Abstract
Over the last 50 years, sequencing, structural biology and bioinformatics have completely revolutionised biomolecular science, with millions of sequences and tens of thousands of three dimensional structures becoming available. The bioinformatics of enzymes is well served by, mostly free, online databases. BRENDA describes the chemistry, substrate specificity, kinetics, preparation and biological sources of enzymes, while KEGG is valuable for understanding enzymes and metabolic pathways. EzCatDB, SFLD and MACiE are key repositories for data on the chemical mechanisms by which enzymes operate. At the current rate of genome sequencing and manual annotation, human curation will never finish the functional annotation of the ever-expanding list of known enzymes. Hence there is an increasing need for automated annotation, though it is not yet widespread for enzyme data. In contrast, functional ontologies such as the Gene Ontology already profit from automation. Despite our growing understanding of enzyme structure and dynamics, we are only beginning to be able to design novel enzymes. One can now begin to trace the functional evolution of enzymes using phylogenetics. The ability of enzymes to perform secondary functions, albeit relatively inefficiently, gives clues as to how enzyme function evolves. Substrate promiscuity in enzymes is one example of imperfect specificity in protein-ligand interactions. Similarly, most drugs bind to more than one protein target. This may sometimes result in helpful polypharmacology as a drug modulates plural targets, but also often leads to adverse side-effects. Many cheminformatics approaches can be used to model the interactions between druglike molecules and proteins in silico. We can even use quantum chemical techniques like DFT and QM/MM to compute the structural and energetic course of enzyme catalysed chemical reaction mechanisms, including a full description of bond making and breaking.
1. INTRODUCTION
Thousands of enzymes play essential roles in catalysing chemical reactions critical for the survival of living organisms [1]. Often enzymes are very precisely selective for their substrate and the chemistry and stereochemistry of the reaction they catalyse; however, some enzymes carry out a variety of different functions with a range of efficiencies [2]. Our understanding of systems biology and metabolism depends on a detailed knowledge of enzyme function [3].
For five decades, the community has been annotating enzymes with Enzyme Commission (EC) numbers, a simple hierarchical description of their function [4]. Over that time, sequencing, structural biology and bioinformatics have completely revolutionised biomolecular science, with millions of sequences in UniProt [5] and tens of thousands of three dimensional (3D) structures in the PDB [6]. Beyond the sheer quantity of data, a plethora of software, much of it freely available, allows sophisticated computations to be carried out on large datasets. Ever increasing hardware capabilities and network capacities ensure that computations can be carried out rapidly either locally or over the internet.
Since around half of all known drugs act by inhibiting or modulating enzymes [7], understanding the structures and functions of enzymes is important for drug design. While the design of small molecules to inhibit or modulate enzymes has been a mainstay of molecular design, more ambitious goals now appear achievable. The design of enzymes themselves is progressing from site-directed re-engineering of natural proteins towards de novo design and assembly of active enzymes. Jiang et al. created from scratch novel active sites capable of catalytic activity as retro-aldol enzymes, basing their design process on a careful analysis of the catalytic requirements of the chemical mechanism and inserting their designs in scaffolds borrowed from naturally occurring proteins [8]. The same level of understanding of enzyme function is also used to predict the unknown functions of protein structures [9]. Function prediction from sequence can also achieve high accuracy if a suitable database including close evolutionary homologues of the query sequence, expected usually to share similar function, is available [10]. However, the high throughput and difficulty of curation of automated function prediction carry a risk of propagating many mis-annotations through the bioinformatics databases [11].
Depending on the context, enzyme function can be defined at different levels. Here, we are chiefly concerned with the molecular function, the chemist’s perspective. For example, to the chemist, angiotensin converting enzyme (ACE) is an exopeptidase catalysing the conversion of angiotensin I to angiotensin II by the hydrolytic removal of two C-terminal residues (EC 3.4.15.1), as well as degrading bradykinin. To the biologist, ACE brings about vasoconstriction and thereby raises blood pressure [12]. Both the chemical and biological descriptions are equally correct, but see the enzyme’s function from different perspectives.
2. TYPES AND SOURCES OF DATA
BRENDA [13, 14] contains copious information about the chemistry, substrate specificity, kinetics, preparation and biological sources of enzymes. Kinetic parameters will be especially valuable in the future, with Bar-Even et al. having demonstrated that the rate acceleration and efficiency of real enzymes often falls far below those of idealised textbook case studies [15]. KEGG (Kyoto Encyclopaedia of Genes and Genomes) is a valuable resource, especially for understanding enzymes in the context of metabolic pathways [16, 17]. The ExplorEnz database is useful for searching and browsing the EC classification, and is also a convenient source of up-to-date statistical information, from which we learn that there are (August 2012) 4867 current EC numbers [18, 19]. Both IntEnz [20, 21] and the official website of the NC-IUBMB [4, 22] also provide gateways to enzyme classification and nomenclature information. The Catalytic Site Atlas (CSA) describes the residues which have catalytic functionality in the active sites of enzymes, based on almost 1,000 papers in the primary literature, with the corresponding information also inferred for about 27,000 homologs. The CSA provides 3D templates which can be used to search a protein structure for spatial signatures of enzymatic activity [23, 24]. These can be searched, for instance, to find evidence of convergent evolution [25]. The major enzyme databases are thoroughly hyperlinked together, so that the sequences from UniProt [5] and X-ray or Nuclear Magnetic Resonance (NMR) structures in the PDB [6] are easily available when browsing any of the major sources of data.
Some of the most valuable data on enzymes relate to the chemical mechanisms by which they operate. EzCatDB contains mechanistic data on 828 enzyme reactions (August 2012) [26, 27]. These data are classified according to EzCatDB’s own RLCP classification system of mechanisms. The Structure-Function Linkage Database (SFLD) concentrates on superfamilies of evolutionarily related, divergently evolved enzymes [28, 29]. The SFLD facilitates study of the evolutionary paths through which related enzyme mechanisms diverge [30]. MACiE combines extensive coverage of the world of enzyme reactions with a very detailed stepwise description of the mechanism of each one, including every chemical bond made or broken [31, 32]. MACiE has been used to show that convergently evolved enzymes typically carry out similar chemical transformations by very different mechanisms [33], for validating atom-to-atom matching between the chemical structures substrates and products [34], and for understanding the complexity of enzymes [35].
3. ENZYME ANNOTATION AND FUNCTION PREDICTION
Sequence based methods are more widely applicable to newly sequenced proteins compared to methods requiring a 3D protein structure. We now consider the kinds of data and methods used to predict enzymatic function from gene or protein sequences.
3.1 Curation of enzyme function
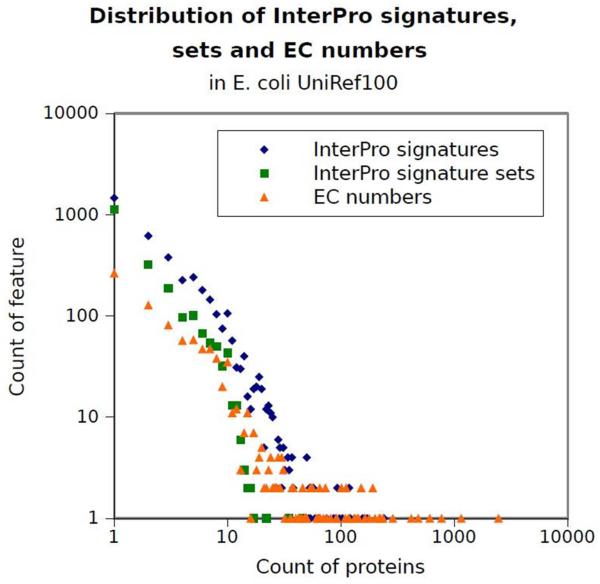
Despite some known limitations, such as some inconsistencies between the rules set by the nomenclature committee and the actual EC number definitions [36], the NC-IUBMB Enzyme Commission (EC) nomenclature [4] is widely used to define enzymatic reactions, and is the current standard for enzyme function classification. The EC nomenclature uses a four digit code, such as EC 1.2.3.4, to represent an enzymatic reaction. Enzymatic classes are long-tail distributed, that is, some EC numbers are very frequent among proteins while most EC numbers only rarely occur, as shown in Figure 1. 80% of EC numbers annotate only about 10% of UniProt [5] enzymes, while the remaining 20% most common EC classes annotate 90% of UniProt enzymes. 731 EC numbers have fewer than five protein examples in UniProt; 277 EC numbers only have one protein example in UniProt.
Figure 1.
Distribution of InterPro signatures, InterPro signature sets and EC numbers in the E. coli UniRef100 data of UniProt [5]. Both axes are logarithmic.
Enzyme resources can be divided into archives, closed and open databases. In archives, only the submitting authors have the right to assign an annotation. Examples of archives are the Protein Data Bank [6] or the EMBL (European Molecular Biology Laboratory) Nucleotide Sequence Database [37]. In closed databases, a privileged group of curators is responsible for the manual annotation of enyzmes. Examples of closed databases include UniProt Swiss-Prot [5], KEGG [38, 39] and Reactome [40]. In contrast, open databases allow any non-malicious user to edit the collected annotations. An example is PDBWiki [41], which allows the community to annotate the protein structure entries deposited in the Protein Data Bank (PDB) [6]. In practice, no current wiki is entirely dedicated to enzyme annotation.
