Abstract
Methylation of the fragile X-related epigenetic element 2 (FREE2) located on the exon 1/intron 1 boundary of the FMR1 gene is related to FMRP expression and cognitive impairment in full mutation (FM; CGG>200) individuals. We examined the relationship between age, the size of the FMR1 CGG expansion and the methylation output ratio (MOR) at 12 CpG sites proximal to the exon 1/intron 1 boundary using FREE2 MALDI-TOF MS. The patient cohort included 119 males and 368 females, i.e. 121 healthy controls (CGG<40), 176 premutation (CGG 55–170) and 190 FM (CGG 213–2000). For all CpG units examined, FM males showed a significantly elevated MOR compared with that in hypermethylated FM females. In FM males the MOR for most CpG units significantly positively correlated with both age and CGG size (P< 0.05). In FM females the skewing towards the unmethylated state was significant for half of the units between birth and puberty (P < 0.05). The methylation status of intron 1 CpG10–12 that was most significantly related to cognitive impairment in our earlier study, did not change significantly with age in FM females. These results challenge the concept of fragile X syndrome (FXS)-related methylation being static over time, and suggest that due to the preference for the unmethylated state in FM females, X-inactivation at this locus is not random. The findings also highlight that the prognostic value of FXS methylation testing is not uniform between all CpG sites, and thus may need to be evaluated on a site-by-site basis.
INTRODUCTION
Fragile X syndrome (FXS) is the most common heritable form of intellectual disability with the frequency of 1 in 3600 in males and 1 in 6000 in females in the general population (1). It is also the major genetic cause of autism, accounting for between 2 and 6.5% of all individuals diagnosed with co-morbid autism disorder (AD) (reviewed in 2). FXS usually results from the lack of the expression of the FMR1 gene due to methylation of its promoter and loss of its protein product (FMRP), which is essential for normal neurodevelopment (3–5). Large expansions (>200) of a trinucleotide CGG repeat located in the 5′ untranslated region of FMR1 are termed full mutation (FM), and are usually associated with the FXS phenotype (6).
Premutation (PM) alleles (55–199 repeats) are much more common (1 in 600 males and 1 in 300 females) than FM alleles, and have been primarily linked to late onset disorders including the fragile X-associated tremor/ataxia syndrome (FXTAS) (7) and fragile X-associated primary ovarian insufficiency (FXPOI) (8). Although PM alleles do not cause FXS, there is also some evidence to suggest that these alleles are associated with attention deficit-hyperactivity disorder, autism and learning deficits (9,10). These neurodevelopmental conditions may be related to a small decrease in FMRP which has been reported in the PM carriers (11). A toxic effect of the elevated expanded FMR1 mRNA observed in PM carriers has also been suggested to contribute to some of these phenotypes (7,12,13) as well as to the PM-related late onset disorders (7,14,15). Thus, the combination of RNA toxicity and decrease in FMRP is thought to contribute to clinical heterogeneity, particularly in PM/FM mosaics and can confound the predictive value of CGG-based testing in molecular diagnosis of FXS.
Methylation-sensitive Southern blot analysis is the current ‘gold standard’ test that provides information on the number of the CGG repeats and methylation status for all types of FMR1 alleles. One of its limitations is that it targets methylation of only a few CpG sites within the FMR1 CpG island located 5′ of the expansion (16) and the methylation status of these few sites does not necessarily represent that of the entire promoter region (17,18). We have recently developed a test for FXS targeting methylation of specific biomarker CpG sites located 3′ of the CGG expansion, predominantly within the FMR1 intron 1 region named fragile X-related epigenetic element 2 (FREE2). In a pilot cohort, methylation of the FREE2 intron 1 sites could be used to identify cognitively impaired FM males and females with specificity and sensitivity approaching 100%, but could not distinguish between PM carriers and healthy controls or between PM carriers and high functioning males with unmethylated FM alleles (17,19). This involvement of intronic sites suggests that the FMR1 promoter is in fact much larger than the CpG island and extends intragenically 3′ of the CGG expansion into exon 1 and the non-coding portion of the gene. In this study, we have performed FREE2 methylation analysis in a much larger cohort of FMR1 expansion carrier males and females, and have examined the relationships of FMR1 exon 1 and intron 1 methylation, the size of the FMR1 CGG expansion and age in both sexes.
RESULTS
We performed FREE2 methylation analysis using the MALDI-TOF MS EpiTYPER system in 119 males including 17 healthy controls (CGG<40), 38 PM (CGG 55-170), 64 FM (213-1500) and in 368 females including 104 healthy controls (CGG<40), 138 PM (CGG 55-170) and 126 FM (CGG 213-200). We have analysed 5 CpG units representing 12 CpG sites (Fig. 1A). It is important to note that methylation analysis of CpG 6/7, 8/9 and 10–12 cannot be separated into single CpG resolution. This is because the fragments generated for these are of the same size (generated through T cleave) as previously described (17,18).
