Abstract
In human beings, Parkinson’s disease (PD) is associated with the oligomerization and amyloid formation of α-synuclein (α-Syn). The polyphenolic Asian food ingredient curcumin has proven to be effective against a wide range of human diseases including cancers and neurological disorders. While curcumin has been shown to significantly reduce cell toxicity of α-Syn aggregates, its mechanism of action remains unexplored. Here, using a series of biophysical techniques, we demonstrate that curcumin reduces toxicity by binding to preformed oligomers and fibrils and altering their hydrophobic surface exposure. Further, our fluorescence and two-dimensional nuclear magnetic resonance (2D-NMR) data indicate that curcumin does not bind to monomeric α-Syn but binds specifically to oligomeric intermediates. The degree of curcumin binding correlates with the extent of α-Syn oligomerization, suggesting that the ordered structure of protein is required for effective curcumin binding. The acceleration of aggregation by curcumin may decrease the population of toxic oligomeric intermediates of α-Syn. Collectively; our results suggest that curcumin and related polyphenolic compounds can be pursued as candidate drug targets for treatment of PD and other neurological diseases.
Keywords: Curcumin, α-synuclein, amyloid, oligomers, toxicity, Parkinson’s disease
The major pathological hallmark of Parkinson’s disease (PD) is the presence of insoluble, fibrous aggregates, composed of α-Syn in intraneuronal inclusions of Lewy bodies (LBs) and Lewy neuritis (LNs).1 The animal models that overexpress human α-Syn indicate the direct involvement of α-Syn aggregation in PD pathogenesis.2,3 Moreover, the discovery of three disease-related substitution mutations (A30P, A53T, and E46K) in the SNCA gene that encodes α-Syn and their effects on the aggregation kinetics in vitro further support the central role of α-Syn aggregation in PD pathogenesis.1,4,5 The monomeric α-Syn is a natively unfolded protein, which transforms into cross-β-sheet rich amyloid by self-assembly at physiological conditions via partially folded intermediates and soluble oligomers.6 Recently, evidence have emerged from both in vitro and in vivo studies that soluble, oligomeric forms of α-Syn have potent neurotoxic activities and may cause the neuronal injury and death in PD.7−11 The α-Syn mutants that preferentially formed oligomers when expressed in the rat brain showed more neurotoxicity and cell death compared to the mutants that mostly formed amyloid fibrils.9 Molecules that inhibit the toxicity of oligomers and/or fibrils either by reducing their formation or by converting their toxic state to nontoxic state would be an immediate step for the development of effective therapeutics against PD.12−15 Guided by this concept, many investigators have either searched for existing small molecules or synthesized inhibitors against α-Syn fibrillogenesis.15−23 Several small polyphenolic molecules such as epigallocatechin gallate and curcumin have been shown to modulate the assembly and/or toxicity of many amyloidogenic protein/peptides such as Aβ, α-Syn, and prion.19,23−26 It has been proposed that the antioxidative properties of these polyphenols along with their structural constraints might be responsible for their efficacy in amyloid inhibition.24,27
Curcumin (diferuloylmethane) (Supporting Information Figure S1) is a well-known polyphenolic in Asian food ingredient turmeric and has been shown to exhibit anti-inflammatory, antimicrobial, and anticarcinogenic activities.28 Due to its low cost, blood brain barrier crossing ability and its pharmacological safety as evident from preclinical studies have suggested the potential therapeutic role of curcumin in neurological disorders including Alzheimer’s, Parkinson’s, and Huntington’s disease.28−33 For example, curcumin binds to amyloid β protein (Aβ) oligomers/fibrils, alters the Aβ aggregation, and reduces the toxicity in AD.31,34
In PD, curcumin has been shown to inhibit the α-Syn aggregation in vitro35−37 and attenuate the α-Syn oligomer toxicity in cells.17,38 However, the reduction of toxicity by curcumin and its effect on the pathway of α-Syn aggregation in physiological conditions is not clearly understood. Our work focuses on studying the effect of curcumin on the morphology and toxicity of oligomeric and fibrillar assemblies of α-Syn. We suggest that curcumin preferentially binds to the preformed α-Syn aggregates, modulates the morphology, and reduces their cellular toxicity by minimizing their hydrophobic surface exposure. In addition, the data reveals that curcumin accelerates α-Syn aggregation in vitro and could reduce the population of soluble oligomers, which are cytotoxic. Thus, curcumin and related polyphenolic compounds could be used for the development of potential drugs against PD.
Results and Discussion
Curcumin Binds to α-Syn Oligomers and Reduces Their Toxicity
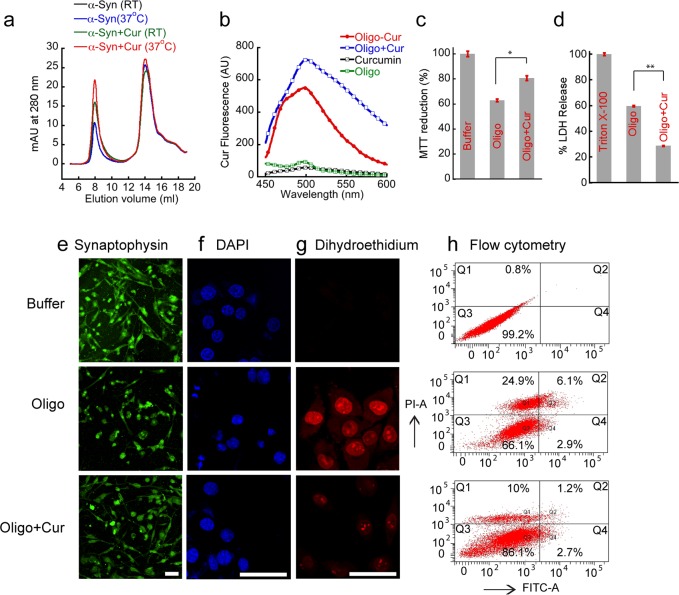
It has been suggested that α-Syn oligomers are much more toxic species compared to mature fibrils,7−9 and curcumin has been reported to attenuate the toxicity of the oligomers.39 Here, we studied the interaction and the effect of curcumin on preformed α-Syn oligomers using size exclusion chromatography (SEC) and fluorescence assay (Figure 1). When freshly solubilized protein in 20 mM MES buffer, pH 6.0, 0.01% sodium azide was injected in SEC, two major species were eluted; protofibrillar oligomers eluted close to void volume (∼8 mL) and monomers at ∼15 mL (Figure 1a). For simplicity, we use the term “oligomers” for protofibrillar oligomers isolated from SEC at ∼8 mL fraction in subsequent sections. Using this SEC profile, the binding of curcumin to oligomers can be studied. To do that, two different sets of protein preparation (5 mg/mL in 20 mM MES, pH 6.0) were incubated in dark with and without 100 μM curcumin: one at room temperature (RT) and other at 37 °C for 30 min. The solutions were then injected into the SEC column. The SEC profile of monomeric α-Syn remained unaltered in the presence and absence of curcumin. However, the oligomeric fraction in the presence of curcumin showed significantly higher absorbance at 280 nm in both RT and 37 °C incubations. The constant absorbance of monomeric fractions ruled out the possibility of more oligomerization of α-Syn in the presence of curcumin in this condition, which may be due to very short incubation time and mild conditions (without agitation) used for the incubation. However, these observations raised the possibility of interaction of curcumin to the preformed oligomeric protein, where curcumin might form an oligomer–curcumin complex, which possesses higher absorbance compared to oligomers alone. However, the probability of increased light scattering at 280 nm of this oligomeric assembly in the presence of curcumin cannot be ruled out. The higher absorbance of oligomers at 280 nm in the SEC profile of α-Syn incubated with curcumin at 37 °C suggests increased binding of curcumin to the preformed oligomers at this temperature (Figure 1a). As the curcumin was dissolved in DMSO, the effect of residual 0.05% DMSO on the size exclusion profile of α-Syn was also monitored and found insignificant. The size exclusion of 100 μM curcumin did not show any significant absorbance at 280 nm at ∼8 mL elution volume (data not shown).
Figure 1.
Curcumin binds α-Syn oligomers and reduces their toxicity. (a) SEC profiles of α-Syn incubated with and without curcumin at room temperature (RT) and 37 °C. (b) Curcumin fluorescence emission spectra of oligomer–Cur complex and preformed oligomers incubated with curcumin (Oligo+Cur). Only curcumin (Cur) and oligomers (Oligo) were used as control. (c) MTT reduction by SH-SY5Y neuronal cell in the presence of 5 μM preformed oligomers in presence and absence of 3 μM curcumin. 20 mM MES buffer, pH 6.0 was used as a control. (d) LDH release assay in SH-SY5Y neuronal cells using 5 μM preformed oligomers in presence and absence of 3 μM curcumin. 0.5% triton X-100 was used as positive control, and the buffer was used as negative control. Differentiated SH-SY5Y cells were treated with buffer, oligo and oligo+Cur for 40 h. (e) Oligo treated cells showed more neurite damage and reduced synaptophysin staining compared to oligo+Cur. Scale bars are 50 μm. (f) DAPI staining indicates that the nuclear morphology of oligo+ Cur treated samples were more intact compared to oligo treated cells. Scale bars are 50 μm. (g) The fluorescence intensity of the oxidized 2-hydroethidium showing significantly high intensity in oligomers treated cells compared to control and cells treated with oligomers+curcumin. Scale bars are 50 μm. (h) Flow cytometry analysis showing that reduction of oligomers toxicity in presence of curcumin. Quadrants Q1, Q2, Q3, and Q4 represent dead cells, late apoptotic cells/necrosis, live cells, and early apoptotic cells, respectively. Statistical significance: *P < 0.05, **P < 0.01.
To further evaluate the interactions of curcumin with α-Syn oligomers, we performed a curcumin fluorescence study. Curcumin is a weak fluorophore in water and produces low intensity fluorescence at ∼540 nm when excited ∼426 nm. However, if it binds to the hydrophobic surface of a protein such as amyloids, its fluorescence quantum yield increases with a blue shift in the fluorescence maximum (λmax).40,41 The curcumin fluorescence of the oligomers–curcumin complex isolated from SEC (Oligo-Cur) showed high fluorescence intensity with blue-shifted λmax ∼ 500 nm (Figure 1b). The fluorescence intensity ∼500 nm was ∼50-fold higher compared to fluorescence intensities of either oligomers or curcumin alone. Similarly, curcumin fluorescence was ∼70-fold more intense when preformed oligomers were isolated from SEC and incubated with curcumin for 30 min at 37 °C prior to fluorescence study (Oligo+Cur) (Figure 1b). These observations support the idea that curcumin strongly binds to the preformed oligomeric α-Syn.
The toxicity of the α-Syn oligomers was evaluated using dopaminergic neuronal cell line SH-SY5Y. To evaluate the toxicity of oligomers in presence and absence of curcumin, MTT assay42 was carried out (Figure 1c). In the presence of 5 μM preformed α-Syn oligomers isolated from SEC, MTT reduction was decreased to 60% (Figure 1c). However, when preformed oligomers were incubated in the presence of curcumin for 30 min before adding to the cells, MTT reduction was increased to 80%. The final curcumin concentration was 3 μM, which alone did not show any toxicity. For further confirmation of toxicity, LDH assay was performed. The LDH release assay is also widely used to determine the cytotoxicity of chemicals or environmental toxic factors.43 LDH is a soluble cytosolic enzyme that is released into the culture medium following the loss of membrane integrity.43 Figure 1d shows that, in the presence of oligomers, the LDH release was 60%, whereas in the presence of oligomers pretreated with curcumin LDH release was decreased to 29%. Both MTT and LDH assays therefore suggest that curcumin is able to reduce the toxicity of α-Syn oligomers.
