Abstract

We utilized three independent techniques, immunocytochemistry (ICC), single cell mass spectrometry (MS), and in situ hybridization (ISH), to localize neuropeptides and their transcripts in the nervous system of the nematode Ascaris suum. AF11 (SDIGISEPNFLRFa) is an endogenous peptide with potent paralytic effects on A. suum locomotory behavior. A highly specific antibody to AF11 showed robust immunostaining for AF11 in the paired AVK neurons in the ventral ganglion. We traced the processes from the AVK neurons into the ventral nerve cord and identified them as ventral cord interneurons. MS and MS/MS of single dissected AVKs detected AF11, two previously characterized peptides (AF25 and AF26), seven novel sequence-related peptides, including several sharing a PNFLRFamide C-terminus, and peptide NY, a peptide with an unrelated sequence. Also present in a subset of AVKs was AF2, a peptide encoded by the afp-4 transcript. By sequencing the afp-11 transcript, we discovered that it encodes AF11, all the AF11-related peptides detected by MS in AVK, and peptide NY. ISH detected the afp-11 transcript in AVK neurons, consistent with other techniques. ISH did not detect afp-11 in the ALA neuron, although both ICC and MS found AF11 in ca. 30% of ALAs. All 10 AF11-related peptides reduced acetylcholine-induced muscle contraction, but they differed in their rate of reversal of inhibition after removal of the peptide.
Keywords: Neuropeptide, mass spectrometry, specific antibody, in situ hybridization, single neuron, nematode
The nervous systems of nematodes appear to be remarkably simple, both numerically and morphologically. The parasite Ascaris suum and the free-living Caenorhabditis elegans each have a total of approximately 300 neurons, 298 in adult female A. suum(1) or 302 in C. elegans hermaphrodites.2 These neurons are very simple morphologically; each neuron has a distinctive morphology that is conserved between the two species, despite their enormous discrepancy in size and their evolutionary distance (their latest common ancestor is estimated to have existed ca. 500 million years ago3). At the level of neuronal morphology, C. elegans looks like a miniature version of A. suum.
Despite this apparent simplicity, obtaining a detailed functional understanding of neuronal circuitry (how the nervous system controls behavior) has been surprisingly slow and difficult. In A. suum, even a detailed anatomical wiring diagram and the description of the electrical properties of individual neurons and their synapses was insufficient to explain adequately the circuit properties of the motor nervous system and the way it controls locomotory behavior.4 At least one part of what was missing from the description was the role of the rich world of modulatory signaling molecules, of which neuropeptides appear to be the most diverse. At least 250 neuropeptides have been predicted in A. suum,5,6 and a similar number was predicted in C. elegans.7 Peptides are known to have potent activities on neurons in nematodes, including the neurons in the motor nervous system.8,9 Although there appears to be some functional redundancy among these peptides, the number of classes of signaling molecules is still large, and they need to be characterized in detail at both the molecular and the physiological levels before we can attempt to understand neural circuits.
Among nematode species, the structure of neuropeptides is also highly conserved. From EST libraries, predicted peptides with identical sequences or very closely related sequences are found across many different species of nematodes,10,11 implying strong positive selection pressure on these sequences. However, it has emerged recently that the particular cellular expression of neuropeptides is not well conserved;12−15 this may explain, at least in part, how it is that structurally similar nervous systems can generate the different behaviors that have contributed to the enormous radiation of nematodes and the success of this phylum in populating such a wide variety of habitats, including, most notoriously, their success as parasites of both animals and plants.16
In an effort to understand the role of neuropeptides in A. suum, this laboratory has characterized and localized an increasing number of Ascaris FMRFamide-like peptides (AF peptides). One, AF11 (SDIGISEPNFLRFamide), is of particular interest, being highly bioactive. When injected into intact worms, it produces a dramatic paralysis, abolishing all locomotory movements.8 Peptide AF11 was originally isolated and sequenced from A. suum in a large-scale fractionation and direct sequencing project that led to the description of the first 19 AF peptides.4,17−19 The C-terminal PNFLRFamide sequence is shared with peptides encoded by the flp-1 gene of C. elegans,20 peptides isolated from Panagrellus redivivus,21 and predicted peptides in several other nematode species.10 Both the early peptide sequencing project19 and initial mass spectrometry (MS) of ganglia22 had shown that there were other peptides with C-terminal PNFLRFa in A. suum, but characterizing them proved difficult. Several attempts to isolate the transcript that encodes these peptides by degenerate PCR with primers based on the AF11 sequence were unsuccessful. A combination of our recently developed technique for dissecting single neurons for MS and sequence determination by tandem MS, together with the recently available A. suum genome survey sequences database (GSS, available on the NCBI database) allowed us to determine the sequence of a family of related peptides, and then to clone and localize the AF11-encoding transcript afp-11, as reported in this paper. Mass spectrometry (MS) has emerged as a particularly powerful technique to determine the peptide complement of organisms. Specifically in nematodes, peptide extraction from whole animals and identification by MS led to the first characterizations of many peptides in A. suum,6C. elegans,23 and C. briggsae.24 In addition, direct MALDI-TOF (matrix assisted laser desorption/ionization–time-of-flight) MS and MS/MS analysis, first of whole ganglia and then single dissected neurons, identified many novel peptides as well as localizing the cellular expression of these peptides.5,14
We developed a highly specific antibody to AF11, which stained only three cells in A. suum (Figure 1). The AVK neurons, a bilateral pair in the most posterior region of the ventral ganglion (VG), were stained consistently and robustly. ALA, a single neuron in the dorsal ganglion (DG), was stained inconsistently and with variable intensity. Application of MALDI-TOF MS and MS/MS to single dissected AVK neurons confirmed that they contain AF11, as well as several other peptides, including a total of 10 sequence-related AF peptides, 6 of which shared the C-terminal PNFLRFamide. Searches of the A. suum genome survey sequences (GSS) revealed three GSS sequences that together encoded all of the AF11-related peptides found in AVK neurons. This led to the cloning of afp-11, the transcript that encodes these peptides, and synthesis of a riboprobe specific to afp-11. In situ hybridization with this riboprobe confirmed that the afp-11 transcript is present in AVK. The afp-11 transcript is not detected in ALA neurons, although single cell MS analysis detected some of the AF11-related peptides at low levels in some samples of ALA.14 This is consistent with the variability seen by ICC.
Figure 1.
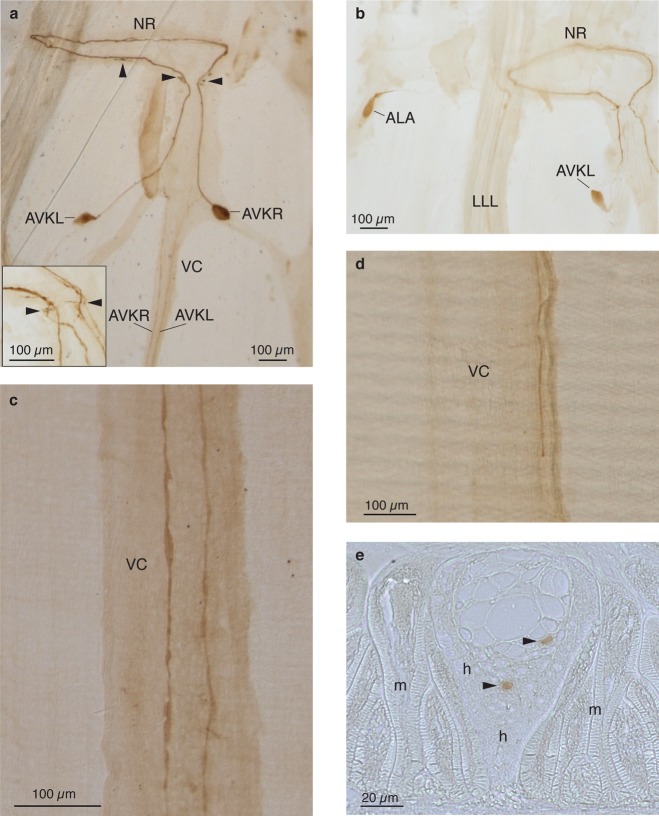
Neurons in the cephalic region of A. suum. (a) Diagram of the head ganglia of A. suum modified from Goldschmidt.56 The worm has been cut longitudinally near the DC and opened flat. Neuronal cell bodies and commissural processes are shown. (b) Neurons expressing AF11 in A. suum. Diagram shows the cell bodies of AVKR (green), AVKL (red) of the VG, and ALA (blue) of the DG. The AVKs each send a process anteriorly toward the midline, turn ipsilaterally into the NR, traverse the NR, and then proceed posteriorly down the contralateral side of the VC. The ALA sends two processes anteriorly into the NR, where they diverge. One goes left, enters the LLL and travels posteriorly; the other goes right and travels down the RLL. (c) Manually dissected AVKR neuron for direct MS analysis. Nerve ring (NR), ventral ganglion (VG), dorsal ganglion (DG),lateral ganglia (LG), retrovesicular ganglion (RVG), left lateral line (LLL), right lateral line (RLL), ventral nerve cord (VC), dorsal nerve cord (DC), deirid commissures (DeC), amphidial commissures (AC).
Thus, the use of three independent techniques showed consistent expression of AF11 and the other peptides encoded by afp-11 in AVK neurons. Because peptides are usually expressed in both cell bodies and neurites of cells in which they are synthesized, ICC allowed us to describe the cellular morphology of the AVK neurons (Figure 1). From their cell bodies in the VG, processes project anteriorly and enter the nerve ring (NR) close to the ventral midline; each process turns ipsilaterally, runs completely around the NR, and then enters the ventral nerve cord (VC) and projects posteriorly. Examination of immunostained transverse sections showed AF11 immunoreactivity in two small ventral nerve cord interneurons.
Many of the peptides encoded by afp-11 affect locomotory behavior, producing paralysis and a significant lengthening of the body,8,25 correlating with muscle relaxation, as reported here. Since the VC projections of the AVK are classified as VC interneurons, we surmise that it is these processes that are responsible for the behavioral effects of these peptides; that is, the peptides are secreted from these VC processes and affect other neurons in the ventral nerve cord and/or nearby muscle.
Results and Discussion
Cellular localization of a neuropeptide is important since it identifies one-half of the intercellular signaling dyad mediated by the peptide, namely, the cell that synthesizes, stores, and presumably releases the peptide. We used three independent techniques, single cell MS, ICC, and ISH, for localizing AF11, a highly bioactive peptide, and the transcript that encodes it. MS and MS/MS are important techniques, since they unambiguously identify the peptide within the dissected cell, whereas ICC is always limited by lack of complete knowledge of cross-reactivity of the antibody with epitopes on presently unknown peptides (or proteins) in the organism. In addition, MS detects unknown peptides within the dissected cell and is therefore an ideal method for characterization (and simultaneous cellular localization) of novel peptides, with no prior sequence information. The new sequences then can be synthesized as peptides for assays of biological activity. ICC, on the other hand, allows the determination of the morphology of the neurites of the cell, which is important for neuronal identification. ISH and ICC both extend the localization information to the entire nervous system, allowing other cells that express the peptide(s) to be identified. Comparison with two other nematodes that encode a sequence-related family of peptides shows that they are expressed in different neurons; this probably contributes to species-specific differences in behavior.