Enzyme annotation is rarely changed or curated in most secondary databases, and is usually represented as a link to a read-only entry in one of the main data sources mentioned above. Enzyme annotation tends to cascade down from archives to other databases unchallenged. For example, in UniProt TrEMBL (the non-manually annotated section of UniProt), EC numbers are usually taken directly from the corresponding EMBL Nucleotide Sequence Database or PDB entry. UniProt Swiss-Prot is, together with KEGG, one of the richest collections of manually curated protein entries. In Swiss-Prot, the justification for a protein annotation is provided as a list of literature entries. However, no specific paper is directly linked to the EC annotation, making it difficult to identify the specific evidence the curator used to assign a certain enzymatic function.
3.2 Automated prediction of enzyme function
At the current rates of genome sequencing and expert annotation, manual curation will never complete the functional annotation of all available enzymes [42], hence the need for automated annotation. We concentrate on methods that use sequence signatures, evolutionarily conserved patterns derived from known genetic sequences, as attributes (features) for machine learning prediction. A signature is usually annotated with its function, derived from the literature, structures or mutant information available for the underlying sequences containing that signature. One of the main databases of signatures is InterPro [43], an umbrella database including twelve other sources of signatures: Pfam, PRINTS, PROSITE, SMART, ProDom, PIRSF, SUPERFAMILY, PANTHER, CATH-Gene3D, TIGRFAMs and HAMAP. Out of all their signatures, InterPro curators manually condense, give unique names and integrate the most reliable signatures. Sequence signatures can have different lengths and tolerance for mutations, ranging from short catalytic sites with a stringent requirement in terms of amino acid type to entire protein domains composed of hundreds of amino acids. InterPro also provides a browser-based web service and offline software to match signatures to any sequence (InterProScan) [44].
Automated prediction is not widespread for EC annotations. In contrast, functional ontologies such as the Gene Ontology already profit from established automated prediction, such as the InterPro2GO method that assigns Gene Ontology terms depending on matches to InterPro signatures [45]. Tetko et al. [46] used principal component analysis to show that the highest contributors to the performance of various protein function prediction methods were InterPro signatures. Hence, in theory, EC labels too could be directly assigned from InterPro domains using the InterPro2GO method and then the EC2GO lists. However, as shown in [10], this method would have much lower accuracy (80%) than more recent machine learning methods (EnzML 97%).
Sequence based methods for predicting enzyme function, in the guise of EC numbers, include EFICAz [47], ModEnzA [48] and PRIAM [49]. However, some enzymes can perform different reactions, either due to the presence of multiple catalytic sites, or because of substrate promiscuity, or by regulation of a single site. Hence, one enzyme can be associated with multiple EC numbers (multi-label), while these methods can only predict one EC number per protein (multi-class). Regarding multi-label prediction, support vector machines were used to predict EC numbers up to the second digit (e.g., EC 1.2.-.-), on 8,291 enzymes [50]. Hierarchical classification was also applied to about 6,000 enzymes from KEGG, obtaining over 85% accuracy in predicting full four-digit EC numbers [51]. EnzML [10] also used InterPro signatures to predict multiple four-digit EC numbers, and reached an accuracy of over 95% on about 300,000 Swiss-Prot proteins using a K-Nearest Neighbours multi-label algorithm [52].
3.3 The Nature and Limitations of Automated Prediction
The overall success of these methods is partly due to InterPro signatures providing a compact representation of protein functionality. The 13.5 million proteins in UniProt are described by only 154,583 unordered sets of InterPro signatures (attributes). Many of these sets are very similar, only differing by one signature. The success of bioinformatics-based prediction methods clearly owes much to the extent to which protein sequence space has already been explored and annotated. For any given query sequence it is very likely that, amongst the hundreds of thousands of sequences with existing functional annotations, there will be some containing the same sequence signatures - indicative of evolutionary relatedness and probably shared function. These methods, which effectively transfer functional annotations amongst evolutionary neighbours, seem to reproduce well the human curation process, including - in all probability - the same mistakes. Propagation of annotations demands trust that the original is correct. Automated methods can only be evaluated against current manual annotation, while they cannot be easily validated against extensive experimental evidence; biochemical characterisation of protein function has a much lower throughput than sequencing [11].
Current manual annotation of enzymes differs substantially between databases such as Swiss-Prot, KEGG and Reactome. In July 2011, up to a third of Swiss-Prot and KEGG enzyme annotations were different. Excluding disagreeing annotations substantially reduces the available data. Creating a gold standard of enzymatic function for automated prediction is also made difficult by the peculiar distribution of EC numbers; using only annotations agreeing in two or more data sources (as done in [10] for Swiss-Prot and KEGG) causes the loss of many rare EC classes. At the same time, if too many redundant sequences are included in the test set, automated methods’ accuracy can easily be overestimated. In the future, enzymatic annotation could usefully profit from the same curation workflow established for Ontology terms [53]. A GO functional class is attached to a protein together with a standardised evidence code (derived from evidence, from literature, automatically predicted etc.) and a link to the available evidence, usually in the form of literature or other supporting data. This would allow more transparent presentation of both manual and automated methods to predict enzymatic function.
4. DESIGNING ENZYMES WITH NOVEL MOLECULAR FUNCTIONS
Nature doesn’t have a problem inventing novel enzymes - but we do. Although our understanding of enzyme structure and dynamics has improved impressively, still we are only on the cusp of being able to properly design a novel enzyme. Here, we discuss examples of computational enzyme design, its limitations and applications. Enzyme design challenges our fundamental grasp of structure and dynamics, and how they enhance catalysis: the structure-function relationship.
4.1 Computational enzyme design
Enzymes are very efficient catalysts, enhancing the rate of reaction many times over. Enzymes are by nature exquisitely selective, capable of selecting amongst closely related substrates to perform controlled reactions [54]. During enzyme catalysis, a substrate binds to the active site, which then stabilizes the transition state, releases the product, and finally the enzyme is restored to its native form, ready for another round of catalysis [55]. The fundamental properties of enzymes are represented in many ways, via sequence, structure and catalytic characteristics [56]. An excellent review by Lipscomb [57] points out the important features of enzyme structure and catalysis that could be used in enzyme design.
Albery et al. [58] considered the evolutionary improvement of enzyme catalysis in nature in terms of molecular structure and catalytic factors, suggesting the importance of the relationship between structure and function in the course of evolution. Eisenmesser et al. [59] showed the fluctuations of the active site’s conformation during catalysis using a nuclear magnetic resonance relaxation method. This study reveals enzyme dynamics with respect to substrate turnover, without including the side-chain dynamics. In contrast, Tousignant et al. [60] have considered the fluctuation of the entire protein to show the modification of catalysis. These analyses add up to a better understanding of the structural dynamics of the entire protein during catalysis. All these sources of evidence show important features needing to be incorporated in enzyme design.
The goal of enzyme design is to reconstruct an enzyme that is capable of performing catalysis efficiently, bearing in mind the properties of enzyme structure and chemical reactions which closely resemble the enzyme reaction in nature. This tests our understanding of many aspects of enzyme function. For example, a study by Kaplan et al. [61] designed an O2-dependent phenol oxidase from scratch, illustrating the powerful role of enzyme selectivity. With progress in designing, engineering and testing the function of enzymes, our basic understanding of structure-function relationships is also improving greatly [62]. The applications of enzyme design are widespread through biomedical to synthetic chemistry, and have expanded into the realms of the chemical industry [63].
4.2 Computational methods and strategy: De novo design and redesign
With our current understanding of enzymes, we are able to improve the catalytic power of an artificial enzyme and discern the important properties while designing or redesigning the protein [64, 65]. De novo [55] designed proteins are novel with respect to structure and catalysis, whereas in redesign [54] the natural protein is mimicked with improved catalysis. In the past [64, 65], enzyme redesign was the more tractable approach.
It is a challenging task to use computational protein design to create an enzyme to perform a desired chemical reaction. Studies by Jiang et al. [8] and by Rothlisberger et al. [66] overcame this challenge by designing a novel catalytically active enzyme where no prior information was provided [67]. In the first paper [8], they constructed a novel enzyme catalyst, performing a retro-aldol reaction on the non-natural substrate 4-hydroxy-4-(6-methyoxy-2-napthyl)-2-butanone. In the second paper [66], they demonstrated a novel computational strategy, combined with molecular evolution, to create novel enzyme catalysts for a reaction with no natural enzyme; the Kemp elimination [68]. Both of these studies used a Rosetta match algorithm [69], rapidly comparing the geometry of active sites in the structure. This method has been used for enzyme catalysis for a variety of chemical reactions [70]. These studies showed an impressive understanding of the relationship between structure and reactivity.
4.3 Challenges remaining in enzyme engineering
Computational enzyme design [67, 71, 72] has achieved significant breakthroughs, with profound biotechnological, pharmacological and biomedical implications: design of new biocatalysts [8, 61, 66] and improving enzymatic activity [73, 74].
The studies by Baker and colleagues [8, 66] demonstrate significant improvement in our fundamental understanding of enzymatic reactions. Kiss et al. [75] evaluated the enzymes designed in [8, 66], suggesting that there is room for improvement in the utilisation of underlying physical principles for enzyme catalysis. They aimed firstly to develop and improve a fast algorithm by filtering out inactive designs, and secondly to reveal any limitations of the earlier studies [8, 66]. Kiss et al. demonstrated that the relative efficiencies of the designed enzymes could be predictively ranked using a MD-based procedure.