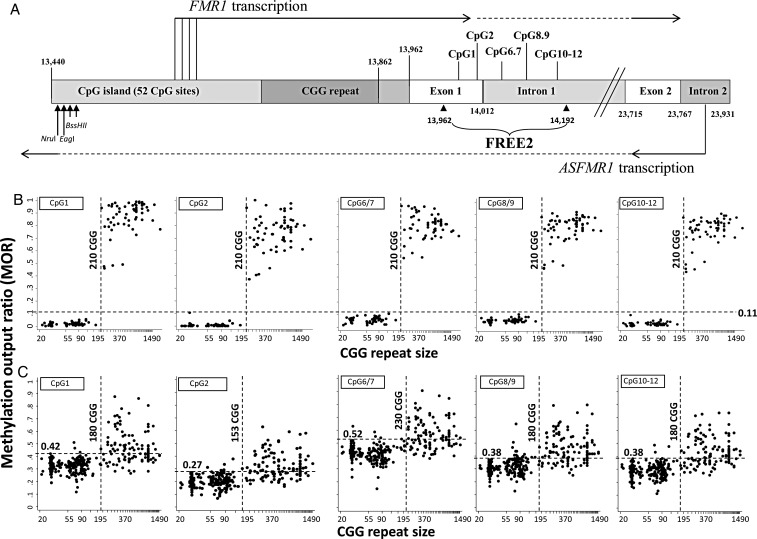
Figure 1.
Comparison between CGG size (x-axis) and methylation output ratio (y-axis) within the FREE2 region. (A) Representation of the intron and exon regions 3′ of the FMR1 CGG expansion (sequence numbering from GenBank L29074 L38501) in relation to FMR1 and ASFMR1 transcription start sites (the broken lines indicated spliced out regions), FREE2 and the FMR1 CpG island and methylation-sensitive restriction sites (NruI, EagI and BssHII) analysed using routine fragile X Southern blot testing. A CGG repeat is located within the 5’ (UTR) of the FMR1 gene. ASFMR1 spans the CGG expansion in the antisense direction and is also regulated by another promoter located in the exon 2 of FMR1. The FREE2 region is located downstream of the CGG expansion, with CpG1 and 2 located within the 3′ end of FMR1 exon 1; CpG6/7, 8/9 and 10–12 located within the 5′ end of FMR1 intron 1. (B) 119 males with CGG size ranging between 22 and 2000 repeats. (C) 368 females with CGG size ranging between 21 and 1500 repeats. Note: PM/FM mosaic individuals and ‘high functioning’ unmethylated FM males as determined by methylation-sensitive Southern blot were not included. For females only the size of the smallest size expanded allele is presented on the x-axis. If a FM allele was identified as a smear; the lowest CGG size expanded allele was presented on the x-axis for both FM males and females. The horizontal-broken line represents the maximum MOR value for the control group with CGG<40, with this value indicated above this line, with an exception of CpG6/7 where one control outlier at the MOR of 0.63 was not considered as the maximum control value. The perpendicular-broken line represents the minimum MOR value between 100 and 300 CGG repeats which is above the maximum value of the control group.
For the male controls, the maximum methylation output ratio (MOR) for 5 CpG units was <0.11 (Fig. 1B, horizontal line). For female controls, the maximum MOR varied particularly at the exon 1/intron 1 boundary, between 0.27 and 0.52 (Fig. 1C, horizontal line). None of the PM samples from males showed a MOR above the maximum value of the control range, while for females, between 2 (3 out of 138) and 12% (17 out of 138) of PM samples showed a MOR above the maximum control value. The number of these hypermethylated PM samples varied between CpG units, with CpGs 1, 6/7 and 10–12 ∼2% and CpGs 2 and 8/9 ∼10%, of all PM female samples examined.
We also determined the MOR in samples with 100–250 CGG triplet repeats that had a MOR above the maximum value of the controls to identify the ‘borderline repeat range’ of the expansion that leads to FMR1 exon 1 and intron 1 hypermethylation in FM males and females. While for males there was a sharp separation in the MOR at 210 CGG repeats for all CpG units examined (Fig. 1B, vertical line), for females this separation varied between CpG units particularly at the exon 1/intron 1 boundary, between 153 and 230 CGGs (Fig. 1C, vertical line).
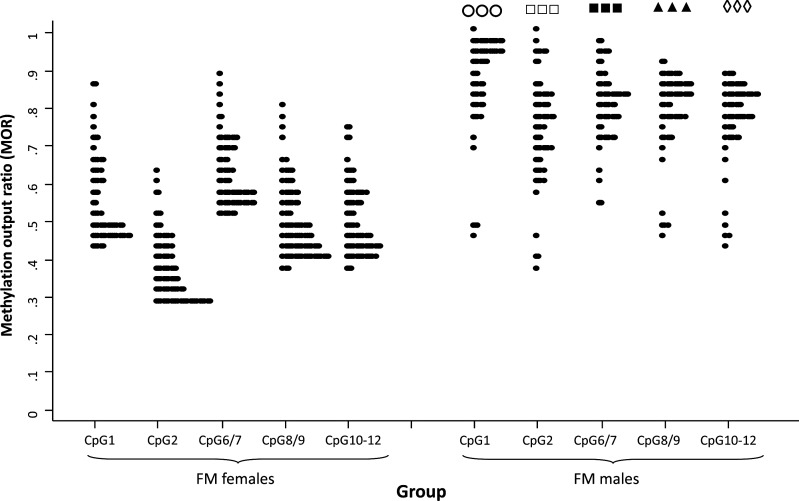
The Shaprico–Wilk test showed that, for all FM males and females with the MOR above the maximum value of the control group, the distribution was not normal (P < 0.05). We therefore used the non-parametric Mann–Whitney test to examine the difference in the median of MOR values between hypermethylated FM males and females. Results presented in the Table 1 and Figure 2 show that methylation for all CpG units in FM males was significantly elevated (one-side P-value) compared with hypermethylated FM females. While for FM males the MOR for most units was between 0.7 and 1, for females this MOR range was generally between 0.4 and 0.7, with distribution skewed towards the maximum control value, with no values reaching 1. Although methylation-sensitive Southern blot analysis of the NruI restriction site within the CpG island (Fig. 1A) indicated that all the samples from FM males were fully methylated, analysis of the FREE2 sites (located >1 kb downstream) indicated that at least half of these were methylation mosaics, within exon 1 and intron 1 sequences.