Characterization of Specific Antibody to AF11
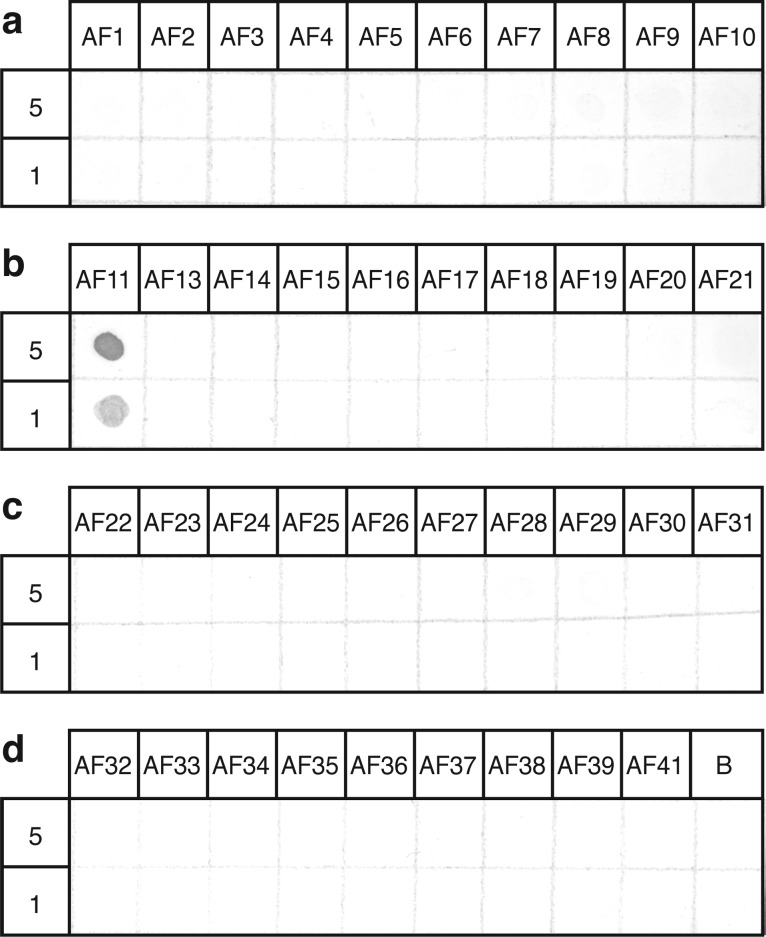
We generated and affinity-purified a polyclonal antibody against AF11 (SDIGISEPNFLRFa). This antibody is highly specific: a dot-enzyme linked immunosorbent assay (dot-ELISA)26 against 39 known AF peptides (sequences given in Supporting Information Table S1) was used to test antibody specificity at three different antibody concentrations (1:1000, 1:7000, and 1:13 000). At the lowest antibody concentration, only AF11 was recognized by the antibody (Figure 2). The medium antibody concentration produced only a faint trace of cross-reactivity for AF28 and 29 (not shown). At the highest concentration, the antibody displayed faint cross-reactivity with all tested AF peptides and slightly stronger cross-reactivity for AF5, 7, 25, 27, 28, 29, and 41 (not shown); these peptides all share a (P/N)XF(L/I)RFamide motif and except for AF5 and 7 are encoded by the same transcript as AF11 (afp-11, described below).
Figure 2.
Dot-ELISA showing specificity of anti-AF11 antibody. At a dilution of 1:13 000, the anti-AF11 antibody showed robust staining for AF11 and no cross-reactivity with BSA or the other peptides (AF1–10, 13–39, and 41). Top rows indicate AF peptide number (see Supporting Information Table S1 for sequences). Peptides were spotted at 5 μg/mL (middle rows) and 1 μg/mL (bottom rows) in 1 mg/mL BSA. BSA was used as a negative control. (a) AF1–10; (b) AF11, AF13–21; (c) AF22–31; (d) AF32–39, AF41, B = BSA.
Localization of AF11 Immunoreactivity in A. suum Head Neurons
The anti-AF11 antibody recognized only three neurons in the head of A. suum (Figure 3a, b). The most posterior pair of neurons in the VG, the left and right AVK neurons (AVKL and AVKR), and their processes were the strongest and most consistently stained. This staining was observed in 110 out of 113 head preparations (98%). In most preparations, the two AVK cells are equally stained, but occasionally one is darker than the other. It is unclear whether such differences in staining intensity are due to variability in tissue fixation, antibody penetration, or peptide expression levels at the instant of fixation.
Figure 3.
AF11 immunoreactivity in AVK neurons and their processes. (a–d) Whole mount preparations; (e) cross-section of plastic-embedded VC. (a) Stained AVK neurons each send a process anteriorly toward the midline, turn ipsilaterally into the NR, proceed all the way around the NR, and then proceed posteriorly down the contralateral side of the VC. The AVK processes usually extend fine, short projections within the NR as well as in the area where they enter and exit the NR into the VC (arrowheads). Inset: higher magnification of another preparation where the AVK processes enter and exit the NR; arrowheads indicate fine extensions. (b) AF11-immunoreactivity in ALA neuron cell body in the dorsal ganglion. ALA sends two processes into the NR with one traveling down the left lateral line (LLL) (faint line) and the other traveling down the right lateral line (not shown). Note the stained AVKL neuron in the lower right. The NR is intact in this preparation, but the processes extending from ALA to the lateral lines were broken during dissection. (c) Stained AVK processes traveling separately down the VC. Note varicosities. (d) Stained AVK process ending in an apparent terminus in the region of the gonopore. (e) In transverse section, the stained AVK axons (arrowheads) travel at different levels in the VC. Their size and position identify them as being in the class of small VC interneurons. Muscle (m), hypodermis (h). The hypodermis that underlies the cuticle is near the bottom edge of this panel; near the top edge is the region where muscle arms divide into fine interdigitating fingers that receive synapses from motor neurons in the VC. Nerve ring (NR), ventral nerve cord (VC), AVK left (AVKL), AVK right (AVKR).
A significant subset of preparations also showed staining of ALA, an unpaired neuron located in the dorsal ganglion.14 In the 83 head preparations where the DG was intact and visible, there was variable staining in ALA. In 50 preparations (60%), there was no AF11-immunoreactivity in the ALA neurons. In 24 preparations (29%), staining in ALA was obvious; the intensity of staining ranged from slightly above background to as dark as in AVK (as in Figure 3b). In the remaining 9 preparations (11%), the difference between faint specific staining and background was ambiguous. The variability in ALA can be contrasted with both the consistency and intensity of positive staining seen in AVK and the absolute lack of staining in RID, the only other cell in the DG, suggesting a biological rather than a procedural basis for this variability. Negative controls (no anti-AF11 antibody) and blocking controls (antibody blocked with AF11) showed no specific staining (not shown).
Determination of AVK Morphology by ICC
Nerve Ring
The AVKs are monopolar cells which each send a process that nearly reaches the ventral midline at the NR, but then reflects ipsilaterally into the NR (Figure 1). Previously in serial sections of unstained preparations, it was not possible to trace their axons any further because of the density of intertwined processes entering and exiting the NR. The anti-AF11 staining allows us to trace the AVK processes as they enter, traverse, and exit the NR and then proceed posteriorly down the VC (Figure 3a–d). Examination of immunostained transverse sections of the VC shows that the VC processes have no contact with muscle (Figure 3e). These features identify AVK neurons as ventral cord interneurons (VC INs), suggesting that they may play a role in the control of locomotory behavior.4
As they enter and exit the ring, the AVK processes usually extend fine, short projections both within the NR and, more prominently, near the ventral midline (Figure 3a, insert). This suggests that the AVKs are making and/or receiving synapses to and from other cells in the NR and possibly communicating with each other at the neck of the ring via these extensions. In A. suum, the hypodermis is thicker where the AVK processes exit the NR, and restricts antibody access. To better characterize the AVK processes in this region, 18 head preparations were treated with a higher concentration of anti-AF11 antibody, which made the AVK processes easy to trace from the NR to the VC. In 17 of these preparations, the VC processes of AVK appeared to have a left-right asymmetry with respect to position and relative intensity of AF11-immunoreactivity, with the process on the left, which comes from AVKR, being slightly thicker and more darkly stained (Figure 3c), especially as it travels posteriorly (in the 18th preparation, this difference was not obvious). The AVKL process is higher in the cord (more medial, in the intact worm) and the AVKR process is lower (closer to the cuticle: see Figure 3e). Asymmetry in the AVKs could have functional implications with respect to modulating locomotory coordination of the worm, with each AVK cell sending a separate, and different, signal to its target(s). Because ICC is not a quantitative method, it is not possible to make a statement about the level of peptide present in the AVK processes other than the observations made above.
VC Processes
In order to describe the morphology of the AVK processes throughout the VC, ICC preparations were made of eight full-length worms, six female (23.5–36.5 cm in length) and two male (21–22 cm). In female worms, staining of the AVK VC processes terminated in the region of the gonopore, which is located approximately one-third of the body length from the head. The two processes were varicose, often with a beaded appearance (Figure 3c), and appeared to taper as they proceeded down the VC. In the region of the gonopore (within 2 cm anterior or posterior to the gonopore), staining either ended in apparent axonal termini (n = 1, Figure 3d) or appeared narrower, and became fainter and fainter until it was no longer visible (n = 5). There was no obvious interaction with the cells of the vulva. In the two male worms, staining could be traced to approximately the same relative position along the length of the worm as in females. In one male worm, each of the two AVK processes terminated in large varicosities, and in the other the processes tapered and became invisible. In both sexes, the AVK processes appear to terminate in the same relative position in the body; no staining was seen in the posterior portions of the worm. This is interesting because the region of the worm where the processes terminate is the region where the anteriorly propagating locomotory waves are initiated in both males and females.