Further, we have analyzed whether curcumin based neuronal cell protection involves changes in hallmarks of neuronal cell death markers like nuclear morphology and syanptophysin staining.44 The SH-SY5Y cells were differentiated using retinoic acid for 5 days (Supporting Information Figure S2a) and subsequently treated with α-Syn oligomers in the presence and absence of curcumin, and only buffer was used as a control. After 40 h of incubation, the attached cells in each culture dish were fixed and immunostained with synaptophysin (Figure 1e). Aβ(25–35) fibril was used as a positive control (Supporting Information Figure S2b). Similar experiments also were done for observing the nuclear morphology using DAPI staining (Figure 1f). The synaptophysin expression was significantly less in the cells treated with oligomers (Figure 1e). Moreover, more fragmented nuclei in cells treated with oligomers were observed (Figure 1f). Conversely, cells treated with buffer and oligomers+curcumin showed mostly normal synaptophysin staining and nuclear morphology (Figure 1e and f, respectively). This observation clearly indicates that curcumin interacts and in turn detoxifies the α-Syn oligomers. Since curcumin is well-known as an antioxidant,29 we studied whether curcumin may reduce the generation of reactive oxygen species (ROS) in SH-SY5Y cells in the presence of oligomers. To do that, 5 days differentiated cells were treated with oligomers in presence and absence of curcumin and with buffer only for 40 h. After incubation, 2-hydroethidium was used to measure the level of superoxide anions generated in the cells. In the presence of superoxide anions, the 2-hydroethidium gets oxidized into 2-hydroxyethidium that binds to DNA and becomes highly fluorescent.45,46 By measuring the fluorescence intensity of 2-hydroxyethidium in cells using confocal microscopy, we analyzed ROS generation in cells. The fluorescence microscopy images (Figure 1g) show a high amount of superoxide generated in cells treated with α-Syn oligomers compared to control (buffer) and cells treated with oligomers+curcumin. The fluorescence intensity quantification suggests that more than 2-fold reduction in 2-hydroxyethidium fluorescence intensity occurred when cells were treated with oligomers+curcumin (Supporting Information Figure S2c). The data further supports that curcumin can act as an antioxidant and reduces the cytotoxicity by minimizing generation and/or scavenging of reactive oxygen species. Further, the toxicity of oligomers in the presence and absence of curcumin was studied by Annexin V and PI binding with undifferentiated cells and subsequently using flow cytometry analysis. It was suggested that Annexin V binds the phospholipid phosphatidylserine (PS) that is translocated from the inner to outer leaflet of plasma membrane during the early apoptosis event in cells (Annexin V+).47,48 However, staining with Annexin V-FITC along with a live/dead dye propidium iodide (PI) allows identification of cells undergoing late apoptosis and/or necrosis (Annexin V+, PI+). In contrast, completely viable cells would not bind either Annexin V or PI (Annexin V–, PI−) and completely dead cells would bind mostly with PI (Annexin V–, PI+).47,48 Our data (Figure 1h) suggest that 34% cells undergo apoptosis and death in oligomer treated cells whereas curcumin treated oligomers showed lesser amount of apoptosis and death (14%). The control of cells treated with buffer only showed only 1% cell death (Figure 1h and Supporting Information Table 1). The data clearly suggest that curcumin reduces the toxicity of oligomers.
Curcumin Alters Morphology of the α-Syn Oligomers
To evaluate the morphological changes of α-Syn oligomers in the presence of curcumin, atomic force microscopy (AFM) experiments were performed. AFM images of oligomers (Oligo) and oligomers–curcumin complex (Oligo-Cur) that were directly isolated from SEC showed numerous globular oligomers (Figure 2a and Supporting Information Figure S3). The oligomers in absence of curcumin mostly were of ∼2–5 nm in height. Whereas the Oligo-Cur complex showed larger sized particles of ∼7–10 nm in height and most often elongated protofibrils were also observed. When preformed oligomers were incubated in the presence of curcumin at 37 °C, large oligomers and elongated protofibrils were observed (Figure 2a and Supporting Information Figure S3, Oligo+Cur), where height of these particles were more than 12 nm. However, incubated oligomers in the absence of curcumin mostly showed round shaped particles with average height of 8 nm. The AFM results demonstrated significant differences in the morphology, distribution, and heights of oligomeric species in the presence and absence of curcumin. The average height of these oligomers increased remarkably in presence of curcumin indicates that curcumin interacts with oligomeric species and transforms them to higher order oligomers with altered morphology. It has been shown previously that, depending upon the extent of proteinase K digestion, the toxicity of oligomers varies.46 Since curcumin alters the morphology and reduces the toxicity of oligomers, we tested the effect of curcumin in proteinase K digestion profile of oligomers (Figure 2b). Our data suggest that the extent of proteinase K digestions were similar when oligomers were treated in presence and absence of curcumin for 30 min, indicating curcumin does not protect oligomers structure from digestion.
Figure 2.
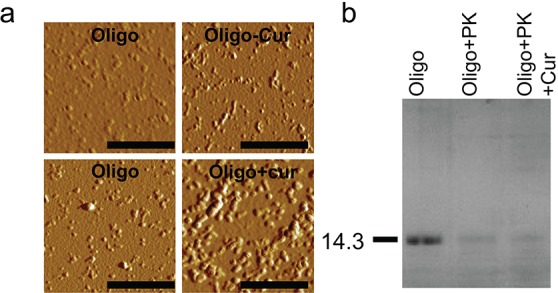
Curcumin modulates morphology of α-Syn oligomers. (a) AFM images of preformed oligomers isolated from SEC. Top left panel showing oligomers directly isolated from SEC. Top right panel showing oligomers morphology of α-Syn–curcumin complex isolated from SEC. Bottom panels showing 30 min incubated α-Syn oligomers in the presence (bottom right) and absence (bottom left) of curcumin. Scale bars are 500 nm. (b) Proteinase K digestion profile of oligomers in the presence and absence of curcumin showing similar extent of proteinase K digestion.
Curcumin Binds to α-Syn Fibrils and Reduces Its Toxicity
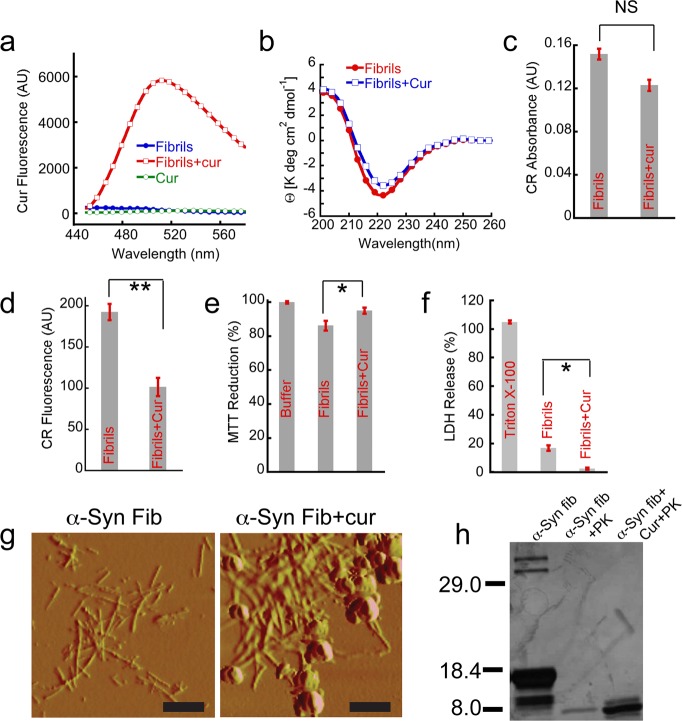
To examine the effect of curcumin on α-Syn fibrils, the preformed fibrils were centrifuged and redissolved in 20 mM MES buffer, pH 6.0, 0.01% sodium azide. The curcumin solution was added to the α-Syn fibrils so that the concentration of both α-Syn and curcumin was 100 μM. These samples were incubated for 20 h at RT and curcumin fluorescence was recorded (Figure 3). Significantly higher fluorescence intensity with prominent blue shift of λmax was observed when α-Syn fibrils were incubated with curcumin (Figure 3a). The results suggest the binding of curcumin to α-Syn fibrils. To study the effect of curcumin on the secondary structure of α-Syn fibrils, circular dichroism (CD) study was performed. The CD spectra of fibrils showed mostly β-sheet structure (with minima at ∼218 nm), and the β-sheet content was little decrease in presence of curcumin (Figure 3b). Congo Red (CR) binding assay49 was used to study the effect of curcumin on preformed amyloid fibrils. Thioflavin T (ThT) is not suitable for this purpose as curcumin decreases the fluorescence quantum yield of ThT.40 CR binding was measured by an increase of the dye’s molar absorptivity at 540 nm. In presence of curcumin, α-Syn amyloid showed slightly lesser binding to CR (Figure 3c). To further confirm the results, CR fluorescence was also performed. Similar to CR absorbance study, if CR binds to amyloid, it gives higher fluorescence at 595 nm when excited at 550 nm.50 The data showed that, in the presence of curcumin, CR fluorescence of fibrils decreased (Figure 3d), suggesting that curcumin may decrease the amyloid content of preformed fibrils. The apparent conflicting data of CD and CR binding suggest that, in the presence of curcumin, the β-sheet content of the amyloids might remain intact while altering their internal structure with lesser binding affinity to CR.
Figure 3.
Curcumin binds to α-Syn fibrils and reduces their toxicity. 100 μM preformed α-Syn fibrils were incubated with and without 100 μM curcumin for 20 h, and these samples were used for all the assays. (a) Significant increase in curcumin fluorescence at ∼500 nm was observed after binding to fibrils when excited at 426 nm. (b) CD spectra of preformed α-Syn fibrils without (red) and with curcumin (blue). Both the spectra showed mostly β-sheet conformation without any significant change. (c) CR absorbance at 540 nm after binding to α-Syn fibrils in the presence and absence of curcumin. (d) CR fluorescence at 595 nm showing decreased in CR binding when preformed α-Syn fibrils were incubated in the presence of curcumin. (e) MTT reduction assay using SH-SY5Y cell line by preformed α-Syn fibrils in the presence and absence of curcumin. (f) LDH release assay using SH-SY5Y cells showing less LDH release by fibrils incubated in the presence of curcumin. 0.5% Triton X-100 used as a positive control and showed 100% cell death. (g) AFM morphology of α-Syn fibrils in the presence and absence of curcumin. Left panel shows the distinct fibrillar morphology of α-Syn amyloids, whereas the right panel shows clustered and clumped fibrillar aggregates along with some amorphous species when α-Syn fibrils were incubated with curcumin. Scale bars are 500 nm. (h) Altered proteinase K digestion profile evident from SDS-PAGE analysis showing α-Syn fibrils in the presence of curcumin is more resistant to proteolytic degradation. Statistical significance *P < 0.05; **P < 0.01; NS P > 0.05.
Although recent studies suggest that α-Syn oligomers are potential neurotoxic species, primarily responsible for PD pathogenesis,7−9 earlier studies showed that matured fibrils are also cytotoxic.51−53 To study the toxicity of amyloid in the presence and absence of curcumin, MTT reduction and LDH release assays were performed using the neuronal cell line SH-SY5Y. The incubation of 5 μM preformed fibrils to the cells for 40 h decreased the MTT reduction by ∼15% (Figure 3e), whereas in fibrils in presence of 3 μM curcumin the MTT reduction decreased ∼6%. A concentration of 3 μM curcumin alone did not show any toxicity to the cells. To further confirm the toxicity of amyloid in the presence and absence of curcumin, LDH assay was performed. Triton X-100 (0.5%) was used as a positive control and considered to release ∼100% of LDH (Figure 3f). The exposure of 5 μM fibrils released only ∼18% of LDH. However, fibrils preincubated with curcumin releases only ∼2% LDH (Figure 3f). The data suggest that toxicity of fibrils could be decreased by curcumin. Our oligomers (Figure 1) and fibrils toxicity (Figure 3) data further suggest that fibrils possess lesser toxicity compared to oligomers, which is consistent with the hypothesis that oligomers are potential neurotoxin in PD pathogenesis.8,9
Curcumin Modifies the α-Syn Fibrils Morphology without Disintegrating Them to Monomers
To study the effect of curcumin on the morphology of α-Syn fibrils, preformed fibrils (100 μM) were incubated in the presence and absence of 100 μM curcumin for 20 h at RT. After incubation, the morphology of the samples was studied by AFM. The AFM data showed numerous fibrillar structures (Figure 3g and Supporting Information Figure S4a) in α-Syn aggregates. However, AFM analysis of incubated samples in the presence of curcumin showed significant population of fibrils along with clumped and amorphous aggregates (Figure 3g and Supporting Information Figure S4b). The data suggest that curcumin may convert the fraction of amyloids into other kind of aggregates. To analyze whether curcumin can dissociate the preformed amyloids into monomeric protein, 100 μM of α-Syn fibrils was incubated in presence and absence of 100, 200, and 300 μM curcumin at 37 °C for 20 h. After incubation, samples were centrifuged at 18 000g for 40 min. The resulting supernatant was collected and analyzed by SDS-PAGE. The SDS-PAGE analysis suggested that, in the absence and presence of varying concentrations of curcumin, similar amounts of soluble fractions were observed (Supporting Information Figure S4c). This analysis suggests that curcumin did not significantly dissociate the preformed α-Syn fibrils into soluble protein up to 20 h of incubation. To further investigate whether curcumin modified the α-Syn fibrils, proteinase K digestion experiments were performed. The undigested α-Syn fibrils showed four different bands in SDS-PAGE with a major band at ∼14 kDa for monomer (Figure 3h). In addition to monomer and core, SDS stable dimers and trimers were also observed, which are consistent with previous reports.54−57 However, after 1 h of proteinase K digestion, only one faint band was observed at ∼8 kDa, representing protease resistant core of fibrils.58 The 1 h of proteinase K digestion of fibrils pretreated with curcumin showed a more intense band at ∼8 kDa and another faint band above ∼8 kDa, suggesting that curcumin may inhibit the protease digestion and/or protect the core of α-Syn fibrils.
