Abstract
Cytoplasmic accumulation and nuclear clearance of TDP-43 characterize familial and sporadic forms of amyotrophic lateral sclerosis and frontotemporal lobar degeneration, suggesting that either loss or gain of TDP-43 function, or both, cause disease formation. Here we have systematically compared loss- and gain-of-function of Drosophila TDP-43, TAR DNA Binding Protein Homolog (TBPH), in synaptic function and morphology, motor control, and age-related neuronal survival. Both loss and gain of TBPH severely affect development and result in premature lethality. TBPH dysfunction caused impaired synaptic transmission at the larval neuromuscular junction (NMJ) and in the adult. Tissue-specific knockdown together with electrophysiological recordings at the larval NMJ also revealed that alterations of TBPH function predominantly affect pre-synaptic efficacy, suggesting that impaired pre-synaptic transmission is one of the earliest events in TDP-43-related pathogenesis. Prolonged loss and gain of TBPH in adults resulted in synaptic defects and age-related, progressive degeneration of neurons involved in motor control. Toxic gain of TBPH did not downregulate or mislocalize its own expression, indicating that a dominant-negative effect leads to progressive neurodegeneration also seen with mutational inactivation of TBPH. Together these data suggest that dysfunction of Drosophila TDP-43 triggers a cascade of events leading to loss-of-function phenotypes whereby impaired synaptic transmission results in defective motor behavior and progressive deconstruction of neuronal connections, ultimately causing age-related neurodegeneration.
INTRODUCTION
Tar DNA binding protein of 43 kDa (TDP-43) has been identified as the major disease protein present in cytoplasmic inclusions in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). Full length TDP-43 protein as well as 25 kDa, C-terminal fragments were found to be hyper-phosphorylated and ubiquitinated in aggregates present in affected brain and spinal cord of ALS and FTLD cases (1,2). TDP-43 inclusions are found as secondary histopathological feature in other neurodegenerative disorders including Alzheimer's, Parkinson's and Huntington's disease, thereby defining a novel proteinopathy (3,4). Dominant missense mutations in the TARDBP gene have been identified in familial and sporadic cases of ALS and FTLD (5–8), indicating that TDP-43 dysfunction may cause disease formation.
TDP-43 encodes an evolutionarily conserved protein comprising a bipartite nuclear localization signal, two RNA recognition motifs (RRMs), a nuclear export signal, a glycine and glycine/glutamine/asparagine-rich C-terminal region. The C-terminus resembles a prion-like domain (9,10) predominantly mutated in TDP-43-related ALS and FTLD cases (11,12). In vitro studies showed that TDP-43 is involved in transcriptional regulation, RNA biogenesis and splicing (13). The C-terminal domain of TDP-43 regulates tissue-specific gene expression, transcriptional repression, and alternative splicing, and is essential for binding to heterogeneous nuclear ribonucleoproteins involved in microRNA and mRNA biogenesis, as well as RNA turnover (12,13).
The pathophysiological role of TDP-43 has been addressed in cell culture and animal models using mutation, alteration or expression of disease-specific variants of TDP-43 and its homologs in worms, flies, zebrafish, mice and rats (11,12,14). These studies revealed that post-translational modification (phosphorylation, ubiquitylation), truncation, mislocalization, nuclear clearance, cytoplasmic accumulation and defective autoregulation can all impact either directly or indirectly onto TDP-43 toxicity (12). The resulting phenotypes include lethality or reduced lifespan, defects in axogenesis and neurite branching, altered synaptic morphology, axon and neuron degeneration, astrogliosis, defective fat metabolism, impaired motor behavior and cognitive deficits (11,14). The available data suggest that TDP-43 toxicity is either directly or indirectly related to disease formation. So far, however, no consensus has emerged as to whether loss (i.e. nuclear clearance) or toxic gain (i.e. cytoplasmic accumulation) of TDP-43, or both, are causally related to disease onset and progression. Moreover, despite the fact that a large number of TDP-43 RNA targets have been identified (15–18), the initiating events and pathogenic pathways underlying TDP-43-mediated neurodegeneration remain to be identified.
Here we show that similar to its human homolog, Drosophila TDP-43, TAR DNA binding protein homolog (TBPH), is expressed in neurons, glia and muscle cells. Both loss of function and overexpression of TBPH result in decreased viability, impaired synaptic transmission, defective locomotion and age-related progressive neurodegeneration. Together these data suggest that both increased and decreased levels of TDP-43 initiate a loss-of-function phenotype, resulting in decreased neuronal viability and subsequent degenerative cell loss that characterize motor neuron diseases like ALS and FTLD.
RESULTS
Drosophila TDP-43, TBPH, is expressed in neurons, glia and muscle cells
BLAST search of the annotated Drosophila melanogaster genome identified two genes with sequence similarities to TARDBP/TDP-43, namely TBPH (19) and CG7804. Genomic sequence analysis revealed that the predicted six isoforms of TBPH are encoded by 5–6 exons, whereas the coding region of CG7804 is intronless with sequence similarity to exon 1–4 of TBPH. The differences in exon–intron structure and coding sequence similarities between CG7804 and TBPH suggest that CG7804 is paralogous to TBPH and likely to be a newly evolved retrogene that derived from TBPH rather than TARDBP (20). Moreover, in contrast to TBPH, CG7804 is not expressed in CNS and lacks a prion-like C-terminus that is predominantly mutated in ALS and FTLD (Supplementary Material, Fig. S1). We therefore focused on TBPH.
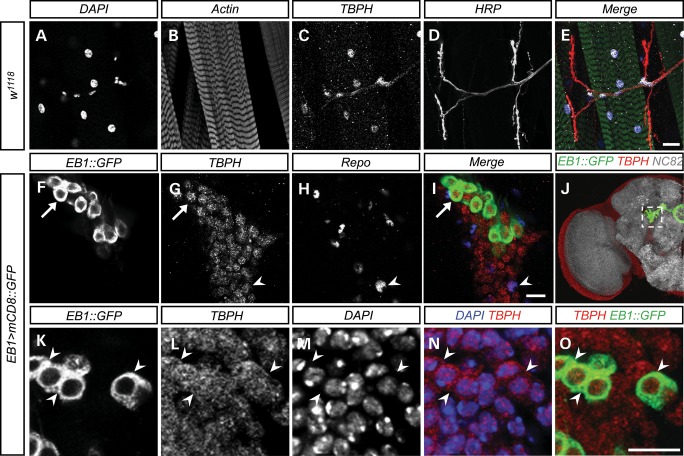
We generated polyclonal antibodies against TBPH-specific peptide sequences. Western blot analysis of control tissue identified a 58 kDa protein encoded by TBPH which was absent in deletions uncovering the genomic locus (Supplementary Material, Fig. S2); we did not detect any cleavage products, TBPH fragments, dimers nor band that indicates post-translational modifications (Supplementary Material, Fig. S2). Whole-mount immunocytochemistry revealed that, similar to human TDP-43, TBPH is expressed throughout development and adulthood in the nucleus of neurons, glia and muscle cells (Fig. 1). Perinuclear, cytoplasmic TBPH expression was detectable in post-mitotic neurons (Fig. 1L–O), although immunoreactivity was less intense than nuclear anti-TBPH labeling. These data suggest that similar to TDP-43, and predicted by nuclear localization and export signals (21,22), TBPH is predominantly localized to the nucleus but also shuttles into the cytoplasm.
Figure 1.
TBPH is expressed in neurons, glia and muscle cells. (A–E) Endogenous TBPH expression was examined at the neuromuscular junction of muscle group 6/7 of abdominal segment II in w1118 wandering third instar larvae. TBPH is expressed in the nucleus of muscle cells (C–E). Images are z-projections of a 10 μm area scanned in 2 μm steps. (F–J) Endogenous TBPH expression was examined in the adult brain of flies expressing membrane-bound GFP in ellipsoid body neurons of the central brain that are considered to be upper motor neurons [EB1>mCD8::GFP; CNS hemisphere shown in (J), the dotted white box corresponds to (F–I)]. TBPH is expressed in neurons (F and G, arrow) and glia (H, arrowhead) throughout the adult brain, including ellipsoid body neurons. Images are single z-slices taken in 1 μm steps. (K–O) TBPH expression is also seen in perinuclear regions of ellipsoid body neurons as shown by its co-localization with membrane bound mCD8::GFP (white arrowheads). Images are single z-sections taken in 1.5 μm steps. Scale bars; 50 μm (A–E); 10 μm (F–I, K–O).
Both loss and gain of TBPH affect survival and motor behavior
To study TDP-43 dysfunction in Drosophila, we generated specific loss- and gain-of-function alleles of TBPH. Imprecise P-element excision resulted in two independent deletions that removed the promoter region and start codon, or the promoter region, start codon, exons 1–3 and parts of exon 4 coding for the TBPH RRMs (Fig. 2A). RT–PCR and western blot analyses determined the absence of TBPH RNA and protein (Fig. 2B and C). For tissue-specific TBPH LOF, we generated two independent hairpin-loop UAS-RNAi lines that target exon 5 of all predicted TBPH isoforms (Supplementary Material, Fig. S3). BLAST search confirmed that our chosen target sequences do not detect off-targets, in contrast to other available TBPH RNAi lines, and western blot analysis revealed that Tub-Gal4 driven UAS-TBPH-RNAi is able to knock down endogenous TBPH protein expression below detection level (Supplementary Material, Fig. S3).
Figure 2.
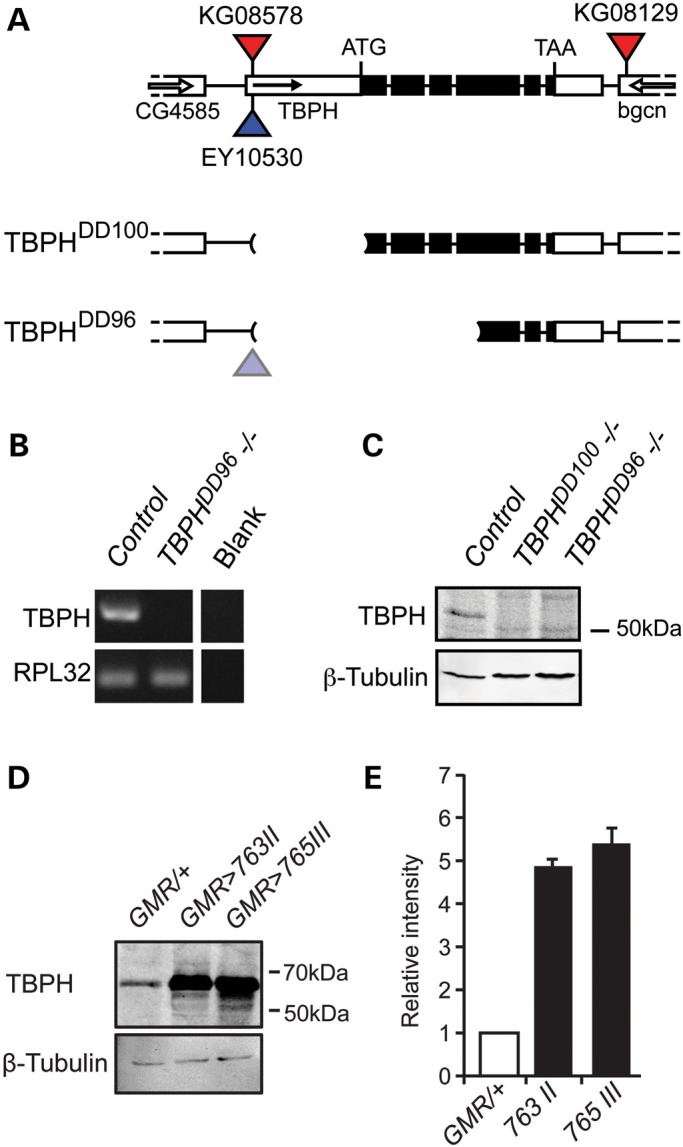
Loss- and gain-of-function alleles of the Drosophila TDP-43 homolog TBPH. (A–C) TBPH null mutant alleles and (D, E) UAS-TBPH overexpression alleles. (A) TBPH null mutant alleles (TBPHDD100 and TBPHDD96) were generated by imprecise P-element excision. Two types of deletion mutant were generated lacking distinct sections of the TBPH genomic region. The TBPHDD100 allele is lacking the promoter region of TBPH, including the start codon. The TBPHDD96 allele is lacking a larger amount of gDNA including the promoter region, the start codon and bases encoding the RNA recognition motifs (RRMs). (B) RT–PCR revealed lack of TBPH mRNA in homozygous TBPH mutants. Ribosomal protein L32 (RPL32) was used as loading control. Control genotype: w1118. (C) TBPHDD100 and TBPHDD96 mutant alleles are protein null, as revealed by the absence of the TBPH-specific 58 kDa protein band present in w1118 flies (control); shown is a western blot using protein extracts from six fly heads blotted with anti-TBPH antibody. (D) Western blot analysis of GMR-Gal4 driven UAS-TBPH shows excess TBPH protein expression in adult heads. Two different UAS-TBPH lines with insertion on different chromosomes (#763 on II, #765 on III) show comparable expression levels. (E) Quantification of GMR>TBPH shows a dramatic increase in TBPH protein levels. Western blot TBPH signal intensities were normalized against β-Tubulin. Mean and SEM are shown.
