Abstract
Background and Purpose
Famitinib is a novel multi-targeted receptor tyrosine kinase inhibitor under development for cancer treatment. This study aims to characterize the metabolic and bioactivation pathways of famitinib.
Experimental Approach
The metabolites in human plasma, urine and feces were identified via ultra-high performance liquid chromatography-quadrupole-time of flight-mass spectrometry and confirmed using synthetic standards. Biotransformation and bioactivation mechanisms were investigated using microsomes, recombinant metabolic enzymes and hepatocytes.
Key Results
Famitinib was extensively metabolized after repeated oral administrations. Unchanged famitinib was the major circulating material, followed by N-desethylfaminitib (M3), whose steady-state exposure represented 7.2 to 7.5% that of the parent drug. Metabolites in the excreta were mainly from oxidative deamination (M1), N-desethylation (M3), oxidative defluorination (M7), indolylidene hydroxylation (M9-1 and M9-5) and secondary phase-II conjugations. CYP3A4/5 was the major contributor to M3 formation, CYP3A4/5 and aldehyde dehydrogenase to M1 formation and CYP1A1/2 to M7, M9-1 and M9-5 formations. Minor cysteine conjugates were observed in the plasma, urine and feces, implying the formation of reactive intermediate(s). In vitro microsomal studies proved that famitinib was bioactivated through epoxidation at indolylidene by CYP1A1/2 and spontaneously defluorinated rearrangement to afford a quinone-imine species. A correlation between famitinib hepatotoxicity and its bioactivation was observed in the primary human hepatocytes.
Conclusion and Implications
Famitinib is well absorbed and extensively metabolized in cancer patients. Multiple enzymes, mainly CYP3A4/5 and CYP1A1/2, are involved in famitinib metabolic clearance. The quinone-imine intermediate formed through bioactivation may be associated with famitinib hepatotoxicity. Co-administered CYP1A1/2 inducers or inhibitors may potentiate or suppress its hepatotoxicity.
Keywords: famitinib, metabolism, bioactivation, quinone-imine, hepatotoxicity
Introduction
Receptor tyrosine kinases (RTKs) are a family of cell surface growth factor receptors that are crucial in many cellular biological processes, such as growth, differentiation, migration and apoptosis. These enzymes act by controlling the initiation of signal transduction pathways. RTKs are not only primary regulators in normal cells but are also critically involved in the development and progression of cancers (Zwick et al., 2001; Gschwind et al., 2004). Therefore, RTKs are important therapeutic targets in anti-cancer drug design. Sunitinib, N-(2-diethylaminoethyl)-5-[(Z)-(5-fluoro-2-oxo-1H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carboxamide, marketed as Sutent® by Pfizer, is an oral, small-molecule, multi-targeted RTK inhibitor approved by the US Food and Drug Administration in 2006 for the treatment of renal cell carcinoma and imatinib-resistant gastrointestinal stromal tumour (Cabebe and Wakelee, 2006; Hartmann et al., 2009). Despite the impressive effectiveness of sunitinib treatment, several cases of liver failure and fatality were documented in clinical trials and post-marketing surveys. In 2010, a regulatory black box warning was released on sunitinib for its hepatotoxicity (Mueller et al., 2008; Eisen et al., 2012). In addition, sunitinib is associated with pulmonary toxicity, which has a low incidence rate but can be severe and sometimes be fatal (Goodman et al., 2007; Socinski et al., 2008).
Famitinib (SHR1020), 5-[2-(diethylamino)ethyl]-2-[(Z)-(5-fluoro-2-oxo-1H-indol-3-ylidene)methyl]-3-methyl-6,7-dihydro-1H-pyrrolo[3,2-c]pyridin-4(5H)-one, a structural analogue of sunitinib, is a novel and potent multi-targeted RTK inhibitor that is currently undergoing phase II clinical trials in China for the treatment of renal cell carcinoma, gastrointestinal stromal tumours, pancreatic cancer and nasopharyngeal carcinoma. Compared with sunitinib, famitinib exhibits superior inhibition activities against multiple RTKs, including stem-cell factor receptor, vascular endothelial growth factor receptor 2/3, platelet-derived growth factor receptor β, FMS-like tyrosine kinase-1/3 receptor and proto-oncogene tyrosine-protein kinase receptor RET, on various cell lines. Animal studies also demonstrated the higher potency of famitinib in tumour growth inhibition in human tumour xenograft models (Lou et al., unpubl. data). Current clinical trials showed that famitinib has anti-tumour effects similar to those of sunitinib but at a considerably lower therapeutic dose. Hepatotoxicity was also observed in patients after famitinib treatment (Zhou et al., 2011). This adverse reaction is of low incidence, develops several days after commencement of therapy and is thus considered as an idiosyncratic toxicity.
To understand fully the efficacy and safety of new drugs, the disposition and metabolism of the agent must be determined during the course of clinical development. Metabolites that present in humans can contribute to the pharmacological effect or cause undesired toxicity (Baillie et al., 2002). The pharmacokinetics, metabolism and disposition of famitinib in rats have been investigated (Chen et al., unpubl. data). After oral administration, famitinib is absorbed with a moderate bioavailability of 40.9% and is readily distributed throughout the body. The plasma levels of famitinib reach the peak at approximately 6.3 h post dose and then decrease with a long half-life of 9.5 h. Famitinib elimination is mainly governed by metabolism, with the metabolites secreted primarily in the feces. N-Desethylfaminitib, which is pharmacodynamically active but exhibits a lower potency than the parent drug, is the major metabolite identified in rats. Thiol conjugates were also detected, implying the tendency of famitinib to form a reactive intermediate. This behaviour intensified the concern on the safe use of this drug. These results in rats indicate an extensive metabolism and similar bioactivation of famitinib may also occur in humans. Therefore, the potential links between the hepatotoxicity of famitinib observed in clinical studies and its bioactivation process must be evaluated.
The objectives of the current study are (i) to characterize the metabolic pathways of famitinib in cancer patients at the steady state following repeated oral administration, (ii) to determine the pharmacokinetic parameters and primary elimination route of famitinib, (iii) to explore the underlying biotransformation and bioactivation mechanisms of famitinib using different in vitro systems and (iv) to conduct a preliminary investigation on the correlation between the formation of the reactive metabolite(s) of famitinib and the famitinib-induced cytotoxicity in primary human hepatocytes.
Methods
Chemicals
Famitinib l-malate capsules manufactured by Jiangsu Hengrui Medicine Co. Ltd. (Lianyungang, China) were used for clinical trials. The reference standards of famitinib (purity 98.8%), 5-[2-(diethylamino)ethyl]-2-[(Z)-(5-chloro-2-oxo-1H-indol-3-ylidene)methyl]-3-methyl-6,7-dihydro-1H-pyrrolo[3,2-c]pyridin-4(5H)-one (SHR115692, purity 98.6%), 5-[2-(ethylamino)ethyl]-2-[(Z)-(5-fluoro-2-oxo-1H-indol-3-ylidene)methyl]-3-methyl-6,7-dihydro-1H-pyrrolo[3,2-c]pyridin-4(5H)-one (SHR116637, purity 98.7%), 5-[2-(ethylamino)ethyl]-2-[(Z)-(5-hydroxy-2-oxo-1H-indol-3-ylidene)methyl]-3-methyl-6,7-dihydro-1H-pyrrolo[3,2-c]pyridin-4(5H)-one (SHR141371, purity 90.0%), 5-[2(diethylamino) ethyl]-2-[(Z)-(5-hydroxy-2-oxo-1H-indol-3-ylidene) methyl]-3-methyl-6,7-dihydro-1H-pyrrolo [3,2-c]pyridin-4(5H)-one (SHR141227, purity 84.3%), 5-[2-(diethylamino)ethyl]-2-[(5-fluoro-2-oxo-1H-indol-3-yl)methyl]-3-methyl-6,7-dihydro-1H-pyrrolo[3,2-c]pyridin-4(5H)-one (SHR 139088, purity 91.7%), 5-[2-(diethylamino)ethyl]-2-[(Z)-(5-fluoro-6-hydroxy-2-oxo-1H-indol-3-ylidene)methyl]-3-methyl-6,7-dihydro-1H-pyrrolo[3,2-c]pyridin-4(5H)-one (SHR141226, purity 75.0%) were synthesized by Jiangsu Hengrui Medicine Co. Ltd. The following chemicals were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO): glutathione (GSH), β-nicotinamide adenine dinucleotide 2′-phosphate reduced tetrasodium salt (NADPH), 1-aminobenzotriazole (ABT), α-naphthoflavone (α-NF), sulphaphenazole, ticlopidine, quinidine, chlormethiazole, ketoconazole (KET), 4-methylpyrazole, raloxifene, omepreazole, l-buthionine-sulphoximine (BSO), (−)borneol and leucine enkephalin. All other reagents and solvents were either analytical or HPLC grade.
Enzyme sources
Microsomal epoxide hydrolase (MEH) and recombinant human NAD(P)H: quinone oxidoreductase 1 (NQO1) were purchased from Sigma-Aldrich Chemical Co. Human liver microsomes (HLM), recombinant cytochrome P450 isozymes (CYP1A1, CYP1A2, CYP1B1, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5 and CYP4A11) and recombinant flavin mono-oxygenases (FMO1, FMO3 and FMO5) were purchased from BD Biosciences (Woburn, MA). Human intestinal microsomes (HIM), human pulmonary microsomes [HPM(NS), non-smoker; HPM(S), smoker], human renal microsomes (HRM) and cryopreserved primary human hepatocytes were obtained from XenoTech LLC (Lenexa, KS).
Human study and sample collection
The mass balance and pharmacokinetics of famitinib in patients were determined in the present open-label and single-centre study. The protocol was approved by the Ethics Committee of the Cancer Institute and Hospital, Chinese Academy of Medical Sciences. The study was conducted in compliance with Good Clinical Practice and the principles of the Declaration of Helsinki. All participants signed informed consent forms after being advised of the nature and risks associated with the study.
Fifteen subjects (eight male and seven female) with solid tumours (Supporting Information Table S1) were enrolled in the study. The mean age of the subjects was 48.7 years (range 39 to 64 years), and weight was 64 kg (range 40 to 79 kg). Each subject received repeated oral administration of 20 mg (group 1, eight subjects) or 27 mg (group 2, seven subjects) famitinib l-malate capsules q.d. for 28 consecutive days. Three subjects from each group participated in the mass balance study. Serial blood samples were collected and placed in heparinized tubes prior to famitinib administration; at 1, 2, 3, 4, 6, 8, 12 and 24 h post dose on days 1 and 28; and at pre-dose on days 6 to 14. Plasma was harvested via centrifugation. Fecal and urine samples were separately collected daily from days 15 to 17. At the end of each collection interval, the total urine volume and the total fecal weight were recorded. Each fecal sample was extracted with 1 L of ethanol, ultrasonicated and then centrifuged at 2000× g. All samples were stored at −20°C until analysis.