Reduction of Solvent Exposed Hydrophobic Surface of Oligomers and Fibrillar α-Syn by Curcumin
It has been suggested that the extent of hydrophobic surface exposure may play a significant role in cellular toxicity of protein aggregates.59,60 We hypothesize that, along with structural and morphological changes, curcumin binding to the preformed α-Syn aggregates may also alter their exposed hydrophobic surfaces and in turn reduce their toxicity. ANS is routinely used as fluorescent dye to detect the solvent-exposed hydrophobic surfaces of the protein.59 However, the ANS and bis-ANS fluorescence quantum yield decreases in presence of curcumin as emission of ANS/bis-ANS falls in the range of excitation of curcumin and energy transfer from ANS/bis-ANS to curcumin may occur (Supporting Information Figure S5). Therefore, we suggest that ANS/bis-ANS is not a suitable fluorescent dye to detect exposed hydrophobic surfaces in presence of curcumin. We used Nile Red (NR), another sensitive dye for this purpose, which shows several fold increase in its fluorescence quantum yield with blue shift of λmax upon binding to the hydrophobic exposed surfaces of the protein.61,62 Moreover, the presence of curcumin did not affect the NR fluorescence significantly (Figure 4). For the analysis of exposed hydrophobic surfaces of the oligomers, we isolated the oligomers in 20 mM MES buffer, pH 6.0, 0.01% sodium azide using SEC. The oligomers were incubated in the presence and absence of curcumin for 30 min at 37 °C, and NR fluorescence was performed. The data showed a significantly high NR fluorescence intensity and blue shift of λmax after binding to α-Syn oligomers (Figure 4a) compared to control. However, NR fluorescence decreased to almost half when similar experiments were performed in presence of curcumin (Oligo+Cur). Control experiments were done only with curcumin, NR and NR+Cur, which did not show any significant fluorescence (Figure 4a). Similarly, NR binding study of Oligo-Cur complex isolated from SEC in 20 mM MES buffer, pH 6, 0.01% sodium azide showed a marked reduction in NR fluorescence compared to oligomers alone (Figure 4b). These results indicate that curcumin reduces the solvent exposed hydrophobic surface of α-Syn oligomers.
Figure 4.
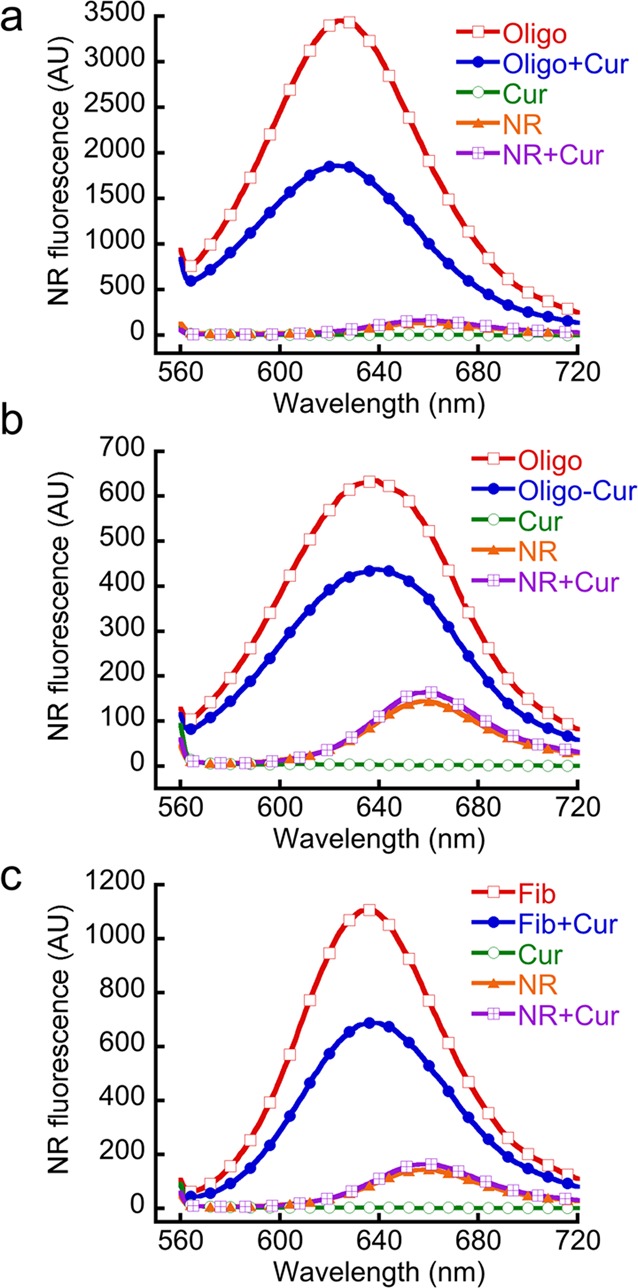
Curcumin reduces the exposed hydrophobic surfaces of oligomeric and fibrilar α-Syn. NR binding to oligomers and fibrils in presence and absence of curcumin was measured by fluorescence. (a) NR binding of α-Syn oligomers obtained from SEC and incubated in presence (Oligo+Cur) and absence (Oligo) of curcumin. The NR fluorescence of curcumin (Cur), NR, and NR+Cur was taken as control. (b) NR binding of Oligomer–curcumin complex (Oligo-Cur) isolated from SEC. Only NR was used as a control. (c) NR binding of α-Syn fibrils in presence and absence curcumin. Curcumin and NR alone showed insignificant fluorescence. All spectra were measured by exciting the solution at 550 nm and emission in the range of 560–720 nm.
To determine the effect of curcumin on hydrophobic surface exposure of α-Syn fibrils, fibril solution in 20 mM MES buffer, pH 6.0, 0.01% sodium azide was mixed with curcumin so that the final concentration ratio of fibrils and curcumin was 1:1 and the mixture was incubated at RT for 20 h. The incubated α-Syn fibril in buffer was used as a control. High NR binding to fibrils was observed (Figure 4c), whereas curcumin treated fibrils showed significantly less NR binding. The data suggests that curcumin may modify the fibril structure by reducing its exposed hydrophobic surface.
Curcumin Binding Correlates with the Extent of α-Syn Oligomerization
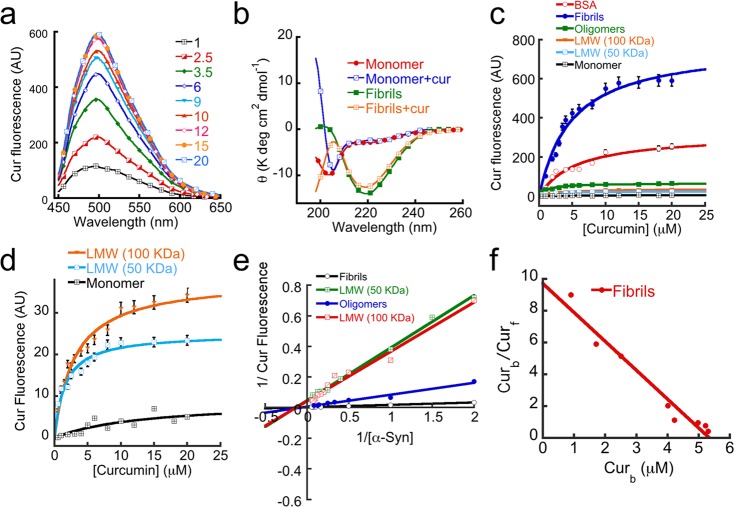
To evaluate the relative binding affinity of curcumin to the different α-Syn species (LMW 50 kDa, LMW 100 kDa, oligomers and fibrils), curcumin fluorescence studies were performed with each species (Figure 5). To do that 5 μM concentration of different protein species were prepared in 20 mM MES buffer, pH 6.0, 0.01% sodium azide and curcumin fluorescence was recorded in presence of varying concentration of curcumin (1–20 μM). A concentration of 5 μM bovine serum albumin (BSA) was used as a positive control. Curcumin fluorescence was done with excitation at 426 nm and emission in the range of 450–650 nm. The curcumin fluorescence plots for each species, monomers, LMW assembly (50 and 100 kDa), oligomers and fibrils, were obtained (Supporting Information Figure S6a). Figure 5a shows the fluorescence intensity of varying concentrations of curcumin in presence of 5 μM α-Syn fibrils. The fluorescence intensity increased with increasing concentration of curcumin. The curcumin fluorescence intensity was highest at 12 μM curcumin and remained almost same up to 20 μM. When fluorescence intensity at λmax was plotted against different curcumin concentrations for individual protein species, curcumin saturation plots were obtained (Figure 5c and d). To reveal whether curcumin binding to different α-Syn species alters the secondary structure, CD was performed with 5 μM monomeric and fibrillar α-Syn in presence and absence of 20 μM curcumin (Figure 5b). The CD study did not show any significant secondary structural change in the presence of curcumin for both monomeric and fibrillar form of α-Syn. The monomers remained unstructured and the fibrils showed characteristic β-sheet conformation (Figure 5b). The curcumin fluorescence saturation plots of all α-Syn species (Figure 5c) indicate that the increase in fluorescence intensity were according to the oligomer order; fibrils > oligomers > LMW-100 kDa > LMW 50 kDa > monomers (Figure 5c, d and Supporting Information Figure S6b). The curcumin fluorescence with BSA was greater than the oligomers but less than the fibrils. In another experiment, curcumin fluorescence was performed using 5 μM curcumin in the presence of varying concentrations (1–20 μM) of each α-Syn species and the double reciprocal plot (Figure 5e) was derived. Further, from the fluorescence saturation curve and double reciprocal plots (Figure 5c and e), Scatchard plots were obtained (Figure 5f and Supporting Information Figure S7) for each species. Using Scatchard plots, curcumin dissociation constants for each species were calculated using established method.41,63 The order of dissociation constants at RT, in 20 mM MES buffer, pH 6.0 were of fibrils (∼0.5 μM) < oligomers (∼2 μM) < LMW 100 kDa (∼5 μM) < LMW 50 kDa (6.5 μM). In contrast, only small fluctuations in curcumin fluorescence were observed in the presence of monomeric α-Syn even in 20 μM curcumin concentration. This study clearly indicates that curcumin prefers to bind higher order oligomers and fibrils not monomer.
Figure 5.
Relative binding of curcumin to different α-Syn species. (a) Curcumin fluorescence spectra of varying concentrations of curcumin (1–20 μM) in the presence of 5 μM preformed α-Syn fibrils. (b) CD spectra of 5 μM α-Syn monomers and fibrils in the presence and absence of 20 μM curcumin. (c) Curcumin fluorescence value at λmax (500 nm) in the presence of different α-Syn species with varying concentrations of curcumin showing maximum curcumin binding for fibrils. (d) Comparative increase in curcumin fluorescence at 500 nm in the presence of 5 μM each of the LMW 50 kDa, LMW 100 kDa and monomer showing increase in curcumin binding according to the oligomer order. (e) Double-reciprocal plots of various α-Syn species. (f) Scatchard plot of the α-Syn fibrils–curcumin complex. Curb and Curf indicate bound and free curcumin, respectively.