Previously, electron microscopical studies of serial sections of the VC determined two classes of INs: the large INs (15–30 μm diameter), which synapse directly onto MNs, and small INs (2–8 μm diameter), which synapse onto each other and onto large INs.27 ICC (with the anti-AF11 antibody) of transverse sections of the VC clearly identifies the AVKs as small INs (Figure 3e). The process with slightly larger diameter (identified as AVKR from its position in the whole mounts and sections) is located within the hypodermis between the bundle of VC neural processes and the cuticle and is relatively isolated from the other neural processes in the VC (Figure 3d). The other process (AVKL) appears to contact the VC neurite bundle, but is distinct from most of the other small interneurons, which are located in a region between the motorneurons and the large INs.27
Localization of AF11 in AVK Neurons by MS
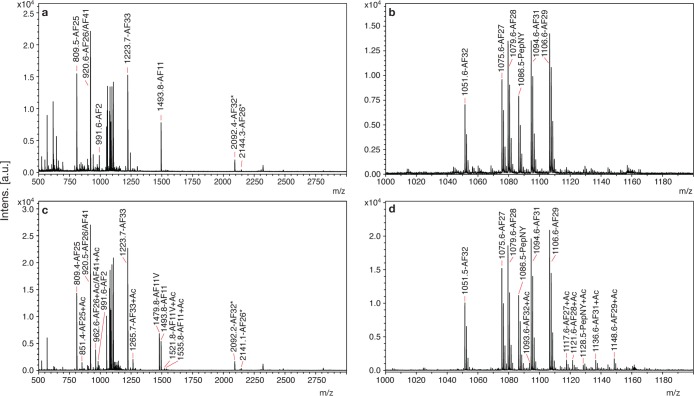
Individual AVK neurons were manually dissected from the ventral ganglion (Figure 1c), and placed on a MALDI-TOF MS target. The paired AVK neurons are recognizable in the dissecting microscope as the most posterior neurons in the VG, each with a monopolar process that projects anteriorly toward the midline and into the nerve ring (Figure 1b). This “balloon-on-a-string” morphology together with their location makes them easy to identify for dissection. Isolated AVK neurons were subjected to MALDI-TOF-MS analysis, which revealed a highly reproducible spectral pattern of nine intense peaks and a variable number of minor peaks (Figure 4a, b; Table 1; Supporting Information Table S2). On-target acetylation produced m/z shifts of 42 Da, consistent with acetylation of single amino groups (Figure 4c, d). Spectra from cells exposed to methylene blue, which we have previously shown to oxidize methionine to its sulfoxide in single cell preparations,14 showed no oxidation peaks (mass shifts of 16 Da), suggesting that none of these peptides contain methionine (Supporting Information Figure S1 and Table S2). The mass to charge ratio (m/z) of three of the nine high-intensity peaks matched the m/z of three previously sequenced peptides, for which the gene sequences were not yet known (AF11-SDIGISEPNFLRFa, 1493.8 Da; KPNF[L/I]RFa, 920.5 Da; NNF[L/I]RFa, 809.4 Da19,22). MS/MS of these peptides confirmed their identities, although it could not resolve the L/I ambiguities (Supporting Information Figure S2a, b). Unlike the anatomical asymmetry seen in the AVK processes, we did not see any obvious differences in spectra between AVKL and AVKR cell bodies, in either peak m/z or relative peak size. Because both MALDI-TOF MS and ICC are nonquantitative methods, we are not able to draw conclusions about relative quantities of peptides in AVKL versus AVKR.
Figure 4.
Neuropeptides from single AVK neurons in native and acetylated forms. (a) Mass spectrum from an individual AVK with no chemical modifications. (b) Expansion of (a) from m/z 1000–1200. (c) Spectrum from an individual AVK treated with acetic anhydride, which acetylates free amines (N-terminus, K) and to a lesser extent Y, and adds 42 mass units. (d) Expansion of (c) from m/z 1000–1200. X axis: m/z is mass-to-charge ratio. Y axis is intensity of MS signal in arbitrary units, a.u. Asterisk (*) indicates extended peptide due to incomplete cleavage.
Table 1. Peptides Expressed in A. suum AVK As Detected by MALDI-TOF MSa.
| transcript | peptide | sequence | M + H |
|---|---|---|---|
| afp-11 | AF25 | NNFLRFa | 809.4 |
| AF31– | SNNFLRFa | 896.5 | |
| AF26 | KPNFLRFa | 920.5 | |
| AF41 | KPNFIRFa | 920.5 | |
| AF32 | GSDPNFLRFa | 1051.5 | |
| AF27 | PADPNFLRFa | 1075.6 | |
| AF28 | SAEPNFLRFa | 1079.6 | |
| PepNY | NDQFDREY | 1086.4 | |
| AF31 | TPSNNFLRFa | 1094.6 | |
| AF29 | NAEPNFLRFa | 1106.6 | |
| AF33 | SNQAQNFLRFa | 1223.6 | |
| AF11-I3V | SDVGISEPNFLRFa | 1479.8 | |
| AF11 | SDIGISEPNFLRFa | 1493.8 | |
| AF32* | TAPPLTFGKKGSDPNFLRFa | 2092.1 | |
| AF26* | NDQFDREYRKPNFLRFa | 2144.1 | |
| afp-4 | AF2 | KHEYLRFa | 991.5 |
Asterisk (*) indicates extended form. Superscript dash (−) indicates truncated form of peptide.
PNFLRFa-Encoding Gene Sequences
Searches of an A. suum GSS database for the three previously reported peptides (AF11, AF25, and AF26)19,22 returned two genomic sequences, GSS 2 and GSS 4 (Figure 5b, d and Supporting Information Figure S2). GSS 2 encoded KPNFIRFa (AF41), and GSS 4 encoded KPNFLRFa (AF26, isobaric with AF41), NNFLRFa (AF25), and an alternative form of AF11 with valine in place of isoleucine at position 3 (SDVGISEPNFLRFa, AF11-I3V). Unique PCR primers from two portions of GSS 4 were used to clone the sequence from isolated total RNA. The cloned product encoded the previously identified form of AF11 (Figure 5e), suggesting that the AF11-I3V is either a mutant form due to an adenosine to guanine transition or is due to a sequencing error. We favor the first explanation since while all individual AVKs analyzed by MALDI-TOF-MS contained a peak corresponding to AF11, one acetylated cell (acetic anhydride exposure adds 42 to free amines within the peptide22) contained an additional peak corresponding to AF11-I3V (Figure 4c). MS/MS of this second peak confirmed this sequence (Figure 6), so we conclude that this individual worm was heterozygous for a mutant allele of this gene, and expressed both AF11 and AF11-I3V in AVK. We do not have an accurate estimate of the frequency of this allele in the population, but it appears to be relatively infrequent, as we have seen only the canonical AF11 in all other MS spectra of AVK neurons, and in all of our cloned afp-11 sequences.
Figure 5.

Nucleotide sequence of afp-11, including GSS, afp-11 mRNA riboprobe sequence, and deduced amino acid sequence of afp-11. Blue indicates encoded peptide. (a–d) Gray indicates predicted intronic region. (b, c) Pink indicates predicted peptide not detected by MS. Bold indicates basic cleavage sites. (a) Sequence from GSS 1 (ED050610) including most of the signal peptide of the afp-11 transcript. (b) Sequence from GSS 2 (ED376227.1) encoding AF41, a portion of PepTF, and part of the signal peptide. c) GSS 3 (ED095318.1) encoding AF29, AF31, AF32, AF33, and the remaining portion of PepTF. (d) GSS 4 (ED362933.1) encoding AF11-I3V, AF25, AF26, AF27, AF28, and PepNY. (e) Cloned portion of GSS 4 from unique primers. (f) Complete afp-11 consensus sequence deduced by 3′ and 5′ RACE and confirmed by unique primers (underlined purple). Green indicates signal peptide. The polyadenylation signal is italicized.
Figure 6.
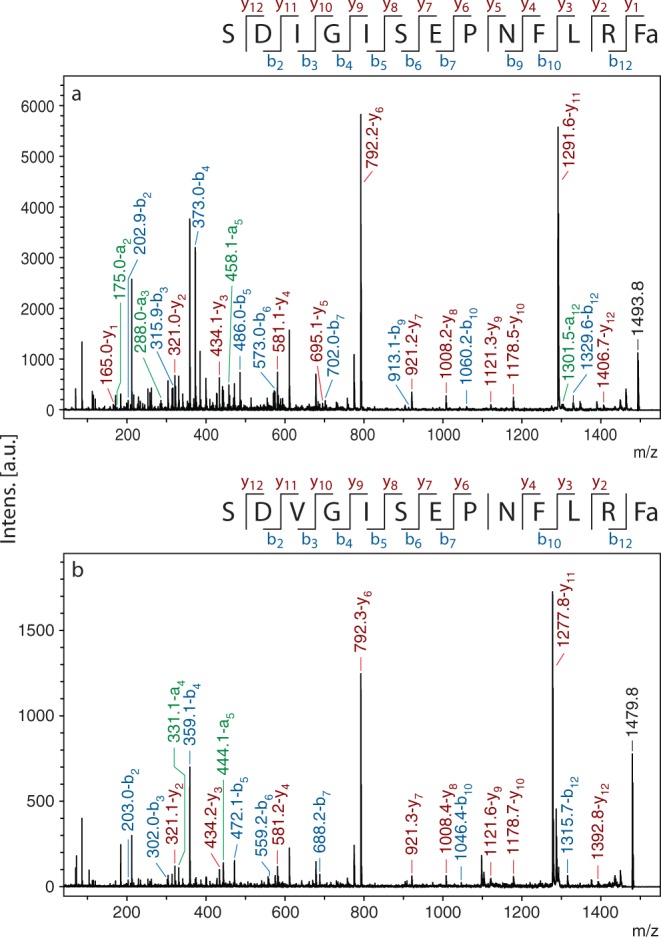
MS/MS spectra of AF11 and AF11-I3V from AVK. Peaks representing a (green), b (blue), and y (red) ions are labeled, and b and y ions are summarized in the sequence at the top of each spectrum. (a) MS/MS spectrum of AF11, m/z 1493.8. (b) MS/MS spectrum AF11-I3V, m/z 1479.8.
Cloning the afp-11 Transcript
Typically, one should be able to clone most transcripts in A. suum by designing unique primers based on GSS information and using as reverse primer SL1 (a splice leader sequence found in 80% of known A. suum transcripts28). However, this was unsuccessful for cloning afp-11. Instead, both 3′ and 5′ rapid amplification of cDNA ends (RACE) was required (Figure 5f). The RACE products produced two overlapping sequences that included both GSS 2 and GSS 4 as well as a sequence that corresponded to an additional GSS (GSS 3, Figure 5c), which encoded several additional highly sequence-related peptides (Figure 5c, d, see below). It is likely that the entire open reading frame was represented, as the 5′ RACE product encoded an initiating methionine and a signal peptide, and the 3′ RACE product terminated in a poly-A tail (Figure 5f). However, we cannot exclude the possibility that the 5′-untranslated region might be more extensive than the sequences we report. There is no evidence that this transcript is trans-spliced to SL1. To confirm the transcript sequence predicted from the overlapping RACE reaction products, PCR was performed with unique primers just inboard from the 5′ and 3′ ends, and resulted in a 843 nucleotide sequence that confirmed the sequence of the transcript deduced from the RACE sequences (Figure 5f; accession number JN547404). A GSS sequence encoding the N-terminal region of the signal peptide was also found (GSS 1, Figure 5a). The recently available draft genome sequence of A. suum(29) included all the sequences reported above, separated by introns with typical 5′ and 3′ splice sites. Most of the predicted signal peptide-encoding region is on a separate exon (Supporting Information Figure S3).