For gain of TBPH function (GOF), several independent UAS lines were generated with single inserts on different chromosomal locations. Expression efficacy was tested using eye-specific GMR-Gal4 activation. Western blot analysis revealed that GMR>TBPH showed increased amounts of TBPH protein (Fig. 2D) comparable among different insertion lines (Fig. 2E). Targeted misexpression resulted in excessive accumulation of TBPH both in the nucleus and cytoplasm (Supplementary Material, Fig. S4).
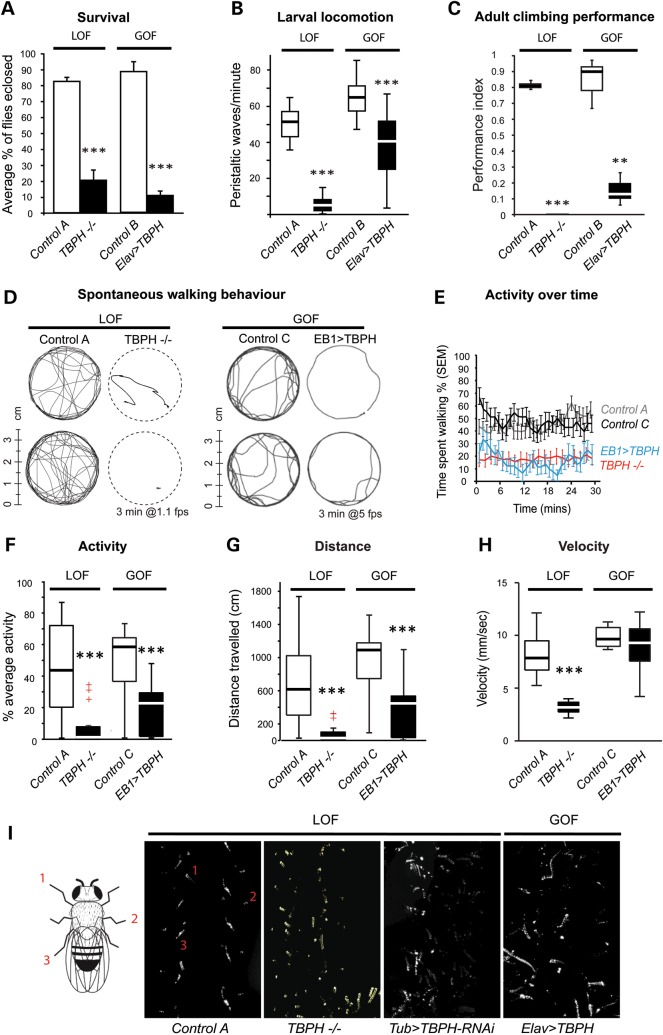
Analysis of zygotic TBPH LOF (TBPH−/−) and Gal4-mediated GOF mutants revealed that both affect development and lifespan: only 10–20% of progeny eclosed as adult while the majority of mutant cases died during late larval/pupal stages (Fig. 3A). Adult TBPH−/− mutant and pan-neuronal Gal4-specific ELAV>TBPH GOF flies were characterized by shortened lifespan, with TBPH−/− adults dying around day 7 and pan-neuronal Gal4-specific ELAV>TBPH flies dying around day 35, when compared with wild-type controls with an average lifespan of 80 days (23,24) (data not shown). Affected larvae exhibited impaired peristalsis and defective locomotion, which was more severe in TBPH−/− LOF mutants compared with pan-neuronal Gal4-specific ELAV>TBPH GOF (Fig. 3B). Adult TBPH−/− LOF mutant escapers and pan-neuronal Gal4-specific ELAV>TBPH GOF flies were characterized by inexistent or severely impaired innate escape and climbing behaviors (Fig. 3C).
Figure 3.
Both loss and gain of TBPH affect survival and motor behavior. (A) Survival analysis quantified the number of larvae that survived to adulthood. The number of fully eclosed adults was significantly reduced in loss-of-function (LOF) TBPH null mutants and gain-of-function (GOF) ELAV>TBPH flies, compared with respective control (w1118/+, control A; Elav/+, control B). (B) Both TBPH LOF and GOF show impaired larval locomotion with a reduction in the number of peristaltic waves per minute compared with controls (w1118/+, control A; Elav/+, control B). (C) TBPH LOF and GOF adults also show poor climbing performance in a startle-induced climbing assay (n = 15) compared with controls (w1118/+, control A; Elav/+, control B). (D–H) Video-assisted motion tracking of TBPHDD96−/− LOF and cell type-specific EB1>TBPH GOF targeted to upper motor neurons. (D) Representative walking tracks recorded over 3 min for TBPHDD96−/− and EB1>TBPH flies, together with their respective controls (w1118/+, control A; EB1/+, control C). (E–G) TBPHDD96−/− LOF and EB1>TBPH GOF flies show a reduction in walking activity over time, activity and total distance traveled compared with the respective controls (w1118/+, control A; EB1/+, control C). (H) TBPHDD96−/− null mutants show reduced walking velocity compared with controls, whereas EB1-specific overexpression of TBPH does not affect the mean velocity (w1118/+, control A; EB1/+, control C). (I) TBPHDD96−/− LOF, Tub>TBPH-RNAi downregulation and ELAV>TBPH GOF flies display disturbed tripod gait. Left cartoon shows tripod gait with left foreleg (1), right middle leg (2) and left hind leg (3). Control flies (w1118) walk in a stereotyped alternating tripod gait pattern. This pattern is disrupted in TBPHDD96−/− null mutants (TBPH−/−), ubiquitous RNAi-mediated TBPH knockdown flies (Tub>TBPH-RNAi) and pan-neuronal Elav-Gal4-mediated TBPH overexpression flies (ELAV>TBPH). Box-plots show median, upper and lower quartiles (box); whiskers contain data 1.5× the interquartile range; + indicates a data point within 3× the interquartile range (outliers). **P < 0.01; ***P < 0.001. Mean and SEM are shown (A, E).
To further investigate motor behavior, we used an open-field paradigm together with video-assisted motion tracking (23,24) (Fig. 3D) and compared adult TBPH−/− mutant LOF escapers with EB1-Gal4-mediated UAS-TBPH GOF flies targeting TBPH to ellipsoid body neurons of the central complex that are upper motor neurons due to their prominent role in the higher control of locomotion (25). Analysis of adult TBPH−/− mutant LOF escapers and EB1>TBPH GOF flies revealed severely reduced walking activity and distance traveled; motor activity remained low over time with only TBPH−/− flies walking slower (Fig. 3D–H). In addition, both LOF and GOF flies were characterized by severe gait abnormalities (Fig. 3I). These data suggest that TBPH dysfunction affects lifespan and motor behavior.
To confirm that the phenotypes observed in homozygous TBPH−/− LOF flies were indeed due to the absence of TBPH, we carried out a complementation test using a deficiency line uncovering the genomic region of chromosome 2R (Df(2R)106) including the TBPH locus, which revealed early larval lethality suggesting non-complementation. In addition and to rule out hidden second-site mutations, we first analyzed heteroallelic combinations using our TBPHDD96 and TBPHDD100 alleles together with a recently reported deletion (26), which also revealed non-complementation. Secondly, we generated a genomic TBPH construct covering the entire coding region of TBPH as well as 3′- and 5′- regions upstream of the TBPH locus (Supplementary Material, Fig. S5). To analyze its rescue potential, we integrated TBPHgenomic on 3R using the attP86Fb landing site and crossed it into the TBPHDD96 mutant background. Analysis of homozygous TBPHDD96−/− flies carrying one or two copies of TBPHgenomic revealed full rescue. Thus, development, larval locomotion, eclosion, adult climbing, walking and tripod gait phenotypes of homozygous TBPHDD96 mutants were all restored in TBPHDD96−/−; TBPHgenomic flies (Supplementary Material, Fig. S6). Together, these data suggest that the observed loss-of-function phenotypes are indeed due to the loss of TBPH, and hence our TBPH −/− mutants represent bona fide TBPH null alleles.
TBPH dysfunction does not affect synapse morphology but function
To determine the cause of TBPH-mediated behavioral phenotypes, we analyzed whether TBPH dysfunction results in muscle defects. Phalloidin staining of either larval L3 or adult day 2 TBPH−/− muscle preparations revealed a muscle pattern indistinguishable of wild-type or heterozygous TBPH+/− controls (data not shown). Next we investigated whether TBPH dysfunction may cause alterations of NMJ synapse structure and morphology, as previously suggested (27–30). We used a collection of synaptic markers (31) including Fasciclin II (FasII), involved in synapse development and plasticity; Bruchpilot (NC82), a component of the synaptic active zone of neurotransmitter release; synaptotagmin (Syt) involved in membrane traffic and calcium-dependent neurotransmitter release; Futsch, a pre-synaptic cytoskeletal protein that binds microtubles and regulates synaptic growth and dendritic morphology; and Neuroglian (Nrg), a cell surface, transmembrane adhesion molecule involved in motor neuron pathfinding. Larval NMJ preparations were double-labeled for synaptic proteins and horse radish peroxidase (HRP) that visualizes axons, dendrites and synaptic boutons in Drosophila. However, for all of the synaptic proteins examined, none were found to be mislocalized or absent in TBPH−/− LOF larvae compared with age-matched w1118 controls (Supplementary Material, Fig. S7). A quantitative analysis of HRP-labeled synaptic boutons and active zone puncta immunolabeled by anti-Bruchpilot, did not reveal any indication for alterations in the number of synaptic boutons or anti-Bruchpilot positive active zones in homozygous TBPHDD96−/− LOF larvae compared with age-matched w1118 controls (Supplementary Material, Fig. S8). These data suggest that the observed TBPH-related larval motor phenotypes are not caused by morphological defects, mislocalization or loss of selected proteins involved in synapse formation and maintenance.