Table 1.
FREE2 methylation comparisons between FM males and females with methylation above the maximum MOR of the control group
| Variable | FM males |
FM females |
P-value | ||||
|---|---|---|---|---|---|---|---|
| n | Median | IQR | n | Median | IQR | ||
| CpG 1 | 62 | 0.918 | 0.136 | 66 | 0.501 | 0.175 | <0.0001 |
| CpG 2 | 63 | 0.775 | 0.165 | 85 | 0.340 | 0.125 | <0.0001 |
| CpG 6/7 | 60 | 0.819 | 0.088 | 74 | 0.590 | 0.140 | <0.0001 |
| CpG 8/9 | 63 | 0.825 | 0.085 | 98 | 0.470 | 0.130 | <0.0001 |
| CpG 10–12 | 64 | 0.803 | 0.090 | 80 | 0.469 | 0.138 | <0.0001 |
IQR, inter-quartile range.
Figure 2.
FREE2 methylation comparisons between FM males and females with the methylation output ratio above the maximum MOR of the control group. CpG1 and 2 are located within the 3′ end of FMR1 exon 1; CpG6/7, 8/9 and 10–12 are located within the 5′ end of FMR1 intron 1. Note: All comparison showed P < 0.001. CpG1—open circles; CpG2—open squares; CpG6/7—closed squares; CpG8/9—closed triangles; CpG10–12—open diamonds.
We have also explored the contribution of CGG size and age to differences in MOR distribution in FM males and females. In FM males, there was a significant increase in the MOR with an increase in CGG size (Table 2), and age (Table 3). In FM females, the CGG size was not significantly associated with the MOR for any of the CpG units (for all CpG units P > 0.2).
Table 2.
Relationship between log transformed CGG size (predictor) and FREE2 methylation in FM males by fitting univariate robust linear regression model for the MOR of each CpG unit
| Outcome variable | n | Coef (β) | SE | P-value | R2 |
|---|---|---|---|---|---|
| CpG 1 | 60 | 0.067 | 0.023 | 0.005 | 0.130 |
| CpG 2 | 61 | 0.044 | 0.035 | 0.212 | 0.026 |
| CpG 6/7 | 58 | −0.022 | 0.025 | 0.369 | 0.014 |
| CpG 8/9 | 61 | 0.043 | 0.020 | 0.034 | 0.074 |
| CpG 10–12 | 42 | 0.050 | 0.018 | 0.008 | 0.111 |
P values less than 0.05 are highlighted in bold.
Table 3.
Relationship between FREE2 methylation in FM males and age (predictor) by fitting univariate robust regression model for the MOR of each CpG unit
| Outcome variable | n | Coef (β) | SE | P-value | R2 |
|---|---|---|---|---|---|
| CpG 1 | 60 | 0.144 | 0.048 | 0.004 | 0.135 |
| CpG 2 | 61 | 0.121 | 0.080 | 0.137 | 0.037 |
| CpG 6/7 | 58 | 0.047 | 0.053 | 0.373 | 0.014 |
| CpG 8/9 | 61 | 0.085 | 0.039 | 0.034 | 0.074 |
| CpG 10–12 | 62 | 0.091 | 0.038 | 0.020 | 0.087 |
Coefficient (Coef) and standard error (SE) were multiplied by 100.
P values less than 0.05 are highlighted in bold.
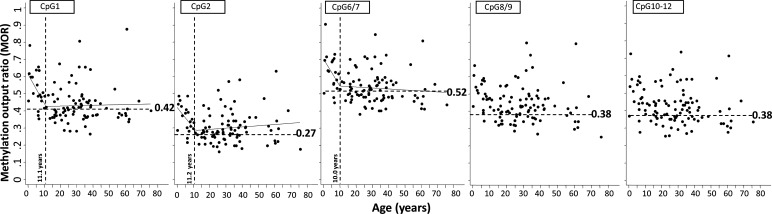
However, the MOR for CpG unit 1 (in exon 1) and CpG unit 6/7 (in intron 1) showed significant negative association with age from birth to puberty (with break points between 11.1 and 10 years; with P = 0.025 and 0.030, respectively). There was no significant relationship with age for MOR of these CpG units after puberty to 80 years of age (Fig. 3 and Table 4).
Figure 3.
Segmented linear regression model comparison between age (x-axis) and methylation output ratio (y-axis) within the FREE2 region of 104 FM females with CGG size ranging between 21 and 1500 repeats. The perpendicular-broken line represents the ‘break point’ age estimated using the segmented linear regression model as detailed in Table 3. From birth to the ‘break point’ age there was significant inverse relationship with MOR for CpG 1, 2 and 6/7, but not CpG8/9 and 10–12. At the ages past the ‘break point’ there was no significant relationship with MOR observed for any of the CpG units. The horizontal-broken line represents the maximum MOR value for the control group with CGG<40, with this value indicated next to this line, with an exception of CpG6/7 where one control outlier at the MOR of 0.63 was not considered as the maximum control value. Note: PM/FM mosaic individuals and FM methylation mosaics as determined by methylation-sensitive Southern blot were not included.