We are yet to achieve the dream of routine enzyme design, engineering of de novo biocatalysis with catalytic power comparable to natural enzymes. Protein design faces major challenges, concerning issues such as computational complexity, energy structure-function dynamics and precise atomic-level simulations. Glowacki et al. [76] designed a simple model using transition-state theory based on the realistic physical parameters embracing the structural conformational change involved in the reaction. A study by McGeagh et al. [77] also favours using the transition state theory for understanding the role of structural changes during enzyme catalysis. It would be beneficial in the future to consider together the dynamics of enzyme structure and function [78, 79] for modelling enzyme catalysis, which is closely related to understanding and predicting natural enzymes’ functionality [9].
5. THE EVOLUTION OF ENZYMES
5.1 The evolution of novel functionality in enzymes
It is thought that the most ancient enzymes showed weak specificity and were able to carry out multiple reactions, but with relatively weak catalytic efficiency [80]. Over the course of evolution these enzymes have become optimised to carry out specific reactions and have hence increased in catalytic efficiency [80].
Broadly speaking, two phenomena displayed by modern day enzymes exemplify how novel functionality may have arisen more recently from already highly evolved enzymes. O’Brien & Herschlag developed the ideas of Jensen, who noted that some modern enzymes showed ‘substrate ambiguity’ [80] and subsequently termed enzymes using an active site for an alternative reaction as ‘catalytically promiscuous’ [81], a description later elaborated on by Khersonsky & Tawfik [82].
In order for a new function in an enzyme to evolve, the gene must be able to accumulate new mutations. As such, it is thought that gene duplication has allowed genes to evolve without affecting the function of the ‘master’ copy [83]. However, the point at which duplication comes into play, i.e., whether duplication allows novel function to manifest in the first place or whether duplication merely permits optimisation of an enzyme already performing a second function, has been under debate since its first proposal; examples of recent reviews include [82, 84]. The idea of enzyme promiscuity has been applied to specific cases of divergence of enzyme function, for example by Russell et al. who discuss enzyme promiscuity in terms of evolution of insecticide resistance as compared to xenobiotic metabolism [85].
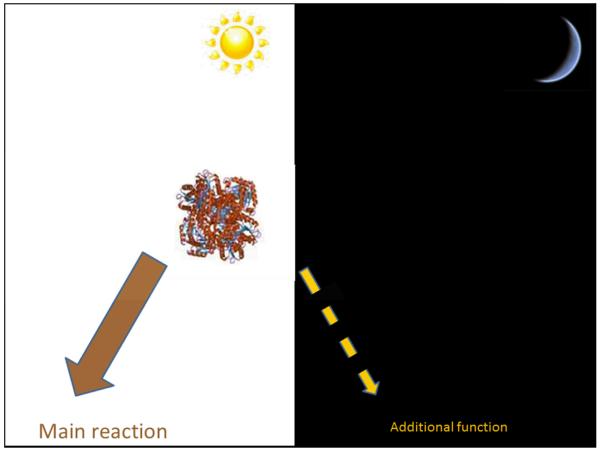
In addition to catalytic promiscuity at the active site, some enzymes are able to ‘moonlight’ [86]. According to Huberts and van der Klei, in moonlighting, the enzyme performs an additional mechanistically different and independent function (Figure 2), using the same domain as its principal function [87]. Moonlighting functions can be structural, regulatory or enzymatic, and tend to occur in locations other than the active site, although this is not always the case as discussed by Copley [88]. Often this additional functionality is context dependent - the dependency upon cellular or extracellular location or other factors is discussed by Jeffery [86]. The ‘classic’ example of moonlighting was discovered by Piatigorsky et al. in 1988 in which the δ-crystallin gene was found to have different roles, one structural, one metabolic, depending on expression levels in the duck [89]. Numerous reviews give examples of moonlighting proteins, we highlight some more recent works that explore the evolutionary mechanisms behind the emergence of novel moonlighting functions [87, 88]. Very recently, a study by Rodríguez Plaza et al., which used protein engineering of ‘hunter-killer peptides’, revealed the possibility that moonlighting proteins are maintained in evolution because the combination of two pre-existing functions in one domain can lead to emergent and novel enzyme functionality [90].
Figure 2.
In moonlighting, the enzyme performs an additional mechanistically different and independent function. This may serve as a starting point for evolving a new principal function.
5.2 Tracing the evolution of function in enzymes
One way to trace back the emergence of function in evolutionary history is by the use of phylogenetics. However, innovation of novel functionality typically represents an ancient event from the perspective of a modern day enzyme. In order to build such a ‘functional phylogeny’, the members chosen must be homologous and their resulting alignment should accurately reflect this homology. This involves discerning which parts of a sequence are most conserved and therefore more likely to have retained some phylogenetic signal.
One way in which to achieve this is by the use of databases such as CATH [91] and SCOP [92], in which enzymes are clustered into homologous groups aided by the conservation of protein structure, which is thought to be more conserved than sequence. Recent studies which have utilised structure to aid inference of distant evolutionary relationships include the exploration of the strictosidine synthase-like proteins [93] and the ferritin-like superfamily [94].
The FunTree online application, released in 2011, allows for the evolution of CATH defined superfamilies (at the ‘Homologous Superfamily’ level) to be explored [95, 96, 97]. The application uses structural alignments to define ‘structurally similar groups’ (SSGs) within the superfamily. Sequences within each SSG are aligned using a structurally informed method. The resulting alignment is then used to build phylogenetic trees that integrate data from the CSA [23, 24] and MACiE [31, 32] and allow for structural superimpositions of structures at each node to be visualised using Jmol [98]. Thus, the evolution of functions in diverse superfamilies can be seen in terms of changes in structure and active site residues.
An application such as FunTree represents an interesting and exciting way to explore evolution. However, one only has to examine the ‘example’ phylogenetic trees on the FunTree homepage [97] to see that in many cases the bifurcations at some nodes, especially more ancient ones, show a low level of bootstrap support. This is unsurprising, given the low level of homologous signal so far back in evolutionary history. The problem of low signal to noise ratio in studies of highly divergent enzymes is on-going and seemingly unavoidable; it will be of much interest to see what strategies will be developed in the future to try and overcome this.
6. PREDICTING DRUG-PROTEIN INTERACTIONS
6.1 On and off-target prediction
It is well known that most drugs bind to more than one protein target. This may sometimes result in polypharmacology, beneficial pharmacological effects from a drug modulating more than one target, or more frequently in adverse side-effects. A drug discovery program takes on average around 6 years before a drug candidate is ready for clinical trials and another 6-7 for the three clinical phases. Hence it is very important to be able to identify adverse effects as soon as possible, in order to save time and money as well as for patient safety.
A number of cheminformatics approaches that model in silico the interactions between druglike molecules and proteins have been proposed. They use different molecular representations including two-dimensional fingerprints (2D), three-dimensional fingerprints (3D), 3D pharmacophore models, and shape based approaches (such as Spherical Harmonic representations, Gaussian maps etc.) [99]. These methods can predict interactions with the primary pharmaceutical target, leading to bioactivity, and also off-target interactions that may lead to side-effects. The use of these techniques for virtual screening in the first stages of drug discovery allows up to millions of compounds to be assessed for bioactivity, ADME and toxicity without the need to spend time or money synthesising them.
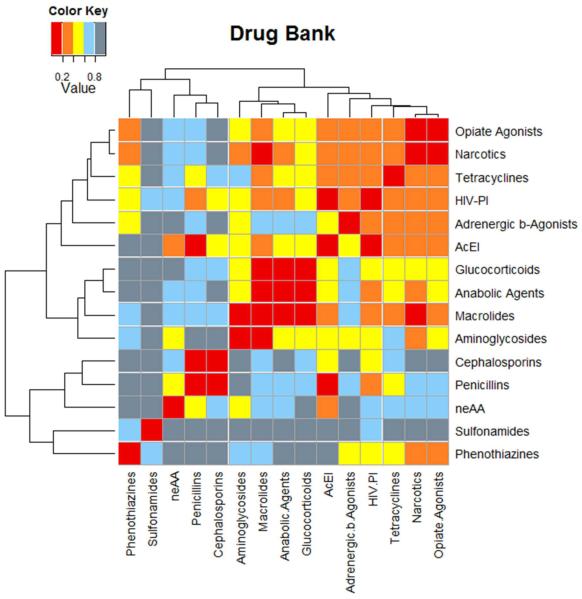
Recent years have seen rapid growth of publically available databases, such as ChEMBL [100], BindingDB [101], PubChem [102] and DrugBank [103], and commercial products like the MDDR [104] and WOMBAT [105]. These databases typically list affinities or activities for molecules against targets, grouping compounds according to their functionality against a given enzyme, protein or disease. Ideally, using those data, one would like to identify all possible drug-to-target interactions or possible target-to-target connections between different proteins and enzymes, as seen in Figure 3. To exploit this vast amount of information, data mining has been combined with many predictive methods such as machine learning, Bayesian and other statistically-based approaches, kernel density estimation and Inductive Logic Programming [106, 107, 108, 109, 110, 111]. Most of these methods use 2D fingerprints to represent and compare molecules, since those methods still outperform the more sophisticated three-dimensional (3D) techniques [112].
Figure 3.
Using 2D fingerprints and a Kernel density estimation function the family associations from the Drug Bank families are predicted. Abbreviations: HIV protease inhibitors (HIV-PI), Angiotensin-converting Enzyme Inhibitors (AcEI), and Non-Essential Amino Acids (neAA).