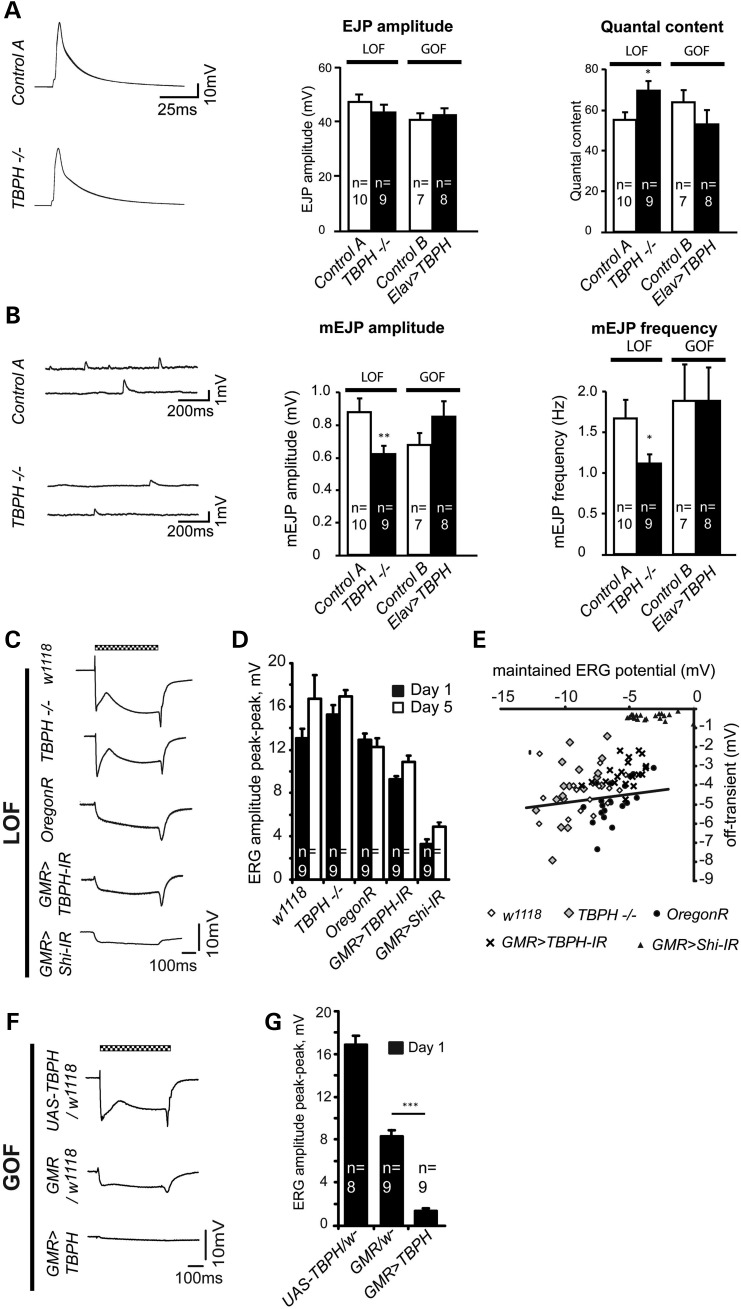
We then carried out electrophysiological recordings of cases and controls to address whether TBPH-mediated behavioral phenotypes might be caused by defects in synaptic efficacy and function. Electrophysiological recordings made at the NMJ of homozygous TBPHDD96−/− LOF and pan-neuronal Gal4-mediated ELAV>TBPH GOF L3 larvae revealed a decrease in both the amplitude and frequency of miniature excitatory junction potentials (mEJPs) in the homozygous loss-of-function background. Low-frequency (0.2 Hz) stimulation of the NMJ showed no difference in evoked excitatory junction potential (EJP) amplitudes. As a consequence of the changed mini amplitude but unaltered EJP amplitude there was an increase in the quantal content in TBPH LOF but not in ELAV>TBPH GOF (Fig. 4A and B). No differences were seen during high-frequency stimulation between cases and controls (data not shown).
Figure 4.
TBPH dysfunction affects synaptic efficacy. (A) Representative excitatory junction potential (EJP) traces are shown for LOF. EJP amplitudes for TBPHDD96−/− LOF and pan-neuronal ELAV>TBPH GOF flies are not significantly different from the respective controls (control A, w1118/+ and control B, Elav/+), however, quantal content is significantly increased in TBPH mutant larvae. (B) Representative traces of spontaneous neurotransmitter release (mEJP) shown for TBPHDD96−/− LOF. Both the mEJP amplitude and frequency are significantly reduced in TBPH mutant larvae. The mEJP is unaffected in ELAV>TBPH gain-of-function flies (w1118/+, control A; Elav/+, control B). (C) Representative ERG traces of the indicated TBPH LOF conditions. The white-eyed TBPHDD96−/− mutants and w1118 flies show a strong, habituating response, steady maintained response and off-transient. The red-eyed OregonR, GMR>Shibire-IR and GMR>TBPH-IR show a smaller amplitude response; GMR>Shibire-IR flies lack on- and off transients. Chequered bar indicates duration of light pulse. (D) Mean peak–peak amplitude of the ERG from the indicated conditions at day 1 and day 5 timepoints. (E) TBPHDD96−/− mutant and GMR>TBPH-IR have significantly smaller off-transients than the respective eye color-matched controls. The negative GMR>Shibire-IR controls, in which synaptic transmission is blocked, show a severely reduced off-transient. The solid line shows the regression line between off-transient and maintained response for wild-type flies. (F) Representative ERG traces of the indicated TBPH GOF conditions. The UAS-TBPH/+ and GMR/+ flies show a normal amplitude response, the smaller response seen in the GMR/+ flies is due to the darker red pigment. GMR>UAS-TBPH flies have a very small response, with no off/on transients visible. (G) Mean peak–peak amplitude of the ERG from the indicated GOF conditions at day 1 is significantly decreased in GMR>TBPH flies compared with the controls (D, E, n = 9; *P < 0.05, **P < 0.01, ***P < 0.001). Mean and SEM are shown (A, B, D, G).
We also recorded electroretinograms (ERGs) from adult TBPH LOF and GOF cases compared with appropriate controls. Both white-eyed TBPH−/− LOF and w1118 control revealed similar responses to blue light pulses with no differences in peak–peak amplitude of the ERG. Moreover, the ERG amplitude of red-eyed GMR>TBPH-RNAi and OregonR were indistinguishable, also when compared between 1- and 5-day-old flies (Fig. 4C and D). We then examined the off-transient and plotted their size as a function of the maintained photoreceptor response. On average, data points from both TBPH LOF strains, TBPH−/− and GMR>TBPH-RNAi flies, were above those from controls (Fig. 4E). We tested the null hypothesis that data points were distributed evenly above and below the control regression line between maintained ERG potential and off-transient, which revealed significant deviation for both TBPH−/− and GMR>TBPH-RNAi. Moreover, we compared the size of the off-transient recorded in both TBPH LOF lines with that estimated from the control regression line which revealed 15 and 25% reduction in the off-transient in TBPH−/− and GMR>TBPH-RNAi, respectively. As positive control, we blocked synaptic transmission in the retina using GMR>Shibire-RNAi, the Drosophila homolog of Dynamin involved in synaptic transmission (32), which resulted in completely abolished off-transient (Fig. 4C) with data points significantly above the wild-type regression line (Fig. 4E).
We also recorded ERGs from TBPH GOF flies where Drosophila TDP-43 was over-expressed in the eye. TBPH/w- controls showed an ERG in response to blue light pulses like wild-type, whereas GMR/w- flies showed a smaller response. In contrast, GMR>TBPH GOF flies showed a very small response, with no off/on transients visible (Fig. 4F and G), suggesting reduced synaptic signaling. Taken together these data suggest that both TBPH LOF and GOF affect synaptic efficacy and function, which is also supported by an impaired innate escape response (Fig. 3C and Supplementary Material, Fig. S9). The observed differential effects on synaptic efficacy and function in TBPH LOF and GOF suggest that Drosophila TDP-43 dysfunction either impairs post-synaptic efficacy or alters the size or loading of synaptic vesicles, which is consistent with the observed behavioral phenotypes of TBPH LOF and GOF (Fig. 3).
Pre-synaptic deficits are early phenotypes of dysfunctional TBPH
To gain further insights into TBPH-related synaptic transmission defects, we wondered whether TBPH dysfunction at the larval NMJ affects pre- or post-synaptic efficiency, or both. To address this question, we manipulated TBPH levels in a tissue-specific manner and focused on Gal4-mediated TBPH knockdown by using UAS-TBPH-RNAi in combination with UAS-Dicer-2 expression, which enhances knockdown efficiency (33). For pre-synapse-specific knockdown at the larval NMJ, we carried out pan-neuronal Gal4-mediated ELAV>Dcr2, TBPH-IR knockdown, and for post-synaptic TBPH knockdown, we used muscle-specific, Gal4-mediated BG57>Dcr2, TBPH-IR knockdown, in both cases utilizing reportedly strong and specific Gal4 drivers (34,35). Effects of pre- or post-synaptic TBPH-RNAi were analyzed using electrophysiological recordings at the L3 larval NMJ (Supplementary Material, Fig. S10).
Recordings at the NMJ of pan-neuronal Gal4-mediated ELAV>Dcr2, TBPH-IR knockdown larvae revealed a decrease in both the amplitude and frequency of mEJPs. Low frequency (0.2 Hz) stimulation of the NMJ showed no difference in evoked EJP amplitudes. As a consequence of the changed mini amplitude but unaltered EJP amplitude, we observed an increase in the quantal content in ELAV>Dcr2, TBPH-IR larvae (Supplementary Material, Fig. S10A and B). These data suggest that, similar to recordings at the NMJ of homozygous TBPHDD96−/− L3 loss-of-function larvae, pan-neuronal Gal4-mediated ELAV>Dcr2, TBPH-IR knockdown leads to impaired synaptic transmission (compare Supplementary Material, Fig. S10A and B with Fig. 4A, B).
In contrast to pre-synaptic knockdown of TBPH, recordings at the NMJ of muscle-specific Gal4-mediated BG57>Dcr2, TBPH-IR L3 knockdown larvae did not reveal any significant alterations in either amplitude or frequency of mEJPs, when compared with controls (Supplementary Material, Fig. S10C and D). Low frequency (0.2 Hz) stimulation of the NMJ revealed a small but significant difference in evoked EJP amplitudes. However, we did not detect any significant differences in quantal content in BG57>Dcr2, TBPH-IR L3 knockdown larvae (Supplementary Material, Fig. S10C and D). Together, these data demonstrate that altered TBPH function causes defective pre-synaptic transmission at the larval NMJ, suggesting that impaired pre-synaptic efficacy is one of the earliest events in the etiology of Drosophila TDP-43-related pathogenesis.