Sequencing of Novel afp-11 Peptides
The translated sequence from afp-11 that corresponds with GSS 3 and GSS 4 predicted 7 novel peptides: PADPNFLRFa (AF27), SAEPNFLRFa (AF28), NAEPNFLRFa (AF29), TPSNNFLRFa (AF31), GSDPNFLRFa (AF32), SNQAQNFLRFa (AF33), and NDQFDREY (PepNY) (Figure 5c, d). Intense peaks with the corresponding m/z of each of these peptides were present in the MS spectra from AVK (Figure 4). MS/MS confirmed all seven peptide sequences (Figure 7, Supporting Information Figure S4a). The MS/MS spectra of AF27, AF28, AF29, and AF32 contained a diagnostically intense peak at 792.4 (PNFLRFa) due to the labile D-P and E-P bonds. MS/MS analysis of synthetic peptides provided additional confirmation of all peptide sequences (Supporting Information Figure S5). The full afp-11 sequence (Figure 5f) contains one additional predicted peptide (TAPPLTFa, PepTF) that is not represented in our spectra. One possible explanation is that it lacks a basic amino acid. In order to be detected by MALDI-TOF, a molecule needs to accept a proton to become charged, and in the −RFamide peptides, the basic amino acid arginine fulfills this role. Synthetic PepTF was only detected by MALDI-TOF MS as a sodium adduct ion (data not shown), but we did not detect the sodium adduct form in spectra from AVK neurons (Figure 4).
Figure 7.
MS/MS of novel AF peptides encoded by afp-11. Peaks representing a (green), b (blue), and y (red) ions are labeled, and b and y ions are summarized in the sequence at the top of each spectrum. Insets in (a), (b), (d), and (e) are at a reduced intensity scale to show the relative intensity of the peaks that are off-scale, particularly the peaks at m/z 792.4 in (a), (b), and (e). (a) MS/MS spectrum of AF27, m/z 1075.6. (b) MS/MS spectrum of AF28, m/z 1079.6. (c) MS/MS spectrum of AF29, m/z 1106.6. (d) MS/MS spectrum of AF31, m/z 1094.6. (e) MS/MS spectrum of AF32, m/z 1051.5. (f) MS/MS spectrum of AF33, m/z 1223.6.
Bioactivity
The relationship between sequence and behavioral effects of these neuropeptides is crucial in understanding the neural control of A. suum behavior. We, therefore, submitted AF11 and its related peptides AF25–29, AF31–33, AF41, and peptides NY and TF to functional analysis using A. suum muscle strip preparations
The effects of peptides on acetylcholine (ACh)-induced contraction of dorsal muscle strips were measured. Single doses of each of the peptides encoded by afp-11 produced a reduction in the contraction and variable recovery over the course of 50 min after removal of the peptide (Figure 8a, b). There was no detected effect on background muscle tension. Control experiments show that ACh-induced contraction declines moderately over 50 min in the absence of peptide (from 100% of initial contraction to 78%).
Figure 8.
Effects of peptides on ACh-induced muscle contraction. ACh (5 μM) was applied until the contraction reached its peak and then was washed out with saline. Contraction strength is normalized to the initial contraction prior to the application of peptide for each individual worm. Error bars show standard error. (a, b) Effects due to single exposure to peptides. 10 μM peptide was introduced at −10 min and washed out at time 0 (time when peptide present indicated by gray shading). Responses to 5 μM ACh were tested every 10 min after removal of peptide. (c, d) Effects during extended exposure to peptide. 10 μM peptide was initially introduced at time 0 (arrow), and 5 μM ACh was applied every 10 min; in each case, ACh was washed out with Ascaris saline containing 10 μM peptide.
For the afp-11 peptides, initial activity is inhibitory, but potency and time course of activity varies. Peptides AF26 (KPNFLRFa) and AF41 (KPNFIRFa) were the most potent, and the effects of AF41 were fully reversible, while those of AF26 were only partially reversible. The effects of AF25, AF27, and AF31 were also fully reversible, whereas the effects of AF11, AF28, AF29, AF32, and AF33 were not fully reversible. The similarity in sequence and activity for AF26 and AF41 compared to other PNFLRFa-ending AF peptides suggests that the N-terminal lysine preceding the stereotypical PNF(L/I)RFa C-terminal sequence results in greater inhibition of ACh-induced contraction. The L/I difference in the third position from the C-terminus was not a factor in initial potency but may alter the time course of neuropeptide activity. No sequence-activity patterns emerged in the reversibility of the effects of the other afp-11 peptides.
The effects of prolonged exposure to the peptides were also examined. Peptide AF26 caused a sustained reduction in the ACh response, but the responses to AF11, AF25, AF28, AF29, AF31, AF32 and AF33 were biphasic, consisting of a rapid inhibition of the response followed by a reduced inhibition (Figure 8c, d).
These results are compatible with previously reported effects on locomotory behavior and on body length when peptides AF25, 26, 29, 31, and 41 were injected into intact worms.25 These peptides caused a cessation of propagating locomotory waveforms and an increase in body length. Peptide-induced changes in cyclic AMP and cyclic GMP in A. suum body wall were also measured in these experiments;25 such changes were small compared with the dramatic increase in cAMP produced by AF1 and AF2, but it is striking that AF26 and AF41 were the only peptides that produced reductions in both cAMP and cGMP levels. The differences in activity of the peptides encoded by the same transcript show that it is necessary to investigate each peptide product individually. Previous experiments have shown that peptide products of the same transcript, afp-6, produced totally opposing effects on behavior and on the physiological responses of individual motor neurons.9
Each of the AF peptides encoded by afp-11 strongly or severely inhibits ACh-induced contraction. These findings are consistent with the findings that in A. suum most AF peptides have potent, though diverse, physiological and behavioral effects.4,8,9,30
Patterns between neuropeptide sequence information and the behavioral effects of neuropeptides are important in understanding neural control of behavior in A. suum. The activity of the endogenous afp-11 peptides demonstrates that A. suum uses multiple neuropeptides encoded from the same gene to inhibit muscle contraction. Numerous peptides, all with inhibitory activity, but with varying potency and reversibility may be used in combination to optimally control locomotory activity.
Minor Peptide Peaks in AVK
In addition to the nine intense peaks characterized above, the MS spectra for individual AVKs also contained a variable number of minor peaks (Figure 4, Supporting Information Table S2). The m/z of one minor peak, found in 8 of 12 neurons, including cells that were chemically modified as well as those that were not, corresponds to the known peptide AF2 (KHEYLRFa). MS/MS of this peak confirmed this identification (Supporting Information Figure S2c). MS/MS of a different minor peak at m/z 896.5 produced the sequence SNNF[L/I]RFa (Supporting Information Figure S4b). This appears to be a truncated form of AF31 (AF31–), although it is unclear if this is an endogenous peptide produced by an unusual proteolytic cleavage or an artifact of sample preparation or MALDI-MS. MS/MS of two other minor peaks with the larger m/z of 2144.1 and 2092.1 suggests that these peptides are NDQFDREYRKPNFLRFa and TAPPLTFGKKGSDPNFLRFa (Supporting Information Figure S4c, d), likely products of incomplete precursor processing, the first linking peptide NY with AF26, and the second peptide PepTF with AF32. The presence of both TAPPLTFGKKGSDPNFLRFa and AF32 suggests that PepTF may indeed be present in AVK neurons. Another minor peak with m/z 2485.3 corresponds to that predicted for TPSNNFLRFGKSNQAQNFLRFa, another product of incomplete precursor processing in which AF31 and AF33 remain uncleaved; attempts to analyze the sequence of this peptide by MS/MS were not successful.
Riboprobe Synthesis and in Situ Hybridization
In order to localize afp-11 by ISH, we used primers that flanked the peptide-encoding region of GSS 4 (AFP11IntF1, ccttcgtttcggtaaatcagc, and AFP11IntR1, tgttgctcctctccatcctc) to synthesize a riboprobe complementary to a 211 nucleotide segment of the transcript (Figure 5e). The antisense strand was generated as the experimental probe, and the sense strand as a negative control. In the 12 whole mount head preparations treated with the antisense probe, we saw robust staining of the two AVK cells (Figure 9) while no staining was seen in any other cells, including ALA. The two negative control preparations did not have any staining (not shown). The ISH staining in AVK shows that the afp-11 transcript is present in AVK. ICC and MS data show that afp-11 is translated and its protein product further processed in AVK.
Figure 9.

Expression of afp-11 in AVK neurons in the ventral ganglion (VG) of A. suum by in situ hybridization. Nerve ring (NR), ventral cord (VC), left lateral line (LLL), right lateral line (RLL), AVK left (AVKL), AVK right (AVKR).
Variability in ALA
Previous direct MALDI-TOF MS of single ALA neurons detected AF11 in three out of nine cells,14 confirming our finding of anti-AF11 immunoreactivity in ca. 30–40% of ALA neurons, and supporting our conclusion that there is biological variability in afp-11-encoded peptide expression in ALA. In contrast, the lack of ISH staining in ALA with the afp-11 riboprobe may be attributable to limitations of the technique. We do not know if afp-11 transcript levels in ALA are too low to be detected by ISH or whether this riboprobe was not able to penetrate ALA as easily as it could AVK because of the surrounding hypodermal tissue that ensheathes the cells in the DG; we consider this possibility less likely, since riboprobes for afp-3 and afp-4 readily penetrate the DG and label ALA or RID.15 Because none of the three methods we used is quantitative, it is impossible to distinguish whether lighter staining in ALA in ICC preparations or lower intensity peaks in the ALA MS spectra are due to lower levels of peptide expression or are due to limitations in the techniques of detection. Another possibility is that ALA expresses an alternatively spliced transcript from the afp-11 gene and that the riboprobe we used for ISH did not detect it efficiently. A further possible explanation is that the lifetime of expressed peptides is longer than that of the mRNA that encodes them, and that different ages or life experiences of individual worms, over which we have no control, account for the observed differences.