6.2 Applications of Predictive Models
One important pharmacological side-effect is phospholipidosis, the excess accumulation of phospholipids within cells, which is often triggered by cationic amphiphilic drugs. This affects many different types of cells and can cause significant delay in the drug development process [113]. Recently, machine learning algorithms and kernel density estimation methods have been used in order to predict the propensity of a drug to induce phospholipidosis, with very good results [106, 108]. Such studies show how important computational methods are and how much effort, time and money can be saved with minimal costs.
Another emerging trend is drug repositioning, where drugs are redirected from one specific therapeutic area to another [114, 115]. Computational methods have succeeded in identifying new drug-target associations. These encouraging results provide more motivation for collaboration between experimental and computational laboratories, such as those supported by the European Cooperation in Science and Technology (COST).
7. QM/MM AND DFT METHODS FOR ENZYME REACTIONS
7.1 Theoretical Chemistry Approaches to Enzyme Function
Computational chemistry can be used to elucidate the complex mechanisms of enzymatic function. Chemical kinetics and thermodynamics are fundamental to our understanding, and ultimately control, of enzymatic reactions. These factors are governed by the changes that occur on the molecular and atomic length scales; hence consideration must be given to quantum mechanical (QM) effects. Methods such as density functional theory (DFT) and partitioned quantum mechanics and molecular mechanics (QM/MM) are therefore employed. Results from calculations of this nature can stand alone, or be used to supplement or replace some traditional descriptors in informatics models, such as quantitative structure-property relationships (QSPR) and quantitative structure-activity relationships (QSAR) [116, 117]. Whatever quantum mechanical methods are used to study an enzymatic pathway, the potential energy surface (PES) of the system must be explored, and the lowest energy, and therefore most probable, path through which the mechanism can proceed should be located.
Conventional quantum chemical methods, like Hartree–Fock (HF), are wave function based methods, and focus their effort on finding an appropriate wave function that is a solution of the Schrödinger equation. HF methods can be systematically improved by adding additional terms accounting for electron correlation, leading to the post-HF methods required for an accurate description of correlation effects such as dispersion.
DFT is a set of quantum chemical methods used in electronic structure calculations, generally for small to medium sized molecules of up to a few hundred atoms [118]. DFT finds its roots in the Thomas-Fermi model of the 1920s [119, 120], although most modern methods are based on the work of Hohenberg and Kohn [121] and Kohn and Sham [122] in the 1960s. DFT, unlike the conventional wave function based quantum chemical methods such as HF, derives a system’s electronic energy using density functionals applied to the electron density [123, 118]. DFT is in principle a more efficient method of QM calculation [123, 124], one for which the limiting factor is again the description of electron correlation and also electron exchange. DFT computing time scales approximately as N3, where N is the number of basis functions [118]. DFT has been described as a ladder [125] in which each rung represents a more sophisticated class of functionals and hence an improved approximation. These approximations, unfortunately, are not necessarily systematic improvements in the same sense as for post-HF methods. DFT functionals continue to advance in complexity and accuracy. Some newer functionals, such as the M06 family of functionals, contain empirical data to account for electron correlation effects [126]. Correction schemes have also been derived to overcome some of the deficiencies, such as dispersion correction schemes [127]. For an overview of DFT methods, we refer the reader to a review by Parr and Yang [128].
7.2 Kohn-Sham DFT applied to enzymes
DFT has been applied to small biomolecules such as peptides in the past, sometimes highlighting the shortcomings of certain DFT functionals [129, 130]. DFT has also been applied to small molecule catalysis [131], making DFT a good starting point for looking at larger and more complex catalytic systems. Applications of conventional Kohn-Sham DFT to enzymatic reaction mechanisms have been generally to cut-down model systems, typically containing the active site region of the enzyme and the substrate. These cut down models are sometimes referred to as mimics, biomimics [132], or theozymes [133]. By cutting a system down to a size at which DFT is useable, one has the advantage of being able to treat the interesting portion of the system at the QM level, in reasonable computer time. However, neglecting a large portion of the system can lead to loss of accuracy.
Work by Georgiev et al. [132] exemplifies such DFT methods. The study looks at the enzyme catalysed ring cleavage of catechol derivatives by the iron-containing dioxygenase enzymes, of which there are two groups: intradiol-cleaving (e.g., catechol 1,2-dioxygenase, EC 1.13.11.1) and extradiol-cleaving dioxygenases (e.g., catechol 1,3-dioxygenase, EC 1.13.11.2). A representative theozyme is used in place of the full enzyme. The study proceeds using the hybrid DFT functional B3LYP [134] and an altered form B3LYP* (adjusted to contain 15% HF exchange instead of 20%). B3LYP* had been previously found to produce better results for systems containing transition metals [135]. Georgiev et al. were able to make a reasoned deduction as to why the catalytic process goes selectively via the intradiol-cleaving dioxygenase mechanism. The study shows that both possible mechanisms produce an alkoxy radical via oxygen-oxygen bond homolysis. At this point, the intradiol-cleaved substrate undergoes a barrier free carbon-carbon bond cleavage. In contrast, the extradiol-cleavage mechanism incurs a second barrier to reaction in the formation of an epoxide. Recent similar studies have provided insights into the mechanistic pathway of N-O bond cleavage in nitrous oxide reductase [136], while Blomberg et al. used DFT to model the catalytic mechanism of type II dehydroquinase [137]. A more comprehensive review of these methods and applications can be found in [138].
7.3 QM/MM
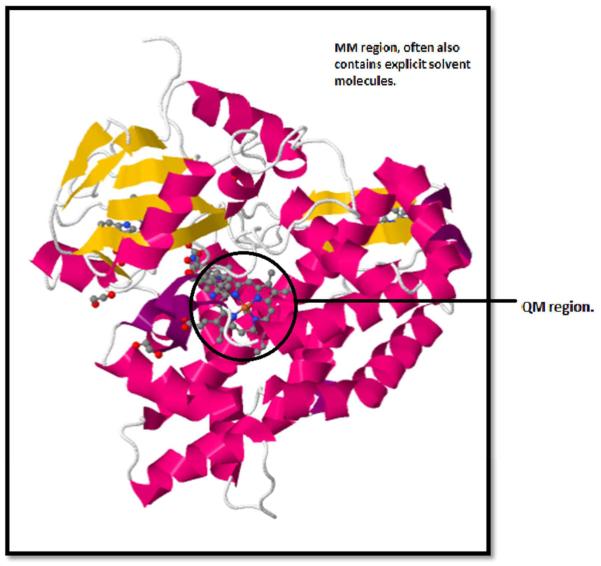
Molecular mechanics (MM) is a level of theory based on classical mechanics, though including non-classical components of the potential energy, such as exchange-repulsion and dispersion. Its force fields have been used for many different applications in chemistry and biochemistry, especially molecular dynamics (MD). One of the major drawbacks of MM is that it cannot be used directly to model systems in which bond breaking and formation occurs, which is often precisely the chemistry we want to study [118]. QM/MM is a method that allows the user to incorporate chemical reactions into MM and also to consider a larger system than allowed by traditional QM methods, such as an enzyme-substrate complex in its entirety. QM/MM involves splitting a calculation into sections; the regions of most interest (typically where bond formation and breaking occur) are treated as QM bubbles, with molecular mechanics (MM) used for the rest of the system, see Figure 4. QM/MM has the advantage over cut-down DFT theozyme models of accounting, at least classically, for the whole macromolecular system. However, the method leads to a complex boundary area between QM and MM regions, and still lacks a QM description of the full system. QM/MM typically uses semi-empirical wave function methods or DFT to describe the QM region, although other methods are possible. In the MM region, a variety of force fields are available, including some polarisable force fields [118, 139, 140]. ChemShell [141] can be used to couple together many popular programs in order to perform QM/MM calculations, while some quantum chemistry packages have self-contained QM/MM functionality. Interesting reviews of QM/MM and quantum chemical methods for studying enzymatic catalysis have been written by Friesner and Guallar [140] and by Lonsdale et al. [142].
Figure 4.
Separation of QM/MM model into QM (inside the black circle) MM (outside the black circle) regions for chromopyrrolic acid bound cytochrome P450 StaP (PDB 2Z3U) [155].
Alhambra et al. [143] studied the proton transfer reaction catalysed by the enzyme enolase, with consideration of QM proton tunnelling. The reaction is the conversion of 2-phospo-D-glycerate (2-PGA) to phosphoenolpyruvate (PEP) (EC 4.2.1.11), an important step in glucose fermentation. A crystal structure was used as an initial starting point along with additional solvent water molecules, equilibrated using a classical MD simulation. A second shorter MD simulation was then run with constraints on the length of the breaking and forming bonds. The system was then split into QM and MM portions. The QM portion, comprising 25 atoms including the substrate and part of the lysine 345 residue, contained the region of bond forming and breaking and was treated at the AM1 semi-empirical level. The rest of the system was frozen at acceptable random geometries from the second MD simulation, and was treated with MM. Geometry optimisation from these random geometries gave the reactants, transition state and products. The study concluded, by consideration of QM effects, that tunnelling was a plausible mechanism. The free energies of activation calculated from the optimised structures were also very similar to those obtained by experiment [143]. Correction schemes using QM/MM to adjust classical simulation results have also been proposed. Beierlein et al. [144] used single point B3LYP DFT-QM/MM calculations at specific geometries to correct their classical simulations of protein-ligand binding free energy via a single step free energy perturbation from MM to DFT-QM/MM using the Zwanzig equation [145].