Metabolite profiling and identification
The pooled plasma samples at the trough (pre-dose on days 11 to 13) and peak (4 and 6 h post dose on day 28) steady-state levels from each group were separately prepared by mixing an equal volume of plasma sample across individuals. Two grand pools of urine samples (steady state on days 15 to 17) from groups 1 and 2 were separately prepared by combining volumes that are proportional to the total volume at each urine collection from all subjects. Two grand pools of fecal extract samples (steady state on days 15 to 17) from groups 1 and 2 were separately prepared by combining volumes that are proportional to the total volume of each fecal extract from all subjects. A 300 μL aliquot of each pooled sample was precipitated with acetonitrile (600 μL) and then centrifuged at 11 000× g. The supernate was collected, evaporated to dryness under a nitrogen stream at 40°C and then reconstituted in 100 μL water/acetonitrile (90:10, v/v) solution. A 10 μL aliquot of the reconstituted solution was injected into the ultra-high performance liquid chromatogram coupled with a quadrupole-time of flight-mass spectrometer (UHPLC/Q-TOF MS) system for analysis.
A 150 μL aliquot of pooled urine sample was incubated with 150 μL β-glucuronidase (2000 units of Type HA-4; Sigma-Aldrich) in 1 M citrate buffer solution (pH 5.0) at 37°C for 16 h. The samples were then prepared as described above.
Metabolite profiling and identification were performed on an Acquity UHPLC/Synapt Q-TOF MS (Waters Corp., Milford, MA) with an electrospray ionization source. Separation was achieved on an Acquity UPLC® HSS T3 column (100 mm × 2.1 mm i.d., 1.8 μm; Waters Corp.). The mobile phase was a mixture of 5 mM ammonium acetate solution with 0.1% formic acid (A) and acetonitrile (B). The gradient elution program consisted of 5% B held for 2 min, a 13 min linear gradient to 36% B and maintained for 1 min, a linear increase to 99% B for the succeeding 1 min and finally a decrease to 5% B to equilibrate the column. The column temperature was 40°C, and the flow rate was 0.45 mL·min−1. The UHPLC effluent was monitored via UV detection at 424 nm. MS detection was conducted in the positive electrospray ionization (ESI+) mode. The main operating parameters for the Q-TOF MS were set as follows: capillary voltage, 3000 V; cone voltage, 40 V; source temperature, 120°C; desolvation temperature, 350°C; collision gas, argon; desolvation gas (nitrogen) flow rate, 700 L·h−1; data acquisition range, m/z 80 to 1000; and data format, centroid. The lock mass solution was leucine enkephalin with a reference mass at m/z 556.2771. Data analysis and instrument control were performed using the MassLynx 4.1 software (Waters Corp.). Metabolite screening was performed using the MetaboLynx software, a subroutine of the MassLynx software, on the basis of accurate mass measurements. The structures of famitinib and its metabolites were elucidated via MSE fragmentation, in which two separate scan functions were programmed with independently low and high collision energies (CEs). Authentic standards, when available, were used to compare the chromatographic retention times and fragmentation patterns.
Determination of famitinib and N-desethylfamitinib (M3, SHR116637) concentrations in plasma, urine and feces
The concentrations of famitinib and M3 in plasma, urine and feces were determined via a validated LC-MS/MS method. A 50 μL aliquot of SHR115692 solution [internal standard (IS), 100 ng·mL−1 for plasma, 500 ng·mL−1 for urine and feces] and 300 μL of 0.1 mmol·L−1 NaOH solution were added to 200 μL of plasma, urine or fecal extract sample. The mixture was extracted with 3 mL ethylether-dichloromethane (3:2, v/v) via vortex mixing for 5 min, followed by centrifugation at 2000× g for 5 min. The upper organic layer was then transferred to another tube and evaporated to dryness at 40°C under a nitrogen stream. The residue was reconstituted in 200 μL of the mobile phase, and a 10 μL aliquot was injected into the LC−MS/MS system for analysis. The LC system consisted of two LC-20AD pumps and a SILHTA autosampler (Shimadzu, Kyoto, Japan). An Ultimate XB-C18 (150 mm × 4.6 mm i.d., 5 μm; Welch Materials, Ellicott, MD) with a SecurityGuard C18 column (4.0 mm × 4.6 mm i.d., 5 μm; Phenomenex, Torrance, CA) was used for the chromatographic analysis. A mixture of 5 mM ammonium acetate/acetonitrile/acetic acid (40:60:0.1, v/v/v) was used as the mobile phase at a flow rate of 0.6 mL·min−1. For the MS detection, an API 4000 triple-quadrupole mass spectrometer (Applied Biosystems, Concord, Ontario, Canada) and multiple reaction monitoring (m/z 411 → 338 for famitinib, m/z 383 → 338 for N-desethylfamitinib and m/z 427 → 354 for SHR115692) were applied in an ESI+ mode. The Analyst 1.4.1 software (Applied Biosystems) was used for data acquisition and processing. Calibration curves were constructed via a linear-weighed least-squares (1/×2) regression method. The standard curve ranges for both famitinib and N-desethylfamitinib were 0.050 to 100 ng·mL−1 in plasma, 1.00 to 2000 ng·mL−1 in urine and 30.0 to 10 000 ng·mL−1 in feces.
The pharmacokinetic parameters of famitinib and M3 were calculated via non-compartmental analysis using the WinNonlin 5.3 software (Pharsight Corp., Mountain View, CA, USA). The peak plasma concentration (Cmax), trough plasma concentration (Cmin) and the time of occurrence of Cmax (Tmax) were directly obtained from the plasma concentration–time profiles. The area under the concentration–time curve up to the last sampling time (AUC0–24h) was calculated using the linear trapezoidal method for ascending concentrations and the log trapezoidal method for descending concentrations over the dosing interval (24 h). The degree of fluctuation values (DF) were estimated as (Cmax − Cmin)/Cav × 100%, where Cav = AUC0–24h/24 h. The accumulation ratios (AR, day 28/day 1) for AUC0–24h were calculated for each subject.
Semi-quantification of famitinib metabolites in urine and feces
Generally, common metabolites such as dealkylated and hydroxylated metabolites share similar UV spectra with the parent compound. Thus, the UV absorbance can roughly reflect the relative quantities of metabolites. The excellent chromatographic resolution of UHPLC allows the separation of metabolites from one other, thereby enabling the semi-quantification of the major metabolites via a UHPLC-UV method. The famitinib metabolites in urine (M0-1, M1, M2, M3-1, M4, M5-2, M7, M9-1, M9-2, M11, M12-1, M15-1, M15-2, M16, M17-1, M17-3, M18, M20-2, M20-3 and M20-4) and feces (M0-1, M1, M2, M3-1, M5-1, M5-3, M7, M9-1, M9-2, M9-4, M9-7, M11, M13, M14-2, M15-1 and M15-2) were semi-quantified. The urine and fecal extract samples (300 μL) were processed via acetonitrile precipitation, evaporation and reconstitution. The chromatographic conditions for semi-quantification were the same as those in Metabolite identification and profiling. The concentration of each analyte was calculated by dividing the UV peak area of the analyte by that of famitinib and then multiplying the result by the concentration of famitinib determined by the LC-MS/MS method.
Microsomal incubations
Stock solutions of famitinib and 5-hydroxyl-defluorofamitinib (M7, SHR141227) were prepared in DMSO. A typical incubation mixture containing the substrate [famitinib (3 μM), M7 (100 nM or 1 μM)] was mixed with microsomes [HLM, HIM, HRM, HPM(NS), HPM(S), 1 mg·mL−1], magnesium chloride (5 mM), EDTA (0.1 mM) and PBS (100 mM, pH 7.4) to a final volume of 250 μL. The final DMSO concentration during the incubation was 0.1%. The mixture was pre-incubated at 37°C for 3 min and then initiated by adding NADPH (final concentration of 1 mM). The reaction was terminated after 60 min of incubation by adding two volumes of ice-cold acetonitrile. To trap the reactive metabolites, separate incubations were performed in the presence of GSH at a final concentration of 2 mM. Control samples without NADPH or the substrate were also prepared. Each incubation was performed in duplicate. The samples were centrifuged at 11 000× g. A 600 μL aliquot of the supernate was collected, evaporated to dryness under a nitrogen stream at 40°C and then reconstituted in 100 μL of a water/acetonitrile (90:10, v/v) solution. A 10 μL aliquot of the reconstituted solution was injected into the UHPLC/Q-TOF MS system for analysis.
To assess the relative contribution of the different microsomal enzyme systems to the metabolism of famitinib by HLM, different chemical inhibitors or enzymes were added into the incubation mixtures, including the non-specific CYP inhibitor ABT (1 mM), the specific CYP1A1/2 inhibitor α-NF (2 μM), CYP2C9 inhibitor sulphaphenazole (6 μM), CYP2B6/2C19 inhibitor ticlopidine (0.4 μM), CYP2D6 inhibitor quinidine (2 μM), CYP2E1 inhibitor chlormethiazole (0.1 μM), CYP3A4/5 inhibitor KET (1 μM), alcohol dehydrogenase inhibitor 4-methylpyrazole (10 μM), aldehyde dehydrogenase inhibitor disulphiram (10 μM), aldehyde oxidase inhibitor raloxifene (100 nM), MEH (1 mg·mL−1) and NQO1 (20 units·mL−1). Each incubation was performed in triplicate.
Recombinant human CYP and FMO incubations
The incubations were performed as previously described, except that the microsomes were replaced by recombinant cDNA-expressed CYP1A1, CYP1A2, CYP1B1, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5, CYP4A11, FMO1, FMO3 and FMO5. The concentrations of enzymes were 50 nM. Control samples without NADPH were also prepared. Each incubation was performed in duplicate. The MS peak area ratios of the detected metabolites to the IS in each incubation system were recorded to determine the contributions of the selected P450/FMO isozymes to the formation of the famitinib metabolites. The highest ratio of each metabolite detected in the incubations was normalized to 100%. The ratios of the specific metabolites in the other enzyme incubation samples were then expressed as percentages relative to this highest level.