Curcumin Does Not Interact with Monomer but Shows a Weak Interaction with 100 kDa LMW α-Syn
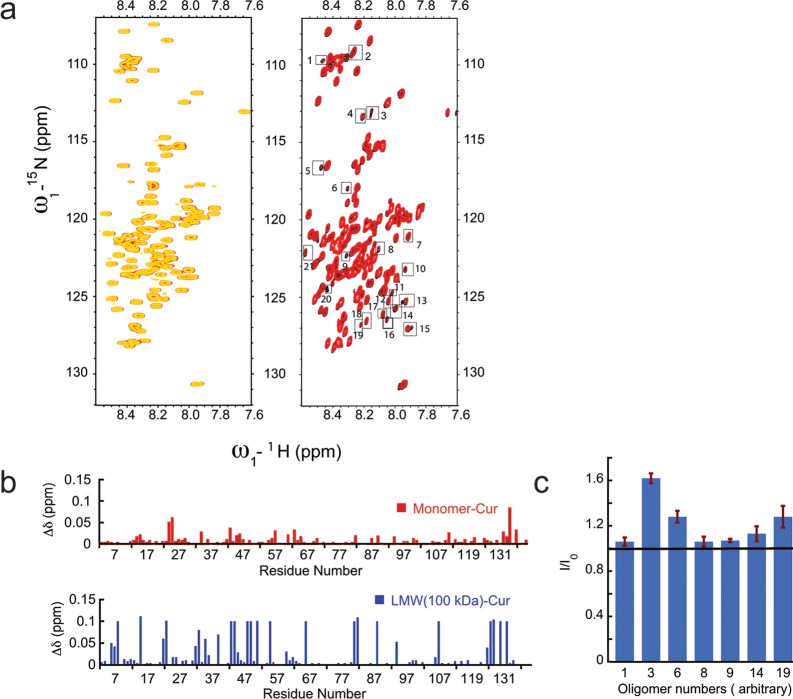
To study the interaction between curcumin and oligomeric species of α-Syn in a residue specific manner, we used (1H–15N) heteronuclear single quantum coherence (HSQC) NMR spectroscopy (Figure 6). The 15N labeled monomeric α-Syn was isolated from SEC, and 150 μM samples in the presence and absence of 75 μM curcumin were used for acquiring the NMR spectra at 25 °C. Figure 6a shows well-resolved HSQC spectra of monomeric α-Syn with sharp peaks and relatively less dispersion in proton chemical shifts at both conditions. The data indicate the unstructured state of the protein. We used published 1H and 15N chemical shift values44,64 for assigning those spectra. The spectra of α-Syn in the presence (Figure 6a, left panel, yellow spectrum) and absence (Figure 6a, left panel, red spectrum) of curcumin were overlaid. Spectra of α-Syn in the presence of curcumin did not show any significant chemical shift perturbation and retained the same resolution as of α-Syn in solution. We calculated the change in chemical shifts and did not observe any significant perturbation in the presence and absence of curcumin (Figure 6b, top panel). Further, the intensity ratios (I/I0) for all nonoverlapping peaks were calculated and values were found to be ∼1.0 (data not shown). The data indicate that curcumin does not interact with the monomeric α-Syn, which lacks any ordered structure.
Figure 6.
Probing interaction of curcumin with monomer and LMW 100 kDa α-Syn by NMR spectroscopy. (a) Overlay of 1H–15N HSQC spectra of 150 μM monomeric α-Syn in the absence (red spectrum) and in the presence of 75 μM curcumin (yellow spectrum, top left). Top right panel shows the overlay of the 1H–15N HSQC spectra of 150 μM 100 kDa LMW α-Syn in the absence (black spectrum) and in the presence of 75 μM curcumin (red spectrum, top right). In the HSQC spectrum of 100 kDa LMW, many additional peaks were observed compared to the HSQC spectrum of monomers and were numbered arbitrarily. (b) Difference in chemical shifts (Δδ) of individual amino acids in monomer (red) and 100 kDa LMW (blue) of α-Syn in the presence and absence of curcumin were calculated. The Δδ (in ppm) was plotted against individual amino acid residues. (c) Intensities of all extra peaks in HSQC spectrum of oligomers in presence (I) and absence (I0) of curcumin were calculated, and relative intensities I/I0 were plotted against oligomer numbers (assigned arbitrary).
NMR experiments were also performed with 15N-labeled LMW 100 kDa α-Syn in the presence and absence of curcumin, and both spectra were overlaid (Figure 6a, right panel). NMR peaks in the 100 kDa LMW spectra both in the presence (Figure 6a, right panel, black spectrum) and absence (Figure 6a, right panel, red spectrum) of curcumin appeared to be broader than those of the monomeric protein and number of peaks in the regions of 8.3–8.5 and 120–125 ppm overlapped with each other. Although most of the peaks had similar resonance frequencies in comparison to the monomer spectrum, ∼21 additional peaks were identified in the spectrum of LMW (arbitrary numbers were given to assign these additional peaks). These peaks were of comparatively low intensity and did not have any corresponding peak in the monomer spectrum, suggesting that such resonances might be arising from oligomer population. We calculated the changes in the resonance frequencies of LMW peaks in the presence and absence of curcumin (Figure 6b, lower panel), which did not show any definite pattern. However, chemical shift changes were relatively larger compared to the monomer. The data suggest that curcumin may transiently interact with LMW α-Syn. To gain further insight, we performed intensity ratio analysis for nonoverlapping peaks (Figure 6c and Supporting Information Figure S8). No clear pattern emerged, suggesting that oligomers do not have preferential binding sites for curcumin. Interestingly, we noticed an increase in intensities for additional peaks in the presence of curcumin shown in Figure 6c and Supporting Information Figure S8, suggesting a possible role of curcumin in stabilizing higher order species of α-Syn.
Curcumin Accelerates α-Syn Aggregation to Produce Morphologically Different Amyloid Fibrils in Vitro
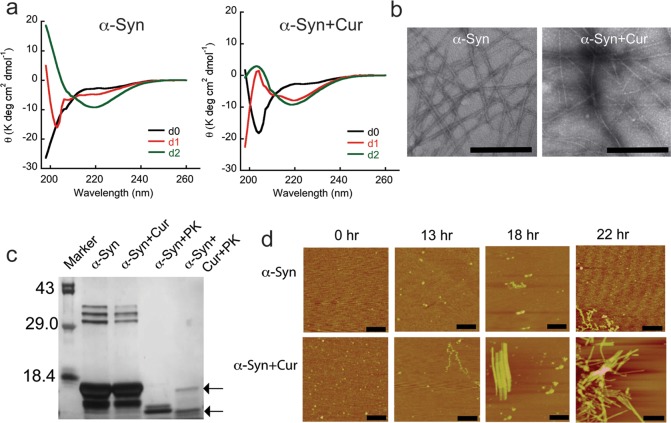
α-Syn is a natively unstructured protein, which gets converted to β-sheet-rich fibrils during incubation in vitro.6 To explore the effect of curcumin on in vitro aggregation, 150 μM 100 kDa LMW α-Syn in 20 mM MES buffer, pH 6.0, 0.01% sodium azide was incubated in the presence and absence of 75 μM curcumin at 37 °C with slight rotation. It is important to note that 100 kDa LMW α-Syn was used for this study as our NMR data suggests that curcumin may weakly interact with this species but not monomers. The curcumin was diluted from DMSO stock solution such that the final DMSO concentration was 0.075% (v/v) in the solution. For control, 150 μM LMW α-Syn in the presence of 0.075% DMSO was also incubated. On each day of incubation, CD was performed to determine the conformational transition of α-Syn in the presence and absence of curcumin. When β-sheets were formed as measured by CD, EM was used for morphological study (Figure 7). Immediately after addition of curcumin, no significant changes in CD spectra of α-Syn were observed. Both in the presence and absence of curcumin, α-Syn showed mostly unstructured conformation on day 0 (Figure 7a). However, CD study has showed that α-Syn was converted mostly to β-sheet conformation at day 1 in the presence of curcumin. On the other hand, α-Syn in the absence of curcumin was mostly unstructured until day 1 and showed β-sheet structure only on day 2. The data suggests that curcumin accelerates the conformational transition from random coil (RC) to β-sheet during amyloid formation (Figure 7a). The EM study after 3 days of incubation suggested that α-Syn alone formed highly ordered amyloid fibrils composed of ∼2–3 individual filaments (Figure 7b). These fibrils were helically twisted to each other with varying degrees of lateral association. The diameter of these fibrils was ∼10–30 nm. In contrast, α-Syn in the presence of curcumin showed narrow fibrils of ∼10–16 nm in diameter with little discernible substructure. To evaluate whether fibrils formed in the absence and presence of curcumin possess different stability/internal structure, a proteinase K digestion experiment was performed (Figure 7c). The SDS-PAGE analysis showed five different bands from undigested fibrils formed either in the presence or absence of curcumin. The three bands of more than 29 kDa were observed possibly due to the presence of higher order SDS stable oligomers. The two bands below 18.4 kDa were observed due to monomers and fragments of α-Syn, which may be the protease stable α-Syn core. Further, the SDS-PAGE analysis of proteinase K digested α-Syn fibrils formed in the absence of curcumin showed significant digestion, indicated by only one band at ∼8 kDa, which may be the protease resistant core of α-Syn fibrils50 (Figure 7c). However, proteinase K digestion of fibrils formed in the presence of curcumin showed two bands below 18.4 kDa, in which one is monomeric and another could be the protease resistant core. The different proteinase K digestion profile suggests that fibrils formed in the presence and absence of curcumin possess different stability against proteases.
Figure 7.
Curcumin accelerates α-Syn aggregation. (a) Time dependent CD spectra of α-Syn in the presence and absence of curcumin. α-Syn in the presence of curcumin showing accelerated conversion of random coil to β-sheet conformation during aggregation. (b) Electron micrographs of the aggregates after 3 days of incubation showing fibrillar morphology of α-Syn in the absence (left) and presence (right) of curcumin. Altered morphology of α-Syn aggregates were seen in the presence of curcumin. Scale bars are 500 nm. (c) SDS-PAGE analysis showing different proteinase K digestion pattern by α-Syn fibrils formed in the presence and absence of curcumin. (d) Time dependent AFM analysis of α-Syn aggregation in the presence and absence of curcumin showing accelerated conversion of oligomers to fibrils in the presence of curcumin. Scale bars are 700 nm.
Further, to determine the detailed kinetics and confirm our observation that curcumin accelerates the α-Syn aggregation, we performed time dependent AFM study during aggregation (Figure 7d). The data suggests that, immediately after preparation, LMW α-Syn in the presence and absence of curcumin formed small globular oligomers and during incubation they gradually transformed to higher order structures. AFM imaging of 18 h incubated α-Syn in the presence of curcumin showed some fibrils along with globular oligomers, which was converted to higher order fibrils at 22 h. In contrast, α-Syn in the absence of curcumin only showed small protofilament-like structures at 22 h without formation of higher order fibrils. The data clearly suggests that curcumin accelerates the aggregation of α-Syn in vitro.
The Kd determination and NMR studies showed that curcumin binds to early oligomers and not to monomers. Moreover, we find that curcumin accelerates the structural conversion and amyloid formation of LMW 100 kDa α-Syn, suggesting that curcumin may effectively reduce the population of toxic oligomeric intermediates. In contrast, previous data suggested that curcumin binds to monomeric α-Syn35 and inhibits its aggregation and amyloid formation.35−37 However, these previous studies were performed in the presence of either 10% 2,2,2-trifluoroethanol (TFE) or 1 mM Fe3+. TFE and Fe3+ are known to modulate the protein aggregation65−67 and complex formation with curcumin,68 respectively. Moreover, previous studies have used ThT for monitoring the aggregation kinetics, which is not a suitable dye for monitoring amyloid aggregation in the presence of polyphenols such as curcumin.40 Interestingly, the modulation of curcumin mediated Aβ aggregation in AD has also been reported to be conflicting, as both acceleration and inhibition of Aβ aggregation were evident in the presence of curcumin.31,69,70 Cole and co-workers have suggested that curcumin inhibits Aβ aggregation and reduces toxicity.31 Further, it has been shown that curcumin reverses Alzheimer’s disease pathology and reduces the plaque load in a mouse model of AD.34 Whereas recent studies suggested that curcumin accelerates the aggregation of Aβ both in vitro and in vivo.70 Moreover, two previous reports also have indicated that curcumin might inhibit the conversion of Aβ monomers to oligomers but accelerates the conversion of preformed oligomers to fibrils with effective reduction of toxic oligomeric population.71,72 However, all these previous studies have suggested a common theme that irrespective of whether curcumin accelerates or delays the protein aggregation, it reduces the toxicity of protein aggregates in vitro and in vivo.