Table 4.
Relationship between FREE2 methylation in FM females and age (predictor) by fitting segmented linear regression model of each CpG on age for the MOR of each CpG unit (plotted in Fig. 3)
| Outcome variable | n | Break point |
Break point Age (years) | Segmented linear regression |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | 95% CI | Coef (β) | SE | P-value | R2a | R2b | AICa | AICb | |||
| CpG 1 | 103 | 11.1 | 5.13–17.0 | <11.1 | −1.68 | 0.74 | 0.025 | 0.144 | 0.041 | −163.9 | −156.2 |
| ≥11.1 | 0.02 | 0.08 | 0.792 | ||||||||
| CpG 2 | 103 | 11.2 | 4.30–18.0 | <11.2 | −1.21 | 0.63 | 0.057 | 0.094 | 0.007 | −198.7 | −193.3 |
| ≥11.2 | 0.07 | 0.06 | 0.310 | ||||||||
| CpG 6/7 | 104 | 10.0 | 4.27–15.8 | <10.0 | −1.56 | 0.70 | 0.030 | 0.160 | 0.082 | −191.4 | −186.2 |
| ≥10.0 | −0.06 | 0.07 | 0.427 | ||||||||
| CpG 8/9 | 104 | 12.2 | 1.86–22.5 | <12.2 | −1.05 | 0.70 | 0.138 | 0.091 | 0.033 | −176.5 | −174.1 |
| ≥12.2 | 0.002 | 0.08 | 0.976 | ||||||||
| CpG 10–12 | 104 | 10.1 | 2.53–17.7 | <10.1 | −1.26 | 0.75 | 0.096 | 0.096 | 0.046 | −179.2 | −177.6 |
| ≥10.1 | −0.04 | 0.08 | 0.632 | ||||||||
P values less than 0.05 are highlighted in bold.
Coefficient (Coef) and standard error (SE) were multiplied by 100. Coefficient of determination (R2) and Akaike information criteria (AIC) were determined for asegmented linear regression models and bLinear regression models.
Comparison between different regression models showed that the segmented linear regression models had a higher R2 values and lower Akaike information criterion (AIC) values than linear regression models, and hence were considered as better fitted models (Table 4). Furthermore, we did not find relationships between CGG size and age to be significant in either FM males or females, suggesting that the differences in the MOR observed in FM groups are not due to somatic stability of the repeat size over time.
DISCUSSION
Relationship between FREE2 methylation and CGG size in males and females
Our results from this and previous studies (19) show that similar to Southern blot, FREE2 methylation can be used to differentiate hypermethylated FM from large unmethylated PM alleles, notably in both sexes. In this study, methylation of FMR1 exon 1 and intron 1 sequences in males showed a sharp increase in the MOR at ∼200 CGG repeats for all CpG units examined. This increase, however, was more gradual in females, occurring between 153 and 230 repeats and varying among different CpG units. Consistent with these findings, using methylation-sensitive Southern blot analysis, Rousseau et al. (20) have previously identified individuals with hypermethylated 176 repeat CGG alleles as well as 243 repeat unmethylated alleles and have thus suggested that methylation of expanded alleles occurs across a ‘borderline range’ rather than at a single size threshold. Furthermore, they have indicated that the CpG island methylation predicted the FXS phenotype more accurately than the size of the CGG expansion within this ‘borderline range’ (20). Our data show a PM/FM ‘borderline range’ (153–230 CGG repeats) in female samples that is most notable at CpG units 2 and 6/7, which are located on either side of the FMR1 exon 1/intron 1 boundary (Fig. 1A). Because this unusual PM/FM ‘borderline range’ is not found in males, it suggests association between abnormal methylation at the boundary and skewing of X inactivation in FMR1 expansion carrier females.
FREE2 methylation and skewed X-inactivation in FM females
Previous studies have shown the FMR1 locus to be methylated as part of X-inactivation and for the gene not to be expressed from the allele on the inactive X chromosome (21). Thus, healthy females who have CGG repeats within the normal range (<40 repeats) should have random methylation of either one of the FMR1 alleles through X-inactivation, with total FMR1 promoter methylation of ∼50%. For asymptomatic FM females who also express FMR1 at normal levels, the normal unmethylated FMR1 allele would be expected to be on the active X chromosome with the methylated FM allele on the inactive X for most of the cells. These females would also have total FMR1 promoter methylation of ∼50%. In contrast, for severely affected FM females with a completely silenced FMR1, the fully methylated allele would be expected to be on the active X and the normal size allele would be on the inactive X. For most of their cells, the total FMR1 promoter methylation would be expected to reach 100% since the normal allele would be also fully methylated due to X-inactivation. However, if X-inactivation was random, for most FXS affected FM females methylation would be expected to be 75%, with half of the cells having methylation equivalent to that of asymptomatic FM females (25% contribution to methylation), while the other half would have methylation equivalent to that of severely affected FM females (50% contribution to methylation). This would mean that there would be a normal distribution of methylation values for FM females, with a mean of 75% and with lower and higher tails of the distribution approaching 50 and 100%, respectively.
