-mediated hydrolysis of K48NC (green) and K48NCI44E (red). Errors represent standard deviation of triplicate measurements.
Interestingly, titration of K48NC with USP21i resulted in depletion of high-FRET and increase in low-FRET populations, but also gave rise to a non-FRET population of K48NC (Fig. 4d,e). The appearance of open, non-FRET K48NC species can be rationalized structurally, as USP21 stretches the linkage across the active site to form a catalytically competent conformation (Fig. 3f). However, the increase in low-FRET populations suggests that USP21i binds semi-open conformations directly (Fig. 4d,e). Estimation of binding constants for the low- and non-FRET species indicated a slightly higher affinity of USP21i for the open non-FRET conformation (Supplementary Fig. 9).
Our data result in two models for DUB interactions with Lys48-linked Ub chains (Fig. 4f). DUBs such as OTUB1 may directly recognize and hydrolyze low-FRET semi-open conformations of Lys48-linked diUb. Other enzymes such as USP21 recognize semi-open and open (i.e. low-FRET and non-FRET) conformations. Binding of semi-open conformations could lead to remodeling to open conformations that are compatible with catalysis (Fig. 4f). Alternatively, dissociation of low-FRET, and re-binding of non-FRET diUb could account for generating a catalytically competent enzyme-substrate complex (Fig. 4f). Both models imply that recognition of Lys48-linkages is governed initially by conformational selection.
Importantly, our data indicate that the compact, high-FRET population of K48NC is not recognized directly. Access to Ub HPs is obstructed in the prevalent compact structure of Lys48-linked diUb (Fig. 1a8-10) that would correlate with a high-FRET population. Since all known DUBs bind the HP of Ub7, interconversion from compact high-FRET to semi-open low-FRET conformations could be crucial for DUB activity (Fig. 4f). To test this hypothesis, we mutated Ile44 in the HP of the proximal Ub of K48NC (K48NCI44E) to disrupt a HP-based interface. We observed a reduction in the high- and low-FRET populations and a shift in their peak positions to lower FRET efficiencies as compared to K48NC (Supplementary Fig. 10), suggesting that the mutation changes the conformational equilibria such that open conformations, as well as alternative compact forms of this chain type may now be populated (e.g. Fig. 1b11). Importantly, kinetic assessment of USP21-mediated cleavage shows that K48NCI44E is hydrolyzed significantly faster than wild-type K48NC (Fig. 4g). We propose that Ub chain conformation and dynamics directly affect the rate of DUB hydrolysis.
In summary, we show here how FRET/TCCD measurements can be used to probe the complex conformational dynamics of Ub chains in solution, thereby identifying novel conformations not detected using NMR or X-ray crystallography. Our results establish that distinct conformational populations of flexible two-domain Ub chains are recognized by UbIPs. In the case of USP21, we suggest that subsequent remodeling may take place to achieve active conformations. Such a combination of ‘conformational selection’ and remodeling would be consistent with the most recent models of macromolecular recognition24, and may further extend these models, since the remodeling step here involves significant translation and presumably rotation of flexibly-linked domains with respect to each other. Hence, conformational selection is an important mechanism of Ub chain recognition. This highlights the importance of understanding the conformational space for the eight different Ub chain types, as this holds the key to linkage-dependent regulation within the Ub system.
Ub chain recognition further depends on the dynamic interconversion of chain conformations. Interfering with conformational dynamics by mutating Ub can directly affect chain hydrolysis by DUBs, providing the first evidence that conformational rearrangements in the chains may govern the speed at which the chains are hydrolyzed. An alternative mechanism to interfere with the conformational dynamics is chain length. Lys48-tetraUb forms a compact structure25 in which all Ub moieties interact with each other through their HPs and secondary interaction sites (Supplementary Fig. 11). This likely alters chain dynamics (‘breathing’ of the chains), which we have here shown to be essential for recognition by DUBs. Consistent with this are recent data showing that some DUBs remove mono- and diUb but not tetraUb or longer polymers from substrates26. It is tempting to speculate that the reported requirement for Lys48-linked tetraUb to trigger proteasomal degradation27 may have originated partly from improved stability of this length/linkage combination towards DUB action. Therefore, factors affecting Ub chain conformation and dynamics (linkage, length, binding partners) may be key regulators of the ubiquitin system.