7.4 Linear Scaling DFT
The essence of linear scaling methods for ab initio and DFT calculations is the ability to localise regions and apply QM methods regionally. The first linear scaling algorithm used for QM calculation was the “divide and conquer” method (DAC) [128, 146, 147], based on DFT. The idea is that, as the electron density is a local property, the system can be decomposed into smaller subsystems, which are calculable separately and later combined together to represent the whole system. This method does not follow the conventional Kohn–Sham equations and instead aims to compute the electron density directly [146, 148]. A number of linear scaling methods have been developed and implemented [148, 149], and reviewed [150]. Linear scaling algorithms are now available in ChemShell [141] and also in stand-alone programs such as ONETEP [151, 152], a linear scaling DFT code.
Cole et al. [153] used ONETEP to study the binding of CO and O2to myoglobin (Mb). The study uses a DFT energy functional corrected for dispersion (DFT+D) and for self-interaction (DFT+U), which corrects the function locally. This is coupled with a basis set achieving accuracy equivalent to that of a plane wave basis set. The heme binding site plus a substantial portion of the surrounding protein structure were used in the study, comprising over 1000 atoms. Elsewhere, an implicit solvation model incorporated into ONETEP was used to test the effects of solvation on protein-ligand binding for T4 lysozyme, containing more than 2000 atoms [154].
CONCLUSIONS
Efforts to understand, predict, control and even redesign the function of enzymes lie at the heart of contemporary bioinformatics and molecular biology, underpinned by the ever-growing fund of genomic data and the free availability of most key databases. These data are enriched by expert curation and annotation that allow associations to be made between sequences, structures, enzyme classification, chemical reactions, mechanisms, cellular functions, protein-ligand interactions, protein expression, metabolism, disease, and pharmacology. Since the volume of data now overwhelms manual annotation, computer algorithms will in future play a critical role in reliably deepening and broadening this knowledge though automated annotation, updating and linking of databases. Phylogenetic reconstructions of evolutionary history will hopefully help us to keep pace with the development of antibiotic resistance, and to understand how novel enzyme functions evolve. With the variety of DFT, ab initio, semi-empirical and QM/MM computational methods now at our disposal, quantum chemical techniques will become increasingly useful for elucidating and understanding enzyme mechanisms. In summary, a wide range of computational techniques from machine learning, through bioinformatics and chemoinformatics, to quantum chemistry will be required to progress the vast quantity of available data on enzymes into useful knowledge, and to translate that knowledge into technological, pharmaceutical and medical applications.
Acknowledgements
We thank the BBSRC for grant BB/I00596X/1 to JBOM which supports LDF’s research, and for studentship funding for RGA. Research by JBOM and LM is supported by WADA. We thank the Scottish Universities Life Sciences Alliance (SULSA) for supporting JBOM, JLMcD and NN, and we also thank the Scottish Overseas Research Student Awards Scheme of the Scottish Funding Council (SFC) for financial support of NN’s studentship. We thank Leo Holroyd and Luke Crawford for helpful discussions.
List of abbreviations
- 2D
Two Dimensional
- 3D
Three Dimensional
- ACE
Angiotensin Converting Enzyme
- ADME
Absorption, Distribution, Metabolism, and Excretion
- BRENDA
BRaunschweig ENzyme Database
- COST
(European) Cooperation in Science and Technology
- CSA
Catalytic Site Atlas
- DFT
Density Functional Theory
- EC
Enzyme Commission
- EMBL
European Molecular Biology Laboratory
- MACiE
Mechanism, Annotation and Classification in Enzymes
- Mb
Myoglobin
- HF
Hartree–Fock
- KEGG
Kyoto Encyclopaedia of Genes and Genomes
- MDDR MDL
Drug Data Report
- MM
Molecular Mechanics
- NC-IUBMB
Nomenclature Committee of the International Union of Biochemistry and Molecular Biology
- NMR
Nuclear Magnetic Resonance
- PES
Potential Energy Surface
- QM
Quantum Mechanics
- QM/MM
Quantum Mechanics / Molecular Mechanics
- QSPR
Quantitative Structure-Property Relationship
- QSAR
Quantitative Structure-Activity Relationship
- RLCP
Reaction, Ligand, Catalysis, Residues
- SFLD
Structure-Function Linkage Database
- SSG
Structurally Similar Group
Footnotes
Conflict of interest
The authors declare no conflict of interest.
REFERENCES
- [1].Bairoch A. The ENZYME database in 2000. Nucleic Acids Research. 2000;28(1):304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Copley S. Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Current Opinion in Chemical Biology. 2003;7(2):265–272. doi: 10.1016/s1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- [3].Li P, Dada JO, Jameson D, Spasic I, Swainston N, Carroll K, Dunn W, Khan F, Malys N, Messiha HL, Simeonidis E, Weichart D, Winder C, Wishart J, Broomhead DS, Goble CA, Gaskell SJ, Kell DB, Westerhoff HV, Mendes P, Paton NW. Systematic integration of experimental data and models in systems biology. BMC Bioinformatics. 2010;11(1):582. doi: 10.1186/1471-2105-11-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].IUBMB . Enzyme Nomenclature 1992: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes. Academic Press; London: 1992. [Google Scholar]
- [5].The UniProt Consortium Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40(D1):D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Research. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hopkins AL, Groom CR. The druggable genome. Nature reviews. Drug discovery. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- [8].Jiang L, Althoff EA, Clemente FR, Doyle L, Röthlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of Retro-Aldol enzymes. Science. 2008;319(5868):1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448(7155):775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].De Ferrari L, Aitken S, van Hemert J, Goryanin I. EnzML: Multi-label prediction of enzyme classes using InterPro signatures. BMC Bioinformatics. 2012;13(1):61. doi: 10.1186/1471-2105-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Furnham N, Garavelli JS, Apweiler R, Thornton JM. Missing in action: enzyme functional annotations in biological databases. Nature Chemical Biology. 2009;5(8):521–525. doi: 10.1038/nchembio0809-521. [DOI] [PubMed] [Google Scholar]
- [12].Fleming I. Signaling by the Angiotensin-Converting enzyme. Circulation Research. 2006;98(7):887–896. doi: 10.1161/01.RES.0000217340.40936.53. [DOI] [PubMed] [Google Scholar]
- [13].Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Söhngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Research. 2011;39(suppl 1):D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].BRENDA [Accessed 23 July 2012];The Comprehensive Enzyme Information System. http://www.brenda-enzymes.info/
- [15].Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, Tawfik DS, Milo R. The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50(21):4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- [16].Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2011;40(D1):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].KEGG Kyoto Encyclopedia of Genes and Genomes. http://www.kegg.jp/ Accessed 23 July 2012.