However, our results show that, in contrast to FM males, for hypermethylated FM females none of the FREE2 CpG units have a MOR approaching 1 (∼100% methylation) (Fig. 2). For FM females, the MOR values of the FREE2 sites which are methylated above the normal range are skewed towards the 40% methylation mark (the normal methylation range in a cohort of 368 females). In contrast, the distribution of abnormally methylated alleles in FM males is skewed towards 100% methylation. This suggests that X-inactivation is not random in FM females and that there is always some residual FMR1 activity, which may explain why FM females with FXS are generally less severely affected than FM males (20).
The main limitation of the current study is that while we report significant skewing towards the unmethylated state in the blood of FM females, it is not clear to what degree FREE2 methylation is or is not skewed in the neurons that are relevant to the underlying pathology of FXS. While this should be explored in future studies, the main line of evidence that already suggests that these findings are relevant to what is happening in the brains of FM females is our recent observation of a significant association between FREE2 methylation in the blood and the type and severity of cognitive impairment in a pilot cohort of 18 FM females (11).
The proposed influence of the FM allele on random X-chromosome inactivation in multiple cell types is consistent with the earlier proposal by Laird (22). Mechanistically, this might be explained by selection for FMRP in a proportion of cells (20,23). The FMRP selection hypothesis is supported by the recently described role of FMRP in cell-cycle control during development, cell proliferation (24), inhibition of apoptosis, particularly in neurones (25) and increase in the number FMRP positive cells over time in long-term cultures of PM/FM mosaic fibroblasts (23).
The primary role of FMRP in neurons is in maintenance of synaptic function (26). This is clearly not its role in the blood as FM males with severe neurodevelopmental changes and silenced FMRP expression show no obvious impairment in blood cell function (27). An alternative hypothesis could be that FM alleles in a proportion of cells promote ‘escape from methylation’ related to X-inactivation in a cell type-independent manner. This could be effected through the expression of ASFMR1/FMR4 long-non-coding RNA at the locus (28,29). This second postulate is supported by previous studies describing ‘escape from X-inactivation’ which was specific to X chromosome loci expressing long-non-coding RNA (30).
Non-random X-inactivation and relationship with age and CGG size in FM females
A number of studies utilizing methylation-sensitive Southern blot analysis have also provided evidence suggesting that X-inactivation skewing towards the normal allele (cells with the expanded allele predominantly found on the inactive X) is significantly associated with age in FM, but not in PM females (27,31). Others have found skewing of 'X-chromosome inactivation towards the normal allele unmethylated state in higher end CGG size PM females (100–180 repeats) (32), but its relationship with age was not examined. We have found no significant relationship of FREE2 methylation with age in control and PM female groups, and no evidence for significant X-chromosome inactivation skewing associated with an increasing CGG size in the PM range in our current and previous studies (19). While there was also no relationship found with CGG size in the FM female group, we did find significant skewing towards the unmethylated state in CpG units most proximal to the exon 1/intron 1 boundary (the MOR values of the control range, Fig. 3). This skewing in FM females was most significant from birth to puberty, after which there was no relationship between methylation and age. It is also notable that these significant relationships with age in FM females were not observed for FREE2 CpG8/9 and 10–12, which are located within intron 1, most distant from the CGG expansion. Together these findings suggest that differences in methylation values increase with distance from the CGG expansion. The inference here is that in FM females, depending on the CpG sites analysed, prognostic value may change, particularly if these analyses are performed in early childhood.
Relationships between FREE2 methylation, age and CGG size in FM males
The age-dependent mechanism of epigenetic modification appears to be different in FM males compared with females. First, it is evident in FM males that methylation of 3 CpG units, representing methylation of 8 out of 12 CpG sites examined (Table 3), significantly increases steadily over the lifetime. This is consistent with our observation that at least half of all FM males with 100% methylated NruI restriction site at the CpG island (according to Southern blot) are partially methylated within FREE2 (Figs. 1 and 2). Since males with the 100% methylated CpG island should not express FMRP, the selection for FMRP positive cells that we suggest contributes to methylation skewing in FM females would not apply to males. Secondly, the direction of the correlation between age and FREE2 methylation is different between males and females. Thirdly, correlations of the CGG size beyond 200 repeats and FREE2 methylation are significant in males but not in females. Together these observations suggest that in FM males with the FMR1 promotor methylated <100%, the CGG expansion is associated with slow accumulation of methylation over time at the exon 1/intron 1 boundary. In contrast, there are no significant changes in methylation after puberty in FM females. Further functional studies are required to clarify if the point of epigenetic differentiation between FM males and females is the somatic contribution of X-inactivation over time.
In summary, while it is widely accepted that methylation analysis is of fundamental importance in diagnostic testing for FXS and other FMR1-related disorders, there are gaps in understanding the relationship between the changes in the epigenetic status and the expansion size on the larger portion of the FMR1 promoter beyond the FMR1 CpG island. Here, we examined the relationship between age, CGG size and methylation of the FREE2 sites at the FMR1 exon 1 and intron 1 boundary. By comparing the methylation of these sites in a large cohort of expansion carriers, we determined that the ‘borderline range’ of CGG triplet expansion that leads to FMR1 exon 1 and intron 1 hypermethylation is different in males and females. We have also shown that, in contrast to males, the FMR1 exon 1 and intron 1 DNA sequences downstream of the CGG expansion in FM females are never fully methylated, demonstrating that the methylation resulting from X-inactivation in FM females at this locus is not random. This observed preference for the unmethylated state in FM females was found to be most significant from birth to puberty, but only for the CpG units most proximal to the CGG expansion. In FM males, however, there was a steady significant increase in methylation of all CpG units over the life time, which may explain the more severe phenotype in FM males and the greater variability in the phenotype of FXS females. The methylation status of intron 1 CpG10-12 most significantly related to cognitive impairment in our earlier study (19), and most distant from the CGG expansion, did not change significantly with age in FM females. This suggests that the prognostic value of FXS methylation at different CpG sites is not uniform, and may need to be evaluated for each targeted biomarker site taking age-related changes into consideration.