Primary human hepatocyte metabolism and cytotoxicity
Cryopreserved primary human hepatocytes were recovered according to the manufacturer's protocol and then were seeded into 96-well collagen I-coated plates (BD Biosciences) at a density of 3 × 105 cells·mL−1. The cells were then cultured for 24 h prior to the addition of agents. Several inhibitors were used to determine the relationship between famitinib bioactivation and cytotoxicity. These inhibitors include the non-specific CYP inhibitor ABT (1 mM), specific CYP1A1/2 inhibitor α-NF (2 μM), GSH depletor BSO (100 μM) and glucuronidation inhibitor (−)borneol (500 μM). Each inhibitor was pre-incubated with the cells 30 min prior to the addition of the drug.
Famitinib (5, 10 and 15 μM) was then incubated with the treated cells at 37°C in a humidified incubator containing 95% air/5% CO2. Each incubation was performed in triplicate. After 24 h of incubation, the mediums from triplicate incubations were collected and pooled to improve the UPLC/Q-TOF MS sensitivity. A 250 μL aliquot of the pooled sample was precipitated with acetonitrile (with IS), evaporated to dryness and then reconstituted in 70 μL of a water/acetonitrile (90:10, v/v) solution. A 10 μL aliquot of the reconstituted solution was injected into the UHPLC/Q-TOF MS system for metabolite profiling.
Cell viability was determined using the Cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies, Gaithersburg, MD) assay. A 100 μL aliquot of serum-free medium containing 10% CCK-8 was added to each well. The plates were incubated at 37°C for 120 min. The absorbance was then measured at 450 nm. The absorbance of the treated cells was compared with that of the controls, whose cells were exposed only to the vehicle and thus were considered as 100% viable.
Statistical analysis
Data are presented as means ± SD values. Differences between means were tested by one-way anova with Dunnet's post hoc test using the Statistical Package for the Social Sciences (SPSS) version 20.0 software (SPSS Inc., Chicago, IL). Statistical significance was defined as P < 0.05. Graphic representations of the results were created using GraphPad Prism version 5.0 software (GraphPad Software Inc., San Diego, CA). In the hepatocyte metabolism experiment, the mediums from triplicate incubations were pooled; thus, the data obtained represent the mean of triplicate incubations. The difference in metabolite formation among different treatment groups was only assessed by direct comparison of the determined absolute values, but not by statistical significance tests.
Results
Metabolite profiling and identification in humans
The identities of the metabolites were elucidated based on their accurate mass, as well as on chromatographic and MS fragmentation comparisons with the parent compound and available reference standards. The MS fragmentation behaviour of the reference famitinib was first investigated. Famitinib exhibited a protonated molecule [M + H]+ at m/z 411.2181 in the positive ion mode. In the high CE spectrum (Figure 1A), famitinib gave a predominant ion at m/z 338.1303, which was formed by the cleavage of the alkyl C–N bond and the loss of the diethylamine moiety. Low-abundance product ions at m/z 296.1194, 294.1025 and 269.1069 were formed from the cleavage of the methylpyrrolo-dihydropyridone. Based on the MS fragmentation pattern, the structure of famitinib was divided into two parts: A and B (Figure 1B). The structures of the metabolites were tentatively characterized by determining the changes in the m/z-values of these two segments.
Figure 1.

Q-TOF mass spectrum of famitinib under high collision energy in the ESI+ mode (A) and tentative structures of the most informative fragment ions for famitinib (B). The structure of famitinib could be divided into two parts according to the fragment pattern.
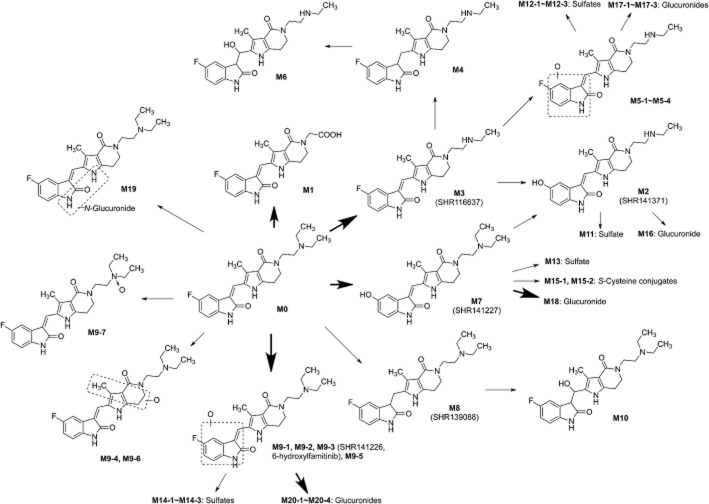
Figures 2 and 3 show a total of 25, 40 and 27 metabolites in human plasma, urine and feces after repeated oral administration of 27 mg famitinib l-malate capsules q.d. for 28 consecutive days respectively. Similar results have also been observed in 20 mg treatment group. These metabolites were designated as M0 to M20 according to their m/z-values. According to the UV chromatograms, M0 was the predominant component in plasma; whereas M3, M18 and M20-2 were relatively minor (<10%). The major components in urine were M0, M3, M7, M16, M17-1, M18, M20-2 and M20-4; whereas those in feces were M0, M0-1, M1, M3, M7, M9-1, M9-5 and M13. Detailed information on these metabolites is summarized in Table 1. The proposed metabolic pathways of famitinib in humans are depicted in Figure 4.
Figure 2.

Metabolic profiles of the pooled plasma samples at the (A) trough (pre-dose on days 11 to 13) and (B) peak (4 h and 6 h post dose on day 28) steady-state level of after oral administration of 27 mg famitinib once daily for 28 days to patients. Upper trace, UHPLC-UV chromatogram. Lower trace, MDF processed chromatogram. The inset is the expanded MDF processed chromatogram. The asterisk (*) indicates the E-isomer of M0.
Figure 3.

Metabolic profiles of the pooled (A) urine and (B) fecal samples after oral administration of 27 mg famitinib once daily for 28 days to patients at the steady-state level (on days 15 to 17). Upper trace, UHPLC-UV chromatogram. Lower trace, MDF-processed chromatogram. The inset is the expanded MDF-processed chromatogram. The asterisk (*) and diamond (♦) indicate the E-isomers of M0 and M3 respectively.
Table 1.
UPLC-Q/TOF MS data for famitinib metabolites detected in human plasma, urine and feces
| No. | Description | Rt (min) | Formula | Measured M + H+ | Error (in ppm) | Fragment ions | Matrices |
|---|---|---|---|---|---|---|---|
| M0 | Parent | 14.4 | C23H27FN4O2 | 411.2202 | 1.5 | 338.1298, 296.1176, 294.1030, 269.1067 | P, U, F |
| M1 | Oxidative deamination to carboxylic acid | 14.7 | C19H16FN3O4 | 370.1212 | 2.5 | 324.1148, 296.1219, 269.1069 | U, F |
| M2 | N-Desethylation + oxidative defluorination | 9.2 | C21H24N4O3 | 381.1911 | −4.1 | 336.1332, 294.1246, 292.1068, 267.1173 | P, U, F |
| M3 | N-Desethylation | 13.3 | C21H23FN4O2 | 383.1892 | 2.3 | 338.1287, 296.1203, 294.1045, 269.1071 | P, U, F |
| M4 | N-Desethylation + hydrogenation | 8.9 | C21H25FN4O2 | 385.2028 | −3.0 | 340.1447, 190.1067, 189.0993 | U, F |
| M5-1 | N-Desethylation + aryl hydroxylation | 9.9 | C21H23FN4O3 | 399.1838 | 1.5 | 354.1237, 312.1097, 310.0985, 285.1002 | U, F |
| M5-2 | N-Desethylation + aryl hydroxylation | 10.1 | C21H23FN4O3 | 399.1830 | −0.5 | 354.1231, 312.1088, 310.0924, 285.1074 | F |
| M5-3 | N-Desethylation + aryl hydroxylation | 10.5 | C21H23FN4O3 | 399.1841 | 2.2 | 354.1234, 312.1155, 310.1021, 285.1064 | P, U, F |
| M5-4 | N-Desethylation + aryl hydroxylation | 11.7 | C21H23FN4O3 | 399.1814 | −4.6 | 354.1243, 312.1100, 310.0994, 285.1034 | P, U, F |
| M6 | N-Desethylation + hydrogenation + aliphatic hydroxylation | 6.9 | C21H25FN4O3 | 401.1984 | −1.2 | 356.1405, 338.1302, 190.1079, 189.0992 | U |
| M7 | Oxidative defluorination | 10.0 | C23H28N4O3 | 409.2224 | −6.2 | 336.1344, 294.1231, 292.1082, 267.1140 | P, U, F |
| M8 | Hydrogenation | 10.0 | C23H29FN4O2 | 413.2344 | −2.1 | 340.1446, 190.1095, 189.1011 | U, F |
| M9-1 | Aryl hydroxylation | 10.8 | C23H27FN4O3 | 427.2141 | −1.0 | 354.1266, 312.1160, 310.1012, 285.0986 | P, U, F |
| M9-2 | Aryl hydroxylation | 11.0 | C23H27FN4O3 | 427.2144 | −0.3 | 354.1240, 312.1134, 310.1019, 285.1035 | F |
| M9-3 | Aryl hydroxylation | 11.6 | C23H27FN4O3 | 427.2152 | 1.6 | 354.1238, 312.1152, 310.0990, 285.1047 | P, U, F |
| M9-4 | Aliphatic hydroxylation | 12.3 | C23H27FN4O3 | 427.2149 | 0.9 | 354.1288, 336.1161, 310.0973, 292.0896, 267.0896 | U |
| M9-5 | Aryl hydroxylation | 12.8 | C23H27FN4O3 | 427.2145 | 0 | 354.1245, 312.1145, 310.0974, 285.1050 | P, U, F |
| M9-6 | Aliphatic hydroxylation | 13.3 | C23H27FN4O3 | 427.2128 | −4.0 | 354.1254, 336.1149, 310.0969, 292.0896 | P, U, F |
| M9-7 | N-Oxidation | 14.6 | C23H27FN4O3 | 427.2129 | −3.8 | 338.1301, 296.1216, 294.1033, 269.1101 | P, U, F |
| M10 | Hydrogenation + aliphatic hydroxylation | 8.1 | C23H29FN4O3 | 429.2299 | −0.6 | 356.1412, 338.1278, 190.1080, 189.0973 | U |
| M11 | N-Desethylation + oxidative defluorination + sulphation | 8.4 | C21H24N4O6S | 461.1483 | −2.5 | 416.0910, 336.1321, 294.1253, 292.1098, 267.1130 | P, U, F |
| M12-1 | N-Desethylation + aryl hydroxylation + sulphation | 9.1 | C21H23FN4O6S | 479.1407 | 1.4 | 434.0811, 354.1241, 312.1086, 310.0989, 285.1051 | P, U, F |
| M12-2 | N-Desethylation + aryl hydroxylation + sulphation | 9.5 | C21H23FN4O6S | 479.1376 | −5.1 | 434.0826, 354.1246, 312.1132, 310.0958, 285.1047 | U |
| M12-3 | N-Desethylation + aryl hydroxylation + sulphation | 10.8 | C21H23FN4O6S | 479.1395 | −1.1 | 434.0845, 354.1253, 312.1097, 310.0998, 285.0997 | U |
| M13 | Oxidative defluorination + sulphation | 9.0 | C23H28N4O6S | 489.1805 | −2.0 | 416.0910, 336.1328, 294.1257, 292.1104, 267.1116 | P, U, F |
| M14-1 | Aryl hydroxylation + sulphation | 8.8 | C23H27FN4O6S | 507.1754 | 8.0 | 354.1266 | U, F |
| M14-2 | Aryl hydroxylation + sulphation | 9.7 | C23H27FN4O6S | 507.1696 | −3.4 | 434.0819, 354.1228, 312.1140, 310.0984, 285.1038 | P, U, F |
| M14-3 | Aryl hydroxylation + sulphation | 10.4 | C23H27FN4O6S | 507.1713 | −0.1 | 434.0917, 354.1275, 312.1255, 310.0955, 285.1071 | U |