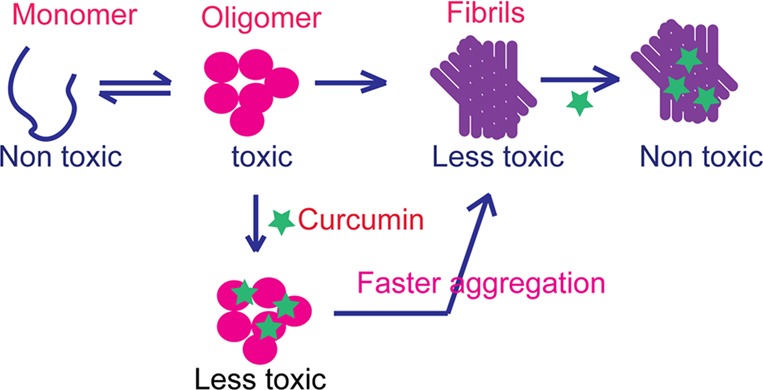
In summary, our present data showed that curcumin binds to preformed oligomers and fibrils and reduces their toxicity by modifying their morphology and hydrophobic surface exposure. Further, curcumin binds to early oligomers and accelerates their conversion to fibrils so that the effective population of toxic oligomeric intermediates could be reduced (Figure 8). This study suggests that curcumin or curcumin related compounds73 could be important small molecules that modulate and/or halt the toxicity of α-Syn aggregates in PD.
Figure 8.
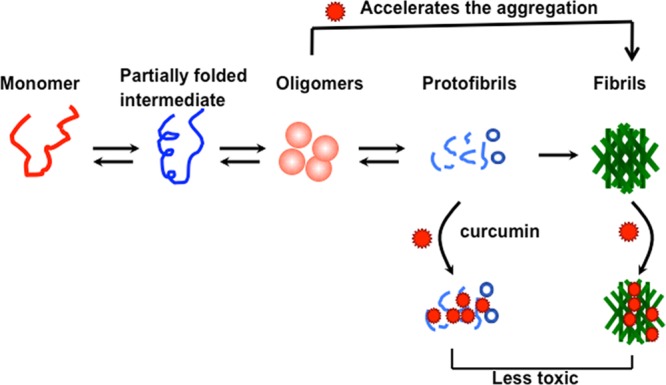
Schematic representation showing the effect of curcumin on α-Syn aggregation and toxicity. Curcumin may interact with various oligomeric intermediates of α-Syn and accelerate their conversion to fibrils. Curcumin also binds to preformed oligomers and fibrils and reduces their toxicity.
Methods
Chemicals and Reagent
Curcumin and other chemicals were purchased from Sigma (St. Louis, MO) except Nile Red dye, which was purchased from Invitrogen (Carlsbad, CA). Water was double distilled and deionized using a Milli-Q system (Millipore Corp., Bedford, MA). WT α-Syn plasmid (pRK172) was a kind gift of Prof. Roland Riek, ETH Zurich, Switzerland.
Preparation of Curcumin Stock Solution
Curcumin stock solution of 100 mM was made in DMSO. This stock solution was further diluted in 20 mM MES buffer, pH 6.0, 0.01% sodium azide to 57 mM. The working solution of curcumin was freshly prepared for each use by dilution of 57 mM curcumin in same buffer.
Protein Purification
α-Syn was expressed in Escherichia coli BL21 (DE3) strain according to the protocol described by Volles and Lansbury.74 For 15N labeled α-Syn expression, minimal media containing 15N-labeled ammonium chloride was used as a nitrogen source. Briefly, the IPTG induced bacterial cells were pelleted down by centrifugation. The pellet was resuspended in buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 150 mM NaCl) with protease inhibitor cocktail (Roche), sonicated followed by heating in boiling water bath for 20 min. Supernatant was collected after centrifugation (14 000g, 30 min). Streptomycin sulfate (10%; 136 μL/mL supernatant) and glacial acetic acid (228 μL/mL supernatant) were added to the supernatant followed by centrifugation (14 000g, 4 °C, 10 min). The resulting supernatant was precipitated by equal volume of saturated ammonium sulfate, prepared at 4 °C. Precipitated protein was washed with a solution of ammonium sulfate (saturated ammonium sulfate and water, 1:1 v/v at 4 °C). The washed pellet was resuspended in 100 mM ammonium acetate and stirred for 10 min. The α-Syn was precipitated, adding an equal volume of absolute ethanol. Ethanol precipitation was repeated twice at RT. Protein was again resuspended in 100 mM ammonium acetate, lyophilized, and stored at −20 °C for further use.
Preparation of Low Molecular Weight (LMW) α-Syn
For binding and fibrillization studies, LMW (50 kDa and 100 kDa) of α-Syn was prepared from lyophilized protein. Briefly, protein was dissolved in 20 mM MES buffer, pH 6.0, 0.01% sodium azide at a concentration of 15 mg/mL. α-Syn is acidic in nature, the pH of resulting solution was ∼5.0 and was sparingly soluble. To solubilize the protein, few μL of 2 M NaOH solution was added until protein was dissolved completely and clear solution was appeared. Then, the pH was adjusted to 6.0 by adding few μL of 2 M HCl solutions. The protein solution was dialyzed against the same buffer overnight at 4 °C using a 10 kDa MWCO mini-dialysis unit (Millipore). This process was carried out to remove salts and fragmented peptides. Further to remove any larger aggregates, the resulting solution was filtered by centricon YM-50 MWCO or YM-100 MWCO filter (Millipore) to obtain LMW preparation of 50 kDa and 100 kDa, respectively. The supernatant was collected and used for the further study.
Amyloid Fibril Formation
The assembly reaction was initiated with 100 kDa LMW α-Syn at a concentration of ∼400 μM in 1.5 mL eppendorf tube in 20 mM MES buffer, pH 6.0, 0.01% sodium azide. The eppendorf tubes containing protein solutions were placed into an EchoTherm model RT11 rotating mixture (Torrey Pines Scientific) with a speed corresponding to 50 r.p.m. inside a 37 °C incubator. The fibril formation was monitored by CD, ThT binding and confirmed by AFM/EM at the end of assembly reaction. For measuring α-Syn aggregation kinetics in presence of curcumin, 100 kDa LMW α-Syn and curcumin in 20 mM MES buffer, pH 6.0, 0.01% sodium azide were mixed such a way that the final concentrations of α-Syn and curcumin was 150 μM and 75 μM, respectively. The DMSO concentration was 0.075% (v/v). As a control, 150 μM of α-Syn was also incubated in presence of 0.075% of DMSO. Three independent experiments were performed for each sample.
Size Exclusion Chromatography (SEC)
Purified α-Syn was dissolved in 20 mM MES buffer, pH 6.0, 0.01% sodium azide as described above. The protein solution was then centrifuged for 30 min at 14 000g using a benchtop microcentrifuge (HITACHI, himac CT15RE, Japan). The resulting solution was clear and free of any larger aggregates. To study the curcumin interactions, protein solution was incubated with and without 100 μM curcumin at room temperature as well as at 37 °C for 30 min. Then 200 μL solutions were loaded on a S200-Superdex gel filtration column attached to an AKTA purifier (GE Healthcare) and eluted isocratically at 4 °C in the same buffer with a flow rate of 0.4 mL/min, and 200 μL fractions were collected.
Circular Dichroism Spectroscopy (CD)
Ten microliters of protein solution was diluted to 200 μL in 20 mM MES buffer, pH 6.0, 0.01% sodium azide. The solution was placed into a 0.1 cm path length quartz cell (Hellma, Forest Hills, NY). Spectra were acquired using a JASCO-810 instrument. All measurements were done at 25 °C. Spectra were recorded over the wavelength range of 198–260 nm. Three independent experiments were performed with each sample. Raw data were processed by smoothing and subtraction of buffer spectra, according to the manufacturer’s instructions.
Curcumin Fluorescence
The curcumin fluorescence spectra were acquired in the emission range of 450–650 nm by exciting the samples at 426 nm. The experiments were performed using a HITACHI spectrofluorometer (model F-2500) with excitation and emission slit widths of 2.5 and 5 nm, respectively. To study the interaction of curcumin with preformed α-Syn oligomers, the solid protein was dissolved in 20 mM MES buffer, pH 6.0, 0.01% sodium azide at a concentration of 5 mg/mL and incubated with 100 μM curcumin at 37 °C. After 30 min of incubation, SEC was performed as described earlier to obtain the preformed oligomers. The curcumin fluorescence was performed with this preformed oligomer–curcumin complex. The fluorescence of 100 μM curcumin in identical conditions was also measured as a control. In another set of experiments, the preformed α-Syn oligomers from ∼8 mL fractions were collected from SEC by injecting 5 mg/mL protein dissolved in 20 mM MES buffer, pH 6.0, 0.01% sodium azide. The preformed oligomers with and without 40 μM curcumin were incubated at 37 °C for 30 min and curcumin fluorescence recorded. Curcumin of 40 μM in identical conditions was also measured for control.
Congo Red Binding Assay
α-Syn preformed fibrils of 100 μM were incubated with and without 100 μM curcumin for 20 h at RT without agitations. A 20 μL aliquot of sample was then mixed with 180 μL of 20 mM MES buffer, pH 6.0, 0.01% sodium azide. Then 0.4 μL of 10 mM CR solution (filtered through 0.2 μM filter) in 20 mM MES buffer, pH 6.0, with 0.01% sodium azide was added such that the final protein concentration became 10 μM and CR 20 μM. After 10 min of incubation at RT, absorbance was measured in the range of 400–600 nm. For the measurement of the CR-only spectrum, 0.4 μL of CR was mixed with 200 μL of 20 mM MES buffer, pH 6.0, with 0.01% sodium azide and absorbance was measured as described. As a control, a 20 μL aliquot of fibrils with 180.4 μL of MES buffer was also measured. Three independent experiments were performed for each sample. For Congo Red fluorescence studies, the samples were prepared in similar way as described for Congo Red absorbance. The Congo Red fluorescence spectra were acquired on a HITACHI fluorescence spectrophotometer (model F-2500). The samples were excited at a wavelength of 550 nm, and emission was recorded in the wavelength range of 560–650 nm. The excitation and emission slit widths were 2.5 and 5.0 nm, respectively. The fluorescence spectrum of 20 μM Congo Red in 20 mM MES buffer, pH 6.0, 0.01% sodium azide was also recorded as a control.
Determination of Kd by Curcumin Fluorescence
Dissociation constants of curcumin to different species of α-Syn were determined based on curcumin fluorescence. For this study, different species of α-Syn were prepared as described earlier. For determining curcumin binding to different species of α-Syn, 5 μM α-Syn species were incubated in the presence and absence of varying concentrations of curcumin (1–20 μM) for 30 min at RT in the dark. BSA was used as a control for this study. Immediate after incubation curcumin fluorescence was measured with excitation at 426 nm and emission in the range 450–650 nm. Experiments were performed using Shimadzu RF-5301PC spectrofluorometer with excitation and emission slit widths of 5 and 5 nm, respectively. The curcumin fluorescence saturation curves were obtained for each α-Syn species, and BSA by plotting curcumin fluorescence value at 500 nm against different concentration of curcumin.