Comparison with Other Nematodes
Sequences and Cellular Expression Patterns
Despite dramatic differences in size and environment, nematodes have been shown to have remarkably similar nervous systems in terms of their neuronal morphology. Neurons in A. suum and C. elegans are so similar that the same nomenclature can be used in both species. In addition, neuropeptide sequences are highly conserved throughout the nematodes. In particular, AF11 belongs to a large family of sequence-related peptides found in many other nematodes. It shares the C-terminal PNFLRFa sequence with the seven peptides encoded by the flp-1 gene of C. elegans.20 As we report here, afp-11, the homologous gene from A. suum, encodes 11 AF peptides, including AF26 (KPNFLRFa), which was predicted from flp-1 (but not yet isolated from C. elegans), and 5 other peptides with the characteristic PNFLRFa sequence; 3 peptides have −NNFLRFa C-termini, and 1 peptide has a QNFLRFa sequence. AF41 (KPNFIRFa) is identical in sequence to peptide PF4, first isolated from Panagrellus redivivus.31
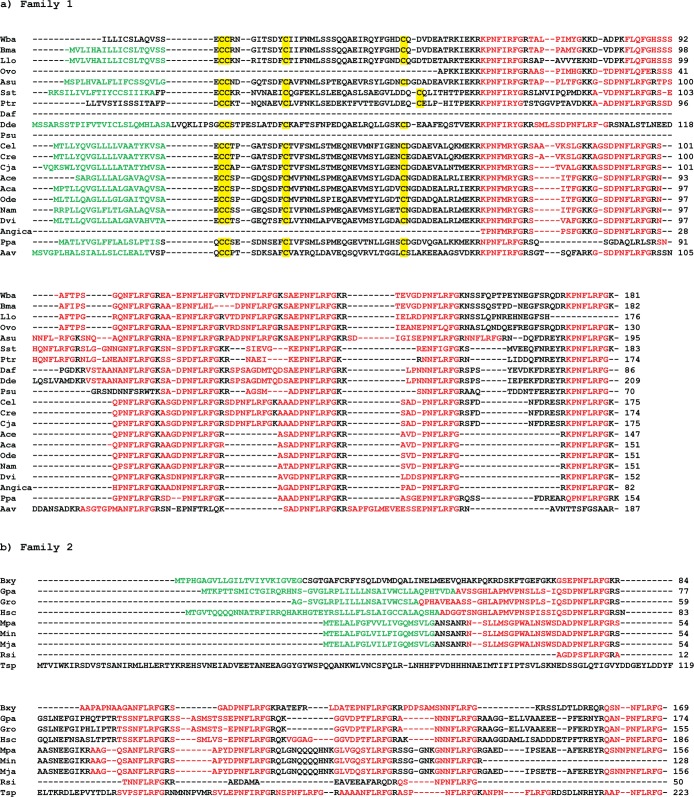
Earlier BLAST searches of nematode ESTs predicted related peptides from 16 species of plant and animal parasitic nematodes.10 We have further searched the more recently available EST and genomic sequences available on the NCBI Web site, using both the A. suumafp-11 sequence and the C. elegansflp-1 sequence as queries. We found many sequences that encode precursor proteins that include predicted homologues of the peptides found in AVK neurons. The majority came from EST libraries; almost all had predicted signal peptides (SignalP32). When the sequences were aligned, they fell into two distinct families (Figure 10). Family 1 was typified by several conserved features: (1) following the signal peptide, an N-terminal region with fairly high sequence similarity that included a highly conserved set of 4 cysteines, the first two being immediate neighbors in the sequence; (2) multiple predicted peptides bounded by basic or dibasic amino acids, with the first peptide typically being KPNFIRFa or KPNFMRYa and the last usually being KPNFLRFa, together with a variable number of other peptides with C-terminal (D/E)PNFLRFa or NNFLRFa sequences. Family 2 lacked the conserved cysteines and the two flanking KPNF(I/M)R(F/Y)a and KPNFLRFa peptides, but included several peptides with PNFLRFa or NNFLRFa sequences. Only one family was found in each nematode species. Since the majority of the sequences are from EST libraries, the relative abundance of transcripts might affect their representation in the library. However, searches of the genomes of A. suum, C. elegans, Brugia malayi, and Loa loa showed sequences from only one of the two groups. The distinction of two classes of genes nicely follows phylogeny, with the Family 2 transcripts being found in clades I and IVb nematodes,33 whereas Family 1 transcripts are found in clades III, IVa, and V.
Figure 10.
CLUSTAL multiple sequence alignments of related predicted precursor proteins that include AF11-related peptides. Most of the data are from ESTs, and some are not full-length. (a) Family 1, (b) Family 2. Red, predicted processed peptides, with emphasis on RFamide peptides; green, predicted signal peptide; yellow, conserved cysteines in Family 1. Accession numbers: Aav, Aphelenchus avenae, GO479351; Aca, Ancylostoma caninum, AW627172; Ace, Ancylostoma ceylanicum, CB276900; Angica, Angiostrongylus cantonensis, JK314934; Asu, Ascaris suum, JN547404; Bma, Brugia malayi, BH006937; Bxy, Bursaphelenchus xylophilus, CJ983516; Cel, Caenorhabditis elegans, BJ108988; Cja, Caenorhabditis japonica, FF133133; Cre, Caenorhabditis remanei, DY525954; Daf, Ditylenchus africanus, FE921558; Dde, Ditylenchus destructor, GU198750; Dvi, Dictyocaulus viviparus, EV852328; Gpa, Globodera pallida, AJ310913; Gro, Globodera rostochiensis, BM354407; Hsc, Heterodera schachtii, CF587423; Llo, Loa loa, XM003243156; Min, Meloidogyne incognita, BQ519755; Mja, Meloidogyne javanica, CF350781; Mpa, Meloidogyne paranaensis, CK426955; Nam, Necator americanus, BG467519; Ode, Esophagostomum dentatum, AY172997; Ovo, Onchocerca volvulis, BF918235; Ppa, Pristionchus pacificus, FE933932; Ptr, Parastrongyloides trichosuri, BI451336; Psu, Panagrolaimus superbus, GW4099937; Rsi, Radolphus similis, EY195794; Sst, Strongyloides stercoralis, BG224354; Tsp, Trichinella spiralis, XM003379676; Wba, Wuchereria bancrofti, CK850238.
Variability of Expression
Despite the strong sequence conservation among peptides from different species, recent evidence suggests that cellular expression of homologous peptides can be dramatically different between nematode species.14,15,34 The results of the present study reinforce this conclusion. In C. elegans, there are several reports of the cellular expression of flp-1 as determined by GFP reporter constructs.13,35 However, there are discrepancies in the reported expression patterns of flp-1: in all cases, there is agreement that the AVK neurons express flp-1, but some reports also record flp-1 expression in AIA, AIY, AVA, AVE, RIG, and RMG neurons.35−37 We have seen no expression of afp-11 in any of these neurons in A. suum. We found variable expression of afp-11 in ALA by ICC and MS,14 but there is no reported expression of flp-1 in the ALA neuron of C. elegans.13 Our data on the cellular localization of AF11 in A. suum reinforce our previous conclusions that the specific cells containing related or identical peptides can be different in the two species. The potato cyst nematode Globodera pallida also expresses a transcript (gpflp-4) encoding similar peptides (Table 2).12 ISH with this transcript found expression only in the RVG and VC motorneurons of G. pallida, not in the VG where the AVK neurons are located.12 Again, this suggests species-specific expression of these related families of peptides.
Table 2. Comparison of Analogous Peptides Localized within the AVK in A. suum, C. elegans, and G. pallidaa.
|
A. suum |
C. elegans |
G. pallida |
||||||
|---|---|---|---|---|---|---|---|---|
| gene | peptide sequence | AVK | gene | peptide sequence | AVK | gene | peptide sequence | AVK |
| afp-11 | KPNFIRFab | yes | flp-1 | SADPNFLRFab | yes | gpflp-4 | SDPNFLRFa | no |
| TAPPLTFa | SQPNFLRFab | TSSNFLRFa | ||||||
| (TAPPLTFGKK)GSDPNFLRFac | ASGDPNFLRFab | SSASMSTSSEPNFLRFa | ||||||
| GSDPNFLRFa | SDPNFLRFab | GGVDPTFLRFa | ||||||
| TPSNNFLRFab | AAADPNFLRFab | ANNNFLRFa | ||||||
| SNQAQNFLRFab | (K)PNFLRFa | QANPNFLRFa | ||||||
| NAEPNFLRFab | AGSDPNFLRFa | |||||||
| PADPNFLRFab | (K)PNFMRYa | |||||||
| SAEPNFLRFab | ||||||||
| SD(I/V)GISEPNFLRFad,e | ||||||||
| NNFLRFab | ||||||||
| NDQFDREYb | ||||||||
| (NDQFDREYR)KPNFLRFac | ||||||||
| KPNFLRFab | ||||||||
| afp-4 | KHEYLRFab | yes | flp-14 | KHEYLRFa | NA | gpflp-3 | KHEYLRFa | no |
In A. suum, afp-11 was localized by MALDI-TOF MS and in situ hybridization. afp-4 has been localized in AVK by MALDI-TOF MS. In C. elegans, flp-1 was localized using GFP constructs.13,35 In G. pallida, in situ hybridization did not detect gpflp-1 and gpflp-14 in AVK.12
Peptides isolated and sequenced.
Incompletely processed peptide.
Located in AVK by immunocytochemistry.
Both I and V forms have been detected.
We need to be cautious about the conclusions we draw from peptide localization comparisons between A. suum and C. elegans, because different techniques were applied to describe the expression patterns of peptide-encoding genes. In A. suum, we have used three independent techniques, ISH, ICC, and single-cell MS, and all detect either the peptides themselves or the transcripts that encode them. In C. elegans, GFP constructs that include upstream 5′ DNA sequences for individual peptide-encoding genes in C. elegans were used.13 Application of SAGE (Serial Analysis of Gene Expression, which detects mRNA) to populations of individual C. elegans neurons obtained by cell-sorting has shown significant differences from the observations of strains with GFP reporters.38 The transcription factors that regulate gene expression are numerous and have complex interactions; they have DNA binding sites that may be located at long distances from the promoter, including in 3′ regions and introns,39 so it is extremely difficult to be sure that the complete set of appropriate control elements are included in GFP constructs. In C. elegans, ISH of the flp-1 transcript has not yet been reported, and the use of antineuropeptide antibodies is so far very limited.
AF2 (KHEYLRFa), occasionally detected in AVK spectra in A. suum (Figure 4a, c), is encoded by the flp-14 and gpflp-3 transcripts in C. elegans and G. pallida, respectively.12,40 GFP reporter constructs in C. elegans for flp-14 failed to show expression in any cells.7 In G. pallida, gpflp-3 was not detected by ISH in the VG. In 10 preparations of A. suum, ISH with an afp-4 antisense probe (afp-4 encodes AF2) showed faint staining of ALA in only one preparation.15 This probe gave faint positive staining of between one and five VG neurons, but they were not identified in these preparations, although their reported position in the ganglion suggests that they include the AVK neurons.
In this and previous reports,14,15,34 we have shown other examples of variability of cellular expression patterns of different gene products. In A. suum, it is unclear how much of this variability is due to genetic differences, since the worms are not isogenic (in fact we report a case of genetic heterogeneity in this paper), and how much are due to nongenetic causes. The variability of expression seen in individual worms reinforces the important and interesting issue of how such individual differences in peptide expression affect individual behavior. Perhaps, as suggested by Marder and colleagues,41 circuits of neurons are intrinsically robust, despite being made up of neurons with variable properties.