Methods summary
DiUb molecules were assembled with Alexa488 and Alexa647 fluorophores as detailed in Supplementary Fig. 1. Single molecule measurements were performed at 20 °C in PBST buffer (150 mM NaCl, 18 mM Na2HPO4, 7 mM NaH2PO4, 0.01% (v/v) Tween20, pH 7.4). UbIPs were incubated with labeled chains for 5 min. The single molecule instrument was described previously28. Recording times depended on the amount of FRET-labeled diUb and the signal-to-noise ratio, and were typically between 30 min and 3 h. Both TCCD and FRET data were collected for all measurements, and were analyzed using a coincidence criterion. Coincident events in both channels15 are only selected when the counts in each channel are above its threshold count value, which was selected automatically as described previously29. For experiments in which TCCD data were compared with FRET data, a common threshold was used for the donor channels (determined by maximizing the association quotient in the TCCD experiment), whereas the acceptor channel thresholds were independently determined for FRET and TCCD (see Methods). This ensured that both the FRET and TCCD measurements of each sample resulted in approximately equal burst rates in the donor channel. Bulk FRET measurements, dye lifetime measurements and PAX experiments were performed as described in Supplementary Methods. Further details can be found in the Methods online and Supplementary Methods.
Methods
Constructs
Ub constructs with K48R or K63R mutations (for distal Ub moiety in the diUb) and N-terminal Ala-Cys sequence were prepared in pOPIN-S (His6-SUMO-tag)30. Ub constructs with mutation G76C (for proximal Ub) were prepared in pOPIN-E (C-terminal His6-tag) 30. Met1-linked diUb in pRS was mutated to append an N-terminal Met-Ala-Cys sequence, and to change C-terminal Gly152 to Cys.
The following UbIP constructs were used: USP21 (aa 196-565) in pOPIN-S21, human NEMO UBAN (aa 257-350) in pGEX-6P13 (GST-tag), OTUB1 (aa 40-271) in pET28a-LIC31 (GST-tag, kind gift of B. Kessler, Oxford) and full-length AMSH in pGEX-6P20 (GST-tag, kind gift of Sylvie Urbe, Liverpool). Inactive DUBs were generated by site-directed mutagenesis using described protocols and primers introducing the following mutations of catalytic residues: C221A (USP21i), C91A (OTUB1i) and E280A (AMSHi). All constructs were verified by sequencing. The Lys63-specific antibody17 was purchased from Millipore.
Labeling, chain ligation and purification of FRET diUb
Alexa Fluor 488 C5 maleimide (A488) and Alexa Fluor 647 C2 maleimide (A647) were purchased from Invitrogen, dissolved in DMSO (1 mg/300 μL), snap-frozen in 20 μL aliquots and stored at −80 °C. Labeling of Ub Cys mutants was achieved by reacting ~80 μM Ub in labeling buffer (see Supplementary Methods) with 1.2-fold molar excess of fluorophore dissolved in DMSO. Met1-linked diUb was labeled in the same manner using equimolar ratios of A488 and A647. The reaction mixture was agitated in the dark at room temperature for three hours. Unreacted dye was removed by size-exclusion chromatography (HiLoad S26/10 column, GE Healthcare) using Ub buffer (see Supplementary Methods). Unlabeled protein was separated from labeled protein using anion-exchange chromatography (MonoQ 5/50, GE Healthcare) in Ub buffer and eluted with a NaCl gradient from 0 to 1 M. Labeling of Ub Cys mutants was confirmed by LCT mass spectrometry (Supplementary Fig. 1a). Enzymatic ligation of Ub chains was performed according to published protocols21 (see Supplementary Methods). DiUb was separated from monoUb by multiple runs of anion-exchange chromatography (MonoQ).
Single-molecule TCCD and FRET instrumentation
The same instrumentation was used for both TCCD and FRET measurements. This has been reported in detail previously28. For TCCD experiments, two lasers at 488 nm and 633 nm illuminated the sample containing labeled diUb at ~25 pM concentration, and emission of fluorescence signals was recorded at 512-557 nm (donor channel) and 667-722 nm (acceptor channel), respectively. Only excitation at 488 nm was used for FRET measurements. All measurements were performed at 20 °C in PBS buffer (150 mM NaCl, 18 mM Na2HPO4, 7 mM NaH2PO4, pH 7.4) including 0.01% (v/v) Tween20 in a total volume of 250 μL.
Prior to the addition of each UbIP such as Lys63-specific antibody, NEMO and inactivated DUBs, TCCD and FRET data were collected for dual-labeled diUb alone. We did not observe significant differences between different batches of FRET-labeled diUb. Inactive DUBs were added to the measured sample, mixed and incubated for > 5 min. Recording times depended on the amount of FRET-labeled diUb and the signal-to-noise ratio, and were typically between 30 min and 3 h. Both TCCD and FRET data were collected for all measurements. Titration measurements were performed in the same manner on the same day. With high concentrations of UbIP, significant noise was sometimes observed, limiting the dynamic range of the titration measurements. To allow for comparison of each dataset, we normalized the number of events detected in TCCD to a common value (see Supplementary Methods), and applied the same normalization factor to the FRET data for the same sample.