In order to obtain a double reciprocal plot, 5 μM curcumin in 20 mM MES buffer, pH 6.0, 0.01% sodium azide was incubated with varying concentrations (0–20 μM) of each α-Syn species. The mixture was incubated at RT for 30 min in the dark. After incubation, curcumin fluorescence was performed. Double reciprocal plots were obtained for each α-Syn species by plotting 1/fluorescence intensity in the Y-axis against 1/protein concentrations in the X-axis. From the intercept of the double reciprocal plot, maximum fluorescence intensity was obtained when all curcumin was bound to protein. Using saturation and double reciprocal plots, concentrations of bound as well as free curcumin to each protein species were calculated. Then ratios of bound curcumin [curcuminb] to free curcumin [curcuminf] were plotted against curcuminb (Scatchard plot). This provides a straight line with a negative slope. The dissociation constants were calculated from the formula Kd = −1/slope according to the established methods.41,63
Two Dimensional Nuclear Magnetic Resonance (2D NMR) spectroscopy
All NMR spectra were acquired on a Bruker Avance 800 MHz spectrometer using a cryogenically cooled triple-resonance probe equipped with z-axis gradient coils. Data were acquired and processed using Topspin 2.1 version and analyzed with Sparky 3.114. DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) was used as an internal reference for the calibration of proton chemical shifts, whereas nitrogen chemical shift was calibrated indirectly. Two-dimensional 1H–15N correlation heteronuclear single quantum coherent (HSQC) experiments were performed on 15N-labeled protein samples in phosphate buffer, pH 6.8 with a (90:10) H2O/D2O ratio, at 25 °C. Various sets of 1H–15N HSQC correlation spectra were recorded on 150 μM α-Syn monomer and 150 μM 100 kDa LMW samples in the absence and presence of 75 μM curcumin. 2D HSQC spectra for monomer were recorded for 256 data points in indirect dimension with 16 scans per transient, whereas for 100 kDa LMW 512 data points were collected in the indirect dimension. For mapping the interaction sites between α-Syn and curcumin, we calculated mean weighted chemical shift perturbations and change in intensity ratios (I/I0). Nonoverlapping 1H–15N HSQC amide cross-peaks were considered, and their intensities (I) with those of the same cross-peaks in the data set of free protein (I0) were calculated. The I/I0 ratios of nonoverlapping cross-peaks were plotted as a function of the amino acid sequence to obtain the relative intensity variation across the polypeptide chain. However, chemical shift perturbations were calculated using ((5Δδ1H) 2 + (Δδ15N)2)1/2 to identify the significant perturbations upon interactions.
Nile Red Binding Assay
The stock solution of 1 mM Nile Red (NR) was prepared in DMSO and stored at −20 °C until further use. All the fluorescence studies were performed in 20 mM MES buffer, pH 6.0 with 1 μM working concentration of Nile Red. 200 μL of 15 μM oligomeric α-Syn (isolated from SEC) was diluted to 400 μL in same buffer. In 200 μL of 7.5 μM oligomers, curcumin was added such that the final curcumin concentration became 40 μM. The solution was incubated at 37 °C for 30 min. The 200 μL of 7.5 μM oligomers without curcumin was used as a control. In another experiment, freshly prepared 5 mg/mL α-Syn solution was incubated in the presence of 100 μM curcumin at 37 °C for 30 min and injected for SEC. The oligomers were collected from ∼8 mL peak fractions. Then 0.2 μL of 1 mM NR was added to 200 μL each of the oligomers prepared in the presence and absence of curcumin, where final NR was 1 μM. NR fluorescence was performed immediately by exiting at 550 nm and emission in the range of 560–720 nm. For NR binding study with preformed fibrils, 200 μL of 75 μM α-Syn fibrils was incubated with and without 75 μM curcumin for 20 h. Then 20 μL of α-Syn fibrils was diluted to 200 μL in 20 mM MES buffer, pH 6.0, 0.01% sodium azide. Then 0.2 μL of 1 mM NR was added so that the final NR concentration became 1 μM. The NR fluorescence was performed as described; 1 μM NR in 200 μL of MES, pH 6.0 was used as a NR control. To study whether curcumin can affect the NR fluorescence, fluorescence was measured for 1 μM NR, 7.5 μM curcumin, and the mixture of NR and curcumin.
Atomic Force Microscopy (AFM)
For atomic force microscopy, the oligomeric and fibrillar samples were diluted to ∼5 μM in distilled water and diluted samples were spotted on a freshly cleaved mica sheet followed by washing with double distilled water. The mica was dried under vacuum desiccators. AFM imaging was done in tapping mode under a silicon nitride cantilever using a Veeco Nanoscope IV multimode atomic force microscope. A minimum of five different areas of three independent samples were scanned with a scan rate of 1.5 Hz.
Electron Microscopy (EM)
α-Syn fibrils formed in the presence and absence of curcumin were diluted in distilled water to a concentration of ∼40 μM, spotted on a glow-discharged, carbon-coated Formvar grid (Electron Microscopy Sciences, Fort Washington, PA), incubated for 5 min, washed with distilled water, and then stained with 1% (w/v) aqueous uranyl formate solution. Uranyl formate solution was freshly prepared and filtered through 0.22 μm sterile syringe filters (Milipore). EM analysis was performed using a FEI Tecnai G2 12 electron microscope at 120 kV with nominal magnifications in the range of 26 000–60 000. Images were recorded digitally by using the SIS Megaview III imaging system. At least two independent experiments were carried out for each sample.
MTT Metabolic Assay
The neuronal cell line of SH-SY5Y was cultured in Dulbecco’s modified Eagle's medium (DMEM) (Himedia, India) supplemented with 10% FBS (Invitrogen), 100 units/mL penicillin, and 100 μg/mL streptomycin in a 5% CO2 humidified environment at 37 °C. Cells were seeded in 96-well plates in 100 μL of media at a cell density of ∼10 000 cells/well. After 24 h of incubation, the old media was replaced with fresh media containing either oligomeric or fibrillar α-Syn preincubated with curcumin so that α-Syn and curcumin concentration became 5 and 3 μM, respectively. Cells were further incubated for 40 h at 37 °C. Only buffer and 3 μM curcumin were also tested as a control. After 40 h, 10 μL of 5 mg/mL MTT prepared in PBS was added to each well and the incubation was continued for 4 h. Finally, 100 μL of a solution containing 50% dimethylformamide and 20% SDS (pH 4.7) was added and incubated for overnight. After incubation in a 5% CO2 humidified environment at 37 °C, absorption values at 560 nm were determined with an automatic micro titer plate reader (Thermo Fisher Scientific). The background absorbance was also recorded at 690 nm and subtracted from the absorbance value of 560 nm.
Lactate Dehydrogenase (LDH) Release Assay
Lactate dehydrogenase release assay was performed to further evaluate the neuroprotective effect of curcumin over the toxic oligomeric and fibrillar α-Syn. Identical concentrations of preformed oligomers/fibrils (with and without curcumin) were used as mentioned for the MTT assay. SH-SY5Y neuronal cells were cultured and plated in 96-well plates as described earlier. LDH assay was performed after 40 h of incubation of neuronal cells with test samples (preformed oligomers/fibrils, with and without curcumin) using LDH toxicological kit (TOX-7, Sigma) according to the manufacturer's instructions.
SDS-PAGE Analysis
A concentration of 100 μM preformed α-Syn fibrils was incubated with and without 100, 200, and 300 μM curcumin for 20 h. After incubation, fibrils solutions were centrifuged at 18 000g, 40 min. Supernatants were collected and analyzed via 12% Bis-Tris NUPAGE gel (Invitrogen) electrophoresis.
Proteinase K Digestion Assay
α-Syn (150 μM) aggregated in the presence and absence of curcumin (75 μM) was subjected to proteinase K treatment. For proteinase K digestion assay, samples were half diluted in 20 mM MES, pH 6.0, 0.01% sodium azide and 1 μL aliquot of 500 μg/mL proteinase K was added to 75 μL of α-Syn fibrils formed in the presence and absence of curcumin. The final concentration of proteinase K was 6.5 μg. After 1 h of incubation at 37 °C, the reaction was stopped by adding SDS sample buffer followed by heating to 100 °C for 10 min in a water bath. In another experiment, 150 μM preformed α-Syn fibrils was incubated with and without 75 μM curcumin for 1 h at room temperature and digested with proteinase K as described above. For proteinase K digestion of oligomers, 30 μM oligomers (isolated from SEC) were incubated with and without 30 μM curcumin at 37 °C for 30 min. The samples were treated with and without 0.1 μg proteinase K for 10 min at 37 °C and reaction was stopped as described. The samples were then used for SDS-PAGE using 12% Bis-Tris NuPAGE gel (Invitrogen) and developed with silver stain (Bangalore Genei, India).
SH-SY5Y Cell Culture and Differentiation
The neuronal cell line of SH-SY5Y was cultured in Dulbecco’s modified Eagle's medium (DMEM) (Himedia, India) medium supplemented with 10% FBS (Invitrogen), 100 units/mL penicillin, and 100 μg/mL streptomycin in a 5% CO2 humidified environment at 37 °C. For cell differentiation, 50 μM all-trans retinoic acid (RA) (Sigma) was used.75 To do that, undifferentiated cells were seeded on cover glass in a 24-well plate for 24 h, and after that cells were treated with 50 μM RA in media. At every alternate day, the old media was replaced with fresh media containing RA. After 5 days of incubation, we analyzed the cell morphology using phase contrast microscopy to observe neurite projections of differentiated cells.
Immunocytochemistry
The 5 days differentiated cells were used for synaptophysin staining. To do so, cell culture media was replaced with fresh media containing oligomers, oligomers+curcumin, and buffer; cells were further incubated for 40 h. The final oligomers and curcumin concentrations were 6 and 3 μM, respectively. After incubation, media was discarded and the cells were fixed with 3.5% formaldehyde for 30 min at 37 °C. After fixing, the cells were washed with PBS (1 mL × 2) and permeabilized with chilled methanol for 20 min at −20 °C. Further, the cells were blocked with blocking solution (2% BSA in PBS) for 30 min at RT. Cells were incubated for 3 h at RT with rabbit monoclonal anti-synaptophysin primary antibody diluted in blocking solution (1:350; ABCAM). After incubation, the solution was discarded and cells were washed with 1 mL of PBS (3 × 5 min) with gentle shaking. The cells were then incubated with anti-rabbit AlexaFluor 488 (1:500, Invitrogen) secondary antibody prepared in 2% BSA in PBS and incubated for 2 h in dark. After removing the secondary antibody solution, the cells were mounted on a clean glass slide using mounting media. A positive control for synaptophysin specificity was used where the differentiated SH-SY5Y cells were treated with 6 μM Aβ(25–35) amyloid fibrils.76 Images were acquired using an Olympus IX 81 confocal microscope.
To visualize the morphology of nucleus, the differentiated cells were stained with 4,6- diamidino-2-phenylindole (DAPI; Sigma) (1 μg/mL) for 5 min at RT. After 5 min, the DAPI solution was removed and the cells were washed with PBS, mounted on a clean glass slide using mounting media, and imaged under confocal microscope.
Reactive Oxygen Species (ROS) Measurement
The differentiated SH-SY5Y cells were treated with oligomers, oligomers+curcumin, and buffer only in 24-well plates for 40 h. After incubation, the old media was discarded and cells were very gently washed with 1× PBS buffer. A 3 mM stock of 2-hydroethidium (Sigma) was freshly prepared in DMSO, and 2 μL of this solution in 1 mL of PBS was added to the cells and incubated for 5 min at RT in dark. The final concentration of 2-hydroethidium was 6 μM. The cells were immediately imaged under confocal microscopy with an excitation at 543 nm and emission above 565 nm. The intensities of oxidized hydroethidium (hydroxyethidium) fluorescence in cells treated with buffer, oligomers, and oligomers+curcumin were quantified using FV500 Tiempo (version 4.3) software.