- [18].McDonald AG, Boyce S, Moss GP, Dixon HB, Tipton KF. ExplorEnz: a MySQL database of the IUBMB enzyme nomenclature. BMC Biochemistry. 2007;8:14. doi: 10.1186/1471-2091-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. [Accessed 14 August 2012];ExplorEnz-The Enzyme Database. http://www.enzyme-database.org/
- [20].Fleischmann A, Darsow M, Degtyarenko K, Fleischmann W, Boyce S, Axelsen KB, Bairoch A, Schomburg D, Tipton KF, Apweiler R. IntEnz, the integrated relational enzyme database. Nucleic Acids Research. 2004;32(suppl 1):D434–D437. doi: 10.1093/nar/gkh119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. [Accessed 23 July 2012];IntEnz. http://www.ebi.ac.uk/intenz/
- [22]. [Accessed 23 July 2012];NC-IUBMB. http://www.chem.qmul.ac.uk/iubmb/enzyme/
- [23].Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Research. 2004;32(suppl 1):D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. [Accessed 23 July 2012];Catalytic Site Atlas. http://www.ebi.ac.uk/thornton-srv/databases/CSA/
- [25].Gherardini PFF, Wass MN, Helmer-Citterich M, Sternberg MJ. Convergent evolution of enzyme active sites is not a rare phenomenon. Journal of Molecular Biology. 2007;372(3):817–845. doi: 10.1016/j.jmb.2007.06.017. [DOI] [PubMed] [Google Scholar]
- [26].Nagano N. EzCatDB: the Enzyme Catalytic-mechanism Database. Nucleic Acids Res. 2005;33(suppl 1):D407–D412. doi: 10.1093/nar/gki080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].EzCatDB [Accessed 14 August 2012]; http://mbs.cbrc.jp/EzCatDB/
- [28].Pegg SCH, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme Structure-Function relationships for functional inference and experimental design: the Structure-Function Linkage Database. Biochemistry. 2006;45(8):2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- [29]. [Accessed 23 July 2012];Structure-Function Linkage Database. http://sfld.rbvi.ucsf.edu/django/
- [30].Meng EC, Babbitt PC. Topological variation in the evolution of new reactions in functionally diverse enzyme superfamilies. Current Opinion in Structural Biology. 2011;21(3):391–397. doi: 10.1016/j.sbi.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Holliday GL, Almonacid DE, Bartlett GJ, O’Boyle NM, Torrance JW, Murray-Rust P, Mitchell JBO, Thornton JM. MACiE (Mechanism, Annotation and Classification in Enzymes): Novel tools for searching catalytic mechanisms. Nucleic Acids Research. 2007;35(suppl 1):D515–D520. doi: 10.1093/nar/gkl774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. [Accessed 23 July 2012];MACiE (Mechanism, Annotation and Classification in Enzymes) http://www.ebi.ac.uk/thornton-srv/databases/MACiE/
- [33].Almonacid DE, Yera ER, Mitchell JBO, Babbitt PC. Quantitative comparison of catalytic mechanisms and overall reactions in convergently evolved enzymes: implications for classification of enzyme function. PLoS Computational Biology. 2010;6(3):e1000700. doi: 10.1371/journal.pcbi.1000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rahman SA, Bashton M, Holliday GL, Schrader R, Thornton JM. Small molecule subgraph detector (SMSD) toolkit. Journal of Cheminformatics. 2009;1:12. doi: 10.1186/1758-2946-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Holliday GL, Fischer JD, Mitchell JBO, Thornton JM. Characterizing the complexity of enzymes on the basis of their mechanisms and structures with a bio-computational analysis. FEBS Journal. 2011;278(20):3835–3845. doi: 10.1111/j.1742-4658.2011.08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Egelhofer V, Schomburg I, Schomburg D. Automatic Assignment of EC Numbers. PLoS Comput Biol. 2010;6(1):e1000661. doi: 10.1371/journal.pcbi.1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cochrane G, Akhtar R, Bonfield J, Bower L, Demiralp F, Faruque N, Gibson R, Hoad G, Hubbard T, Hunter C, Jang M, Juhos S, Leinonen R, Leonard S, Lin Q, Lopez R, Lorenc D, McWilliam H, Mukherjee G, Plaister S, Radhakrishnan R, Robinson S, Sobhany S, Hoopen PT, Vaughan R, Zalunin V, Birney E. Petabyte-scale innovations at the European Nucleotide Archive. Nucleic Acids Res. 2009;37(suppl 1):D19–D25. doi: 10.1093/nar/gkn765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kotera M, Hirakawa M, Tokimatsu T, Goto S, Kanehisa M. The KEGG databases and tools facilitating omics analysis: latest developments involving human diseases and pharmaceuticals. Methods Mol Biol. 2012;802(1):19–39. doi: 10.1007/978-1-61779-400-1_2. [DOI] [PubMed] [Google Scholar]
- [39].Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(suppl 1):D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H, D’Eustachio P, Stein L. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39(suppl 1):D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Stehr H, Duarte JM, Lappe M, Bhak J, Bolser DM. PDBWiki: added value through community annotation of the protein data bank. Database. 2010;2010 doi: 10.1093/database/baq009. doi: 10.1093/database/baq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Baumgartner WA, Cohen KB, Fox LM, Acquaah-Mensah G, Hunter L. Manual curation is not sufficient for annotation of genomic databases. Bioinformatics. 2007;23(13):i41–i48. doi: 10.1093/bioinformatics/btm229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJA, Thimma M, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37(suppl 1):D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- [45].Burge S, Kelly E, Lonsdale D, Mutowo-Muellenet P, McAnulla C, Mitchell A, Sangrador-Vegas A, Yong S-Y, Mulder N, Hunter S. Database. 2012;2012 doi: 10.1093/database/bar068. doi: 10.1093/database/bar068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tetko IV, Rodchenkov IV, Walter MC, Rattei T, Mewes H-W. Beyond the ‘best’ match: machine learning annotation of protein sequences by integration of different sources of information. Bioinformatics. 2008;24(5):621–628. doi: 10.1093/bioinformatics/btm633. [DOI] [PubMed] [Google Scholar]
- [47].Tian W, Arakaki AK, Skolnick J. EFICAz: a comprehensive approach for accurate genome-scale enzyme function inference. Nucleic Acids Res. 2004;32(21):6226–6239. doi: 10.1093/nar/gkh956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Desai DK, Nandi S, Srivastava PK, Lynn AM. ModEnzA: Accurate identification of metabolic enzymes using function specific profile HMMs with optimised discrimination threshold and modified emission probabilities. Adv Bioinformatics. 2011;2011:743782. doi: 10.1155/2011/743782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Claudel-Renard C, Chevalet C, Faraut T, Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31(22):6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cai CZ, Han LY, Ji ZL, Chen YZ. Enzyme family classification by support vector machines. Proteins: Structure, Function, and Bioinformatics. 2004;55(1):66–76. doi: 10.1002/prot.20045. [DOI] [PubMed] [Google Scholar]
- [51].Astikainen K, Holm L, Pitkanen E, Szedmak S, Rousu J. Towards structured output prediction of enzyme function. BMC Proceedings. 2008;2(suppl 4):S2. doi: 10.1186/1753-6561-2-s4-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Spyromitros E, Tsoumakas G, Vlahavas I. An empirical study of lazy multilabel classification algorithms; SETN ‘08 Proceedings of the 5th Hellenic conference on Artificial Intelligence: Theories, Models and Applications; 2008.pp. 401–406. [Google Scholar]
- [53].Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R. Nucleic Acids Research. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Penning TM, Jez JM. Enzyme Redesign. Chemical Reviews. 2001;101(10):3027–3046. doi: 10.1021/cr000049n. [DOI] [PubMed] [Google Scholar]
- [55].Baltzer L, Nilsson H, Nilsson J. De Novo Design of Proteins - What Are the Rules? Chemical Reviews. 2001;101(10):3153–3164. doi: 10.1021/cr0000473. [DOI] [PubMed] [Google Scholar]
- [56].McCammon JA. Protein Dynamics. Rep. Prog. Phys. 1984;47(1):1–46. [Google Scholar]
- [57].Lipscomb WN. Structure and catalysis of enzymes. Annu Rev Biochem. 1983;52:17–34. doi: 10.1146/annurev.bi.52.070183.000313. [DOI] [PubMed] [Google Scholar]
- [58].Albery WJ, Knowles JR. Efficiency and Evolution of Enzyme Catalysis. Angewandte Chemie International Edition in English. 1977;16(5):285–293. doi: 10.1002/anie.197702851. [DOI] [PubMed] [Google Scholar]
- [59].Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme Dynamics During Catalysis. Science. 2002;295(5559):1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- [60].Tousignant A, Pelletier JN. Protein Motions Promote Catalysis. Chemistry & Biology. 2004;11(8):1037–1042. doi: 10.1016/j.chembiol.2004.06.007. [DOI] [PubMed] [Google Scholar]
- [61].Kaplan J, DeGrado WF. De novo design of catalytic proteins. Proc Natl Acad Sci USA. 2004;101(32):11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nanda V, Zahid S, Xu F, Levine D. Chapter twenty - Computational Design of Intermolecular Stability and Specificity in Protein Self-assembly. In: Michael LJ, Ludwig B, editors. Methods in Enzymology. Vol. 487. Academic Press; 2011. pp. 575–593. [DOI] [PubMed] [Google Scholar]
- [63].Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. Engineering the third wave of biocatalysis. Nature. 2012;485(7397):185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- [64].Tao H, Cornish VW. Milestones in directed enzyme evolution. Current Opinion in Chemical Biology. 2002;6(6):858–864. doi: 10.1016/s1367-5931(02)00396-4. [DOI] [PubMed] [Google Scholar]
- [65].Gro M, Plaxco KW. Protein engineering: Reading, writing and redesigning. Nature. 1997;388(6641):419–420. doi: 10.1038/41204. [DOI] [PubMed] [Google Scholar]