| M15-1 | Oxidative defluorination + cysteine conjugation | 8.3 | C26H33N5O5S | 528.2280 | -0.1 | 455.1397, 368.1074 | P, U, F |
| M15-2 | Oxidative defluorination + cysteine conjugation | 8.6 | C26H33N5O5S | 528.2296 | 3.0 | 455.1393, 368.1052 | U, F |
| M16 | N-Desethylation + oxidative defluorination + glucuronidation | 6.7 | C27H32N4O9 | 557.2245 | −0.4 | 512.1636, 381.1942, 336.1339 | P, U |
| M17-1 | N-Desethylation + aryl hydroxylation + glucuronidation | 7.2 | C27H31FN4O9 | 575.2128 | −4.4 | 530.1567, 354.1204, 285.1029 | P, U |
| M17-2 | N-Desethylation + aryl hydroxylation + glucuronidation | 7.6 | C27H31FN4O9 | 575.2131 | −3.8 | 530.1570, 354.1263, 285.1036 | U |
| M17-3 | N-Desethylation + aryl hydroxylation + glucuronidation | 8.8 | C27H31FN4O9 | 575.2158 | 0.9 | 530.1566, 354.1230, 285.0958 | P, U |
| M18 | Oxidative defluorination + glucuronidation | 7.3 | C29H36N4O9 | 585.2559 | −0.2 | 512.1672, 409.2227, 336.1313 | P, U |
| M19 | N-Glucuronidation | 9.7 | C29H35FN4O8 | 587.2515 | −0.3 | 514.1600, 338.1313 | P, U, F |
| M20-1 | Aryl hydroxylation + glucuronidation | 7.3 | C29H35FN4O9 | 603.2464 | −0.2 | 530.1556, 427.2138, 354.1244, 285.1056 | U |
| M20-2 | Aryl hydroxylation + glucuronidation | 7.8 | C29H35FN4O9 | 603.2469 | 0.5 | 530.1538, 427.2136, 354.1228, 285.1039 | P, U |
| M20-3 | Aryl hydroxylation + glucuronidation | 8.4 | C29H35FN4O9 | 603.2485 | 3.1 | 530.1555, 427.2127, 354.1253, 285.1082 | P, U |
| M20-4 | Aryl hydroxylation + glucuronidation | 9.7 | C29H35FN4O9 | 603.2463 | −0.5 | 530.1545, 427.2132, 354.1248, 285.1032 | P, U |
P, plasma; U, urine; F, feces.
Figure 4.
Proposed metabolic pathways of famitinib in humans. Famitinib is extensively metabolized in vivo by oxidative deamination and N-desethylation of the triethylamine moiety, oxidative defluorination and hydroxylation of the indolylidene moiety, combinations of above pathways and secondary phase-II conjugation.
M0
The extracted ion chromatograms of m/z 411.2196 showed two peaks at 10.4 and 14.4 min in plasma, urine and feces. Upon further fragmentation, these peaks exhibited product ions that are identical to those of the parent drug. The retention time of the latter peak was the same as that of the reference famitinib, indicating that this component was unmetabolized famitinib, designated as M0. The retention time of the earlier peak was the same as that of the degradation product of famitinib after UV exposure, suggesting that this peak represented the E-isomer of the exocyclic double bond of M0 that probably formed during sample handling. The light-driven E/Z isomerization is not uncommon and has been reported for many compounds that possess a C = C bond (Tamai and Miyasaka, 2000; Xie et al., 2011; Speed et al., 2012). Hence, all experimental operations should be carried out in the dark.
M1
M1 was eluted at 14.7 min and displayed a protonated molecule at m/z 370.1203, which is consistent with the removal of a C4H11N moiety and the addition of O2 to famitinib. The high CE mass spectrum of M1 gave two fragment ions at m/z 296.1219 and 269.1069, reflecting unchanged part B. A prominent fragment ion at m/z 324.1148, which is attributed to the neutral loss of a formic acid from the precursor ion, was observed. This finding implies the presence of a carboxylic acid group in the molecule of M1. Hence, M1 was proposed as an oxidative deaminated metabolite of famitinib. M1 was a major metabolite in the urine and feces, but was not detectable in the plasma.
M3
The extracted ion chromatograms of m/z 383.1883 showed two peaks at 9.5 and 13.3 min in the plasma, urine and feces. The protonated molecules were 28.0313 Da less than the parent drug, which is indicative of N-desethylation. The fragment ions were coincident with those observed in the high CE spectrum of the parent drug. A comparison with a synthetic standard (SHR116637) confirmed that the latter peak was N-desethylfamitinib, designated as M3. The earlier peak was the E-isomer of the exocyclic double bond of M3. M3 was a major component of human plasma, urine and feces.
M2 and M7
M2 and M7, detected in the plasma, urine and feces, were eluted at 9.2 and 10.0 min respectively. The protonated molecules of M2 (m/z 381.1926) and M7 (m/z 409.2249) were 2 Da lower than those of N-desethylfamitinib and famitinib respectively. In addition, their fragment ions at m/z 336.1332, 294.1246, 292.1068 and 267.1173 were also 2 Da less than the corresponding fragment ions of N-desethylfamitinib and famitinib. An analysis of the accurate mass showed that the loss of 2 Da can be attributed to defluorination and the incorporation of a hydroxyl group. The identities of M2 (N-desethyl-5-hydroxyl-defluorofamitinib) and M7 (5-hydroxyl-defluorofamitinib) were further confirmed using synthetic standards (SHR141371 and SHR 141227). M7 was found to be a major metabolite in urine and feces.
M4 and M8
The retention times of M4 (m/z 385.2040) and M8 (m/z 413.2353) were 8.9 and 10.0 min in both urine and feces respectively. An analysis of the accurate mass indicated that the elemental compositions of M4 and M8 were two hydrogen atoms more than those of N-desethylfamitinib and famitinib, respectively, which is indicative of hydrogenation. The high CE mass spectra of M4 and M8 revealed identical fragment ions at m/z 340.1447, 190.1067 and 189.0993. The ion at m/z 340.1447, which is 2 Da higher than those of N-desethylfamitinib and famitinib, was formed by the loss of an ethylamine/diethylamine moiety. Moreover, the two diagnostic ions at m/z 190.1067 and 189.0993 may have been generated by further cleavage of the bond between the 3-C of indolylidene and the exocyclic carbon atom of the ion at m/z 340.1447, with the charge retained on the methylpyrrolo-dihydropyridone part. This fragmentation pathway was specific and was not observed for the parent drug, thus implying that hydrogenation occurred at the exocyclic double bond. Further comparison of the retention time and MS fragmentation patterns with those of the synthetic standard (SHR139088) confirmed the structural assignment of M8. Similarly, M4 was tentatively identified as hydrogenated N-desethylfamitinib.
M5 and M9
M5 and M9 exhibited protonated molecules at m/z 399.1832 and 427.2145, which are 15.9949 Da higher than those of N-desethylfamitinib and famitinib respectively. This result is in line with those of a series of mono-oxygenated metabolites.
Four chromatographic peaks with a pseudo-molecular ion at m/z 399.1832 were detected at 9.9, 10.1, 10.5 and 11.4 min and were designated as M5-1 to M5-4 respectively. Under further fragmentation, the four metabolites showed the same fragment ions at m/z 354.1237, 312.1097, 310.0985 and 285.1002, all of which were ∼16 Da higher than the corresponding fragment ions that originated from part B of famitinib. In addition, no dehydrated ion was observed for these four metabolites, indicating the occurrence of aryl hydroxylation. Based on these results, M5-1 to M5-4 were tentatively identified as the indolylidene-hydroxylated metabolites of N-desethylfamitinib.
Seven chromatographic peaks with a protonated molecule at m/z 427.2145 were detected at 10.8, 11.0, 11.6, 12.3, 12.8, 13.3 and 14.6 min, and were labelled as M9-1 to M9-7 respectively. M9-1 and M9-5 were major components in the feces. M9-1, M9-2, M9-3 and M9-5 gave fragment ions that are identical to those of M5, suggesting that these components were the indolylidene-hydroxylated metabolites of the parent drug famitinib. M9-3 was confirmed as 6-hydroxylfamitinib based on chromatographic and MS fragmentation comparisons with the synthetic standard SHR141226. The fragment ions of M9-1, M9-2, M9-3 and M9-5 were also detected in the mass spectra of M9-4 and M9-6. In addition, the appearance of ions at m/z 336.1149 and 292.0892 due to the dehydration of the fragment ions at m/z 354.1288 and 310.0973, respectively, indicated the presence of an aliphatic hydroxyl group. Thus, the hydroxylation probably occurred at the methylpyrrolo-dihydropyridone moiety. Moreover, M9-7 showed the same fragment ions as the parent compound, which were all derived from part B. This result indicates that the diethylamine group (part A) was the site of oxidation. M9-7 was eluted later than the parent, and no dehydrated ion appeared in the spectrum, favouring the possibility of an N-oxide.
M6 and M10
M6 and M10, which were found only in urine, were eluted at 6.9 and 8.1 min respectively. The protonated molecules of M6 (m/z 401.1989) and M10 (m/z 429.2302) were 15.9949 Da higher than those of M4 and M8, which were assumed to result from a combination of the hydrogenation and mono-oxidation of N-desethylfamitinib and famitinib respectively. M6 and M10 fragmented in a similar manner under the high CE mode. The fragment ion at m/z 356.1405 was 16 Da higher than those of M6 and M10, indicating that the oxygen atom was introduced into part B. The sequential dehydration of this fragment ion yielded an ion at m/z 338.1302, thus indicating that M6 and M10 were aliphatic hydroxyl metabolites. The characteristic fragment ions at m/z 190.1079 and 189.0992 represented that the methylpyrrolo-dihydropyridone moiety was unmodified. These results suggest that the site of hydroxylation was located on the exocyclic methylene of M4 and M8.