Bioactivity
Studies of biological activity suggest that at least some of these PNFLRFa peptides from other nematodes may show similar biological effects to the afp-11 peptides. Peptide PF1 (SDPNFLRFa) from Panagrellus redivivus causes relaxation of A. suum muscle42,43 which is similar to the effects we observed with the afp-11 peptides; similarly, six of the peptides encoded by afp-11 caused flaccid paralysis when injected into intact A. suum.25 In C. elegans, deletion mutants that encompass the flp-1 gene produce an array of behavioral defects, including hyperactive movement, and flp-1 overexpression is reported to have the opposite effect.36 PF4 (AF41) was shown by Maule et al.31 to cause relaxation of A. suum muscle. A thorough investigation of its mode of action was reported by Holden-Dye et al.44 who showed that it produces a rapid chloride-dependent hyperpolarization of A. suum muscle through a mechanism that is independent of the action of GABA, which also causes a rapid chloride-dependent hyperpolarization. In addition, there is a later response to PF4 that is also hyperpolarizing but independent of chloride. The relaxation in A. suum muscle produced by P. redivivus peptides PF1 and PF2 (SDPNFLRFa and SADPNFLRFa)42,43 is associated with a small hyperpolarization that is independent of chloride.44
In this paper, we show that AF41 and the A. suum homologues of PF1 and PF2 are encoded by the same transcript, along with six other sequence-related peptides. Thus, it seems that the peptide products of this gene produce inhibitory responses in muscle but by different mechanisms. Investigations of the A. suum gene afp-6, which encodes peptides AF21, 22, and 23, also showed that peptide products of one gene can have different activity; although all three peptides relaxed and hyperpolarized muscle, they had different effects on motorneurons: AF21 and 23 depolarized three classes of motorneurons (DE1, DE2, and DI), while AF22 hyperpolarized them.9
Conclusions
With all three of our techniques, we found consistent and robust results indicating the presence of afp-11, or the peptides that it encodes, in AVK. Direct MALDI-TOF MS analysis of single neurons continues to be a powerful tool for neuropeptide discovery and localization. When applied to the AVKs of the VG, we discovered eight novel peptides. We also identified a natural heterozygote using this method. ICC identified the AVKs as a pair of small ventral nerve cord interneurons. The AVK VC processes extend down the VC through the anterior third of the worm, which is the region where anteriorly propagating locomotory waveforms are generated. Electrophysiological work on A. suum DE2 (dorsal excitatory type 2) or DI (dorsal inhibitory) MNs showed that AF11 has very little effect on their membrane potential or input resistance.8 Previous reports have shown that the sequence-related peptide SDPNFLRFa, isolated from P. redivivus, acts on A. suum muscle.45 In the intact worm, the effects on motility could also involve other classes of motorneurons and/or interneurons, or involve interactions with other peptides or signaling systems, either from the afp-11 transcript or from other genes.
Methods
Animals
A. suum were collected from a local slaughterhouse. The worms were maintained in buffered saline (PBS; 150 mM NaCl, 8.5 mM sodium phosphate, pH 7.2–7.4) at 37 °C for up to 3 days before use.
Peptide Synthesis
Peptides were synthesized by the University of Wisconsin—Madison Biotechnology Center Peptide Synthesis Facility. They were >95% pure, as determined by HPLC and MALDI-TOF MS.
Antibody Production, Purification, and Testing
Synthetic AF11 (SDIGISEPNFLRFamide) was conjugated via glutaraldehyde to Imject mariculture keyhole limpet hemocyanin, (mcKLH, Pierce Protein Research Products). Polyclonal antibody serum was obtained from rabbits that had been immunized with the AF11-mcKLH conjugate (Panigen Inc., Blanchardville, WI). Serum was purified via affinity chromatography on AF11-conjugated silica bead columns (0.5 mg/mL AF11 in MOPS buffer, pH 7.5–8.0 incubated overnight at 4 °C with 1–2 mL Affigel Beads (Bio-Rad, Hercules, CA) suspended in MOPS) as described in Yew et al.9 After elution with Actisep (Sterogene, Carlsbad, CA), the purified anti-AF11 antibody (primary antibody) was tested for working concentration and specific immunoreactivity on a dot-ELISA spotted with 39 synthetic AF peptides as per Sithigorngul et al.26
Because the dot-ELISA is more sensitive than tissue staining, with antibody-peptide recognition of dot-ELISAs having been recorded at femtomole levels of peptide,26 the antibody was used at an approximately 10-fold higher concentration for the whole mount preparations. This is the standard procedure used in this laboratory for both polyclonal and monoclonal anti-AF peptide antibodies.
Whole Mount Immunocytochemistry (ICC)
Dissections for immunocytochemical study were as in Johnson and Stretton,46 with a brief description of the procedure and any exceptions noted below. A. suum were injected with 5 mg/mL collagenase (Type H, Sigma-Aldrich) in Ascaris saline (4 mM sodium chloride, 125 mM sodium acetate, 24.5 mM potassium chloride, 5.9 mM calcium chloride, 4.9 mM magnesium chloride, 5 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer, pH 6.8) to dissociate muscle tissue. A single injection with 0.1–0.2 mL of collagenase solution was made approximately 3 cm from the tip of the head for head preparations, and injections were made at 3–5 cm intervals along the entire worm’s length for whole mount full-length body dissections. For optimal collagenase activity, the worms were incubated for 1.5–2 h at 37 °C. For the head dissections, the anterior 1.5–2.5 cm of the worms was removed for dissection, and for the whole-length worm dissections each worm was cut transversely into 7–12 pieces ranging from 1.5 to 4.0 cm each. The pieces were then rinsed in PBS, cut longitudinally along the right lateral line (RLL), and pinned flat on a Sylgard-lined dish. The tissue was fixed overnight in 1% or 2% paraformaldehyde in PBS.47 Most preparations were subjected to the anti-AF11 primary antibody at a working concentration of 1:1000 in P1+ (0.1% BSA, 0.5% NP-40, 0.5% TritonX-100, 10% normal calf serum in PBS, pH 7.3–7.4). In some preparations, to better visualize the processes of the stained neurons in the VC, antibody concentration was raised to 1:100, 1:200, or 1:500. Negative controls were treated without primary antibody. For the blocking control preparations, anti-AF11 antibody (1:1,000) was incubated overnight at 4 °C with 1.5 mg/mL AF11 prior to use.
The secondary antibody was a goat-anti-rabbit polyclonal antibody conjugated to horseradish peroxidase (GAR-HRP, Bio-Rad) used at 1:500 in P1+. The preparations were then exposed to staining solution (0.006% H2O2, 0.03% 3–3′-diaminobenzidine-4-HCl in PBS), dehydrated through an ethanol series, and mounted on slides with Permount (Fisher Scientific). The mounted preparations were observed in bright field optics (Zeiss Universal microscope), and photographed with a Zeiss MRc digital camera with Zeiss Axio Vision AC software. Nonuniform illumination of the low power images was corrected by using the software shading correction. The images were not further digitally manipulated.
ICC of Transverse Sections
Stained transverse 6 μm sections of the A. suum head region were prepared as previously described48 except for the following: the sections were treated with the anti-AF11 primary antibody at 1:200 in P1+ and the GAR-HRP secondary antibody at 1:500 in P1+. The glycerin mounting medium consisted of 5% gelatin (w/v) in a 1:1 solution of glycerol and distilled water.
Molecular Biology
Total RNA isolation, cDNA preparation, 3′ and 5′ RACE, PCR, cloning, riboprobe synthesis, and in situ hybridization were performed as previously described.15 A general outline of the procedure and any variations from the published protocol are noted below. All primers were from Integrated DNA Technologies.
Total RNA Isolation and cDNA Preparation
A. suum total RNA isolation and cDNA preparation was performed as previously described.15
RACE and Extended Transcript Sequence Confirmation
3′ and 5′ RACE was performed according to manufacturer’s protocol using Clontech SMART RACE cDNA Amplification kit. Total A. suum RNA was used as the template for making the 5′ and 3′ RACE-ready cDNA. Specific primers were: 5′ RACE primer GSP1 (GTTGCTCCTCTCCATCCTCTTTAGTTCTC) and 3′ RACE primer GSP2 (CCTTCGTTTCGGTAAATCAGCTGAGCC). The RACE products were extracted from a 1% agarose gel and cloned into chemically competent E. coli (TOPO TA Dual Promoter Top10 Cloning kit, Invitrogen). Purified plasmid (QIAprep Spin Miniprep Kit, Qiagen) was sequenced by the UW—Madison Biotech DNA Sequencing Facility. The sequencing results were interpreted using ChromasPro software. To confirm the RACE results, PCR was performed on A. suum cDNA using specific 5′ primer afp11FWD1 (CGCACATATAAGCCATCGAA) and afp11REV1 (CAACAGAGAAACGACGTAACGA). Cloning and sequence confirmation were performed as above.
Synthesis of Riboprobes
PCR was performed with the specific primers AF11IntF1 (CCTTCGTTTCGGTAAATCAGC) and AF11IntR1 (TGTTGCTCCTCTCCATCCTC) with A. suum cDNA template to amplify a 211 base-pair segment of the peptide-encoding region of afp-11 transcript. Cloning and sequence confirmation were performed as above. Purified plasmid containing the confirmed sequence was linearized with Not1 or Spe1 (New England Biolabs) and used as template for riboprobe synthesis. In vitro transcription (Maxiscript SP6/T7 kit, Ambion) was performed to produce both antisense and sense digoxigenin-UTP-labeled riboprobes for the afp-11 transcript.
In Situ Hybridization (ISH)
A. suum head preparations were dissected in 0.1 M phosphate buffer, pH 7.2, and fixed in 1% paraformaldehyde in 0.1 M phosphate buffer overnight at 4 °C. The preparations were dehydrated with an ethanol series. They were rehydrated and treated for 15 min at 37 °C with a proteinase-K solution, refixed, rinsed, and transferred to 12-well plates. The preparations were incubated overnight with riboprobe in a hybridization buffer. Then they were rinsed, treated with RNase A, blocked for nonspecific antibody and endogenous alkaline phosphatase (AP) activity, and incubated with anti-digoxigenin AP-conjugated antibody fragments. The preparations were then stained with NBP/BCIP staining solution (Roche), mounted on slides with Clear Mount (Electron Microscopy Sciences), and viewed and photographed with the same equipment as the ICC whole mounts.
Sample Preparation for Mass Spectrometry
Single neurons were isolated by using slight modifications of the methods described previously.14 Briefly, adult female A. suum were injected with 2 mg/mL collagenase in Ascaris saline (as described above). After 1.5 h at 37 °C, the head was removed and rinsed with water. The head region was then cut longitudinally and pinned flat on a Sylgard-lined dish filled with 170 mM ammonium acetate.49 By using tungsten needles, AVK neurons were manually dissociated from the hypodermis and the other neurons in the VG. In some experiments, cells were stained with methylene blue to aid cell identification: prior to dissection, the preparation was exposed to 16 mM methylene blue in EtOH, and after 1–2 min, the sample was rinsed two to three times with 170 mM ammonium acetate. Dissection then proceeded as described above.