MATERIALS AND METHODS
Participants
The patient cohort consisted of 487 individuals (119 males and 368 females), with samples collected from birth to 82 years of age in males, and birth to 80 years of age in females. Of these, 436 were expansion carriers and control blood DNA samples collected as part of FXS cascade testing and routine molecular microarray testing through Victorian Clinical Genetics Services (VCGS) and the Greenwood Genetic Center as described previously (33). All samples were de-identified before use in this study. Additional 23 PM and 28 FM participants were recruited through the VCGS and the M.I.N.D. Institute, University of California at Davis Medical Center, Sacramento, from families seen at the Fragile X Treatment and Research Center through a collaborative genotype–phenotype NICHD-funded study. The study was approved by the Royal Children's Hospital Human Research Ethics Committee, Victoria, Australia and by the Institutional Review Board of the University of California at Davis.
Molecular studies
Processing of DNA samples and the assessment of the size of CGG repeat from the extracted DNA was conducted using a fully validated PCR amplification assay (34,35). CGG repeat sizing and methylation of the FMR1 CpG island restriction sites of all samples >55 repeats was also performed using a methylation-sensitive Southern blot procedure with appropriate normal and abnormal controls, as previously described (36). Briefly, EcoRI and NruI digestion was performed on 7–9 μg of DNA. The FMR1 alleles were detected using the StB12.3 probe, labelled with Dig-11-dUTP by PCR (PCR Dig Synthesis kit; Roche Diagnostics). Southern blot methylation for the expanded FMR1 alleles was determined as previously described with alleles classified as either unmethylated, partially methylated or fully methylated (16,37). Alleles at CGG sizes >150 repeats that were methylated by Southern blot were classified as FM; alleles between 55 and 200 repeats that were unmethylated by Southern blot were classified as PM. FREE2 methylation was assessed in the same samples using the Sequenom EpiTYPER system, as previously described (17,35,36). FREE2 methylation analysis for each sample was performed in duplicate, giving two separate MORs, averaged to take account of combined technical variation resulting from bisulfite conversion, PCR and mass cleave reactions.
Data analysis
For both FM males and females, the smallest expanded CGG triplet allele was included when examining relationships between methylation and CGG size, as described previously (20). FM/PM mosaic individuals as classified by Southern blot were excluded from these analyses. Testing for normality of the MOR distribution was conducted using the Shaprico–Wilk test at the significance level of P = 0.05. Depending on the results of this test, either the two-sample t-test for the means, if the data were normal, or non-parametric Mann–Whitney test for median, if the data were non-normal, was used.
Non-parametric regression using locally weight smoothing (LOWESS) was used to explore the natural relationship between each CpG MOR (outcome variable) and age. Initial analyses using this method suggested that for FM female data each of outcome variable has a non-linear relationship with age, with the initial linear inverse relationship which then leveled out. For FM male data the relationship was linear from birth to 82 years of age. Therefore, we fitted the robust linear regression models for FM males and segmented linear regression models to FM females. The later method allows for different slopes for each interval of age, while the former method down-weighing the effect of outlier observations. The goodness of fit of a model was assessed using the coefficient of determination (R2), ranging from 0 to 1, and Akaike information criteria (AIC). An R2 value approaching 1, and lower AIC indicated a better predictor. We used these measures to compare the performance of linear and segmented linear regression models for FM female data.
All analyses were conducted using the publicly available R statistical computing package (R Development Core Team 2012. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, ISBN 3-900051-07-0. URL: http://www.r-project.org/). Within R we used ‘segmented’ package to conduct segmented linear regression (Muggeo, V.M.R. 2008. segmented: an R Package to Fit Regression Models with Broken-Line Relationships. R News, 8/1, 20-25. URL: http://cran.r-project.org/doc/Rnews/).
FUNDING
This work was supported by the Victorian Government's Operational Infrastructure Support Program, NHMRC development grant (No 1017263 to H.R.S. and D.E.G.), E.W. Al Thrasher Award, USA (to H.R.S. and D.E.G.), Martin & E.H. Flack Trust, Australia (to H.R.S. and D.E.G.), National Institute of Child Health and Human Development grant, USA (HD36071 to D.Z.L. and R.J.H.) and in part by a grant from the South Carolina Department of Disabilities and Special Needs (SCDDSN). This study was also supported by NHMRC project grant (No 104299 to H.R.S. and D.E.G.).
ACKNOWLEDGEMENTS
We thank the study participants for their contribution and Dr Benjamin Ong from the Sequenom Platform Facility (MCRI). Dedicated to the memory of Ethan Francis Schwartz 1996–1998.
Conflict of Interest statement. D.E.G. is an inventor on a patent related to the technology described in this article. R.J.H. has received grant funding from Roche, Novartis, Seaside Therapeutics, Forest and Curemark for treatment studies in fragile X syndrome or autism. She has also consulted with Novartis regarding treatment of fragile X syndrome. The other authors declare that they have no conflicts of interest.