TBPH is essential for neuronal activity underlying motor behavior
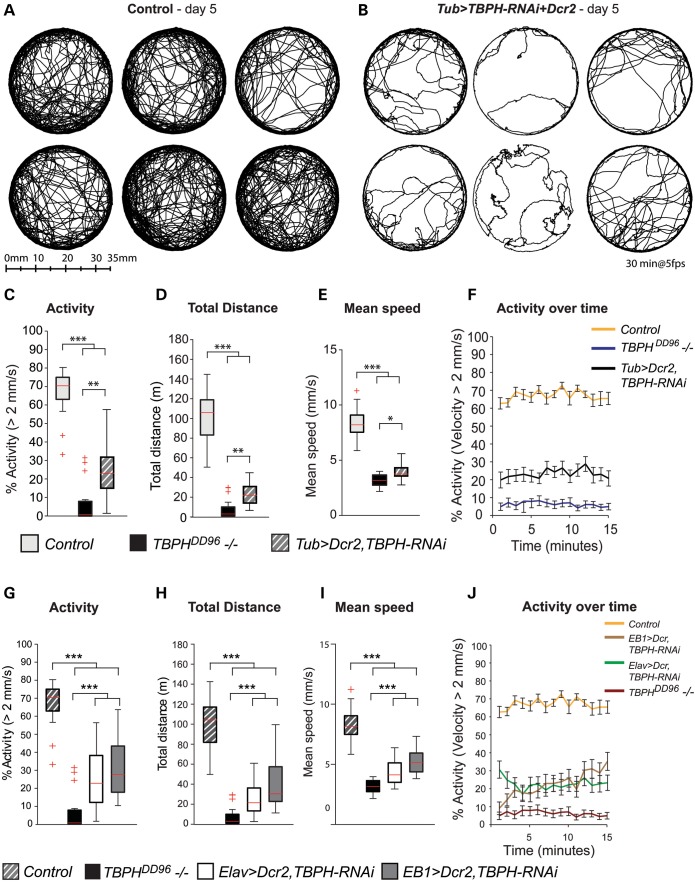
To further dissect TBPH dysfunction, we focused on cell-type-specific LOF and GOF and carried out TBPH-RNAi together with Dcr2 targeted by Tubulin-Gal4, which is ubiquitously and constitutively active during development, adulthood and aging (24), as well as pan-neuronal Elav-Gal4 and EB1-Gal4 specific to upper motor neurons. Tub>Dcr2, TBPH-IR mimicked TBPH mutations, causing larval/pupal lethality with few adult escapers (data not shown) and a disturbed tripod gait (Fig. 3I). To rule out that UAS-Dcr2 itself does cause a walking phenotype, we analyzed Tub>Drc2 flies which revealed no differences in walking activity, distance or activity over time when compared with controls (Supplementary Material, Fig. S11). Adult Tub>Dcr2, TBPH-IR flies displayed impaired motor behavior comparable to TBPHDD96−/− null mutants with severely reduced walking activity and distance traveled; their motor behavior remained limited over time with significantly reduced walking speed (Fig. 5), and walking was characterized by severe gait abnormalities (Fig. 3I). We observed similar motor phenotypes with Gal4-mediated UAS-TBPH-RNAi LOF targeted either to post-mitotic neurons (Elav>Dcr2, TBPH-IR) or to upper motor neurons (EB1>Dcr2, TBPH-IR) (Fig. 5). Moreover, analysis of open-field locomotion revealed that upper motor neuron-specific TBPH GOF (EB1>TBPH) and upper motor neuron-specific TBPH-RNAi knockdown (EB1>Dcr2, TBPH-IR), resulted in similar phenotypes characterized by severely impaired motor behavior (Figs 3E–H and 5). These data suggest that both tissue-specific loss and gain of Drosophila TDP-43 function impair neuronal activity underlying essential motor behavior.
Figure 5.
Similar to TBPH mutants, cell-type specific knockdown of TBPH leads to impaired motor behavior. Gal4-mediated UAS-TBPH-RNAi knockdown targeted either ubiquitously (Tub-Gal4) or to all neurons (Elav-Gal4), or targeted to upper motor neurons (EB1-Gal4). RNAi efficiency was enhanced by co-expressing UAS-Dcr2. (A–J) Motor behavior analysis using video assisted open-field motion tracking. (A) Representative walking tracks recorded over 30 min for 5-day-old Tub>Dcr2, TBPH-RNAi flies and heterozygous Dcr2,TBPH-RNAi controls. (C–F) Tracking analyses reveal that, comparable to TBPHDD96−/− mutants, Tub>Dcr2,TBPH-RNAi flies have a significantly reduced average activity, total distance travelled, mean speed and activity over time when compared with heterozygous Dcr2,TBPH-RNAi controls. (G–J) Dcr2, TBPH-RNAi expression was activated by either the pan-neuronal ELAV-Gal4 driver, or the EB1-Gal4 driver (ELAV>Dcr2,TBPH-RNAi or EB1>Dcr2,TBPH-RNAi, respectively). Tracking analyses reveal that, comparable to TBPHDD96−/− mutants, the walking activity, total distance travelled, mean speed and activity over time of 5-day-old ELAV>Dcr2,TBPH-RNAi and EB1>Dcr2,TBPH-RNAi flies is significantly reduced when compared with that of the control (heterozygous Dcr2,TBPH-RNAi). Box-plots show median, upper and lower quartiles (box); whiskers contain data 1.5× the interquartile range; +indicates a data point within 3× the interquartile range (outliers). (n = 24; *P < 0.05, **P < 0.01, ***P < 0.001). Mean and SEM are shown (F, J).
TBPH dysfunction differentially affects synaptic integrity in an age-related manner
Next we investigated whether impaired synaptic activity caused by TBPH dysfunction may lead to synaptic alterations in aging flies. We therefore analyzed brains of aged flies with or without either UAS-TBPH-IR LOF or UAS-TBPH GOF targeted to EB1-Gal4-specific upper motor neurons. The EB1-Gal4 driver becomes active during late pupal stages and remains active during adulthood and aging in a population of roughly 80 upper motor neurons (ellipsoid body ring neurons). In contrast to severe motor phenotypes (see Figs 3E–H and 5G–J), dysfunction or loss of this small population of upper motor neurons does not significantly alter the lifespan of flies and hence is an ideal assay system to study Drosophila TDP-43 dysfunction at the cellular and synaptic level (see Figs 6 and 7). We used EB1-Gal4 driven UAS-TBPH-IR LOF or UAS-TBPH GOF and co-expressed a membrane-bound form of GFP (UAS-mCD8::GFP) that also visualizes axons and synaptic terminals, and co-labeled whole mount brains with anti-Fasciclin 2 (Fas2) that is also expressed at pre- and post-synaptic sites of the ellipsoid body ring neuropil (Supplementary Material, Fig. S12).
Figure 6.
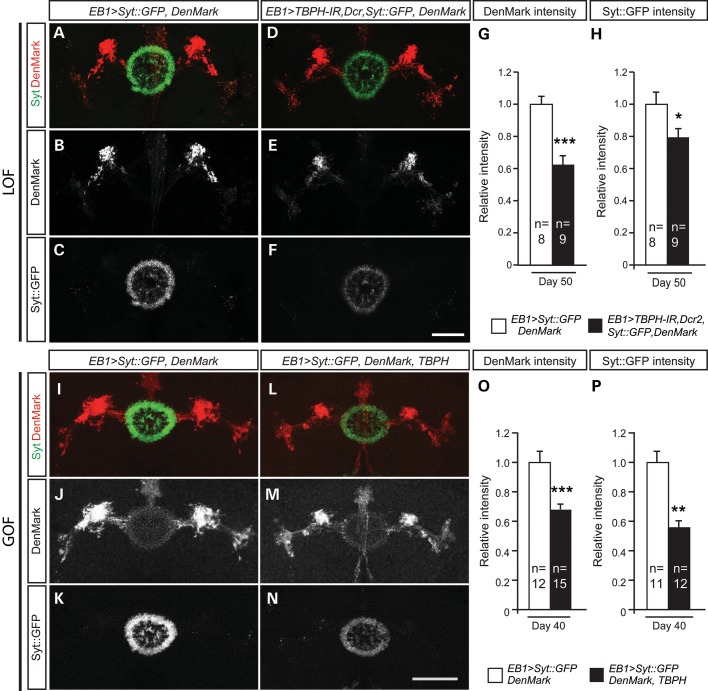
Both cell type-specific TBPH LOF and GOF cause trans-synaptic defects. (A) Expression of pre-synaptic marker Syt::GFP (green) and dendritic marker DenMark (red) in upper motor neurons of the adult central brain of 50-day-old flies using the ellipsoid body neuron-specific EB1-Gal4 driver (EB1>Syt::GFP, DenMark). Syt::GFP (green) expression is seen in the pre-synaptic region, whereas DenMark visualizes post-synaptic dendritic compartment. DenMark signal and Syt::GFP are indicated in single panels (B and C, respectively). (D–F) Driving TBPH-RNAi in EB1 neurons leads to downregulation of DenMark (E) and Syt::GFP (F). (G) DenMark signal is significantly reduced in TBPH LOF (EB1>Syt::GFP, DenMark, TBPH-IR). (H) TBPH knockdown significantly downregulates Syt::GFP. (I–K) DenMark (J) and Syt::GFP (K) expression in EB1 neurons of 40-day-old flies. (L and M) TBPH overexpression in EB1 neurons leads to downregulation of DenMark (M) and Syt::GFP (N). (O) DenMark signal is significantly reduced in TBPH GOF (EB1>Syt::GFP, DenMark, TBPH). (P) TBPH overexpression significantly downregulates Syt::GFP. *P < 0.05, **P < 0.01, ***P < 0.001. Mean and SEM are shown. Scale bar: 50 µm.
Figure 7.
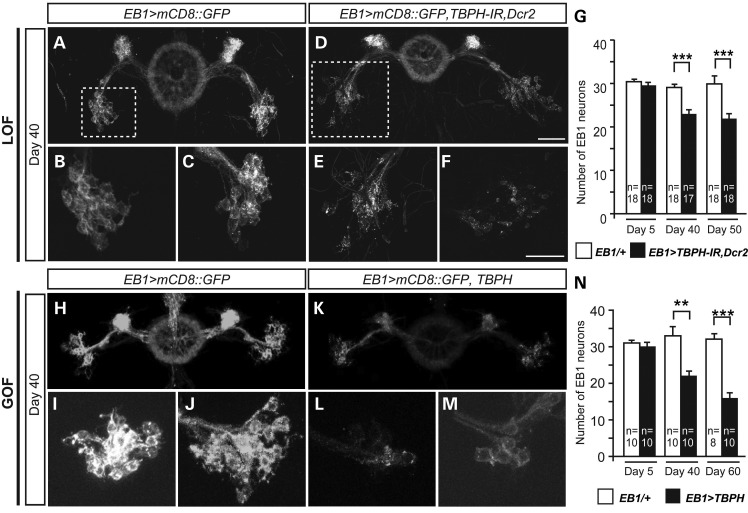
Upper motor neuron-specific RNAi-mediated downregulation and gain of TBPH causes loss of neuronal connections and age-related neurodegeneration. (A–C) Control flies with mCD8::GFP overexpression in upper motor neurons at day 40 (B, enlargement of dashed box in A; C, additional example). (D–F) TBPH LOF flies (EB1>mCD8::GFP, Dcr2, TBPH-IR) with TBPH-RNAi targeted to upper motor neurons at day 40. The GFP expression in the ellipsoid body structure does not diminish in the aged brain, however, dispersed cells with fragmented and granular GFP expression are detectable (E, enlargement of dashed box in D; F, additional example). (G) Quantification of EB1 neuron cell number (GFP positive) in control and TBPH LOF flies at day 5, 40 and 50. A significant decrease in neuron number is observed in aged TBPH LOF flies. (H–J) Control flies with mCD8::GFP overexpression in EB1 ring neurons at day 40 (I–J, enlarged example of cell bodies). (K–M) TBPH and mCD8::GFP overexpression in upper motor neurons leads to an age-related reduction in mCD8::GFP expression (L–M, enlarged example of cell bodies). (N) Quantification of EB1 cell numbers (GFP positive) in control and TBPH GOF flies at day 5, 40 and 60. A significant decrease in neuron number is observed over time in TBPH GOF flies. **P < 0.01, ***P < 0.001. Mean and SEM are indicated. Scale bar: 50 µm.