Single-molecule data analysis
Coincidence criterion
Both FRET and TCCD experiments were analyzed using a coincidence criterion. Coincident events in both channels15 are only selected when the counts in each channel are above its threshold count value. This approach filters out the zero peak, enabling the analysis of only significant FRET events.
The Z-parameter was calculated for each significant event in the FRET and TCCD experiments according to equation 1
| (1) |
where ID and IA are the intensities of the coincident burst in the donor and acceptor channel, respectively, and γ is the instrumental factor to account for differences in detection efficiency and quantum yield between the different channels (see Supplementary Methods). The Z-parameter for each significant event was used to generate histograms for each experiment.
Threshold selection
The association quotient, Q, is the ratio of the rate of coincident events to the sum of the rate of all events in the donor and acceptor channels, with a correction factor to avoid double counting of coincident events15. It is defined as
| (2) |
where the event rates (s−1) in the donor and acceptor channels are A and B, respectively; the observed rate of coincident events is C; and the estimated rate at which coincident events occur by chance is
| (3) |
where τ is the interval time in seconds. The chance coincident event rates were calculated by randomly shuffling the order of the frames in one detection channel and subsequently pairing these with the non-shuffled frames in the other channel and counting the number of coincident events15. We have previously shown that appropriate thresholds can be found automatically by treating the association quotient as a function of the thresholds and finding its maximum29. For experiments in which TCCD data were compared with FRET data, a common threshold was used for the donor channels (determined by maximizing Q in the TCCD experiment), whereas the acceptor channel thresholds were independently determined for FRET and TCCD. This ensured that both the FRET and TCCD measurements of each sample resulted in approximately equal burst rates in the donor channel (see below and Supplementary Fig. 3). Automated threshold selection did not significantly bias experiments where multiple FRET populations co-existed as shown by the good agreement with PAX measurements (see Supplementary Material).
Detector thresholds
The selection of thresholds for analysis of FRET and TCCD experiments is described in the Methods section. Typical thresholds for K48NC were 5-6 kHz in the donor channel and 8-9 kHz in the acceptor channel for both FRET and TCCD measurements. For K63NC, typical thresholds in the donor channel were 6-8 kHz in both FRET and TCCD, and in the acceptor channel 5-6 kHz for FRET and 9-10 kHz for TCCD. For M1NC, typical thresholds in the donor channel were 7-8 kHz in both FRET and TCCD, and in the acceptor channel 5-6 kHz for FRET and 7-9 kHz for TCCD. UbIPs increase the background, and therefore the automated threshold determination software calculates optimized thresholds that were slightly increased as compared to measurements without UbIP.
Identification of non-FRET populations
We investigated how different UbIPs changed the FRET population(s) of diUb samples. For certain UbIPs, the number of events detected in TCCD was significantly higher than from the corresponding FRET measurement (see Supplementary Fig. 12 and 13). We could therefore infer a ‘non-FRET’ population from the differences, and estimate the fraction of these events according to equation 4
| (4) |
where NF is the fraction of molecules displaying no FRET signal, and ATCCD and AFRET are the integrated areas under the fitted curves of the TCCD and FRET Z-parameter histograms, respectively. We tested our ability to recover non-FRET populations by preparing a range of samples made up of two dual-labeled DNA duplexes, one with a high FRET efficiency (duplex 2, see Supplementary Methods) and another with a FRET efficiency of zero (duplex 1). Supplementary Fig. 3 shows that we were able to recover the correct populations at this FRET efficiency.
Periodic Acceptor Excitation (PAX)
We used Periodic Acceptor Excitation (PAX) to confirm the presence or absence of populations with very low FRET efficiencies, which in traditional FRET and coincidence experiments are usually either undetected due to the thresholds applied to the fluorescent events, or hidden below a zero-peak. In PAX experiments, the laser at the acceptor wavelength is modulated, making it possible to determine both the donor:acceptor dye stoichiometry and the FRET efficiency of fluorophore-labeled single molecules transiting the confocal volume32. The criterion for selecting molecules is firstly to determine whether there is an acceptor dye present, and then calculate the stoichiometry of the dyes in the molecule; only if the stoichiometry indicates the presence of a donor label is the event counted. Both thresholds are independent of FRET efficiency, and this therefore leads to the unbiased detection of molecules with different FRET efficiencies, i.e. molecules with very low FRET efficiencies are detected with an equal propensity as those with higher FRET efficiencies.
The PAX instrumentation is described in Supplementary Material. Each single molecule takes approximately 1 ms to transit the confocal volume. During this time, the 633 nm red laser was pulsed 20 times; the data collected when the laser was off were combined, as were those when the laser was on. A 3 μs delay was introduced between each laser-switching event and data collection to allow for instrument response time. The collection regime used, described in detail in Supplementary Material, was able to recover the correct populations of FRET and non-FRET events in pre-determined DNA samples (see Supplementary Material, and Supplementary Fig. 4).