M11, M12, M13 and M14
M11 and M13 were eluted at 8.4 and 9.0 min respectively. The protonated molecules of M11 (m/z 461.1495) and M13 (m/z 489.1805) were 79.9584 Da higher than those of M2 and M7, indicating that these molecules were sulphate conjugates of M2 and M7 respectively. The molecules shared identical fragment ions to M2 and M7, except for the ion at m/z 416.0910 (336 + 80). This pattern suggests that the sulphuric acid was attached to the part B of M2 and M7, specifically to the 5-hydroxyl group produced from oxidative defluorination. M13 was a major metabolite in feces. By using the same strategy, M12-1 to M12-3 (m/z 479.1400) and M14-1 to M14-3 (m/z 507.1713) were tentatively identified as the phenol sulphates of M5 and M9 respectively.
M15
The M15-1 (found in plasma, urine and feces) and M15-2 (found in urine and feces) were eluted at 8.3 and 8.6 min respectively. These metabolites displayed a protonated molecule at m/z 528.2280, with an elemental composition of C26H33N5O5S. These results indicate the addition of C3H5NO2S to M7, which is consistent with the introduction of one molecule of cysteine and the removal of two hydrogen atoms. In the high CE spectra of M15-1 and M15-2, the base peak ion was observed at m/z 455.1382, which corresponded to the neutral loss of the diethylamine moiety. Another diagnostic fragment ion appeared at m/z 368.1006 and was formed by the cleavage of the C–S bond of the cysteinyl moiety. The appearance of this ion implies the presence of an aromatic-oriented thioether motif (Xie et al., 2012). The para-hydroxyaniline moiety of M7 provided an initial structural alert because it can be oxidized to a highly electrophilic quinone-imine species, as indicated by parallel studies on several drugs possessing the para-hydroxyaniline moiety (Dahlin et al., 1984; Kang et al., 2008; Wen and Moore, 2011). Thus, M15-1 and M15-2 may be cysteine conjugates, with the cysteinyl moiety attached to the benzene ring of the quinone-imine intermediate.
M16, M17, M18, M19 and M20
M16 and M18 were eluted at 6.7 and 7.3 min respectively. The protonated molecules of M16 (m/z 557.2247) and M18 (m/z 585.2559) were 176.0321 Da higher than those of M2 and M7 respectively. The neutral loss of 176 Da from the precursor ions yielded fragment ions at m/z 381.1942 and 409.2227, respectively, indicating the presence of glucuronide conjugates. In addition, the loss of the ethylamine/diethylamine moiety and the subsequent neutral loss of glucuronic acid (176 Da) resulted in the appearance of ions at m/z 512.1636 and 336.1339. These results indicate that the glucuronic acid was attached to the part B of M2 and M7, specifically to the 5-hydroxyl group produced from oxidative defluorination. By using the same strategy, M17-1 to M17-3 (m/z 575.2153) and M20-1 to M20-4 (m/z 603.2466) were tentatively identified as the phenol glucuronides of M5 and M9, respectively, whereas M19 was assigned as the N-glucuronide of famitinib. A further attempt to determine the site of glucuronidation was made by subjecting the urine sample to β-glucuronidase. After incubation, the chromatographic peaks of M16, M17, M18 and M20 disappeared, whereas those of M2, M5-1, M5-2, M5-4, M7, M9-1, M9-2, M9-3 and M9-5 markedly increased. M19 was resistant to hydrolysis. These results support our structural assignments. M16, M17-1, M18, M20-2 and M20-4 were observed as major components in urine.
Pharmacokinetics and excretion of famitinib
The plasma concentrations of famitinib and its major circulating metabolite N-desethylfamitinib (M3) were determined during the oral administration of the drug (20 or 27 mg) to patients once daily for 28 days. A summary of the pharmacokinetic parameters for famitinib and M3 in both groups is presented in Table 2. After the first dose, the plasma concentrations of famitinib and M3 reached a maximum (Cmax) between 4.4 and 6.8 h and between 5.3 and 7.8 h respectively. Both famitinib and M3 were slowly removed from circulation, and their concentrations at 24 h were 35.6% to 42.7% and 46.8% to 51.1% of the Cmax respectively. The AUC0–24h of M3 was approximately 4.5% that of its parent drug. After the last dose, the Cmax of famitinib and M3 were observed at 5.1 h to 7.1 h and at 6.1 to 12.8 h respectively. The AUC0–24h of M3 was 7.2% to 7.5% that of its parent drug. After repeated administration, the Cmin (trough concentrations) of famitinib and M3 reached steady-state levels on days 7 to 10 and on days 10 to 14 respectively. The accumulation ratios of famitinib and M3 were 1.80 to 2.73 and 2.68 to 6.03 respectively.
Table 2.
Pharmacokinetic parameters (mean ± SD) of famitinib (M0) and N-desethylfamitinib (M3) following oral administration of 20 or 27 mg famitinib L-malate once daily for 28 days in patients
| tmax (h) | Cmax (ng·mL−1) | Cmin (ng·mL−1) | Cav (ng·mL−1) | AUC0–24h (ng·h·mL−1) | DF (%) | AR | ||
|---|---|---|---|---|---|---|---|---|
| Dose = 20 mg (n = 8) | ||||||||
| Day 1 | M0 | 6.75 ± 2.38 | 21.7 ± 7.78 | N.A. | N.A. | 333 ± 119 | N.A. | N.A. |
| M3 | 7.75 ± 2.71 | 0.963 ± 0.366 | N.A. | N.A. | 14.9 ± 5.85 | N.A. | N.A. | |
| Day 28 | M0 | 7.13 ± 3.36 | 44.2 ± 18.4 | 20.8 ± 8.32 | 33.5 ± 14.5 | 805 ± 348 | 71.0 ± 31.3 | 2.73 ± 1.29 |
| M3 | 12.8 ± 9.68 | 3.12 ± 1.22 | 1.70 ± 0.837 | 2.52 ± 1.18 | 60.5 ± 28.4 | 63.5 ± 31.5 | 6.03 ± 4.82 | |
| Dose = 27 mg (n = 7) | ||||||||
| Day 1 | M0 | 4.44 ± 1.24 | 47.7 ± 11.3 | N.A. | N.A. | 674 ± 221 | N.A. | N.A. |
| M3 | 5.33 ± 2.78 | 1.99 ± 0.490 | N.A. | N.A. | 31.2 ± 8.00 | N.A. | N.A. | |
| Day 28 | M0 | 5.14 ± 1.07 | 73.5 ± 15.9 | 28.2 ± 9.79 | 50.2 ± 14.7 | 1205 ± 352 | 97.5 ± 37.3 | 1.80 ± 0.375 |
| M3 | 6.14 ± 3.39 | 4.57 ± 1.58 | 2.63 ± 1.34 | 3.60 ± 1.57 | 86.5 ± 37.7 | 64.0 ± 40.8 | 2.68 ± 0.860 |
N.A., not applicable.
At steady state, the amount of famitinib and the identified major metabolites that were excreted daily via urine and feces accounted for 14.2% ± 5.1% and 56.3% ± 23.4% of the dose respectively. In the urine samples, a combination of the unchanged famitinib and its E-isomer M0-1 represented 2.4% of the dose. The major metabolites M3, M7, M16, M17-1, M18, M20-2 and M20-4 accounted for 1.1%, 1.0%, 1.1%, 0.6%, 1.6%, 1.6% and 1.6% of the dose respectively. In the fecal samples, a combination of the unchanged famitinib and its E-isomer M0-1 represented 17.6% of the dose. The major metabolites M1, M3, M7, M9-1, M9-5 and M13 accounted for 6.2%, 5.9%, 3.7%, 4.2%, 7.3% and 3.0% of the dose respectively. The combined urinary and fecal excretions of the two cysteine conjugates (M15-1 and M15-2) accounted for 3.3% of the dose.
In vitro metabolism of famitinib by human liver microsomes
To understand the biotransformation mechanisms of famitinib, the metabolism of famitinib was first investigated using HLM, HIM, HRM and HPM. Approximately 70% of the parent compound was consumed in the HLM incubations, whereas the utilized amounts of the parent compound in the HIM, HRM and HPM incubations were only 23.1%, 21.7% and 16.6%, respectively, of what was consumed in the HLM incubations. Therefore, the liver is suspected to be the major biotransformation site for famitinib.
The incubation of famitinib with the NADPH-supplemented HLM generated 12 oxidative metabolites (Figure 5). Based on the peak areas, N-desethylfamitinib (M3) was the main in vitro metabolite with small amounts of oxidatively deaminated carboxylic metabolite (M1), N-desethylated and subsequent oxidative-defluorinated metabolite (M2), two N-desethylated and aryl-hydroxylated metabolites (M5-3 and M5-4), oxidatively defluorinated metabolite (M7), hydrogenated metabolite (M8), three indolylidene-hydroxylated metabolites (M9-1, M9-3 and M9-5) and N-oxidized metabolite (M9-7). In addition, the new metabolite M21, which was not observed in vivo, was detected and tentatively characterized as an alcohol product from oxidative deamination based on its elemental composition and high CE spectrum. The generation of M1 and M21 is supposed to be associated with the same oxidative deamination pathway through an aldehyde intermediate, which could be oxidized to the carboxylic metabolite M1 or could alternatively react with reductase to produce the alcohol metabolite M21. Thus, both M1 and M21 can be monitored as indicators of oxidative deamination to aldehyde pathway in the succeeding mechanistic studies. The properties of the metabolites identified in the HLM incubations are listed in Table 3. The absence of NADPH in the incubations implies the absence of any metabolites.
Figure 5.

Metabolic profiles of famitinib (3 μM) in HLM: (A) without NADPH, (B) with NADPH and (C) with both NADPH and GSH. The inset is the expanded chromatogram.
Table 3.