REFERENCES
- 1.Hagerman R.J., Berry-Kravis E., Kaufmann W.E., Ono M.Y., Tartaglia N., Lachiewicz A., Kronk R., Delahunty C., Hessl D., Visootsak J., et al. Advances in the treatment of fragile X syndrome. Pediatrics. 2009;123:378–390. doi: 10.1542/peds.2008-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagerman R.J., Hagerman R.J., Hagerman P. Fragile X Syndrome: Diagnosis, Treatment and Research. Baltimore: John Hopkins; 2002. pp. 3–109. [Google Scholar]
- 3.Pieretti M., Zhang F.P., Fu Y.H., Warren S.T., Oostra B.A., Caskey C.T., Nelson D.L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 4.Weiler I.J., Greenough W.T. Synaptic synthesis of the Fragile X protein: possible involvement in synapse maturation and elimination. Am. J. Med. Genet. 1999;83:248–252. doi: 10.1002/(sici)1096-8628(19990402)83:4<248::aid-ajmg3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Irwin S.A., Galvez R., Greenough W.T. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb. Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- 6.Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 7.Jacquemont S., Hagerman R.J., Leehey M.A., Hall D.A., Levine R.A., Brunberg J.A., Zhang L., Jardini T., Gane L.W., Harris S.W., et al. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291:460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]
- 8.Hunter J.E., Epstein M.P., Tinker S.W., Charen K.H., Sherman S.L. Fragile X-associated primary ovarian insufficiency: evidence for additional genetic contributions to severity. Genet. Epidemiol. 2008;32:553–559. doi: 10.1002/gepi.20329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farzin F., Perry H., Hessl D., Loesch D., Cohen J., Bacalman S., Gane L., Tassone F., Hagerman P., Hagerman R. Autism spectrum disorders and attention-deficit/hyperactivity disorder in boys with the fragile X premutation. J. Dev. Behav. Pediatr. 2006;27:S137–S144. doi: 10.1097/00004703-200604002-00012. [DOI] [PubMed] [Google Scholar]
- 10.Clifford S., Dissanayake C., Bui Q.M., Huggins R., Taylor A.K., Loesch D.Z. Autism spectrum phenotype in males and females with fragile X full mutation and premutation. J. Autism Dev. Disord. 2007;37:738–747. doi: 10.1007/s10803-006-0205-z. [DOI] [PubMed] [Google Scholar]
- 11.Tassone F., Beilina A., Carosi C., Albertosi S., Bagni C., Li L., Glover K., Bentley D., Hagerman P.J. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA. 2007;13:555–562. doi: 10.1261/rna.280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greco C.M., Hagerman R.J., Tassone F., Chudley A.E., Del Bigio M.R., Jacquemont S., Leehey M., Hagerman P.J. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- 13.Hagerman R.J., Leehey M., Heinrichs W., Tassone F., Wilson R., Hills J., Grigsby J., Gage B., Hagerman P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 14.Jacquemont S., Leehey M.A., Hagerman R.J., Beckett L.A., Hagerman P.J. Size bias of fragile X premutation alleles in late-onset movement disorders. J. Med. Genet. 2006;43:804–809. doi: 10.1136/jmg.2006.042374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddalena A., Richards C.S., McGinniss M.J., Brothman A., Desnick R.J., Grier R.E., Hirsch B., Jacky P., McDowell G.A., Popovich B., et al. Technical standards and guidelines for fragile X: the first of a series of disease-specific supplements to the Standards and Guidelines for Clinical Genetics Laboratories of the American College of Medical Genetics. Quality Assurance Subcommittee of the Laboratory Practice Committee. Genet. Med. 2001;3:200–205. doi: 10.1097/00125817-200105000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries B.B., Wiegers A.M., Smits A.P., Mohkamsing S., Duivenvoorden H.J., Fryns J.P., Curfs L.M., Halley D.J., Oostra B.A., van den Ouweland A.M., et al. Mental status of females with an FMR1 gene full mutation. Am. J. Hum. Genet. 1996;58:1025–1032. [PMC free article] [PubMed] [Google Scholar]
- 17.Godler D.E., Tassone F., Loesch D.Z., Taylor A.K., Gehling F., Hagerman R.J., Burgess T., Ganesamoorthy D., Hennerich D., Gordon L., et al. Methylation of novel markers of fragile X alleles is inversely correlated with FMRP expression and FMR1 activation ratio. Hum. Mol. Genet. 2010;19:1618–1632. doi: 10.1093/hmg/ddq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godler D.E., Slater H.R., Bui Q.M., Ono M., Gehling F., Francis D., Amor D.J., Hopper J.L., Hagerman R., Loesch D.Z. FMR1 intron 1 methylation predicts FMRP expression in blood of female carriers of expanded FMR1 alleles. J. Mol. Diagn. 2011;13:528–536. doi: 10.1016/j.jmoldx.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godler D.E., Slater H.R., Bui Q.M., Storey E., Ono M.Y., Gehling F., Inaba Y., Francis D., Hopper J.L., Kinsella G., et al. Fragile X mental retardation 1 (FMR1) intron 1 methylation in blood predicts verbal cognitive impairment in female carriers of expanded FMR1 alleles: evidence from a pilot study. Clin. Chem. 2012;58:590–598. doi: 10.1373/clinchem.2011.177626. [DOI] [PubMed] [Google Scholar]