Cells were transferred to a Bruker ground stainless steel target plate, rinsed on-target with 0.5 μL isopropanol,50 and allowed to dry. The matrix, α-cyano-4-hydroxycinnamic acid (CHCA; Sigma-Aldrich), was rinsed with 5% acetic acid for one minute, and then dissolved in 100 μL 50% acetonitrile and vortexed for 90 s. Using a Nanoliter Cool Wave Syringe II,51,52 matrix was applied robotically to the sample 1–3 times in 30–100 nL amounts and allowed to dry after each application.
MS Acquisition
A Bruker Ultraflex III MALDI-TOF/TOF MS (Bruker Daltonics, Billerica, MA) equipped with a Smartbeam laser, a reflectron, and a LIFT cell was used to obtain MS and MS/MS spectra. Compass v. 1.2 software was used to control the instrument. Due to the delicacy of the sample, all spectra were obtained from 50 laser shots per acquisition instead of the default 200 shots. MS spectra were obtained in positive ion reflector mode, with an m/z range of 450–4000 Da. Polypropylene glycol (PPG) 1000 or polyethylene glycol (PEG) 1500 in methanol was used for external calibration.
Acetylation of Cells
Prior to application of CHCA, 0.5 μL of 3:1 methanol/acetic anhydride was applied to each cell and allowed to dry.5 After the solution evaporated, the cell was covered with matrix as described above.
Oxidation of Cells
In cells exposed to Methylene Blue, as described above, methionine residues were oxidized to the sulfoxide, with a gain of 16 Da. Methylene Blue was originally used as a method for staining cell bodies to aid the dissection, and it was found that methionine-containing peptides were partially oxidized.14
Assignment of Peaks and Interpretation of Mass Spectra
All spectra were analyzed using Bruker Daltonics flexAnalysis 3.0 software. For all MS spectra, masses were assigned automatically by the software. All MS/MS data underwent background subtraction and smoothing prior to automatic assignment of masses. Occasionally, additional peaks were assigned a mass manually. Peaks were considered to be above noise when the intensity of the peak was at least twice that of the baseline. A peak was initially identified as a known peptide if the observed m/z value was within ±0.2 Da of a previously sequenced A. suum peptide.4,5,9,17−19,22 Molecular masses were calculated by Prospector MS-Product (http://prospector.ucsf.edu). Assignments were confirmed by MS/MS and in some cases by chemical modifications (acetylation and/or oxidation). To identify novel peptides, de novo sequencing was performed by hand, using a mass assignment error of ±0.2 Da. Further verification was obtained by on-target chemical modification, by comparing the MS/MS spectra of synthetic peptides to the spectra obtained from single neurons, and by tBLASTn searches and cloning of gene sequences.
Database Searches
All putative peptide sequence assignments were searched against a catalog of 447 546 A. suum GSSs (available in NCBI). Since the peptide sequences are short, a local tBLASTn search was used with a word size of 2, E value of 20 000, and a PAM30 matrix. Search results were translated using the ExPASy Translate tool (http://au.expasy.org/tools/dna.html) and visually examined for possible peptide cleavage sites, as well as a C-terminal glycine for amidated peptides. Possible splice sites were predicted by GenScan (http://genes.mit.edu/GENSCAN.html53), or were detected directly by PCR cloning of the mRNA sequence and comparison with genomic sequences. Signal peptides were predicted by SignalP (http://www.cbs.dtu.dk/services/SignalP/32).
To search for homologous sequences in other nematodes, BLAST searches were performed on the NCBI EST and genomic sequence databases by using afp-11 or flp-1 as subjects. Sequences were aligned by CLUSTAL-W in MEGA.54
Muscle Tension Measurements
Dissections were made by slitting the worm longitudinally along the lateral lines and isolating a 1.5 cm dorsal strip approximately 1 cm posterior to the head.55 These preparations contained dorsal muscle cells (all longitudinal) and the motor axons of the dorsal nerve cord. Throughout dissection and tension measurements, preparations were maintained in Ascaris saline (described above) at 37 °C. For tension measurements, each end of the muscle strip was ligatured with silk thread; one end was tied to a fixed hook in a 10 mL chamber, and the other was attached to an isometric force transducer. The output from the transducer was fed into a computer for recording and analysis of tension/time relationships using DataTrax software.
The effects of the peptides on acetylcholine (ACh)-induced contraction were measured in two separate procedures. In the first, muscle responses were monitored after a single exposure to peptide, paying attention to the time course and extent of the recovery of tension. In the second procedure, the muscle strip tension was followed during continuous exposure to peptide. Peptides were added by micropipet to give a final concentration of 10 μM. An initial response to 5 μM ACh for each worm was compared to ACh contractions at 10 min intervals after the addition of the peptide. For recovery experiments, each peptide solution was washed out of the chamber 10 min after peptide addition. For continued exposure experiments, muscle strips were washed and then continuously exposed to peptide solution at the same concentration. Solutions were mixed by bubbling with nitrogen gas.
Acknowledgments
We are grateful to Bill Feeny for help with the figures, and Philippa Claude for critically reviewing the manuscript.
Glossary
Abbreviations
- ACh
acetylcholine
- AF
Ascaris FMRFamide-like
- afp
Ascaris FMRFamide-like precursor protein transcript
- BSA
bovine serum albumin
- DG
dorsal ganglion
- dot-ELISA
dot-enzyme linked immunosorbent assay
- EST
expressed sequence tag
- flp
FMRFamide-like peptide gene
- GSS
genomic survey sequence
- ICC
immunocytochemistry
- ISH
in situ hybridization
- MALDI-TOF
matrix assisted laser desorption/ionization - time-of-flight
- MS
mass spectrometry
- m/z
mass-to-charge ratio
- nlp
neuropeptide-like protein gene
- NR
nerve ring
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PEG
polyethylene glycol
- PPG
polypropylene glycol
- RACE
rapid amplification of cDNA ends
- RVG
retrovesicular ganglion
- VC
ventral nerve cord
- VG
ventral ganglion
Supporting Information Available
Amino acid sequence of known AF peptides, peak lists of peaks present in AVK, MS/MS of previously identified peptides, and of synthetic peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
⊥ J.L.J.: Department of Biological Sciences, San Jose State University, San Jose CA, 95192.
Author Contributions
J.L.J. was responsible for MS, bioinformatics, data analysis, and manuscript preparation. I.R.V. was responsible for antibody characterization, ICC, cloning, ISH, and manuscript preparation. K.M.A. was responsible for single cell dissections. A.H.M. was responsible for muscle strip assays. M.A.R. was responsible for antibody characterization, ICC, and ISH. M.M.V. was responsible for MS. A.O.S. was responsible for data analysis, bioinformatics, and manuscript preparation.
This research was supported by the National Institutes of Health (Grants AI15429, T32 GM007507, and NCRR/SIG S10RR024601) and by a John Bascom Professorship, University of Wisconsin—Madison, to A.O.S.
The authors declare no competing financial interest.
Author Contributions
∥ These authors contributed equally to this work.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Stretton A.; Donmoyer J.; Davis R.; Meade J.; Cowden C.; Sithigorngul P. (1992) Motor behavior and motor nervous system function in the nematode Ascaris suum. J. Parasitol. 78, 206–214. [PubMed] [Google Scholar]
- White J.; Southgate E.; Thomson N.; Brenner S. (1986) The structure of the nervous system of the nematode C. elegans. Philos. Trans. R. Soc. London, Ser. B 314, 1–340. [DOI] [PubMed] [Google Scholar]
- Vanfleteren J. R.; Van de Peer Y.; Blaxter M. L.; Tweedie S. A.; Trotman C.; Lu L.; Van Hauwaert M. L.; Moens L. (1994) Molecular genealogy of some nematode taxa as based on cytochrome c and globin amino acid sequences. Mol. Phylogenet. Evol. 3, 92–101. [DOI] [PubMed] [Google Scholar]
- Davis R. E.; Stretton A. O. (1996) The motornervous system of Ascaris: electrophysiology and anatomy of the neurons and their control by neuromodulators. Parasitology 113(Suppl), S97–117. [DOI] [PubMed] [Google Scholar]
- Yew J. Y.; Kutz K. K.; Dikler S.; Messinger L.; Li L.; Stretton A. O. (2005) Mass spectrometric map of neuropeptide expression in Ascaris suum. J. Comp. Neurol. 488, 396–413. [DOI] [PubMed] [Google Scholar]
- Jarecki J. L.; Frey B. L.; Smith L. M.; Stretton A. O. (2011) Discovery of Neuropeptides in the Nematode Ascaris suum by Database Mining and Tandem Mass Spectrometry. J. Proteome Res. 10, 3098–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Kim K. (2008) Neuropeptides. WormBook 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. E.; Stretton A. O. (2001) Structure–activity relationships of 18 endogenous neuropeptides on the motor nervous system of the nematode Ascaris suum. Peptides 22, 7–23. [DOI] [PubMed] [Google Scholar]
- Yew J. Y.; Davis R.; Dikler S.; Nanda J.; Reinders B.; Stretton A. O. (2007) Peptide products of the afp-6 gene of the nematode Ascaris suum have different biological actions. J. Comp. Neurol. 502, 872–882. [DOI] [PubMed] [Google Scholar]
- McVeigh P.; Leech S.; Mair G. R.; Marks N. J.; Geary T. G.; Maule A. G. (2005) Analysis of FMRFamide-like peptide (FLP) diversity in phylum Nematoda. Int. J. Parasitol. 35, 1043–1060. [DOI] [PubMed] [Google Scholar]
- McVeigh P.; Alexander-Bowman S.; Veal E.; Mousley A.; Marks N. J.; Maule A. G. (2008) Neuropeptide-like protein diversity in phylum Nematoda. Int. J. Parasitol. 38, 1493–1503. [DOI] [PubMed] [Google Scholar]