Analysis of brains of Gal4-mediated upper motor neuron-specific EB1>Dcr2, TBPH-IR at day 5, 20 and 40 did not reveal any significant differences at synaptic termini of the EB ring neuropil based on distribution, intensity, and shape of mCD8::GFP or Fas2 expression (Supplementary Material, Fig. S12A–J); however, by day 40, but not at day 5 or day 20, we consistently observed misshapen, fragmented and scattered cell bodies of EB ring neurons, together with a severe reduction in the number and fasciculation of axonal projections, when compared with age-matched controls (Supplementary Material, Fig. S12E–H, compare with Supplementary Material, Fig. S12A–D). In contrast, we observed strong reduction already by day 5 of mCD8::GFP labeling in cell body membranes, axons and synaptic termini of TBPH GOF brains (EB1>mCD8::GFP, TBPH), which was more pronounced by day 40; Fas2 immunoreactivity, however, appeared unaffected (Supplementary Material, Fig. S12K–T).
To determine in more detail whether the age-related synaptic alterations were either pre- or post-synaptic, we made use of the pre-synaptic marker synaptotagmin fused to GFP (UAS-Syt::GFP) in combination with the post-synaptic marker DenMark, a fusion protein of telencephalin and mCherry, which labels the somatodendritic compartment in Drosophila (36) (Fig. 6). Analysis of day 5, day 40 and day 50 adult EB1>Syt::GFP, DenMark controls revealed that expression of these markers alone did not cause obvious phenotypes (Fig. 6A–C, I–K, and data not shown). However, when co-expressed together with UAS-TBPH-IR and UAS-Dcr2 in upper motor neurons, Gal4-mediated upper motor neuron-specific EB1>Dcr2, TBPH-IR RNAi-mediated LOF specifically caused down-regulation of both pre-synaptic Syt::GFP and dendritic DenMark at day 50 (Fig. 6D–F). To rule out preparation or immunolabeling artifacts, independent replicates of fly brains were similarly processed and analyzed with identical confocal microscopy settings, which established significant reduction of reporter gene expression in pre-synaptic (Fig. 6H) and post-synaptic compartments when compared with age-matched controls (Fig. 6G). Similar analyses were carried out on day 5 and day 40 Gal4-mediated upper motor neuron-specific GOF flies, with adult EB1>Syt::GFP, DenMark, TBPH flies showing a down-regulation of both pre-synaptic Syt::GFP and dendritic DenMark at day 5 and day 40 (Fig. 6L–N) compared with EB1>Syt::GFP, DenMark controls (Fig. 6I–K). Independent replicates of brains were similarly analyzed and showed a significant reduction of reporter gene expression in pre-synaptic (Fig. 6P) and post-synaptic compartments when compared with age-matched controls (Fig. 6O and data not shown). Together these findings suggest that upper motor neuron-specific TBPH GOF and RNAi LOF affect synaptic integrity; in both cases, phenotypes occur in a progressive and age-related manner, and are associated with severely impaired motor behavior (Figs 3D–H and 5).
TBPH dysfunction results in age-related and progressive neurodegeneration
Synaptic deficits and subsequent neurodegeneration are considered to be hallmarks of ALS and other neurodegenerative diseases (37,38). We therefore investigated whether TBPH LOF and GOF can lead to degenerative cell loss. To visualize and number targeted neurons, we analyzed brains of aged flies that co-express UAS-mCD8::GFP with or without either UAS-TBPH-IR LOF or UAS-TBPH GOF again targeted to EB1-Gal4 specific upper motor neurons (Fig. 7).
Analysis of EB1>mCD8::GFP, Dcr2, TBPH-IR brains at day 5 and 20 did not reveal any significant differences in neuron numbers; however, by day 40 and 50, we observed a significant reduction of targeted EB neurons (Fig. 7G), showing a worsening of the phenotype over time. For the remaining EB neurons, mCD8::GFP labeling revealed misshapen, fragmented and scattered cell bodies when compared with age-matched controls (Figs 7D–F, compare with 7A–C). A comparable but more severe degenerative phenotype was observed in TBPH GOF. Brains of 5-day-old EB1>mCD8::GFP, TBPH flies showed reduced mCD8::GFP expression in perikarya as well as axonal extensions and synaptic arborizations of ellipsoid body ring neurons. GFP expression was further reduced by day 40 (Fig. 7K–M, compare with 7H–J) and almost undetectable in day 60 old EB1>mCD8::GFP, TBPH flies when compared with age-matched controls. Analysis of independent replicates of brains of age-matched EB1>mCD8::GFP, TBPH and EB1> mCD8::GFP flies that were similarly processed and analyzed with identical confocal microscopy settings, established significant and progressive, age-related loss of upper motor neurons (Fig. 7N).
To independently confirm age-related and progressive loss of neurons, we studied pox neuro (poxn) expression, which is specific to ellipsoid body ring neurons (39). Notably, loss of poxn-expressing cells progressed between day 5 and day 40 in EB1>mCD8::GFP, TBPH flies (Supplementary Material, Fig. S13). In addition to the decrease and subsequent loss of mCD8::GFP and poxn expression, degenerative cell loss was confirmed by the specific and age-related decrease in the number of ellipsoid body ring neurons that expressed excessive amounts of TBPH in EB1>mCD8::GFP, TBPH flies (Supplementary Material, Fig. S4 and data not shown). TUNEL labeling of brains of aged EB1>mCD8::GFP, TBPH flies did not indicate increased apoptotic activity (Supplementary Material, Fig. S14), suggesting that progressive loss of upper motor neurons was not caused by programmed cell death. Together these data suggest that prolonged dysfunction of Drosophila TDP-43 causes age-related and progressive neurodegeneration.
Gain of TBPH does not downregulate or mislocalize endogenous TBPH
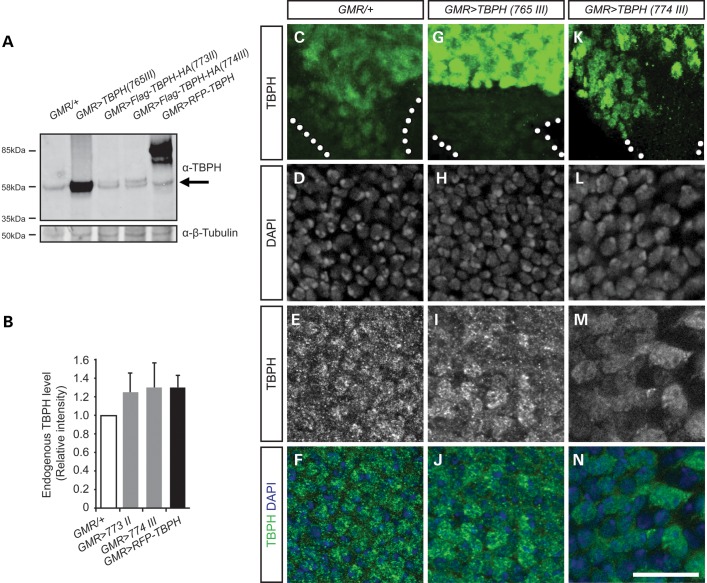
The phenotypic similarities between tissue-specific TBPH LOF and GOF may suggest that the observed synaptic deficits, impaired motor behavior and age-related neurodegeneration might be due to a common pathogenic mechanism. Previous in vitro and in vivo studies indicated that TDP-43 regulates its own expression by a negative feedback loop (17,40,41) that may participate in a feed-forward mechanism whereby cytoplasmic aggregation depletes nuclear TDP-43 function, essentially leading to a loss-of-function phenotype. We tested this hypothesis and over-expressed UAS constructs coding for tagged or untagged full length TBPH (Fig. 8). We utilized GMR-Gal4 and specifically targeted TBPH GOF to differentiating photoreceptor cells which resulted in a rough eye phenotype due to photoreceptor cell degeneration (42) (data not shown). Western blot analysis of heads of 1–3-day-old cases and controls identified a 58 kDa band in each condition similar to the control (Fig. 8A). Quantification of independent replicates confirmed that GMR-Gal4 specific TBPH GOF with C- and/or N-terminal tags did not alter endogenous levels of TBPH expression (Fig. 8B).
Figure 8.
GMR-Gal4-mediated gain of TBPH does not downregulate or mislocalize endogenous TBPH. (A) Western blot analysis of GMR-Gal4-driven untagged (58 kDa) or tagged UAS-TBPH (Flag-TBPH-HA, 60 kDa; RFP-TBPH, 85 kDa). Levels of endogenous TBPH (arrow at 58 kDa) are unaffected in day 1–3 flies overexpressing either form of TBPH. (B) Quantification of endogenous TBPH shows no alteration in TBPH protein levels. Endogenous TBPH signal intensities were normalized against β-Tubulin. Mean and SEM are indicated. (C, G, K) Third instar eye imaginal discs immunolabeled with anti-TBPH does not reveal TBPH-positive aggregates in cell bodies nor in the optic stalk (dotted area). (D–F, H–J, L–N) Third instar eye imaginal discs immunolabeled with anti-TBPH and DAPI identifies that nuclear expression of endogenous TBPH is maintained in gain of TBPH. Note that signal intensity in I, J, M and N is adjusted for overexpressed TBPH to avoid saturation of the signal. Scale bar: 10 um.
We then tested whether GMR-Gal4 specific TBPH GOF may lead to aggregate formation and/or deplete nuclear expression, hence affecting the function of endogenous TBPH without changing overall TBPH protein levels. Third instar eye imaginal discs were immunolabeled with anti-TBPH and anti-FLAG or anti-HA and counterstained with DAPI to determine potential aggregates and nuclear localization. In none of the cases examined did we detect aggregate formation, nor did we observe loss or alteration of nuclear TBPH expression (Fig. 8C–N and data not shown). We also tested whether GMR-Gal4-specific TBPH GOF may result in ubiquitinated TBPH and thus render the endogenous protein non-functional even though expression levels and nuclear localization appeared unaltered. However, immunolabeling with anti-FK2, which recognizes poly-ubiquitination, did not reveal any differences to controls (data not shown). Together these data suggest that gain of Drosophila TDP-43 function does not alter its own expression level or localization, although it results in severe synaptic, behavioral and neurodegenerative phenotypes.
DISCUSSION
Here we have systematically compared the phenotypic consequences of loss and gain of TDP-43 function in Drosophila. Our results demonstrate that deregulation of TBPH affects neuronal function and viability. In either case, deregulated TBPH initiates synaptic deficits and impaired motor behavior, ultimately causing age-related and progressive neurodegeneration. Our findings indicate that common loss-of-function phenotypes underlie TDP-43 dysfunction in Drosophila, which has implications for understanding TDP-43-mediated pathogenesis in ALS and FTLD.