- 20.Rousseau F., Heitz D., Tarleton J., MacPherson J., Malmgren H., Dahl N., Barnicoat A., Mathew C., Mornet E., Tejada I., et al. A multicenter study on genotype-phenotype correlations in the fragile X syndrome, using direct diagnosis with probe StB12.3: the first 2,253 cases. Am. J. Hum. Genet. 1994;55:225–237. [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchgessner C.U., Warren S.T., Willard H.F. X inactivation of the FMR1 fragile X mental retardation gene. J. Med. Genet. 1995;32:925–929. doi: 10.1136/jmg.32.12.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laird C.D. Proposed mechanism of inheritance and expression of the human fragile-X syndrome of mental retardation. Genetics. 1987;117:587–599. doi: 10.1093/genetics/117.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salat U., Bardoni B., Wohrle D., Steinbach P. Increase of FMRP expression, raised levels of FMR1 mRNA, and clonal selection in proliferating cells with unmethylated fragile X repeat expansions: a clue to the sex bias in the transmission of full mutations? J. Med. Genet. 2000;37:842–850. doi: 10.1136/jmg.37.11.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W., Jiang F., Bi X., Zhang Y.Q. Drosophila FMRP participates in the DNA damage response by regulating G2/M cell cycle checkpoint and apoptosis. Hum. Mol. Genet. 2012;21:4655–4668. doi: 10.1093/hmg/dds307. [DOI] [PubMed] [Google Scholar]
- 25.Jeon S.J., Han S.H., Yang S.I., Choi J.W., Kwon K.J., Park S.H., Kim H.Y., Cheong J.H., Ryu J.H., Ko K.H., et al. Positive feedback regulation of Akt-FMRP pathway protects neurons from cell death. J. Neurochem. 2012;123:226–238. doi: 10.1111/j.1471-4159.2012.07886.x. [DOI] [PubMed] [Google Scholar]
- 26.Fatemi S.H., Folsom T.D. The role of fragile X mental retardation protein in major mental disorders. Neuropharmacology. 2011;60:1221–1226. doi: 10.1016/j.neuropharm.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rousseau F., Heitz D., Oberle I., Mandel J.L. Selection in blood cells from female carriers of the fragile X syndrome: inverse correlation between age and proportion of active X chromosomes carrying the full mutation. J. Med. Genet. 1991;28:830–836. doi: 10.1136/jmg.28.12.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ladd P.D., Smith L.E., Rabaia N.A., Moore J.M., Georges S.A., Hansen R.S., Hagerman R.J., Tassone F., Tapscott S.J., Filippova G.N. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 29.Khalil A.M., Faghihi M.A., Modarresi F., Brothers S.P., Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS ONE. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinius B., Shi C., Hengshuo L., Sandhu K.S., Radomska K.J., Rosen G.D., Lu L., Kullander K., Williams R.W., Jazin E. Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics. 2010;11:614. doi: 10.1186/1471-2164-11-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor A.K., Safanda J.F., Fall M.Z., Quince C., Lang K.A., Hull C.E., Carpenter I., Staley L.W., Hagerman R.J. Molecular predictors of cognitive involvement in female carriers of fragile X syndrome. JAMA. 1994;271:507–514. [PubMed] [Google Scholar]
- 32.Garcia-Alegria E., Ibanez B., Minguez M., Poch M., Valiente A., Sanz-Parra A., Martinez-Bouzas C., Beristain E., Tejada M.I. Analysis of FMR1 gene expression in female premutation carriers using robust segmented linear regression models. RNA. 2007;13:756–762. doi: 10.1261/rna.206307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inaba Y., Herlihy A.S., Schwartz C.E., Skinner C., Bui Q.M., Cobb J., Shi E.Z., Francis D., Arvaj A., Amor A.J., et al. Fragile X Related Element 2 methylation analysis may provide a suitable option for inclusion of fragile X syndrome and/or sex chromosome aneuploidy into newborn screening: a technical validation study. Genet. Med. 2012 doi: 10.1038/gim.2012.134. doi:10.1038/gim.2012.134. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 34.Loesch D.Z., Godler D.E., Khaniani M., Gould E., Gehling F., Dissanayake C., Burgess T., Tassone F., Huggins R., Slater H., et al. Linking the FMR1 alleles with small CGG expansions with neurodevelopmental disorders: preliminary data suggest an involvement of epigenetic mechanisms. Am. J. Med. Genet. A. 2009;149A:2306–2310. doi: 10.1002/ajmg.a.32990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khaniani M.S., Kalitsis P., Burgess T., Slater H.R. An improved diagnostic PCR assay for identification of cryptic heterozygosity for CGG triplet repeat alleles in the fragile X gene (FMR1) Mol. Cytogenet. 2008;1:5. doi: 10.1186/1755-8166-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tassone F., Pan R., Amiri K., Taylor A.K., Hagerman P.J. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J. Mol. Diagn. 2008;10:43–49. doi: 10.2353/jmoldx.2008.070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tassone F., Hagerman R.J., Chamberlain W.D., Hagerman P.J. Transcription of the FMR1 gene in individuals with fragile X syndrome. Am. J. Med. Genet. 2000;97:195–203. doi: 10.1002/1096-8628(200023)97:3<195::AID-AJMG1037>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]