Flow Cytometry Analysis
For relative quantification of cell death and apoptosis in oligomers and oligomers+curcumin treated samples, flow cytometry measurement was performed using an Annexin V-FITC apoptosis detection kit (Sigma). To do that, undifferentiated cells were grown in T25 cell culture flasks (Nunc) until ∼70% confluency (∼106 cells). The cells were then treated with oligomers, oligomers+curcumin, and only buffer for 40 h. After incubation, the cells were trypsinized, centrifuged, and used for cell death assay using the Annexin V-FITC apoptosis detection kit (APOAF, Sigma). The cell pellet was washed with 1× PBS and further resuspended in 1× binding buffer (Sigma). Cells were stained with Annexin V-FITC and propidium iodide (PI) according to the manufacturer’s instructions. Unstained cells (without Annexin V-FITC and PI) were used as a control, and cells stained with either Annexin V-FITC or PI were used as fluorescent compensation controls. Annexin V-FITC and PI staining were quantified in a flow cytometer (FACSAria, BDBiosciences, San Jose, CA) and analyzed using the BDFACS Diva software. For each sample, 20 000 cells were analyzed. The upper left quadrant (Q1) represents the dead cell population (only PI positive), the upper right quadrant (Q2) represents the population of cells in the late stage of apoptosis (Annexin V-FITC positive and PI positive), the lower left quadrant (Q3) represents the population of viable/live cells (Annexin V-FITC negative and PI negative), and the lower right quadrant (Q4) represents the population of cells in early stages of apoptosis (only Annexin V-FITC positive). The X-axis represents the area of Annexin V-FITC fluorescence, and the Y-axis represents PI fluorescence intensities.
Statistical Analysis
Statistical significance was determined by using one-way ANOVA followed by Newman-Keuls multiple comparison post hoc test; *P < 0.05; **P < 0.01; NS P > 0.05.
Acknowledgments
The authors wish to acknowledge Central SPM Facility (IRCC, IIT Bombay) for AFM imaging and SAIF (IIT Bombay) for electron microscopy. The authors also thank Dr. Shamik Sen, Reeba S. Jacob, Shruti Sahay, A. Anoop, and Subhadeep Das for critically reading the manuscript and Prem Verma (Department of Physics, IIT Bombay) for the help during AFM imaging. We would also like to thank Ankit Rai, Sudarshan Kini, and Sachin Tawade for their valuable help in confocal microscopy and flow cytometry analysis.
Glossary
Abbreviations
- α-Syn
α-synuclein
- CD
circular dichroism
- EM
electron microscopy
- AFM
atomic force microscopy
- AD
Alzheimer’s disease
- PD
Parkinson’s disease
- NMR
nuclear magnetic resonance
- LMW
low molecular weight
- CR
Congo Red
- NR
Nile Red
- SEC
size exclusion chromatography
Supporting Information Available
Table 1 and Supplementary figures S1 to S8. This materials is available free of charge via the Internet http://pubs.acs.org
Author Contributions
§ Equally contributing authors.
Author Contributions
S.K.M. and P.K.S. conceived and designed the experiments. P.K.S., D.G., V.K., G.M.M., and A.K. performed the experiments: S.K.M., A.K., P.K.S., and D.G. analyzed the data. S.K.M., P.K.S., and A.K. wrote the paper.
The work was supported by Grants from CSIR (37(1404)/10/EMR-11), DST (SR/FR/LS-032/2009), and DBT (BT/PR14344Med/30/501/2010 and BT/PR13359/BRB/10/752/2009), Government of India.
The authors declare no competing financial interest.
Supplementary Material
References
- Cookson M. R. (2005) The biochemistry of Parkinson’s disease. Annu. Rev. Biochem. 74, 29–52. [DOI] [PubMed] [Google Scholar]
- Lansbury P. T.; Brice A. (2002) Genetics of Parkinson’s disease and biochemical studies of implicated gene products - Commentary. Curr. Opin Cell. Biol. 14, 653–660. [DOI] [PubMed] [Google Scholar]
- Feany M. B. (2000) Studying human neurodegenerative diseases in flies and worms. J. Neuropathol. Exp. Neurol. 59, 847–856. [DOI] [PubMed] [Google Scholar]
- Conway K. A.; Harper J. D.; Lansbury P. T. (1998) Accelerated in vitro fibril formation by a mutant α-synuclein linked to early-onset Parkinson disease. Nat. Med. 4, 1318–1320. [DOI] [PubMed] [Google Scholar]
- Conway K. A.; Lee S. J.; Rochet J. C.; Ding T. T.; Williamson R. E.; Lansbury P. T. (2000) Acceleration of oligomerization, not fibrillization, is a shared property of both a-synuclein mutations linked to early-onset Parkinson’s disease: Implications for pathogenesis and therapy. Proc. Natl. Acad. Sci. U.S.A. 97, 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V. N.; Lee H. J.; Li J.; Fink A. L.; Lee S. J. (2001) Stabilization of partially folded conformation during α-synuclein oligomerization in both purified and cytosolic preparations. J. Biol. Chem. 276, 43495–43498. [DOI] [PubMed] [Google Scholar]
- Goldberg M. S.; Lansbury P. T. (2000) Is there a cause-and-effect relationship between α-synuclein fibrillization and Parkinson’s disease?. Nat. Cell Biol. 2, E115–E119. [DOI] [PubMed] [Google Scholar]
- Karpinar D. P.; Balija M. B.; Kugler S.; Opazo F.; Rezaei-Ghaleh N.; Wender N.; Kim H. Y.; Taschenberger G.; Falkenburger B. H.; Heise H.; Kumar A.; Riedel D.; Fichtner L.; Voigt A.; Braus G. H.; Giller K.; Becker S.; Herzig A.; Baldus M.; Jackle H.; Eimer S.; Schulz J. B.; Griesinger C.; Zweckstetter M. (2009) Pre-fibrillar α-synuclein variants with impaired β-structure increase neurotoxicity in Parkinson’s disease models. EMBO J. 28, 3256–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winner B.; Jappelli R.; Maji S. K.; Desplats P. A.; Boyer L.; Aigner S.; Hetzer C.; Loher T.; Vilar M.; Campioni S.; Tzitzilonis C.; Soragni A.; Jessberger S.; Mira H.; Consiglio A.; Pham E.; Masliah E.; Gage F. H.; Riek R. (2011) In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. U.S.A. 108, 4194–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Z. S.; Neugebauer V.; Dineley K. T.; Kayed R.; Zhang W.; Reese L. C.; Taglialatela G. (2012) α-Synuclein oligomers oppose long-term potentiation and impair memory through a calcineurin-dependent mechanism: relevance to human synucleopathic diseases. J. Neurochem. 120, 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzer K. M.; Haasen D.; Karow A. R.; Moussaud S.; Habeck M.; Giese A.; Kretzschmar H.; Hengerer B.; Kostka M. (2007) Different species of α-synuclein oligomers induce calcium influx and seeding. J. Neurosci. 27, 9220–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo N. D., Maji S. K., Fradinger E. A., Bitan G., and Teplow D. B.. (2005) The Amyloid β Protein. In Amyloid proteins: The β-sheet conformation and disease (Sipe J. D., Ed.), pp 384–491, Wiley-VCH Publishers, Weinheim, Germany. [Google Scholar]
- Mason J. M.; Kokkoni N.; Stott K.; Doig A. J. (2003) Design strategies for anti-amyloid agents. Curr. Opin. Struct. Biol. 13, 526–532. [DOI] [PubMed] [Google Scholar]
- Cohen F. E.; Kelly J. W. (2003) Therapeutic approaches to protein-misfolding diseases. Nature 426, 905–909. [DOI] [PubMed] [Google Scholar]
- Hong D. P.; Fink A. L.; Uversky V. N. (2008) Structural characteristics of α-synuclein oligomers stabilized by the flavonoid baicalein. J. Mol. Biol. 383, 214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Agnaf O. M.; Paleologou K. E.; Greer B.; Abogrein A. M.; King J. E.; Salem S. A.; Fullwood N. J.; Benson F. E.; Hewitt R.; Ford K. J.; Martin F. L.; Harriott P.; Cookson M. R.; Allsop D. (2004) A strategy for designing inhibitors of α-synuclein aggregation and toxicity as a novel treatment for Parkinson’s disease and related disorders. FASEB J. 18, 1315–1317. [DOI] [PubMed] [Google Scholar]
- Masuda M.; Suzuki N.; Taniguchi S.; Oikawa T.; Nonaka T.; Iwatsubo T.; Hisanaga S.; Goedert M.; Hasegawa M. (2006) Small molecule inhibitors of α-synuclein filament assembly. Biochemistry 45, 6085–6094. [DOI] [PubMed] [Google Scholar]
- Lamberto G. R.; Binolfi A.; Orcellet M. L.; Bertoncini C. W.; Zweckstetter M.; Griesinger C.; Fernandez C. O. (2009) Structural and mechanistic basis behind the inhibitory interaction of PcTS on α-synuclein amyloid fibril formation. Proc. Natl. Acad. Sci. U.S.A. 106, 21057–21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnhoefer D. E.; Bieschke J.; Boeddrich A.; Herbst M.; Masino L.; Lurz R.; Engemann S.; Pastore A.; Wanker E. E. (2008) EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 15, 558–566. [DOI] [PubMed] [Google Scholar]
- Masuda M.; Hasegawa M.; Nonaka T.; Oikawa T.; Yonetani M.; Yamaguchi Y.; Kato K.; Hisanaga S.; Goedert M. (2009) Inhibition of α-synuclein fibril assembly by small molecules: analysis using epitope-specific antibodies. FEBS Lett. 583, 787–791. [DOI] [PubMed] [Google Scholar]
- Zhu M.; Rajamani S.; Kaylor J.; Han S.; Zhou F.; Fink A. L. (2004) The flavonoid baicalein inhibits fibrillation of α-synuclein and disaggregates existing fibrils. J. Biol. Chem. 279, 26846–26857. [DOI] [PubMed] [Google Scholar]
- Rao J. N.; Dua V.; Ulmer T. S. (2008) Characterization of α-synuclein interactions with selected aggregation-inhibiting small molecules. Biochemistry 47, 4651–4656. [DOI] [PubMed] [Google Scholar]
- Bieschke J.; Russ J.; Friedrich R. P.; Ehrnhoefer D. E.; Wobst H.; Neugebauer K.; Wanker E. E. (2010) EGCG remodels mature α-synuclein and amyloid-beta fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 107, 7710–7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana M.; Hogen T.; Levin J.; Hillmer A.; Giese A.; Vassallo N. (2011) Inhibition and disaggregation of α-synuclein oligomers by natural polyphenolic compounds. FEBS Lett. 585, 1113–1120. [DOI] [PubMed] [Google Scholar]
- Ferreira N.; Saraiva M. J.; Almeida M. R. (2011) Natural polyphenols inhibit different steps of the process of transthyretin (TTR) amyloid fibril formation. FEBS Lett. 585, 2424–2430. [DOI] [PubMed] [Google Scholar]
- Hafner-Bratkovic I.; Gaspersic J.; Smid L. M.; Bresjanac M.; Jerala R. (2008) Curcumin binds to the α-helical intermediate and to the amyloid form of prion protein - a new mechanism for the inhibition of PrP(Sc) accumulation. J. Neurochem. 104, 1553–1564. [DOI] [PubMed] [Google Scholar]
- Porat Y.; Abramowitz A.; Gazit E. (2006) Inhibition of amyloid fibril formation by polyphenols: structural similarity and aromatic interactions as a common inhibition mechanism. Chem. Biol. Drug. Des. 67, 27–37. [DOI] [PubMed] [Google Scholar]
- Goel A.; Kunnumakkara A. B.; Aggarwal B. B. (2008) Curcumin as ″Curecumin″: from kitchen to clinic. Biochem. Pharmacol. 75, 787–809. [DOI] [PubMed] [Google Scholar]
- Cole G. M.; Teter B.; Frautschy S. A. (2007) Neuroprotective effects of curcumin. Adv. Exp. Med. Biol. 595, 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim G. P.; Chu T.; Yang F. S.; Beech W.; Frautschy S. A.; Cole G. M. (2001) The curry spice curcumin reduces oxidative damage and amyloid pathology in an Alzheimer transgenic mouse. J. Neurosci. 21, 8370–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F.; Lim G. P.; Begum A. N.; Ubeda O. J.; Simmons M. R.; Ambegaokar S. S.; Chen P. P.; Kayed R.; Glabe C. G.; Frautschy S. A.; Cole G. M. (2005) Curcumin inhibits formation of amyloid beta oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J. Biol. Chem. 280, 5892–5901. [DOI] [PubMed] [Google Scholar]