- [66].Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453(7192):190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- [67].Nanda V. Do-it-yourself enzymes. Nat Chem Biol. 2008;4(5):273–275. doi: 10.1038/nchembio0508-273. [DOI] [PubMed] [Google Scholar]
- [68].Casey ML, Kemp DS, Paul KG, Cox DD. Physical organic chemistry of benzisoxazoles. I. Mechanism of the base-catalyzed decomposition of benzisoxazoles. Journal of Organic Chemistry. 1973;38(13):2294–2301. [Google Scholar]
- [69].Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Röthlisberger D, Baker D. New algorithms and an in silico benchmark for computational enzyme design. Protein Science. 2006;15(12):2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D. De Novo Enzyme Design Using Rosetta3. PLoS ONE. 2011;6(5):e19230. doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Suárez M, Jaramillo A. Challenges in the computational design of proteins. J. R. Soc. Interface. 2009;6(suppl 4):S477–S491. doi: 10.1098/rsif.2008.0508.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lippow SM, Tidor B. Progress in computational protein design. Current Opinion in Biotechnology. 2007;18(4):305–311. doi: 10.1016/j.copbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Dantas G, Kuhlman B, Callender D, Wong M, Baker D. A Large Scale Test of Computational Protein Design: Folding and Stability of Nine Completely Redesigned Globular Proteins. Journal of Molecular Biology. 2003;332(2):449–460. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- [74].Jaramillo A, Wernisch L, Héry S, Wodak SJ. Folding free energy function selects native-like protein sequences in the core but not on the surface. Proceedings of the National Academy of Sciences. 2002;99(21):13554–13559. doi: 10.1073/pnas.212068599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kiss G, Röthlisberger D, Baker D, Houk KN. Evaluation and ranking of enzyme designs. Protein Science. 2010;19(9):1760–1773. doi: 10.1002/pro.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Glowacki DR, Harvey JN, Mulholland AJ. Taking Ockham’s razor to enzyme dynamics and catalysis. Nature Chemistry. 2012;4(3):169–176. doi: 10.1038/nchem.1244. [DOI] [PubMed] [Google Scholar]
- [77].McGeagh JD, Ranaghan KE, Mulholland AJ. Protein dynamics and enzyme catalysis: Insights from simulations. Biochimica et Biophysica Acta. 2011;1814(8):1077–1092. doi: 10.1016/j.bbapap.2010.12.002. [DOI] [PubMed] [Google Scholar]
- [78].Kamerlin SCL, Warshel A. At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis? Proteins: Structure, Function, and Bioinformatics. 2010;78(6):1339–1375. doi: 10.1002/prot.22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lassila JK. Conformational diversity and computational enzyme design. Current Opinion in Chemical Biology. 2010;14(5):676–682. doi: 10.1016/j.cbpa.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jensen RA. Enzyme recruitment in evolution of new function. Ann. Rev. Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- [81].O’Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chemistry & Biology. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- [82].Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annual Review of Biochemistry. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- [83].Ohno S. Evolution by gene duplication. Springer-Verlag; New York: 1970. p. xv.p. 160. [Google Scholar]
- [84].Copley SD. Toward a systems biology perspective on enzyme evolution. Journal of Biological Chemistry. 2012;287(1):3–10. doi: 10.1074/jbc.R111.254714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Russell RJ, Scott C, Jackson CJ, Pandey R, Pandey G, Taylor MC, Coppin CW, Liu J-W, Oakeshott JG. The evolution of new enzyme function: lessons from xenobiotic metabolizing bacteria versus insecticide-resistant insects. Evolutionary Applications. 2011;4(2):225–248. doi: 10.1111/j.1752-4571.2010.00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Jeffery CJ. Moonlighting proteins. Trends in Biochemical Sciences. 1999;24(1):8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- [87].Huberts DHEW, van der Klei IJ. Moonlighting proteins: An intriguing mode of multitasking. Biochimica et Biophysica Acta. 2010;1803(4):520–525. doi: 10.1016/j.bbamcr.2010.01.022. [DOI] [PubMed] [Google Scholar]
- [88].Copley SD. Moonlighting is mainstream: Paradigm adjustment required. BioEssays. 2012;34(7):578–588. doi: 10.1002/bies.201100191. [DOI] [PubMed] [Google Scholar]
- [89].Piatigorsky J, O’Brien WE, Norman BL, Kalumuck K, Wistow GJ, Borras T, Nickerson JM, Wawrousek EF. Gene sharing by delta-crystallin and argininosuccinate lyase. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(10):3479–3483. doi: 10.1073/pnas.85.10.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Rodríguez Plaza JG, Villalón Rojas A, Herrera S, Garza-Ramos G, Torres Larios A, Amero C, Zarraga Granados G, Gutiérrez Aguilar M, Lara Ortiz MT, Polanco Gonzalez C, Uribe Carvajal S, Coria R, Peña Díaz A, Bredesen DE, Castro-Obregon S, del Rio G. Moonlighting peptides with emerging function. PLoS ONE. 2012;7(7):e40125. doi: 10.1371/journal.pone.0040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Cuff AL, Sillitoe I, Lewis T, Clegg AB, Rentzsch R, Furnham N, Pellegrini-Calace M, Jones D, Thornton J, Orengo CA. Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Research. 2011;39(Database issue):D420–D426. doi: 10.1093/nar/gkq1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. Journal of Molecular Biology. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- [93].Hicks MA, Barber AE, Giddings L-A, Caldwell J, O’Connor SE, Babbitt PC. The evolution of function in strictosidine synthase-like proteins. Proteins. 2011;79(11):3082–3098. doi: 10.1002/prot.23135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lundin D, Poole AM, Sjöberg BM, Högbom M. Use of Structural Phylogenetic Networks for Classification of the Ferritin-like Superfamily. Journal of Biological Chemistry. 2012;287(24):20565–20575. doi: 10.1074/jbc.M112.367458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Furnham N, Sillitoe I, Holliday GL, Cuff AL, Rahman SA, Laskowski RA, Orengo CA, Thornton JM. FunTree: a resource for exploring the functional evolution of structurally defined enzyme superfamilies. Nucleic Acids Research. 2012;40(D1):D776–D782. doi: 10.1093/nar/gkr852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Furnham N, Sillitoe I, Holliday GL, Cuff AL, Laskowski RA, Orengo CA, Thornton JM. Exploring the evolution of novel enzyme functions within structurally defined protein superfamilies. PLoS Computational Biology. 2012;8(3):e1002403. doi: 10.1371/journal.pcbi.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].FunTree [Accessed 8 August 2012]; http://www.ebi.ac.uk/thornton-srv/databases/FunTree/
- [98]. [Accessed 8 August 2012];Jmol: an open-source Java viewer for chemical structures in 3D. http://jmol.sourceforge.net/
- [99].Ghemtio L, Perez-Nueno VI, Leroux V, Asses Y, Souchet M, Mavridis L, Maigret B, Ritchie DW. Recent Trends and Applications in 3D Virtual Screening. Combinatorial Chemistry & High Throughput Screening. 2012;15 doi: 10.2174/138620712803519707. in press. [DOI] [PubMed] [Google Scholar]
- [100].Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Research. 2012;40(D1):D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Research. 2007;35(suppl 1):D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: Integrated platform of small molecules and biological activities. Annual Reports in Computational Chemistry. 2008;4 Chapter 12. [Google Scholar]
- [103].Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39(suppl 1):D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].MDL Drug Data Report, 2010.2. Accelrys, Inc.; San Diego, CA (USA): 2010. [Google Scholar]
- [105].Olah M, Mracec M, Ostopovici L, Rad R, Bora A, Hadaruga N, Olah I, Banda M, Simon Z, Mracec M, Oprea TI. WOMBAT: World of Molecular Bioactivity, in Chemoinformatics in Drug Discovery (ed T. I. Oprea) Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim; Germany: 2005. doi: 10.1002/3527603743.ch9. [Google Scholar]
- [106].Lowe R, Mussa HY, Nigsch F, Glen RC, Mitchell JBO. Predicting the mechanism of phospholipidosis. Journal of Chemoinformatics. 2012;4:2. doi: 10.1186/1758-2946-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Lowe R, Mussa HY, Mitchell JBO, Glen RC. Classifying Molecules Using a Sparse Probabilistic Kernel Binary Classifier. J. Chem. Inf. Model. 2011;51(7):1539–1544. doi: 10.1021/ci200128w. [DOI] [PubMed] [Google Scholar]
- [108].Lowe R, Glen RC, Mitchell JBO. Predicting Phospholipidosis Using Machine Learning. Mol. Pharmaceutics. 2010;7(5):1708–1718. doi: 10.1021/mp100103e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas A, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Whaley R, Glennon RA, Hert J, Thomas KLH, Edwards DD, Shoichet BK, Roth BL. Predicting new molecular targets for known drugs. Nature. 2009;462(7270):175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Chen B, Wild D, Guha R. PubChem as a Source of Polypharmacology. J. Chem. Inf. Model. 2009;49(9):2044–2055. doi: 10.1021/ci9001876. [DOI] [PubMed] [Google Scholar]
- [111].Cannon EO, Amini A, Bender A, Sternberg MJE, Muggleton SH, Glen RC, Mitchell JBO. Support vector inductive logic programming outperforms the naive Bayes classifier and inductive logic programming for the classification of bioactive chemical compounds. Journal of Computer-Aided Molecular Design. 2007;21(5):269–280. doi: 10.1007/s10822-007-9113-3. [DOI] [PubMed] [Google Scholar]