UPLC-Q/TOF MS data for famitinib metabolites detected in human liver microsomes
| No. | Description | Rt (min) | Formula | Measured M + H+ | Error (in ppm) | +NADPH | +NADPH + GSH |
|---|---|---|---|---|---|---|---|
| M0 | Parent | 14.5 | C23H27FN4O2 | 411.2218 | 5.4 | + | + |
| M1 | Oxidative deamination to carboxylic acid | 15.2 | C19H16FN3O4 | 370.1231 | 7.6 | + | + |
| M2 | N-Desethylation + oxidative defluorination | 9.3 | C21H24N4O3 | 381.1953 | 7.0 | + | + |
| M3 | N-Desethylation | 13.3 | C21H23FN4O2 | 383.1891 | 2.7 | + | + |
| M5-3 | N-Desethylation + aryl hydroxylation | 10.6 | C21H23FN4O3 | 399.1859 | 6.7 | + | + |
| M5-4 | N-Desethylation + aryl hydroxylation | 11.7 | C21H23FN4O3 | 399.1870 | 9.5 | + | + |
| M7 | Oxidative defluorination | 10.1 | C23H28N4O3 | 409.2264 | 6.0 | + | + |
| M8 | Hydrogenation | 10.1 | C23H29FN4O2 | 413.2385 | 7.9 | + | + |
| M9-1 | Aryl hydroxylation | 10.9 | C23H27FN4O3 | 427.2141 | 0.9 | + | + |
| M9-3 | Aryl hydroxylation | 11.6 | C23H27FN4O3 | 427.2122 | −5.4 | + | + |
| M9-5 | Aryl hydroxylation | 12.9 | C23H27FN4O3 | 427.2169 | 5.6 | + | + |
| M9-7 | N-Oxidation | 14.7 | C23H27FN4O3 | 427.2170 | 5.8 | + | + |
| M15-1 | Oxidative defluorination + cysteine conjugation | 8.4 | C26H33N5O5S | 528.2305 | 4.7 | − | + |
| M15-2 | Oxidative defluorination + cysteine conjugation | 8.7 | C26H33N5O5S | 528.2310 | 5.7 | − | + |
| M21 | Oxidative deamination to alcohol | 15.0 | C19H18FN3O3 | 356.1408 | −0.6 | + | + |
| M22-1 | Oxidative defluorination + cysteinylglycine conjugation | 8.3 | C28H36N6O6S | 585.2508 | 2.2 | − | + |
| M22-2 | Oxidative defluorination + cysteinylglycine conjugation | 8.6 | C28H36N6O6S | 585.2493 | −0.3 | − | + |
| M23-1 | N-Desethylation + oxidative defluorination + GSH conjugation | 8.2 | C31H39N7O9S | 686.2556 | −7.6 | − | + |
| M23-2 | N-Desethylation + oxidative defluorination + GSH conjugation | 8.7 | C31H39N7O9S | 686.2586 | −3.2 | − | + |
| M24-1 | Oxidative defluorination + GSH conjugation | 8.7 | C33H43N7O9S | 714.2919 | −0.3 | − | + |
| M24-2 | Oxidative defluorination + GSH conjugation | 9.1 | C33H43N7O9S | 714.2958 | 5.2 | − | + |
Identification of the enzymes involved in famitinib biotransformation
The catalytic activities of a panel of human recombinant CYP and FMO isozymes in oxidizing famitinib were then examined. As summarized in Table 4, multiple CYPs and FMOs participated in the oxidative biotransformation of famitinib. All the tested isoforms were capable of famitinib N-desethylation. The highest M3 production was observed in the incubations with CYP3A5, followed by those with CYP2C8, CYP1B1 and CYP3A4. Based on their average hepatic expression, CYP3A4 was identified as the principle isoform that mediates famitinib N-desethylation. Although the relative abundance of CYP3A5 is much lower than CYP3A4 in the liver, the role of CYP3A5 in M3 formation should not be neglected due to its differential expression levels among individuals. Likewise, M21 was predominantly formed by CYP3A4/5; whereas M7, M9-1, M9-3 and M9-5 were catalysed by CYP1A1/2. The highest M9-7 production was measured during incubation with FMO1, followed by that with FMO3. FMO1 was found to be highly expressed in the kidneys but not in the liver, whereas FMO3 was mainly present in the liver. Thus, FMO3 is most likely to be the major enzyme responsible for the formation of M9-7.
Table 4.
Major oxidative metabolites and GSH conjugates of famitinib as catalysed by recombinant human cytochrome P450s and FMOs
| Enzyme | M3 | M7 | M9-1 | M9-3 | M9-5 | M9-7 | M21 | M24-1 |
|---|---|---|---|---|---|---|---|---|
| CYP1A1 | 29.6 | 87.7 | 55.2 | 31.4 | 39.6 | 24.8 | – | 55.0 |
| CYP1A2 | 11.6 | 100 | 100 | 100 | 100 | 1.2 | – | 100 |
| CYP1B1 | 45.0 | 8.9 | 14.6 | 15.2 | 22.6 | 9.5 | – | 7.6 |
| CYP2A6 | 8.0 | – | – | – | – | – | 25.4 | – |
| CYP2B6 | 7.3 | – | – | – | – | 1.7 | 19.6 | – |
| CYP2C8 | 63.8 | – | – | – | – | 1.8 | 44.5 | – |
| CYP2C9 | 9.8 | – | – | – | – | 2.6 | – | – |
| CYP2C19 | 10.4 | – | – | – | – | 0.9 | – | – |
| CYP2D6 | 8.0 | – | – | – | – | 1.7 | – | – |
| CYP2E1 | 7.2 | – | – | – | – | 2.3 | – | – |
| CYP3A4 | 31.3 | 2.0 | 1.9 | 19.5 | 1.2 | 1.5 | 77.7 | 2.0 |
| CYP3A5 | 100 | – | – | – | – | 0.9 | 100 | – |
| CYP4A11 | 6.8 | – | – | – | – | 1.0 | 17.2 | – |
| FMO1 | 2.5 | – | – | – | – | 100.0 | – | – |
| FMO3 | 5.4 | – | – | – | – | 30.6 | – | – |
| FMO5 | 4.9 | – | – | – | – | – | – | – |
Data were presented as mean for two separate experiments. The highest ratio of each metabolite detected in the incubations was neutralized to 100%. The ratios of the specific metabolites in the other enzyme incubation samples were then expressed as percentages relative to this highest level.
To determine further the relative contributions of the different CYPs in famitinib biotransformation, ABT (non-specific CYP inhibitor), α-NF (CYP1A1/2 inhibitor), sulphaphenazole (CYP2C9 inhibitor), ticlopidine (CYP2C19 inhibitor), quinidine (CYP2D6 inhibitor), chlormethiazole (CYP2E1 inhibitor) and KET (CYP3A4/5 inhibitor) were used to monitor the famitinib depletion and metabolite formation in the HLM incubations. The results of these incubation studies are presented in Figure 6. The depletion of famitinib could be exclusively inhibited by the presence of ABT or KET because the concentrations of unchanged famitinib in the ABT and KET incubations were both approximately thrice as much as that of control. These results suggest that CYP3A4/5 is the most important enzyme for famitinib oxidation in HLM. In the presence of KET, the formations of M3, M21 and M1 were inhibited by 70%, 81% and 92% respectively. In the presence of α-NF, the formations of M7 and M9-5 (a major indolylidene-hydroxylated metabolite) were inhibited by 53% and 74% respectively. M9-1 and M9-3 showed similar trends. Other inhibitors did not show any marked inhibition on these pathways. These results verified that CYP3A4/5 was the key enzyme in N-desethylation and oxidative deamination to aldehyde, whereas CYP1A1/2 had the highest efficacy in oxidative defluorination and indolylidene hydroxylation.
Figure 6.

Incubations of famitinib (3 μM) in HLM in the presence of NADPH. Different enzyme inhibitors, including ABT (non-specific CYP inhibitor), α-NF (CYP1A1/2 inhibitor), sulphaphenazole (CYP2C9 inhibitor), ticlopidine (CYP2C19 inhibitor), quinidine (CYP2D6 inhibitor), chlormethiazole (CYP2E1 inhibitor), KET (CYP3A4/5 inhibitor), 4-methylpyrazole (alcohol dehydrogenase inhibitor), disulfiram (aldehyde dehydrogenase inhibitor), raloxifene (aldehyde oxidase inhibitor), were used to assess the relative contribution of different microsomal enzyme systems to the metabolism of famitinib. (A) Effect of CYP inhibitors on the depletion of the parent drug. (B) Effect of CYP inhibitors on the formation of the oxidative-deaminated carboxylic metabolite (M1), oxidative-deaminated alcohol metabolite (M21), N-desethylated metabolite (M3), oxidative-defluorinated metabolite (M7) and indolylidene-hydroxylated metabolite (M9-5). (C) Effect of 4-methylpyrazole, disulfiram, and raloxifene on the formation of M1 and M21. Each column represents the mean ± SD of triplicate incubations. *P < 0.05, **P < 0.01, ***P < 0.001, versus control.
To probe the underlying mechanisms for the oxidation of the aldehyde intermediate to M1, 4-methylpyrazole (alcohol dehydrogenase inhibitor), disulfiram (aldehyde dehydrogenase inhibitor) and raloxifene (aldehyde oxidase inhibitor) were added to the HLM incubations for the measurement of M1 and M21 formations. As shown in Figure 6, the yield of M1 was attenuated by 42%, 94% and 18% in the presence of 4-methylpyrazole, disulfiram and raloxifene respectively. Thus, aldehyde dehydrogenase was implicated as the other pivotal enzyme for M1 formation. Furthermore, the addition of these inhibitors increased M21 formation, thereby demonstrating that M1 and M21 were generated by two competing pathways.
Bioactivation of famitinib and 5-hydroxyl-defluorofamitinib (M7) in HLM
The co-incubation of famitinib with GSH, NADPH and HLM allowed for the detection of four GSH conjugates, namely, M23-1, M23-2, M24-1 and M24-2 (Figure 5), which were eluted at 8.2, 8.7, 8.7 and 9.1 min respectively. The protonated molecules of M23-1 and M23-2 (m/z 686.2608) and M24-1 and M24-2 (m/z 714.2921) were 305.0672 Da higher than that of M2 and M7, thereby indicating the addition of one molecule of GSH and the removal of two hydrogen atoms. Upon further fragmentation, identical product ions were observed at m/z 641.2028, 512.1597 and 368.1035. The base peak ion at m/z 368.1035 was generated via the cleavage of the C–S bond of the glutathionyl moiety, which suggested the existence of an aromatic-orientated thioether motif in the GSH conjugates. The ions at m/z 641.2028 and 512.1597 resulted from the loss of the ethylamine/diethylamine moiety and the subsequent neutral loss of the pyroglutamic acid moiety (129 Da). These results indicated that the GSH molecule was most likely attached to the part B of M2 and M7, specifically to the indolylidene moiety. Additionally, several secondary metabolites of M24-1 and M24-2 were detected, including two cysteinylglycine conjugates (M22-1 and M22-2) and two cysteine conjugates (M15-1 and M15-2). M24-1 was the predominant GSH conjugate. Thus, for simplicity, only M24-1 was shown in the mechanistic studies, but similar trends were observed with all the other thiol conjugates. The identified thiol conjugates are listed in Table 3.