Both loss and gain of TDP-43 trigger disease formation
TDP-43 related ALS and FTLD cases are characterized by both nuclear clearance and the cytoplasmic accumulation of full length, truncated and modified forms of TDP-43 (1). TDP-43 has been shown to autoregulate its own expression (17,41), and its ubiquitylation, phosphorylation, aggregation, truncation, mislocalization, nuclear clearance and defective autoregulation can contribute either directly or indirectly to disease formation (12). However, no consensus has emerged as to whether loss or toxic gain of TDP-43 function, or both, are causally related to disease onset and progression.
In our study, we systematically compared loss- and gain of TBPH, the Drosophila homolog of TDP-43, and analyzed their effect on synaptic function and morphology, motor control and age-related neuronal survival. Our results demonstrate that loss of function, RNAi-mediated downregualtion and overexpression of wild-type Drosophila TDP-43 are sufficient to cause age-related neurodegeneration. In the case of TDP-43 overexpression, we did not detect alterations in expression levels or mislocalization of the endogenous protein; we also found no evidence for ubiquitylation, aggregation, truncation or autoregulation, and based on western blot data, excessive phosphorylation appears unlikely. Our data thus argue that TDP-43 truncation, modification, altered autoregulation or aggregates are not a prerequisite for toxicity and age-related neurodegeneration, which is also supported by data in other model systems (28,41,43–46).
Previous studies showed that TDP-43 can form homodimers (47) and functions in multiprotein/RNA complexes (15,48,49) where multiple TDP-43 molecules are incorporated into each ribonucleoprotein complex (50). These data suggest that equilibrated, physiological levels of TDP-43 and a stoichiometric relationship with ribonucleoprotein complex components are essential for proper biogenesis, spatio-temporal expression and hence, regulation of TDP-43 target genes. Our results establish that deregulation of TDP-43 levels is a core event underlying disease formation, which can be triggered by either decreasing or increasing levels of TDP-43. This suggests that de-regulation of TDP-43 levels, by loss of function, downregulation or overexpression, alters the stoichiometry of participating components and, hence, ribonucleoprotein complex formation required for proper physiological function. In either condition, the net result of de-regulated TDP-43 is impaired physiological function which, when continuing, affects neuronal survival, ultimately causing progressive cell loss. In support of this notion, we determined synaptic deficits as one of the earliest phenotypes of TDP-43 dysfunction.
Pre-synaptic deficits are early, initiating events of TDP-43-related pathogenesis
In our study, we found that both loss and gain of Drosophila TDP-43 result in synaptic deficits and impaired motor behavior, followed by progressive, age-related neurodegeneration. In contrast to previous reports in Drosophila (27,28,29), we did not detect any obvious structural defects at the larval neuromuscular junction or mislocalization of synaptic proteins that might explain the observed behavioral phenotypes. This discrepancy might be attributable to differences in genetic background and/or methods used. Motor terminals are dynamic structures that show developmental and circadian fluctuations in shape and size (51). In our study, we used bona fide TBPH null alleles together with genomic rescue, as well as overexpression alleles. Instead of structural defects at the larval NMJ, our experiments reveal that affected flies are characterized by defective synaptic transmission, a phenotype detectable both at the larval NMJ and in the adult. In addition, our tissue-specific RNAi-mediated knockdown experiments identified pre-synaptic, rather than post-synaptic deficits as the earliest detectable phenotypes at the larval NMJ, suggesting that pre-synaptic deficits are initiating events in disease formation.
Our observations therefore allow for the first time the description of a sequence of events whereby TDP-43 dysfunction causes impaired synaptic transmission and motor abnormalities, followed by the subsequent loss of neuronal connections that precede degenerative cell death in an age-related and progressive manner. Such a sequence of events suggests that TDP-43 related pathogenesis progresses from the synapse and axon to the neuronal cell body, thereby resembling affinities to the ‘dying back’ phenomena, a hallmark of several neurodegenerative diseases (37). Comparable events have been, at least to some extent, reported for Wallerian degeneration (52), and recent genetic evidence indicates that neuronal ‘dying back’ underlies an active process of self-destruction distinct from apoptotic cell death (53). A ‘dying back’ process also characterizes human ALS patients and mouse superoxide dismutase 1 (SOD1) models of ALS (38,54). Like TDP-43 inclusions, SOD1 mutations are commonly identified in familial ALS (55). Although further, conclusive experiments are required, it is tempting to speculate that TDP-43 might not only regulate RNAs related to synaptic function (15,17), but potentially also negatively regulates the transcription or translation of target genes involved in the active self-destruction of neuronal connections.
TDP-43 dysfunction results in loss-of-function phenotypes
In addition to similarities between loss and gain of function phenotypes, we also observed phenotypic differences, including electrophysiology at the larval NMJ but also at the level of synaptic integrity and neurodegeneration in adult brain, where GOF typically caused more severe phenotypes than LOF. While some of these differences might be attributable to differing efficiencies of UAS/Gal4-mediated LOF and GOF, or a possible gain of toxic function, it is striking that both conditions lead to comparable, detrimental phenotypes with similar endpoint: age-related and progressive neurodegeneration.
It should be emphasized, however, that the similar phenotypic outcomes of loss and gain of Drosophila TDP-43 function, including age-related neurodegeneration, do not necessarily implicate similar mechanisms related to TDP-43 mediated pathogenesis. It is conceivable but remains to be shown that loss, downregulation or gain of TDP-43 function could trigger distinct pathogenic mechanisms that all converge on a functional node which when defective initiates disease formation. Previous studies suggest that such a functional node is likely the stochiometric activity of TDP-43 in ribonucleoprotein complexes that are essential for the regulation and correct processing of multiple target genes, including those related to synaptic function (15,17). This model is consistent with familial cases of ALS and FTLD, were TDP-43 is predominantly mutated in the C-terminal, prion-like domain (5–8) that regulates tissue-specific gene expression, transcriptional repression and alternative splicing, and is essential for binding to ribonucleoproteins involved in microRNA and mRNA biogenesis, as well as RNA turnover (9,10,12,13). According to this model, C-terminal, but also RRM mutations may hinder the participation of TDP-43 in ribonucleoprotein complex formation and hence, their function, leading to physiological deficits and eventually neurodegeneration.
In summary, our findings demonstrate that both loss and gain of TDP-43 function result in functional deficits whereby impaired synaptic transmission and defective motor behavior precede progressive deconstruction of neuronal connections, ultimately causing age-related neurodegeneration. Our data therefore provide in vivo evidence for common loss-of-function phenotypes underlying TDP-43 dysfunction that likely also characterize TDP-43-mediated ALS and FTLD.
MATERIALS AND METHODS
Fly stocks
Fly stocks were maintained at 25°C on standard cornmeal food, unless for aging experiments where aged flies were maintained on 15% sugar/yeast medium (23). The following strains were used: Oregon R (wild-type); w1118; w; TBPHnull/CyO,GFP; + (26) Df(2R)106/SM5; w; +; Elav-Gal4; GMR-Gal4; and Tub-Gal4; UAS-mCD8::GFP (56); UAS-Dcr2; UAS-Shibire-RNAi (Bloomington Stock Centre); p{Gawb}C57 (BG57, ref. 34); EB1-Gal4 (57); UAS-DenMark, UAS-Syt::GFP (36); UAS-RFP-TBPH (58).
Transgenic constructs
The TBPH gene was amplified by PCR from cDNA clone GH09868 (DGRC). PCR primers to amplify TBPH in pUAST vector are forward primer: 5′-GGAGATCTATGGATTTCGTTCAAGTGTCGG-3′; reverse primer: 5′-GGCTCGAGTTAAAGAAAGTTTGACTTCTCCGC-3′.
Primers used to generate Flag-TBPH-HA are: forward primer: 5′-GGAGATCTATGGACTACAAGGACGACGATGACAAGGATTTCGTTCAAGTGTCGG-3′; reverse primer: 5′-GGCTCGAGTTAGGCATAGTCTGGGACGTCATATGGATAAAGAAAGTTTGACTTCTCCGC-3′.
BglII and XhoI sites were used to insert TBPH into pUAST vector. DNA sequence was confirmed by sequencing. The UAS-TBPHYA765 line was used for all behavioral, electrophysiology and immunohistochemistry experiments. To specifically knockdown TBPH, the target region was chosen by E-RNAi web service (59). TBPH RNAi construction was carried out as previously described (33). PCR primers with EcoRI (forward) and XbaI (reverse) sites were designed to subclone TBPH RNAi construct into pMF3 vector (33).
TBPH-RNAi: forward primer: 5′-GGGAATTCCACTCATACCACCCACAGGG-3′; reverse primer: 5′-GGTCTAGAGTTATGCGGCTGGTTCATTC-3′.
Insertion of RNAi construct was confirmed by restriction enzyme digestion and sequencing. Plasmid injection and generation of transgenic flies were performed by BestGene Inc (CA, USA).
To generate a TBPH genomic rescue construct, we used TBPH genomic DNA from pacman bac library (CH322-116J04, bacpac.chori.org) covering position 19751906–19744713 on the second chromosome, which encodes the entire TBPH coding sequence. TBPHgenomic was integrated into the pUC 3GLA plasmid (gift from Matthias Soller) by recombination. TBPHgenomic in pUC 3GLA was used to generate transgenic flies by site-specific integration into the attp86Fb fly strain (60) and flies harboring TBPHgenomic pUC 3GLA were selected by compound eye-specific GFP expression.
P-element-mediated mutagenesis
The TBPHDD96 and TBPHDD100 null alleles were generated by imprecise excision of the EY10530 P-element, which is inserted in the promoter region of the TBPH locus. It was mobilized using HoP1 Δ2-3 transposase (gift from Iris Salecker). Four hundred and fifty excision lines were established from individual events and genomic deletions within the TBPH locus were identified by PCR and confirmed as protein null by western blot. TBPHDD96 and TBPHDD100 breakpoints are 19750093 … 19747817 and 19750093 … 19747925, respectively. The primers used to confirm the presence of neighboring gene CG4585 were: forward primer: 5′-CTGAGCATCTTCTGCGAC-3′; reverse primer: 5′-GAAATTAGCTCGCCATGG-3′.
Breakpoints and P-element insertion sites were sequenced using the following primers:TBPHDD100, forward primer: 5′-TTCCATGGCGAGCTAATT-3′; reverse primer: 5′- GTGTCCAGGTTGCGGTACTT-3′ TBPHDD96 forward primer: 5′-CGAGTTGCCGCCGTAAT-3′; reverse primer: 5′-CCTTTCACTCGCACTTAT-3′. Sequencing was carried out at MWG Eurofins.
Generation of TBPH antibody
The anti-TBPH antibody was generated by injecting two rabbits with peptide sequences PQGNHMNPGRNGHHR, corresponding to residues 291–305 and QSSGSQNAAEKSNFL, residues 517–531. Immunization was carried out by Eurogentec.