- Khuwaja G.; Khan M. M.; Ishrat T.; Ahmad A.; Raza S. S.; Ashafaq M.; Javed H.; Khan M. B.; Khan A.; Vaibhav K.; Safhi M. M.; Islam F. (2011) Neuroprotective effects of curcumin on 6-hydroxydopamine-induced Parkinsonism in rats: behavioral, neurochemical and immunohistochemical studies. Brain Res. 1368, 254–263. [DOI] [PubMed] [Google Scholar]
- Hickey M. A.; Zhu C.; Medvedeva V.; Lerner R. P.; Patassini S.; Franich N. R.; Maiti P.; Frautschy S. A.; Zeitlin S.; Levine M. S.; Chesselet M. F. (2012) Improvement of neuropathology and transcriptional deficits in CAG 140 knock-in mice supports a beneficial effect of dietary curcumin in Huntington’s disease. Mol. Neurodegener. 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alloza M.; Borrelli L. A.; Rozkalne A.; Hyman B. T.; Bacskai B. J. (2007) Curcumin labels amyloid pathology in vivo, disrupts existing plaques, and partially restores distorted neurites in an Alzheimer mouse model. J. Neurochem. 102, 1095–1104. [DOI] [PubMed] [Google Scholar]
- Ahmad B.; Lapidus L. J. (2012) Curcumin prevents aggregation in α-synuclein by increasing reconfiguration rate. J. Biol. Chem. 287, 9193–9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K.; Yamada M. (2006) Antioxidant compounds have potent anti-fibrillogenic and fibril-destabilizing effects for α-synuclein fibrils in vitro. J. Neurochem. 97, 105–115. [DOI] [PubMed] [Google Scholar]
- Pandey N.; Strider J.; Nolan W. C.; Yan S. X.; Galvin J. E. (2008) Curcumin inhibits aggregation of α-synuclein. Acta Neuropathol. 115, 479–489. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Yu Y.; Li X.; Ross C. A.; Smith W. W. (2011) Curcumin protects against A53T α-synuclein-induced toxicity in a PC12 inducible cell model for Parkinsonism. Pharmacol. Res. 63, 439–444. [DOI] [PubMed] [Google Scholar]
- Wang M. S.; Boddapati S.; Emadi S.; Sierks M. R. (2010) Curcumin reduces α-synuclein induced cytotoxicity in Parkinson’s disease cell model. BMC Neurosci. 11, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson S. A.; Ecroyd H.; Kee T. W.; Carver J. A. (2009) The thioflavin T fluorescence assay for amyloid fibril detection can be biased by the presence of exogenous compounds. FEBS J. 276, 5960–5972. [DOI] [PubMed] [Google Scholar]
- Rai D.; Singh J. K.; Roy N.; Panda D. (2008) Curcumin inhibits FtsZ assembly: an attractive mechanism for its antibacterial activity. Biochem. J. 410, 147–155. [DOI] [PubMed] [Google Scholar]
- Mosmann T. (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63. [DOI] [PubMed] [Google Scholar]
- Behl C.; Davis J. B.; Lesley R.; Schubert D. (1994) Hydrogen peroxide mediates amyloid beta protein toxicity. Cell 77, 817–827. [DOI] [PubMed] [Google Scholar]
- Braga C. A.; Follmer C.; Palhano F. L.; Khattar E.; Freitas M. S.; Romao L.; Di Giovanni S.; Lashuel H. A.; Silva J. L.; Foguel D. (2011) The anti-Parkinsonian drug selegiline delays the nucleation phase of α-synuclein aggregation leading to the formation of nontoxic species. J. Mol. Biol. 405, 254–273. [DOI] [PubMed] [Google Scholar]
- Zhao H.; Joseph J.; Fales H. M.; Sokoloski E. A.; Levine R. L.; Vasquez-Vivar J.; Kalyanaraman B. (2005) Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence. Proc. Natl. Acad. Sci. U.S.A. 102, 5727–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremades N.; Cohen S. I.; Deas E.; Abramov A. Y.; Chen A. Y.; Orte A.; Sandal M.; Clarke R. W.; Dunne P.; Aprile F. A.; Bertoncini C. W.; Wood N. W.; Knowles T. P.; Dobson C. M.; Klenerman D. (2012) Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell 149, 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimpietri D.; Nico B.; Vacca A.; Mangieri D.; Catarsi P.; Ponzoni M.; Ribatti D. (2005) Synergistic inhibition of human neuroblastoma-related angiogenesis by vinblastine and rapamycin. Oncogene 24, 6785–6795. [DOI] [PubMed] [Google Scholar]
- Vermes I.; Haanen C.; Steffens-Nakken H.; Reutelingsperger C. (1995) A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods 184, 39–51. [DOI] [PubMed] [Google Scholar]
- Klunk W. E.; Jacob R. F.; Mason R. P. (1999) Quantifying amyloid β-peptide (Aβ) aggregation using the Congo Red-Aβ (CR-Aβ) spectrophotometric assay. Anal. Biochem. 266, 66–76. [DOI] [PubMed] [Google Scholar]
- Maji S. K.; Perrin M. H.; Sawaya M. R.; Jessberger S.; Vadodaria K.; Rissman R. A.; Singru P. S.; Nilsson K. P.; Simon R.; Schubert D.; Eisenberg D.; Rivier J.; Sawchenko P.; Vale W.; Riek R. (2009) Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Agnaf O. M. A.; Irvine G. B. (2002) Aggregation and neurotoxicity of α-synuclein and related peptides. Biochem. Soc. Trans. 30, 559–565. [DOI] [PubMed] [Google Scholar]
- Sidhu A.; Wersinger C.; Moussa C. E.; Vernier P. (2004) The role of α-synuclein in both neuroprotection and neurodegeneration. Ann. N.Y. Acad. Sci. 1035, 250–270. [DOI] [PubMed] [Google Scholar]
- Grelle G.; Otto A.; Lorenz M.; Frank R. F.; Wanker E. E.; Bieschke J. (2011) Black tea theaflavins inhibit formation of toxic amyloid-β and α-synuclein fibrils. Biochemistry 50, 10624–10636. [DOI] [PubMed] [Google Scholar]
- Langer F.; Eisele Y. S.; Fritschi S. K.; Staufenbiel M.; Walker L. C.; Jucker M. (2011) Soluble Abeta seeds are potent inducers of cerebral beta-amyloid deposition. J. Neurosci. 31, 14488–14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlisny M. B.; Ostaszewski B. L.; Squazzo S. L.; Koo E. H.; Rydell R. E.; Teplow D. B.; Selkoe D. J. (1995) Aggregation of secreted amyloid beta-protein into sodium dodecyl sulfate-stable oligomers in cell culture. J. Biol. Chem. 270, 9564–9570. [DOI] [PubMed] [Google Scholar]
- Walsh D. M.; Tseng B. P.; Rydel R. E.; Podlisny M. B.; Selkoe D. J. (2000) The oligomerization of amyloid beta-protein begins intracellularly in cells derived from human brain. Biochemistry 39, 10831–10839. [DOI] [PubMed] [Google Scholar]
- Neumann M.; Kahle P. J.; Giasson B. I.; Ozmen L.; Borroni E.; Spooren W.; Muller V.; Odoy S.; Fujiwara H.; Hasegawa M.; Iwatsubo T.; Trojanowski J. Q.; Kretzschmar H. A.; Haass C. (2002) Misfolded proteinase K-resistant hyperphosphorylated α-synuclein in aged transgenic mice with locomotor deterioration and in human α-synucleinopathies. J. Clin. Invest. 110, 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar M.; Chou H. T.; Luhrs T.; Maji S. K.; Riek-Loher D.; Verel R.; Manning G.; Stahlberg H.; Riek R. (2008) The fold of α-synuclein fibrils. Proc. Natl. Acad. Sci. U.S.A. 105, 8637–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi B.; Kumita J. R.; Barros T. P.; Esbjorner E. K.; Luheshi L. M.; Crowther D. C.; Wilson M. R.; Dobson C. M.; Favrin G.; Yerbury J. J. (2010) ANS binding reveals common features of cytotoxic amyloid species. ACS Chem. Biol. 5, 735–740. [DOI] [PubMed] [Google Scholar]
- Campioni S.; Mannini B.; Zampagni M.; Pensalfini A.; Parrini C.; Evangelisti E.; Relini A.; Stefani M.; Dobson C. M.; Cecchi C.; Chiti F. (2010) A causative link between the structure of aberrant protein oligomers and their toxicity. Nat. Chem. Biol. 6, 140–147. [DOI] [PubMed] [Google Scholar]
- Daban J. R.; Samso M.; Bartolome S. (1991) Use of Nile Red as a fluorescent probe for the study of the hydrophobic properties of protein-sodium dodecyl sulfate complexes in solution. Anal. Biochem. 199, 162–168. [DOI] [PubMed] [Google Scholar]
- Sackett D. L.; Wolff J. (1987) Nile Red as a polarity-sensitive fluorescent probe of hydrophobic protein surfaces. Anal. Biochem. 167, 228–234. [DOI] [PubMed] [Google Scholar]
- Fuchs H.; Gessner R. (2001) The result of equilibrium-constant calculations strongly depends on the evaluation method used and on the type of experimental errors. Biochem. J. 359, 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliezer D.; Kutluay E.; Bussell R.; Browne G. (2001) Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 307, 1061–1073. [DOI] [PubMed] [Google Scholar]
- Munishkina L. A.; Phelan C.; Uversky V. N.; Fink A. L. (2003) Conformational behavior and aggregation of α-synuclein in organic solvents: Modeling the effects of membranes. Biochemistry 42, 2720–2730. [DOI] [PubMed] [Google Scholar]
- Anderson V. L.; Ramlall T. F.; Rospigliosi C. C.; Webb W. W.; Eliezer D. (2010) Identification of a helical intermediate in trifluoroethanol-induced α-synuclein aggregation. Proc. Natl. Acad. Sci. U.S.A. 107, 18850–18855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fezoui Y.; Teplow D. B. (2002) Kinetic studies of amyloid β-protein fibril assembly - Differential effects of α-helix stabilization. J. Biol. Chem. 277, 36948–36954. [DOI] [PubMed] [Google Scholar]
- Dairam A.; Fogel R.; Daya S.; Limson J. L. (2008) Antioxidant and iron-binding properties of curcumin, capsaicin, and S-allylcysteine reduce oxidative stress in rat brain homogenate. J. Agric. Food Chem. 56, 3350–3356. [DOI] [PubMed] [Google Scholar]
- Hamaguchi T.; Ono K.; Yamada M. (2010) REVIEW: Curcumin and Alzheimer’s disease. CNS Neurosci. Ther. 16, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar I.; Jonson M.; Nilsson K. P.; Thor S.; Hammarstrom P. (2012) Curcumin promotes Aβ fibrillation and reduces neurotoxicity in transgenic Drosophila. PLoS One 7, e31424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi T.; Ono K.; Murase A.; Yamada M. (2009) Phenolic compounds prevent Alzheimer’s pathology through different effects on the amyloid-beta aggregation pathway. Am. J. Pathol. 175, 2557–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necula M.; Kayed R.; Milton S.; Glabe C. G. (2007) Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. J. Biol. Chem. 282, 10311–10324. [DOI] [PubMed] [Google Scholar]
- Orlando R. A.; Gonzales A. M.; Royer R. E.; Deck L. M.; Vander Jagt D. L. (2012) A chemical analog of curcumin as an improved inhibitor of amyloid β oligomerization. PLoS One 7, e31869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volles M. J.; Lansbury P. T. Jr. (2007) Relationships between the Sequence of α-Synuclein and its Membrane Affinity, Fibrillization Propensity, and Yeast Toxicity. J. Mol. Biol. 366, 1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J.; Khoshaghideh F.; Patel S.; Lee S. J. (2004) Clearance of α-synuclein oligomeric intermediates via the lysosomal degradation pathway. J. Neurosci. 24, 1888–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amijee H.; Bate C.; Williams A.; Virdee J.; Jeggo R.; Spanswick D.; Scopes D. I.; Treherne J. M.; Mazzitelli S.; Chawner R.; Eyers C. E.; Doig A. J. (2012) The N-Methylated Peptide SEN304 Powerfully Inhibits Aβ(1–42) Toxicity by Perturbing Oligomer Formation. Biochemistry 51, 8338–8352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.