Based on the detected GSH conjugates, the bioactivation of famitinib is likely to proceed through the oxidation of M7 to a quinone-imine intermediate. To address this possibility, M7 was incubated in NADPH- and GSH-supplemented HLM at a low substrate concentration (100 nM), which was identical to the yields of M7 in NADPH-supplemented HLM with famitinib. Unexpectedly, no GSH-related conjugate was discernible. When a much higher substrate concentration (1 μM) was applied, M24-1 and M24-2 could be detected and their yields were comparable with those observed in the famitinib incubations. These observations confirmed that the reactive intermediate of famitinib was a quinone imine. This intermediate was oxidized directly from famitinib, rather than from M7, because the GSH conjugation in the M7 incubations was much lower than that in the famitinib incubations.
M7 originated from famitinib oxidative defluorination, whose initial step is reported to be epoxidation (Park et al., 2001). The resulting epoxide or the quinone imine that was rearranged from the epoxide is highly electrophilic. Therefore, we anticipated that famitinib was bioactivated through a pathway that involved epoxidation and defluorinated rearrangement to a quinone imine, which could be trapped by GSH or reduced to M7. To test this hypothesis, MEH and NQO1 were added to the incubations containing HLM with famitinib, GSH and NADPH. As shown in Figure 7, the formation of M24-1 was decreased by 47%, 50% and 71% in the presence of MEH, NQO1 and a combination of both, respectively. MEH likewise showed a moderate inhibitory effect (48%) on the formation of M7, whereas the yield of M7 was slightly increased after adding NQO1. These results collectively supported the abovementioned hypothesis.
Figure 7.

Effect of human MEH and NQO1 on the formation of the oxidative-defluorinated metabolite (M7) and the GSH conjugate (M24-1). Each column represents the mean ± SD of triplicate incubations. ***P < 0.001, versus control.
Identification of CYP isozymes responsible for famitinib bioactivation
To determine which CYP isozymes(s) preferentially bioactivating famitinib, the formation of M24-1 was investigated using a panel of human recombinant CYP isozyme incubations with famitinib. Based on the data presented in Table 4, CYP1A2 had the highest specific activity with respect to the GSH adduct. The specific activity of CYP1A1 was approximately 55% that of CYP1A2. CYP1B1 and CYP3A4 also catalysed M24-1 formation, but the levels of conjugate formation were only 7.9% and 2.0%, respectively, of the levels formed by CYP1A2. Furthermore, the capability of CYP isozymes to oxidize M7 directly into the quinone-imine intermediate was tested. Multiple CYP isozymes, including CYP1A1, CYP1A2, CYP1B1, CYP2A6, CYP2B6, CYP2E1 and CYP3A4 were found to be involved in this bioactivation pathway.
Based on the results from recombinant CYP experiments, the selective inhibitors of CYP1A1/2 and CYP3A4, which are α-NF and KET, respectively, were evaluated in HLM, HIM, HRM, HPM(S) and HPM(NS). As shown in Figure 8, HLM generated almost 12 times the amount of M24-1 as HIM, and hundreds of times more than both HRM and HPM. Although the formation of M24-1 in HPM was low, a striking difference between smokers and non-smokers was observed. The amount of M24-1 in HPM(S) was twice higher than that in HPM(NS). The addition of α-NF attenuated M24-1 formation in HLM, HRM, HPM(NS) and HPM(S) by 57%, 27%, 20% and 52% respectively, but did not alter the formation rate of M24-1 in HIM. The addition of KET suppressed M24-1 formation by 30% in HIM but had minimal inhibitory effects on HLM, HKM and HPM.
Figure 8.

Comparison of the relative amounts of the GSH conjugate M24-1 formed in different human microsomal incubations, including human hepatic, intestinal, renal and pulmonary microsomes (smoker and non-smoker), with or without the addition of CYP inhibitors α-NF (CYP1A1/2 inhibitor) and KET (CYP3A4/5 inhibitor). Data are reported as the mean of duplicate incubations.
Cytotoxicity and metabolism studies in primary human hepatocytes
The clinical hepatotoxicity of famitinib has been documented (Zhou et al., 2011). Thus, the correlation between the formation of the quinone-imine intermediate and famitinib-induced hepatotoxicity was investigated using primary human hepatocytes. The effect of famitinib on cell viability was initially evaluated at various concentrations. The cells were exposed to 0 μM to 15 μM famitinib for 24 h. The treatment caused a dose-dependent cytotoxicity on the hepatocytes. As low as 50% cell viability was observed in the group treated with famitinib at 15 μM, relative to the vehicle treatment (Table 5).
Table 5.
Famitinib-induced cytotoxicity in the primary human hepatocytes
| Famitinib concentration (μM)a | ||||
|---|---|---|---|---|
| Treatment | 0 | 5 | 10 | 15 |
| Hepatocyte | 100 | 87.5 ± 1.1 | 83.2 ± 2.4 | 47.5 ± 7.3 |
| +1 mM ABT | 95.7 ± 5.0 | 101 ± 3.6** | 102 ± 4.1*** | 59.8 ± 0.5* |
| +2 μM α-NF | 99.9 ± 0.2 | 103 ± 3.6** | 99.1 ± 2.7*** | 62.5 ± 0.8* |
| +100 μM BSO | 97.6 ± 4.0 | 76.4 ± 2.7* | 73.7 ± 1.2* | 37.8 ± 6.7 |
| +500 μM (−)-Borneol | 95.5 ± 3.9 | 87.3 ± 5.3 | 82.6 ± 2.0 | 48.6 ± 1.9 |
Data were presented as mean ± SD for the cell viability of three separate experiments.
P < 0.05,
P < 0.01,
P < 0.001, versus respective incubations containing no inhibitors.
To determine the molecular mechanisms of cytotoxicity, different compounds were added to the culture medium to examine their ability to modulate the toxic response. Meanwhile, the culture medium was collected and subjected to metabolite identification to verify if the cytotoxic effects of famitinib were caused by bioactivation. The main metabolites identified in humans were detected in hepatocyte incubations (Supporting Information Table S2). The major thiol conjugates were the cysteine conjugates (M15-1 and M15-2), whereas the GSH conjugates (M24-1 and M24-2) and the cysteinylglycine conjugates (M22-1 and M22-2) were present in trace amounts. After adding BSO (GSH depletor), a 22% to 51% decline in M15-1 formation was observed. This finding indicates that the cysteine conjugates of famitinib in humans were derived from GSH conjugates via the mercapturate pathway. The formation of M15-1 was measured as an indicator of the relative amount of the toxic quinone imine in the hepatocyte incubations.
The co-incubations with ABT (non-specific CYP inhibitor) and α-NF (CYP1A1/2 inhibitor) at all tested concentrations decreased the amount of M15-1 by 76% to 98% and thus evidently increased hepatocyte viability (Table 5 and Figure 9). These results suggest that CYP1A1/2 is involved in the bioactivation of famitinib into a quinone-imine metabolite. By contrast, BSO (GSH depletor) could exert a potentiating effect on famitinib cytotoxicity by increasing the amount of the quinone-imine species. Regardless of the ability of (−)-borneol (glucuronidation inhibitor) to increase the amount of M7 in the incubations by 24% to 40%, the inhibitor did not affect the yield of M15-1 and cytotoxicity, thereby supporting the fact that the toxic quinone imine did not originate from M7. No obvious cytotoxicity was observed when the cells were cultured with the inhibitors alone.
Figure 9.

Formation of the cysteine conjugate M15-1 in primary human hepatocyte incubations with different famitinib concentrations (5, 10 and 15 μM) in the presence of ABT (non-specific CYP inhibitor), α-NF (CYP1A1/2 inhibitor), BSO (GSH depletor) and (−)-borneol (glucuronidation inhibitor).
Discussion and conclusions
This study characterized the metabolic pathways of famitinib in cancer patients, determined its pharmacokinetic parameters and excretion properties, and established its biotransformation and bioactivation mechanisms. The results also clarified that the cytotoxicity of famitinib towards primary human hepatocytes is triggered by the reactive quinone-imine species.
Famitinib was slowly absorbed, with peak plasma levels occurring at 4.4 h to 6.8 h post dose. This finding is similar to that observed in a pre-clinical rat study. Based on the UHPLC-UV chromatograms of the plasma samples, the circulating drug-related materials were largely composed of the unmodified drug. The major circulating metabolite was the pharmacologically active metabolite N-desethylfamitinib (M3), whose steady-state exposure was less than 7.5% of that observed in its parent compound. Likewise, this level was less than that observed in the pre-clinical toxicology species (Chen et al., unpubl. data). Thus, no additional toxicological investigations were necessary. The steady-state levels of famitinib and M3 were achieved at 7 to 10 days and at 10 to 14 days post treatment, respectively, with the respective Cmin values of 20.8 to 28.2 ng·mL−1 and 1.70 to 2.63 ng·mL−1. Relatively small fluctuations were observed in their plasma levels at the steady state. The accumulation degrees of famitinib and M3 were moderate, with mean accumulation ratios ranging from 1.80 to 2.73 and 2.68 to 6.03 respectively. This accumulation may be due to the short dosing interval (24 h) in relation to the long terminal elimination half-lives, which were determined to be 33.5 and 48.4 h for famitinib and M3, respectively, in another pharmacokinetic study after a single 25 mg oral dose of famitinib to eight healthy male volunteers (Chen et al., unpubl. data).
The urine and fecal UHPLC-UV chromatograms indicated that famitinib was extensively metabolized in patients. A total of 40 and 27 metabolites were identified in urine and feces, respectively, by UHPLC/Q-TOF MS. Whenever possible, their proposed structures were supported by a comparison with the synthetic standards. The metabolic scheme of famitinib in humans is presented in Figure 4. Famitinib and its metabolites were eliminated mainly via the fecal route (approximately 56.3% of the daily dose), whereas the urinary clearance was a minor route of elimination (approximately 14.2% of the daily dose). Majority (approximately 70%) of the drug-related compounds recovered in the feces could be attributed to the metabolites, rather than the parent compound, revealing that famitinib was well absorbed from the gut, metabolized in the liver and then cleared by the biliary–fecal route. Moreover, the metabolic patterns in humans were consistent over 28 days of repeated administration. Thus, the metabolic enzymes responsible for famitinib metabolism were neither induced nor inhibited.