Immunohistochemistry
Adult and larval CNS dissection was carried out as previously described (23). Larval NMJ dissections were carried out according to established protocol (61). Primary antibodies used were: mouse anti-Repo (1:20); rat anti-Elav (1:30); mouse anti-Futsch (1:50); mouse 3C11 (1:100); mouse anti-synaptotagmin (1:25); mouse anti-FasII (1:10); mouse NC82 (anti-bruchpilot) (1:100) and mouse anti-Neuroglian (1:4) all obtained from the Developmental Studies Hybridoma Bank (DSHB) under the auspices of the NICHD and maintained by The University of Iowa; rabbit anti-TBPH antibody (1:3000); rabbit anti-Poxn (1:100; Adachi, Ludlow & Hirth, unpublished); mouse anti-polyubiquitin (FK2, 1:200; ENZO); rabbit anti-FLAG (1:500; Cell Signaling Tech); mouse anti-HA (1:40; Roche); goat anti-HRPCy3 (1:100; Stratech), 488-Phalloidin (1:1000; Invitrogen). Secondary antibodies were Alexa fluor 488, 568 and 647 (each 1:150; Invitrogen).
Image acquisition and analysis
Images were obtained either with Motic BA400 or Leica TCS SP5 confocal microscope with Leica Application Suite Advanced Fluorescence (LAS AF) version 2.0.2 software. For confocal images, channels were scanned sequentially. For comparative images, the same microscope settings were used for the control and experimental genotypes. For Figs 6 and 7, confocal z-stacks were rendered as 3-D projections. Signal quantification of digital images was carried out using Fiji (FIJI). Significance of GFP and FasII signal intensity was calculated using unpaired Student's t-test. Equal variance assumptions were based on Levene's test for equality of variance.
NMJ bouton count
Early L1 larvae were picked from 4 h egg collection plates (fruit agar) and placed on cornmeal food. At wandering L3, larvae were dissected according to established protocol (61). All preparations were dissected within a 1.5 h window to avoid differences in circadian rhythm effect on bouton number. Z-stacks were taken of the MN6/7-Ib motor neurons innervating muscle group 6/7 in segment A3. Boutons were counted by hand and the number of NC82 puncta were analyzed using the ITCN plug-in for ImageJ.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
Tissues of interest were fixed in 4% formaldehyde and subsequently permeabilized in 100 mm Citrate/0.1% Triton X-100 for 30 min at 65°C. Tissues were briefly washed with 0.5% Triton-X 100 in PBS and rinsed twice with TUNEL assay buffer (In Situ Cell Death Detection Kit; Roche). After incubating for 90 min at 37°C, TdT enzyme was added and further incubated for 3 h at 37°C. For positive control, freshly dissected tissue was soaked in 2N HCl for 30 min to induce apoptosis.
Western blots
Fly tissue was lysed in RIPA buffer (150 mm NaCl, 1% NP-40, 5 mm EDTA, 0.5% sodium, deoxycholate and 0.1% SDS, 50 mm Tris, pH8.0) containing complete proteinase inhibitor (Roche) and phosstop phosphatase inhibitor (Roche). Samples were centrifuged to take the RIPA-soluble fraction. SDS–PAGE was run in 8% SDS gel. Rabbit anti-TBPH antibody was used at 1:2000–3000 and anti-β-Tub (DSHB) was used at 1:300. Secondary antibodies were IRDye 800 conjugated goat anti-rabbit (1:10000, Rockland Immunochemicals) and Alexa Fluor 680 goat anti-mouse (1:10000, Invitrogen). Membrane images were acquired using Odyssey.
Reverse transcriptase PCR
RNA was extracted by homogenizing whole flies in Trizol, adding chloroform, incubating on ice for 15 min, followed by centrifugation at 16 000g at 4°C. The upper phase was added to isopropanol and incubated and spun as before. The pellet was washed in 70% ethanol and dissolved in nuclease-free H2O. RNA purity was measured using a Nanodrop. DNase treatment was carried out according to manufacturer's instructions (Ambion). cDNA was generated by mixing 1 µg RNA and random hexamers, heating to 70°C and adding 5 × M-MLV reaction buffer, M-MLV-RT, RNAsin and dNTPs according to manufacturer's instructions (Ambion). Samples were incubated at 37°C for 1 h and 70°C for 15 min. PCR products were run on 1% agarose gel. The primers used were as follows; TBPH: forward primer: 5′- ATCTTGGATGGCTCAGAACG-3′, reverse primer: 5′-GTCGGTCTTTATTCCGTTGG-3′ RPL32: forward primer: 5′-CGCCGCTTCAAGGGACAGTATC-3′, reverse primer: 5′-CGACAATCTCCTTGCGCTTCTT-3′.
Footprint/gait analysis
Flies were briefly anaesthetized with CO2 and one-third of the wings were removed to prevent escape. Flies were left to recover in a petri dish for 10–15 min before being tapped onto a soot-covered glass microscope slide (slides were held over a candle flame until covered in an even coating of soot). Footprints were visualized by lighting the slide from below and images were captured with a Q-capture camera.
Startle-induced negative geotaxis
Flies' climbing ability was assayed as described earlier (23). Equal variance assumptions were based on Levene's test for equality of variance.
Eclosion analysis
Embryos were collected on fruit agar plates in 5 h batches and left in a 25°C incubator until larvae had hatched. Fifty first instar larvae were placed in a vial and a tally made of all fully eclosed adult flies, mean values were calculated and plotted as a percentage of total number of larvae picked. Significance was calculated using unpaired t-test. This was carried out in triplicate for each genotype.
Larval motility
Thirty wandering third instar larvae were individually placed on a fruit agar plate and allowed to recover for 30 s. The number of peristaltic waves, travelling in either direction, was scored over 1 min. Significance was calculated using unpaired t-test (two-tailed). Equal variance assumptions were based on Levene's test for equality of variance.
Video-assisted motion tracking
Tracking arenas were modified six-well tissue culture plates (35 mm diameter wells) filled with silicon rubber (Sylguard) to leave a 3 mm space so that flies could walk freely but not hop or fly. 18–24 flies were briefly anaesthetized with CO2, placed in separate arenas and left to recover at 25°C for 45 min before being placed above an array of white LEDs within a temperature-controlled incubator. Tracking was carried out at 25°C. A black and white CCD camera (Hitachi, MP-M1A) positioned above the arenas was connected to a PC via an analog capture card (Integral Technologies, Flashbus MV Lite). Recordings were carried out during the same time slot. Recorded videos were converted to fly movie format using the motmot package (62) and loaded into Ctrax software (63) to analyze the positions of the flies throughout the video. Position data for the 30 min file was exported as a Matlab-compatable (Mathworks) matrix file. Errors in the tracking were fixed using Matlab (Mathworks) as well as FixErrors GUI (63), which is described in further detail at http://ctrax.sourceforge.net/fixerrors.html. Fixed trajectories were analyzed in Matlab using custom scripts (written by D.M.H). This analysis determined the mean velocity, mean activity, activity over time and mean cumulative distance traveled by the population of flies in the arena. Activity was defined as movement per frame above a velocity of 2 mm/s. Average activity was the percentage of frames where the fly was active (>2 mm/s velocity). Mean velocity was the average of velocities in each frame of the recording only when the fly was active. Box-plots were generated in Matlab where the boxes show median, upper and lower quartiles; whiskers contain data 1.5× the interquartile range; + indicates a data point within 3× the interquartile range. Significance was calculated using the Mann–Whitney U-test with a Bonferroni correction to account for multiple comparisons.
Escape response
The amplitude of the escape response was carried out as previously described (64). In summary, anaesthetized flies were fixed to a tungsten pin on the end of a cocktail stick using glue gum. The pin was placed so that it covered the thorax. Flies were left to recover for 20 min before being mounted over the flexible ergometer and lowered gently until the flies reached out and place their legs on the jump platform, bringing it up to a resting position. A stimulating electrode was placed in the eye and one in the cervical connective to stimulate the Giant Fiber System. A supra-threshold 0.8 mV was applied and as the flies jumped, the amount of beam displacement was measured.
Electrophysiology
Wandering third instar larvae were dissected in HL3 buffer (65) containing 1.8 mm CaCl2. Nerves innervating the body muscle walls were cut near the ventral ganglion and stimulated using a suction electrode and isolated pulse stimulator Digitimer DS2A (constant current modification), with a current double that needed to initiate a compound response. All recordings for TBPH LOF/GOF and appropriate controls were made intracellularly in muscle 6, abdominal segment 3, at ambient room temperature using microelectrodes filled with 3 M KCl that had tip resistances of 5–10 MΩ. In 1.8 mm extracellular Ca2+ resting membrane potentials were −61.1 ± 1.0 mV (n = 10) and −55.8 ± 2.5 mV (n = 9, P < 0.05) and input resistances were 10.1 ± 0.7 MΩ (n = 10) and 9.6 ± 0.4 MΩ (n = 9, P > 0.05) for w1118 and TBPH−/− larvae, respectively. Resting membrane potentials were −54.1 ± 1.4 mV (n = 7) and −52.0 ± 1.5 mV (n = 8, P = 0.0018) and input resistances were 5.9 ± 0.2 MΩ (n = 7) and 6.4 ± 0.3 MΩ (n = 8, P < 0.05) for ELAV/+ and ELAV>TBPH, respectively. Data, filtered at 1 kHz and digitized at 10 kHz, were acquired using an Axopatch 200B amplifier and a Digidata 1320A data acquisition board (Molecular Devices Data) were acquired using pClamp8.02 (Molecular Devices) and analyzed with Clampfit8.02 (Molecular Devices) or MiniAnalysis (Synaptosoft). EJP amplitude histograms were constructed by averaging 50 separate events stimulated at 0.2 Hz from an individual muscle cell and then calculating the mean response from at least six larvae per line. Spontaneous release events were recorded for 120 s without any electrical stimulation. All events were analyzed and mean mEJP amplitudes from individual larvae were averaged to generate the histograms. To obtain the frequency of spontaneous release events the number of events for each recording was divided by 120 and then averaged to generate histograms. Significance was calculated using unpaired Students t-test (two-tailed).
Electroretinograms
Female flies were aspirated into a pooter, and then gently blown into a truncated pipette tip. They were restrained with nail varnish (Creative Nail Design). No anaesthesia was used. Blunt glass pipettes, filled with simple Drosophila saline (130 mm NaCl, 4.7 mm KCl, 1.9 mm CaCl2 (66) were used as recording electrodes, placing one tip centrally on the surface of the eye and the other in the mouthparts. Flies were adapted to the dark for 2 min and then presented with a standard blue light stimulus from three Kingbright, KAF-5060PBESEEVGC light-emitting diodes (maximum emission wavelength 465 nm) situated 6 cm away from the eye. Light pulses were monitored with a BPX65 photodiode (Centronics) next to the head of the fly. In the dark, the photodiode current was 0.5 nA; during the stimuli 400 nA. Each fly was tested with at least three blue light stimuli, >10 s apart. Their responses were captured using Dasylab software (measX, Mönchengladbach, Stuttgart) and the average waveform calculated. At least nine flies of each genotype were tested at each timepoint. Significance was calculated using Bonferroni post hoc analysis.
Statistical analysis
Statistical analysis was carried out as previously described (24); for details of the statistical tests used, see Supplementary Material, Table S1.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the UK Medical Research Council (G-0802208 to I.M.R. and G-070149 to F.H.), the Royal Society (Hirth/2007/R2 to F.H.), the Motor Neurone Disease Association (Hirth/Oct07/6233 to F.H., A.A.-C., C.E.S. and Hirth/Mar12/6085 to F.H. and C.E.S.), Parkinson's UK (G-0714 to F.H.), Alzheimer's Research UK (ARUK-PhD2012-18 to F.H.) and the Fondation Thierry Latran (2/2011/DrosAls to F.H.). Funding to pay the Open Access publication charges for this article was provided by the UK Medical Research Council and the Fondation Thierry Latran.