- [112].Venkatraman V, Perez-Nueno VI, Mavridis L, Ritchie DW. Comprehensive Comparison of Ligand-Based Virtual Screening Tools Against the DUD Data set Reveals Limitations of Current 3D Methods. J. Chem. Inf. Model. 2010;50(12):2079–2093. doi: 10.1021/ci100263p. [DOI] [PubMed] [Google Scholar]
- [113].Anderson N, Borlak J. Drug-induced phospholipidosis. FEBS Letters. 2006;580(23):5533–5540. doi: 10.1016/j.febslet.2006.08.061. [DOI] [PubMed] [Google Scholar]
- [114].Chong CR, Sullivan DJ. New uses for old drugs. Nature. 2007;448(7154):645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- [115].Tartaglia LA. Complementary new approaches enable repositioning of failed drug candidates. Expert Opinion on Investigational Drugs. 2006;15(11):1295–1298. doi: 10.1517/13543784.15.11.1295. [DOI] [PubMed] [Google Scholar]
- [116].Palmer DS, Llinas A, Morao I, Day GM, Goodman JM, Glen RC, Mitchell JBO. Predicting Intrinsic Aqueous Solubility by a Thermodynamic Cycle. Molecular Pharmaceutics. 2008;5(2):266–279. doi: 10.1021/mp7000878. [DOI] [PubMed] [Google Scholar]
- [117].Frolov AI, Ratkova EL, Palmer DS, Fedorov MV. Hydration Thermodynamics Using the Reference Interaction Site Model: Speed or Accuracy? The Journal of Physical Chemistry B. 2011;115(19):6011–6022. doi: 10.1021/jp111271c. [DOI] [PubMed] [Google Scholar]
- [118].Cramer CJ. Essentials of Computational Chemistry: Theories and Models. 2nd ed. John Wiley & Sons; Chichester, UK: 2004. [Google Scholar]
- [119].Thomas LH. The calculation of atomic fields. Mathematical Proceedings of the Cambridge Philosophical Society. 1927;23(5):542–548. [Google Scholar]
- [120].Fermi E. Un metodo statistico per la determinazione di alcune priorieta dell’atome. Rend Accad Naz Lincei. 1927;6(6):602–607. [Google Scholar]
- [121].Hohenberg P, Kohn W. Inhomogeneous Electron Gas. Physical Review. 1964;136(3B):B864–B871. [Google Scholar]
- [122].Kohn W, Sham LJ. Self-Consistent Equations Including Exchange and Correlation Effects. Physical Review. 1965;140(4A):A1133–A1138. [Google Scholar]
- [123].Field MJ. Simulating enzyme reactions: Challenges and perspectives. Journal of Computational Chemistry. 2002;23(1):48–58. doi: 10.1002/jcc.1156. [DOI] [PubMed] [Google Scholar]
- [124].Li D, Wang Y, Han K. Recent density functional theory model calculations of drug metabolism by cytochrome P450. Coordination Chemistry Reviews. 2012;256(11-12):1137–1150. [Google Scholar]
- [125].Perdew JP, Schmidt K. Jacob’s ladder of density functional approximations for the exchange-correlation energy. AIP Conference Proceedings. 2001;577(1):1–20. [Google Scholar]
- [126].Zhao Y, Truhlar D. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor Chem Account. 2008;120(1-3):215–241. [Google Scholar]
- [127].Grimme S. Accurate description of van der Waals complexes by density functional theory including empirical corrections. Journal of Computational Chemistry. 2004;25(12):1463–1473. doi: 10.1002/jcc.20078. [DOI] [PubMed] [Google Scholar]
- [128].Parr RG, Yang W. Density-Functional Theory of the Electronic Structure of Molecules. Annual Review of Physical Chemistry. 1995;46(1):701–728. doi: 10.1146/annurev.pc.46.100195.003413. [DOI] [PubMed] [Google Scholar]
- [129].Holroyd LF, van Mourik T. Insufficient description of dispersion in B3LYP and large basis set superposition errors in MP2 calculations can hide peptide conformers. Chemical Physics Letters. 2007;442(1-3):42–46. [Google Scholar]
- [130].Valdes H, Pluhácková K, Pitonák M, Rezác J, Hobza P. Benchmark database on isolated small peptides containing an aromatic side chain: comparison between wave function and density functional theory methods and empirical force field. Physical Chemistry Chemical Physics. 2008;10(19):2747–2757. doi: 10.1039/b719294k. [DOI] [PubMed] [Google Scholar]
- [131].Chass GA, Kantchev EAB, Fang D-C. The fine balance between one cross-coupling and two β-hydride elimination pathways: a DFT mechanistic study of Ni(π-allyl)2-catalyzed cross-coupling of alkyl halides and alkyl Grignard reagents. Chemical Communications. 2010;46(16):2727–2729. doi: 10.1039/b922326f. [DOI] [PubMed] [Google Scholar]
- [132].Georgiev V, Noack H, Borowski T, Blomberg MRA, Siegbahn PEM. DFT Study on the Catalytic Reactivity of a Functional Model Complex for Intradiol-Cleaving Dioxygenases. J. Phys. Chem. B. 2010;114(17):5878–5885. doi: 10.1021/jp911217j. [DOI] [PubMed] [Google Scholar]
- [133].Simón L, Goodman JM. Enzyme Catalysis by Hydrogen Bonds: The Balance between Transition State Binding and Substrate Binding in Oxyanion Holes. The Journal of Organic Chemistry. 2010;75(6):1831–1840. doi: 10.1021/jo901503d. [DOI] [PubMed] [Google Scholar]
- [134].Becke AD. Density-functional thermochemistry. III. The role of exact exchange. The Journal of Chemical Physics. 1993;98(7):5648–5652. [Google Scholar]
- [135].Salomon O, Reiher M, Hess BA. Assertion and validation of the performance of the B3LYP* functional for the first transition metal row and the G2 test set. The Journal of Chemical Physics. 2002;117(10):4729–4737. [Google Scholar]
- [136].Ertem MZ, Cramer CJ, Himo F, Siegbahn PE. N-O bond cleavage mechanism(s) in nitrous oxide reductase. Journal of Biological Inorganic Chemistry. 2012;17(5):687–698. doi: 10.1007/s00775-012-0888-x. [DOI] [PubMed] [Google Scholar]
- [137].Blomberg LM, Mangold M, Mitchell JBO, Blumberger J. Theoretical study of the reaction mechanism of streptomyces coelicolor type II dehydroquinase. Journal of Chemical Theory and Computation. 2009;5(5):1284–1294. doi: 10.1021/ct800480d. [DOI] [PubMed] [Google Scholar]
- [138].Siegbahn PEM, Blomberg MRA. Transition-Metal Systems in Biochemistry Studied by High-Accuracy Quantum Chemical Methods. Chemical Reviews. 2000;100(2):421–438. doi: 10.1021/cr980390w. [DOI] [PubMed] [Google Scholar]
- [139].Ponder JW, Wu C, Ren P, Pande VS, Chodera JD, Schnieders MJ, Haque I, Mobley DL, Lambrecht DS, DiStasio RA, Head-Gordon M, Clark GNI, Johnson ME, Head-Gordon T. Current Status of the AMOEBA Polarizable Force Field. J. Phys. Chem. B. 2010;114(8):2549–2564. doi: 10.1021/jp910674d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Friesner RA, Guallar V. Ab initio quantum chemical and mixed quantum mechanics/molecular mechanics (QM/MM) methods for studying enzymatic catalysis. Annual Review of Physical Chemistry. 2004;56(1):389–427. doi: 10.1146/annurev.physchem.55.091602.094410. [DOI] [PubMed] [Google Scholar]
- [141]. [Accessed 9 August 2012];ChemShell, a Computational Chemistry Shell. Available from: http://www.chemshell.org.
- [142].Lonsdale R, Ranaghan KE, Mulholland A. J. Computational enzymology. Chem. Commun. 2010;46(14):2354–2372. doi: 10.1039/b925647d. [DOI] [PubMed] [Google Scholar]
- [143].Alhambra C, Gao J, Corchado JC, Villà J, Truhlar DG. Quantum Mechanical Dynamical Effects in an Enzyme-Catalyzed Proton Transfer Reaction. J. Am. Chem. Soc. 1999;121(10):2253–2258. [Google Scholar]
- [144].Beierlein FR, Michel J, Essex JW. A Simple QM/MM Approach for Capturing Polarization Effects in Protein-Ligand Binding Free Energy Calculations. The Journal of Physical Chemistry B. 2011;115(17):4911–4926. doi: 10.1021/jp109054j. [DOI] [PubMed] [Google Scholar]
- [145].Zwanzig RW. High-Temperature Equation of State by a Perturbation Method. I. Nonpolar Gases. The Journal of Chemical Physics. 1954;22(8):1420–1426. [Google Scholar]
- [146].Yang W. Direct calculation of electron density in density-functional theory. Physical Review Letters. 1991;66(11):1438–1441. doi: 10.1103/PhysRevLett.66.1438. [DOI] [PubMed] [Google Scholar]
- [147].Zhao Q, Yang W. Analytical energy gradients and geometry optimization in the divide-and-conquer method for large molecules. The Journal of Chemical Physics. 1995;102(24):9598–9603. [Google Scholar]
- [148].Lee T-S, Lewis JP, Yang W. Linear-scaling quantum mechanical calculations of biological molecules: The divide-and-conquer approach. Computational Materials Science. 1998;12(3):259–277. [Google Scholar]
- [149].Goedecker S, Scuserza GE. Linear scaling electronic structure methods in chemistry and physics. Computing in Science & Engineering. 2003;5(4):14–21. [Google Scholar]
- [150].Goedecker S. Linear scaling electronic structure methods. Reviews of Modern Physics. 1999;71(4):1085–1123. [Google Scholar]
- [151].Haynes PD, Mostof AA, Skylaris CK, Payne MC. ONETEP: linear-scaling density-functional theory with plane-waves. Journal of Physics: Conference Series. 2006;26(1):143. [Google Scholar]
- [152].Skylaris C-K, Haynes PD, Mostofi AA, Payne MC. Introducing ONETEP: Linear-scaling density functional simulations on parallel computers. The Journal of Chemical Physics. 2005;122(8):084119. doi: 10.1063/1.1839852. [DOI] [PubMed] [Google Scholar]
- [153].Cole DJ, O’Regan DD, Payne MC. Ligand Discrimination in Myoglobin from Linear-Scaling DFT+U. The Journal of Physical Chemistry Letters. 2012;3(11):1448–1452. doi: 10.1021/jz3004188. [DOI] [PubMed] [Google Scholar]
- [154].Dziedzic J, Fox SJ, Fox T, Tautermann CS, Skylaris C-K. Large-scale DFT calculations in implicit solvent—A case study on the T4 lysozyme L99A/M102Q protein. Int. J. Quantum Chem. 2012 doi: 10.1002/qua.24075. [Google Scholar]
- [155].Makino M, Sugimoto H, Shiro Y, Asamizu S, Onaka H, Nagano S. Crystal structures and catalytic mechanism of cytochrome P450 StaP that produces the indolocarbazole skeleton. Proceedings of the National Academy of Sciences. 2007;104(28):11591–11596. doi: 10.1073/pnas.0702946104. [DOI] [PMC free article] [PubMed] [Google Scholar]