The estimated populations from K48NC agree within 20 % with those estimated using the TCCD/FRET methodology and within 7 % for the K63NC sample (see Supplementary Methods for further details, and Supplementary Fig. 5). Qualitatively both measurements show that that K48NC exists predominantly in closed conformations, whereas K63NC is in equilibrium between open and half-closed. An extra non-FRET population of 2% is observed for K48NC in PAX. This is below the detection limit of TCCD/FRET of about 5% estimated from the intercept in Supplementary Fig. 3b. This suggest there might be a small non-FRET population present, at the limit of detection of both methods, and this has been take into account in the models presented (Fig. 4f). Overall these comparison experiments with PAX confirm that TCCD/FRET measures all the populations present and can be used to follow the relative changes in these populations on addition of binding partner.
Supplementary Material
Acknowledgments
We would like to thank members of the Komander, Jackson and Klenerman labs, Roger Williams, Stefan Freund, Chris Johnson, Stephen McLaughlin and Alan Fersht for helpful discussions. Work in the Komander lab is supported by the Medical Research Council [U105192732] and the EMBO Young Investigator Program. GB and SI were supported by the BBSRC, the Newton Trust and an EMBO YIP small grant to D. Komander. Work in the Klenerman lab is supported by EPSRC.
Footnotes
Author contributions YY, GB and MHH designed and performed the experiments, including single molecule measurements, and analyzed the data. YY and GB generated all proteins used in this study. YY performed kinetic experiments. MHH and SI built the PAX instrument and AZZ programmed the control for PAX measurements. SI performed single molecule experiments and contributed to data analysis. MJRR and AO performed lifetime measurements. DK, SEJ and DK directed the research and analyzed the results. All authors contributed to the writing of the manuscript.
The authors declare no competing financial interest.
References
- 1.Conte Lo, L., Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 2.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 3.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Iwai K. Linear polyubiquitin chains: a new modifier involved in NFκB activation and chronic inflammation, including dermatitis. Cell Cycle. 2011;10:3095–3104. doi: 10.4161/cc.10.18.17437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 8.Cook WJ, Jeffrey LC, Carson M, Chen Z, Pickart CM. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J Biol Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 9.Ryabov Y, Fushman D. Interdomain mobility in di-ubiquitin revealed by NMR. Proteins. 2006;63:787–796. doi: 10.1002/prot.20917. [DOI] [PubMed] [Google Scholar]
- 10.Tenno T, et al. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells. 2004;9:865–875. doi: 10.1111/j.1365-2443.2004.00780.x. [DOI] [PubMed] [Google Scholar]
- 11.Hirano T, et al. Conformational dynamics of wild-type Lys-48-linked diubiquitin in solution. J Biol Chem. 2011;286:37496–37502. doi: 10.1074/jbc.M111.256354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varadan R, et al. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 13.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohaim A, Kawasaki M, Kato R, Dikic I, Wakatsuki S. Structure of a compact conformation of linear diubiquitin. Acta Crystallogr D Biol Crystallogr. 2012;68:102–108. doi: 10.1107/S0907444911051195. [DOI] [PubMed] [Google Scholar]
- 15.Orte A, Clarke R, Balasubramanian S, Klenerman D. Determination of the fraction and stoichiometry of femtomolar levels of biomolecular complexes in an excess of monomer using single-molecule, two-color coincidence detection. Anal Chem. 2006;78:7707–7715. doi: 10.1021/ac061122y. [DOI] [PubMed] [Google Scholar]
- 16.Fraser JS, et al. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462:669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 18.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Sato Y, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 20.McCullough J, et al. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr Biol. 2006;16:160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 21.Ye Y, et al. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011;12:350–357. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483:618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juang Y-C, et al. OTUB1 Co-opts Lys48-Linked Ubiquitin Recognition to Suppress E2 Enzyme Function. Mol Cell. 2012;45:384–397. doi: 10.1016/j.molcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 26.Schaefer JB, Morgan DO. Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J Biol Chem. 2011;286:45186–45196. doi: 10.1074/jbc.M111.310094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orte A, Clarke RW, Klenerman D. Fluorescence coincidence spectroscopy for single-molecule fluorescence resonance energy-transfer measurements. Anal Chem. 2008;80:8389–8397. doi: 10.1021/ac8009092. [DOI] [PubMed] [Google Scholar]
- 29.Clarke RW, Orte A, Klenerman D. Optimized threshold selection for single-molecule two-color fluorescence coincidence spectroscopy. Anal Chem. 2007;79:2771–2777. doi: 10.1021/ac062188w. [DOI] [PubMed] [Google Scholar]
- 30.Berrow NS, et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edelmann MJ, et al. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–390. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 32.Kapanidis AN, et al. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.