- Kimber M. J.; Fleming C. C.; Prior A.; Jones J. T.; Halton D. W.; Maule A. G. (2002) Localisation of Globodera pallida FMRFamide-related peptide encoding genes using in situ hybridisation. Int. J. Parasitol. 32, 1095–1105. [DOI] [PubMed] [Google Scholar]
- Kim K.; Li C. (2004) Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J. Comp. Neurol. 475, 540–550. [DOI] [PubMed] [Google Scholar]
- Jarecki J. L.; Andersen K.; Konop C. J.; Knickelbine J. J.; Vestling M. M.; Stretton A. O. (2010) Mapping neuropeptide expression by mass spectrometry in single dissected identified neurons from the dorsal ganglion of the nematode Ascaris suum. ACS Chem. Neurosci. 1, 505–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda J. C.; Stretton A. O. (2010) In situ hybridization of neuropeptide-encoding transcripts afp-1, afp-3, and afp-4 in neurons of the nematode Ascaris suum. J. Comp. Neurol. 518, 896–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stretton A. O.; Cowden C.; Sithigorngul P.; Davis R. E. (1991) Neuropeptides in the nematode Ascaris suum. Parasitology 102(Suppl), S107–116. [DOI] [PubMed] [Google Scholar]
- Cowden C.; Stretton A. O.; Davis R. E. (1989) AF1, a sequenced bioactive neuropeptide isolated from the nematode Ascaris suum. Neuron 2, 1465–1473. [DOI] [PubMed] [Google Scholar]
- Cowden C.; Stretton A. O. (1993) AF2, an Ascaris neuropeptide: isolation, sequence, and bioactivity. Peptides 14, 423–430. [DOI] [PubMed] [Google Scholar]
- Cowden C.; Stretton A. O. (1995) Eight novel FMRFamide-like neuropeptides isolated from the nematode Ascaris suum. Peptides 16, 491–500. [DOI] [PubMed] [Google Scholar]
- Rosoff M. L.; Burglin T. R.; Li C. (1992) Alternatively spliced transcripts of the flp-1 gene encode distinct FMRFamide-like peptides in Caenorhabditis elegans. J. Neurosci. 12, 2356–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary T. G.; Price D. A.; Bowman J. W.; Winterrowd C. A.; Mackenzie C. D.; Garrison R. D.; Williams J. F.; Friedman A. R. (1992) Two FMRFamide-like peptides from the free-living nematode Panagrellusredivivus. Peptides 13, 209–214. [DOI] [PubMed] [Google Scholar]
- Yew J. Y.; Dikler S.; Stretton A. O. (2003) De novo sequencing of novel neuropeptides directly from Ascaris suum tissue using matrix-assisted laser desorption/ionization time-of-flight/time-of-flight. Rapid Commun. Mass Spectrom. 17, 2693–2698. [DOI] [PubMed] [Google Scholar]
- Husson S. J.; Clynen E.; Baggerman G.; De Loof A.; Schoofs L. (2005) Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem. Biophys. Res. Commun. 335, 76–86. [DOI] [PubMed] [Google Scholar]
- Husson S. J.; Landuyt B.; Nys T.; Baggerman G.; Boonen K.; Clynen E.; Lindemans M.; Janssen T.; Schoofs L. (2009) Comparative peptidomics of Caenorhabditis elegans versus C. briggsae by LC-MALDI-TOF MS. Peptides 30, 449–457. [DOI] [PubMed] [Google Scholar]
- Reinitz C. A.; Pleva A. E.; Stretton A. O. (2011) Changes in cyclic nucleotides, locomotory behavior, and body length produced by novel endogenous neuropeptides in the parasitic nematode Ascaris suum. Mol. Biochem. Parasitol. 180, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sithigorngul P.; Stretton A. O.; Cowden C. (1991) A versatile dot-ELISA method with femtomole sensitivity for detecting small peptides. J. Immunol. Methods. 141, 23–32. [DOI] [PubMed] [Google Scholar]
- Johnson C. D., and Stretton A. O. W. (1980) Neural control of locomotion in Ascaris: anatomy, electrophysiology, and biochemistry. In Nematodes as Biological Models (Zuckerman B., Ed.), pp 159–195, Academic Press, New York. [Google Scholar]
- Maroney P. A.; Denker J. A.; Darzynkiewicz E.; Laneve R.; Nilsen T. W. (1995) Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA 1, 714–723. [PMC free article] [PubMed] [Google Scholar]
- Jex A. R.; Liu S.; Li B.; Young N. D.; Hall R. S.; Li Y.; Yang L.; Zeng N.; Xu X.; Xiong Z.; Chen F.; Wu X.; Zhang G.; Fang X.; Kang Y.; Anderson G. A.; Harris T. W.; Campbell B. E.; Vlaminck J.; Wang T.; Cantacessi C.; Schwarz E. M.; Ranganathan S.; Geldhof P.; Nejsum P.; Sternberg P. W.; Yang H.; Wang J.; Gasser R. B. (2011) Ascaris suum draft genome. Nature 479, 529–533. [DOI] [PubMed] [Google Scholar]
- Reinitz C. A.; Herfel H. G.; Messinger L. A.; Stretton A. O. (2000) Changes in locomotory behavior and cAMP produced in Ascaris suum by neuropeptides from Ascaris suum or Caenorhabditis elegans. Mol. Biochem. Parasitol. 111, 185–197. [DOI] [PubMed] [Google Scholar]
- Maule A. G.; Shaw C.; Bowman J. W.; Halton D. W.; Thompson D. P.; Thim L.; Kubiak T. M.; Martin R. A.; Geary T. G. (1995) Isolation and preliminary biological characterization of KPNFIRFamide, a novel FMRFamide-related peptide from the free-living nematode, Panagrellus redivivus. Peptides 16, 87–93. [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D.; Nielsen H.; von Heijne G.; Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Parkinson J.; Mitreva M.; Whitton C.; Thomson M.; Daub J.; Martin J.; Schmid R.; Hall N.; Barrell B.; Waterston R. H.; McCarter J. P.; Blaxter M. L. (2004) A transcriptomic analysis of the phylum Nematoda. Nat. Genet. 36, 1259–1267. [DOI] [PubMed] [Google Scholar]
- Sithigorngul P.; Jarecki J. L.; Stretton A. O. W. (2011) A specific antibody to neuropeptide AF1 (KNEFIRFamide) recognizes a small subset of neurons in Ascaris suum: differences from Caenorhabditis elegans. J. Comp. Neurol. 519, 1546–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Much J. W.; Slade D. J.; Klampert K.; Garriga G.; Wightman B. (2000) The fax-1 nuclear hormone receptor regulates axon pathfinding and neurotransmitter expression. Development 127, 703–712. [DOI] [PubMed] [Google Scholar]
- Nelson L. S.; Kim K.; Memmott J. E.; Li C. (1998) FMRFamide-related gene family in the nematode, Caenorhabditis elegans. Brain Res. 58, 103–111. [DOI] [PubMed] [Google Scholar]
- Altun-Gultekin Z.; Andachi Y.; Tsalik E. L.; Pilgrim D.; Kohara Y.; Hobert O. (2001) A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128, 1951–1969. [DOI] [PubMed] [Google Scholar]
- Etchberger J. F.; Lorch A.; Sleumer M. C.; Zapf R.; Jones S. J.; Marra M. A.; Holt R. A.; Moerman D. G.; Hobert O. (2007) The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21, 1653–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D.; Hon G. C.; Hawkins R. D.; Kheradpour P.; Stark A.; Harp L. F.; Ye Z.; Lee L. K.; Stuart R. K.; Ching C. W.; Ching K. A.; Antosiewicz-Bourget J. E.; Liu H.; Zhang X.; Green R. D.; Lobanenkov V. V.; Stewart R.; Thomson J. A.; Crawford G. E.; Kellis M.; Ren B. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Nelson L. S.; Kim K.; Nathoo A.; Hart A. C. (1999) Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann. N.Y. Acad. Sci. 897, 239–252. [DOI] [PubMed] [Google Scholar]
- Marder E. (1998) From biophysics to models of network function. Annu. Rev. Neurosci. 21, 25–45. [DOI] [PubMed] [Google Scholar]
- Franks C. J.; Holden-Dye L.; Williams R. G.; Pang F. Y.; Walker R. J. (1994) A nematode FMRFamide-like peptide, SDPNFLRFamide (PF1), relaxes the dorsal muscle strip preparation of Ascaris suum. Parasitology 108(Pt 2), 229–236. [DOI] [PubMed] [Google Scholar]
- Bowman J. W.; Friedman A. R.; Thompson D. P.; Maule A. G.; Alexander-Bowman S. J.; Geary T. G. (2002) Structure-activity relationships of an inhibitory nematode FMRFamide-related peptide, SDPNFLRFamide (PF1), on Ascaris suum muscle. Int J Parasitol 32, 1765–1771. [DOI] [PubMed] [Google Scholar]
- Holden-Dye L.; Brownlee D. J.; Walker R. J. (1997) The effects of the peptide KPNFIRFamide (PF4) on the somatic muscle cells of the parasitic nematode Ascaris suum. Br. J. Pharmacol. 120, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. W.; Winterrowd C. A.; Friedman A. R.; Thompson D. P.; Klein R. D.; Davis J. P.; Maule A. G.; Blair K. L.; Geary T. G. (1995) Nitric oxide mediates the inhibitory effects of SDPNFLRFamide, a nematode FMRFamide-related neuropeptide, in Ascaris suum. J. Neurophysiol. 74, 1880–1888. [DOI] [PubMed] [Google Scholar]
- Johnson C. D.; Stretton A. O. (1987) GABA-immunoreactivity in inhibitory motor neurons of the nematode Ascaris. J. Neurosci. 7, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sithigorngul P.; Stretton A. O.; Cowden C. (1990) Neuropeptide diversity in Ascaris: an immunocytochemical study. J. Comp. Neurol. 294, 362–376. [DOI] [PubMed] [Google Scholar]
- Guastella J.; Johnson C. D.; Stretton A. O. (1991) GABA-immunoreactive neurons in the nematode Ascaris. J. Comp. Neurol. 307, 584–597. [DOI] [PubMed] [Google Scholar]
- Berman E. S.; Fortson S. L.; Checchi K. D.; Wu L.; Felton J. S.; Wu K. J.; Kulp K. S. (2008) Preparation of single cells for imaging/profiling mass spectrometry. J. Am. Soc. Mass Spectrom. 19, 1230–1236. [DOI] [PubMed] [Google Scholar]
- Schwartz S. A.; Reyzer M. L.; Caprioli R. M. (2003) Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass. Spectrom. 38, 699–708. [DOI] [PubMed] [Google Scholar]
- Hilker B.; Clifford K. J.; Sauter A. D. Jr.; Sauter A. D. 3rd; Harmon J. P. (2009) The measurement of charge for induction-based fluidic MALDI dispense event and nanoliter volume verification in real time. J. Am. Soc. Mass Spectrom. 20, 1064–1067. [DOI] [PubMed] [Google Scholar]
- Tu T.; Sauter A. D. Jr.; Sauter A. D. 3rd; Gross M. L. (2008) Improving the signal intensity and sensitivity of MALDI mass spectrometry by using nanoliter spots deposited by induction-based fluidics. J. Am. Soc. Mass Spectrom. 19, 1086–1090. [DOI] [PubMed] [Google Scholar]
- Burge C.; Karlin S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Tamura K.; Peterson D.; Peterson N.; Stecher G.; Nei M.; Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash A. S.; Tucker J. F. (1967) The bioassay of gamma-aminobutyric acid using a muscle preparation from Ascaris lumbricoides. J. Pharm. Pharmacol. 19, 240–245. [DOI] [PubMed] [Google Scholar]
- Goldschmidt R. (1908) Das nervensystem von Ascaris. Z. Wiss. Zool. 90, 73–136. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.