Supplementary Material
ACKNOWLEDGEMENTS
We thank B. Hassan, T. Lee, I. Salecker, A. Voigt, D.C. Zarnescu, the Bloomington Stock Centre, the Vienna Drosophila RNAi Centre and the Developmental Studies Hybridoma Bank at the University of Iowa for stocks and reagents.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 2.Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 3.Forman M.S., Trojanowski J.Q., Lee V.M. TDP-43: a novel neurodegenerative proteinopathy. Curr. Opin. Neurobiol. 2007;17:548–555. doi: 10.1016/j.conb.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen-Plotkin A.S., Trojanowski J.Q., Lee V.M. TAR DNA-binding protein 43 in neurodegenerative disease. Nat. Rev. Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gitcho M.A., Baloh R.H., Chakraverty S., Mayo K., Norton J.B., Levitch D., Hatanpaa K.J., White C.L. 3rd, Bigio E.H., Caselli R., et al. TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabashi E., Valdmanis P.N., Dion P., Spiegelman D., McConkey B.J., Vande Velde C., Bouchard J.P., Lacomblez L., Pochigaeva K., Salachas F., et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 8.Yokoseki A., Shiga A., Tan C.F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A., et al. TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann. Neurol. 2008;63:538–542. doi: 10.1002/ana.21392. [DOI] [PubMed] [Google Scholar]
- 9.Ayala Y.M., Pantano S., D'Ambrogio A., Buratti E., Brindisi A., Marchetti C., Romano M., Baralle F.E. Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J. Mol. Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 10.Gitler A.D., Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion. 2011;5:179–187. doi: 10.4161/pri.5.3.17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Da Cruz S., Cleveland D.W. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr. Opin. Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee E.B., Lee V.M.Y., Trojanowski J.Q. Gains and losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 2012;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buratti E., Baralle F.E. The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biol. 2010;7:420–429. doi: 10.4161/rna.7.4.12205. [DOI] [PubMed] [Google Scholar]
- 14.Wegorzewska I., Baloh R.H. TDP-43 based animal models of neurodegeneration: new insights into ALS pathology and pathophysiology. Neurodegenerat. Dis. 2011;8:262–274. doi: 10.1159/000321547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sephton C.F., Cenik C., Kucukural A., Dammer E.B., Cenik B., Han Y., Dewey C.M., Roth F.P., Herz J., Peng J., et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A.L., Zupunski V., et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.C., Sun E., Wancewicz E., Mazur C., et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagier-Tourenne C., Polymenidou M., Hutt K.R., Vu A.Q., Baughn M., Huelga S.C., Clutario K.M., Ling S.C., Liang T.Y., Mazur C., et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukacsovich T., Asztalos Z., Juni N., Awano W., Yamamoto D. The Drosophila melanogaster 60A chromosomal division is extremely dense with functional genes: their sequences, genomic organization, and expression. Genomics. 1999;57:43–56. doi: 10.1006/geno.1999.5746. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Q., Zhang G., Zhang Y., Xu S., Zhao R., Zhan Z., Li X., Ding Y., Yang S., Wang W. On the origin and evolution of new genes in Drosophila. Genome Res. 2008;18:1446–1455. doi: 10.1101/gr.076588.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strong M.J., Volkening K., Hammond R., Yang W., Strong W., Leystra-Lantz C., Shoesmith C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Ayala Y.M., Zago P., D'Ambrogio A., Xu Y.F., Petrucelli L., Buratti E., Baralle F.E. Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 2008;121:3778–3785. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- 23.White K.E., Humphrey D.M., Hirth F. The dopaminergic system in the aging brain of Drosophila. Front. Neurosci. 2010;4:205–333. doi: 10.3389/fnins.2010.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humphrey D.M., Parsons R.B., Ludlow Z.N., Riemensperger T., Esposito G., Verstreken P., Jacobs H.T., Birman S., Hirth F. Alternative oxidase rescues mitochondria-mediated dopaminergic cell loss in Drosophila. Hum. Mol. Genet. 2012;21:2698–2712. doi: 10.1093/hmg/dds096. [DOI] [PubMed] [Google Scholar]
- 25.Strauss R. The central complex and the genetic dissection of locomotor behaviour. Curr. Opin. Neurobiol. 2002;12:633–638. doi: 10.1016/s0959-4388(02)00385-9. [DOI] [PubMed] [Google Scholar]
- 26.Fiesel F.C., Voigt A., Weber S.S., Van den Haute C., Waldenmaier A., Görner K., Walter M., Anderson M.L., Kern J.V., Rasse T.M., et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 2010;29:209–221. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feiguin F., Godena V.K., Romano G., D'Ambrogio A., Klima R., Baralle F.E. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 28.Lin M.J., Cheng C.W., Shen C.K. Neuronal function and dysfunction of Drosophila dTDP. PLoS One. 2011;6:e20371. doi: 10.1371/journal.pone.0020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godena V.K., Romano G., Romano M., Appocher C., Klima R., Buratti E., Baralle F.E., Feiguin F. TDP-43 regulates Drosophila neuromuscular junctions growth by modulating Futsch/MAP1B levels and synaptic microtubule organization. PLoS One. 2011;6:e17808. doi: 10.1371/journal.pone.0017808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J.W., Brent J.R., Tomlinson A., Shneider N.A., McCabe B.D. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J. Clin. Invest. 2011;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins C.A., DiAntonio A. Synaptic development: insights from Drosophila. Curr. Opin. Neurobiol. 2007;17:35–42. doi: 10.1016/j.conb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Kitamoto T. Conditional modification of behaviour in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 33.Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 34.Budnik V., Koh Y.H., Guan B., Hartmann B., Hough C., Woods D., Gorczyca M. Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron. 1996;17:627–640. doi: 10.1016/s0896-6273(00)80196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirth F. Drosophila melanogaster in the study of human neurodegeneration. CNS Neurol. Disord. Drug Targets. 2010;9:504–523. doi: 10.2174/187152710791556104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicolaï L.J., Ramaekers A., Raemaekers T., Drozdzecki A., Mauss A.S., Yan J., Landgraf M., Annaert W., Hassan B.A. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc. Natl Acad. Sci. USA. 2010;107:20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raff M.C., Whitmore A.V., Finn J.T. Axonal self-destruction and neurodegeneration. Science. 2002;296:868–871. doi: 10.1126/science.1068613. [DOI] [PubMed] [Google Scholar]
- 38.Fischer L.R., Culver D.G., Tennant P., Davis A.A., Wang M., Castellano-Sanchez A., Khan J., Polak M.A., Glass J.D. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Boll W., Noll M. The Drosophila Pox neuro gene: control of male courtship behaviour and fertility by a complete dissection of all enhancers. Development. 2002;129:5667–5681. doi: 10.1242/dev.00157. [DOI] [PubMed] [Google Scholar]
- 40.Ayala Y.M., De Conti L., Avendaño-Vázquez S.E., Dhir A., Romano M., D'Ambrogio A., Tollervey J., Ule J., Baralle M., Buratti E., et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Igaz L.M., Kwong L.K., Lee E.B., Chen-Plotkin A., Swanson E., Unger T., Malunda J., Xu Y., Winton M.J., Trojanowski J.Q., et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J. Clin. Invest. 2011;121:726–738. doi: 10.1172/JCI44867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritson G.P., Custer S.K., Freibaum B.D., Guinto J.B., Geffel D., Moore J., Tang W., Winton M.J., Neumann M., Trojanowski J.Q., et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J. Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miguel L., Frébourg T., Campion D., Lecourtois M. Both cytoplasmic and nuclear accumulations of the protein are neurotoxic in Drosophila models of TDP-43 proteinopathies. Neurobiol. Dis. 2010;41:398–406. doi: 10.1016/j.nbd.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 44.Guo W., Chen Y., Zhou X., Kar A., Ray P., Chen X., Rao E.J., Yang M., Ye H., Zhu L., et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat. Struct. Mol. Biol. 2011;18:822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y.F., Gendron T.F., Zhang Y.J., Lin W.L., D'Alton S., Sheng H., Casey M.C., Tong J., Knight J., Yu X., et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J. Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shan X., Chiang P.M., Price D.L., Wong P.C. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc. Natl Acad. Sci. USA. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuo P.H., Doudeva L.G., Wang Y.T., Shen C.K., Yuan H.S. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim S.H., Shanware N.P., Bowler M.J., Tibbetts R.S. Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J. Biol. Chem. 2010;285:34097–34105. doi: 10.1074/jbc.M110.154831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ling S.C., Albuquerque C.P., Han J.S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D.W. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl Acad. Sci. USA. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehnert K.I., Beramendi A., Elghazali F., Negro P., Kyriacou C.P., Cantera R. Circadian changes in Drosophila motor terminals. Dev. Neurobiol. 2007;67:415–421. doi: 10.1002/dneu.20332. [DOI] [PubMed] [Google Scholar]
- 52.Conforti L., Adalbert R., Coleman M.P. Neuronal death: where does the end begin. Trends Neurosci. 2007;30:159–166. doi: 10.1016/j.tins.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 53.Osterloh J.M., Yang J., Rooney T.M., Fox A.N., Adalbert R., Powell E.H., Sheehan A.E., Avery M.A., Hackett R., Logan M.A., et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azzouz M., Leclerc N., Gurney M., Warter J.M., Poindron P., Borg J. Progressive motor neuron impairment in an animal model of familial amyotrophic lateral sclerosis. Muscle Nerve. 1997;20:45–51. doi: 10.1002/(sici)1097-4598(199701)20:1<45::aid-mus6>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 55.Pasinelli P., Brown R.H. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y.Q., Rodesch C.K., Broadie K. Living synaptic vesicle marker: synaptotagmin-GFP. Genesis. 2002;34:142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- 57.Wang J., Zugates C.T., Liang I.H., Lee C.H., Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- 58.Estes P.S., Boehringer A., Zwick R., Tang J.E., Grigsby B., Zarnescu D.C. Wild-type and A315T mutant TDP-43 exert differential neurotoxicity in a Drosophila model of ALS. Hum. Mol. Genet. 2011;20:2308–2321. doi: 10.1093/hmg/ddr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horn T., Boutros M. E-RNAi: a web application for the multi-species design of RNAi reagents—2010 update. Nucl. Acids Res. 2010;38:W332–W339. doi: 10.1093/nar/gkq317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brent J.R., Werner K.M., McCabe B.D. Drosophila Larval NMJ Dissection. J. Vis. Exp. 2009;24:e1107. doi: 10.3791/1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Straw A.D., Dickinson M.H. Motmot, an open-source toolkit for realtime video acquisition and analysis. Source Code Biol. Med. 2009;4:5. doi: 10.1186/1751-0473-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Branson K., Robie A.A., Bender J.A., Perona P., Dickinson M.H. High-throughput ethomics in large groups of Drosophila. Nat. Methods. 2009;6:451–457. doi: 10.1038/nmeth.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elliott C.J.H., Sparrow J.C. In vivo measurement of muscle output in intact Drosophila. Methods. 2012;56:78–86. doi: 10.1016/j.ymeth.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 65.Stewart B.A., Atwood H.L., Renger J.J., Wang J., Wu C.F. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J. Comp. Physiol. A. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- 66.Heisenberg M. Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. J. Exp. Biol. 1971;55:85–100. doi: 10.1242/jeb.55.1.85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.