Based on the UHPLC-UV chromatographic peak area, the primary routes of famitinib biotransformation involved N-desethylation (M3), oxidative deamination (M1), oxidative defluorination (M7), indolylidene hydroxylation (M9-1 and M9-5) and the subsequent phase-II conjugations of these metabolites. The in vitro metabolism of recombinant human isozymes and the microsomal chemical inhibition studies demonstrated that multiple enzymes, including CYP3A4/5, CYP1A1/2, aldehyde dehydrogenase and FMO3, participated in the oxidative metabolism of famitinib.
Microsomal studies showed that CYP3A4/5-mediated N-desethylation is important to famitinib. Similarly, CYP3A is also reported to be the most effective enzyme in the metabolism of sunitinib, a structural analogue of famitinib (Speed et al., 2008). Drug–drug interactions between sunitinib and the CYP3A inhibitor KET or the inducer rifampicin have been reported. The concurrent administration of sunitinib with KET or rifampin resulted in a 58.3% increase or a fourfold reduction, respectively, in the AUC of sunitinib when compared with dosing healthy volunteers with sunitinib alone (Washington et al., 2003; Bello et al., 2005; Kim et al., 2009). However, the contribution of CYP3A to the overall metabolism and disposition of famitinib in humans was found to be less vital when compared with its effects on sunitinib. The exposure of the N-desethylated metabolite of sunitinib was approximately 55% that of the parent drug, and its fecal excretion (25% of the administered dose) was almost twice that of the parent (Speed et al., 2012). Much lower exposure and excretion of M3 were observed for famitinib, such that the exposure of M3 was less than 7.5% that of famitinib and the daily excretion of M3 (7% of the administered dose) was only one-third that of the parent. Thus, the CYP3A-based drug–drug interaction of famitinib would be less significant than that of sunitinib. The discrepancy between the in vivo and in vitro metabolism data of famitinib might be explained by the rapid phase II conjugations of the hydroxyl metabolites (CYP1A-catalysed) and the sequential rapid excretion by some transporters.
The identification of two cysteine conjugates (M15-1 and M15-2) of famitinib, whose combined urinary and fecal excretions accounted for 3.3% of the administered dose, provided evidence for the formation of potentially harmful reactive metabolites in vivo. HLM and hepatocyte metabolism studies revealed that the cysteine conjugates in humans presumably arose from the processing of GSH conjugates via the mercapturate pathway by γ-glutamyltransferase and cysteinyl-glycine dipeptidase. Moreover, the major reactive intermediate of famitinib was confirmed to be a quinone imine because the same GSH conjugates were detected in both famitinib and 5-hydroxyl-defluorofamitinib (M7) incubations, and the formation of these conjugates could be suppressed by adding NQO1. Quinone imines are known electrophiles that can alkylate cellular nucleophilic macromolecules or alter the redox state of cells (Stepan et al., 2011). Acetaminophen, amodiaquine and diclofenac exert their toxicities by oxidizing the para-hydroxyaniline group into the reactive quinone-imine species (Smolarek et al., 1990; Bort, 1999; Tafazoli and O'Brien, 2009). However, the quinone-imine of famitinib was not mainly formed by M7 oxidation. Instead, the intermediate originated directly from the parent molecule because much lower GSH conjugation was observed in the M7 incubations than in the famitinib incubations. The observed inhibitory effect of MEH on the production of GSH conjugates in famitinib incubations suggests that the first step of bioactivation is the epoxidation of the indolylidene moiety of famitinib to yield an epoxide intermediate, which is defluorinated and converted into the quinone-imine. The resulting quinone imine can react with GSH or can be reduced by NADPH or reductases to form M7. A plausible bioactivation scheme for famitinib is depicted in Figure 10. The present results could not rule out the possibility that GSH may directly react with a small portion of the epoxide intermediate prior to its defluorination and rearrangement.
Figure 10.

Proposed bioactivation mechanism of 5-fluoro-indolylidenes. The first step of bioactivation is the epoxidation of the indolylidene moiety to yield an epoxide intermediate, which is defluorinated and converted into the quinone imine. The resulting quinone imine can react with GSH or can be reduced by NADPH or reductases to form 5-hydroxyl-indolylidenes.
The incubation of famitinib with 12 major human recombinant CYP isoforms supplemented with NADPH and GSH proved that CYP1A1/2 could catalyse famitinib bioactivation. CYP1B1 and CYP3A4 could also catalyse this bioactivation but to a much lesser degree. The CYPs were mainly expressed in the liver, intestine, kidney and lung. Similarly, famitinib was highly distributed in these organs in rats. After a single oral dose (5 mg·kg−1) was given to rats, the famitinib concentrations in the lung, liver, kidney and intestinal were over 30, 15, 10 and 5 times higher than those in the plasma, respectively (Supporting Information Table S3). If the tissue/plasma ratios measured in rats are representative of those in humans, similar tissue concentrations would be expected. Therefore, famitinib bioactivation was investigated in human liver, intestinal, renal and pulmonary microsomes. The GSH conjugates were detected in all the microsomal incubations, among which the highest amount of the GSH conjugate M24-1 was observed in HLM incubations. The high distribution of famitinib to the liver and lung tissues may increase the incidence of toxicity in these tissues. Based on the activity of specific chemical inhibitors and the relative concentrations of CYPs in different tissues, CYP1A1/2 was identified to be the primary enzyme responsible for famitinib bioactivation in the liver, kidney and lung; whereas CYP3A4 was found to be responsible for famitinib bioactivation in the intestine. Additionally, exposure to cigarette smoke increases the mRNA and protein levels of CYP1A1 in human lung and liver tissues (Villard et al., 1998; Hukkanen et al., 2002). In this study, the incubations of famitinib with HPM(S) did show a twofold increase in M24-1 relative to that with HPM(NS). Thus, the augmented expression of CYP1A1 in the lungs and livers of smokers may elevate their risk of associated toxicities.
Idiosyncratic hepatotoxicity is a common adverse reaction of small-molecule RTK inhibitors observed in clinic, such as imatinib, dasatinib, erlotinib, gefitinib, lapatinib and sunitinib (Ayoub et al., 2005; Ho et al., 2005; Liu et al., 2007; Bonvin et al., 2008; Loriot et al., 2008; Mueller et al., 2008; Teo et al., 2012). Some of these RTK inhibitors could undergo bioactivation to form reactive metabolites (Li et al., 2009a, b; 2010; Teng et al., 2010). However, the hepatotoxic mechanisms and the correlation between bioactivation and hepatotoxicity, especially in view of the potential role of CYP in promoting the toxicity, remain unclear. This study proved that famitinib is toxic to primary human hepatocytes with an IC50 of 13.7 μM. Furthermore, the addition of the non-specific CYP inhibitor ABT and the CYP1A1/2 inhibitor α-NF into the hepatocyte incubation could reverse the cytotoxicity induced by famitinib and suppress the formation of the quinone imine. These metabolism-based events may potentially and partially contribute to the clinically observed idiosyncratic hepatotoxicity. Particularly, BSO treatment could potentiate the famitinib-induced cytotoxicity, which is attributed to the BSO-induced GSH depletion. This depletion may intensify the reactions of electrophilic quinone-imine species with nucleophilic centres on critical biological proteins, thereby initiating cytotoxicity. Furthermore, sunitinib also has a 5-fluoroindolylidene moiety in its molecule. Thus, analogous bioactivation process of sunitinib was also observed (Supporting Information Figure S1). In the current study, sunitinib exhibited identical cytotoxicity to famitinib, with an IC50 of 13.2 μM. However, the proposed lower therapeutic dose of famitinib (25 mg) than sunitinib (50 mg) reduces its risk of toxicity because the daily dose seems to be a key factor in drug toxicity (Lammert et al., 2008; Stepan et al., 2011).
In conclusion, famitinib was well absorbed, extensively metabolized and then eliminated primarily via biliary/fecal route in cancer patients. The vast majority of drug-related materials in circulation are represented by the unchanged famitinib. The metabolites in excreta were mainly derived from the CYP3A4/5-mediated N-desethylation, CYP3A4/5- and aldehyde-dehydrogenase-mediated oxidative deamination and subsequent carboxylation, CYP1A1/2-mediated oxidative defluorination and indolylidene hydroxylation, and secondary phase II conjugations. The quinone-imine intermediate formed directly from famitinib via epoxidation by CYP1A1/2 at the indolylidene moiety and sequential defluorination–rearrangement may be responsible for the famitinib-induced cytotoxicity in primary human hepatocytes. Therefore, co-administered drugs that can induce CYP1A1/2 may potentiate the toxicity of famitinib. The metabolic and cytotoxicity data have expanded understanding on the famitinib-induced hepatotoxicity. The potential safety issues regarding famitinib, especially in patients who continue to smoke, need further assessment. Efforts should be made to unveil the underlying molecular mechanisms. This study constitutes the first report confirming that the hepatotoxicity of RTK inhibitors could be initiated by CYP-dependent bioactivation. Understanding the correlation between bioactivation and toxicity will provide an important theoretical basis for the safe clinical use of RTK inhibitors and the development of new therapeutic compounds.
Acknowledgments
This work was partially supported by the National Natural Science Foundation of China [81173115] and the National Basic Research Program of China [2009CB930300].
The authors thank the clinical staff of the Cancer Institute and Hospital, Chinese Academy of Medical Sciences (Beijing, China) for conducting the clinical studies; Jiangsu Hengrui Medicine Co. Ltd. (Lianyungang, China) for synthesizing the standard compounds; Mr Rongwei Shi and Ms Hua Li for their contribution in the LC-MS/MS analysis of famitinib and N-desethylfamitinib; and Dr Liang Li for his helpful discussion.
Glossary
- ABT
1-aminobenzotriazole
- BSO
l-buthionine-sulphoximine
- CCK-8
cell counting kit-8
- CE
collision energy
- CYP
cytochrome P450
- FMO
flavin mono-oxygenases
- GSH
glutathione
- HIM
human intestinal microsomes
- HLM
human liver microsomes
- HPM
human pulmonary microsomes
- HRM
human renal microsomes
- IS
internal standard
- KET
ketoconazole
- MEH
microsomal epoxide hydrolase
- NADPH
β-nicotinamide adenine dinucleotide 2′-phosphate reduced tetrasodium salt
- NQO1
recombinant human NAD(P)H : quinone oxidoreductase 1
- RTK
receptor tyrosine kinase
- UHPLC/Q-TOF MS
ultra-high performance liquid chromatography-quadrupole-time of flight-mass spectrometer
- α-NF
α-naphthoflavone
Conflicts of interest
The authors state no conflicts of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Figure S1 Extracted ion chromatograms of oxidative-defluorinated metabolite (A) and GSH conjugates (B) of sunitinib in HLM incubations.
Table S1 Information on subjects enrolled in the mass balance and pharmacokinetic study.
Table S2 UPLC-Q/TOF MS data for famitinib metabolites detected in primary human hepatocytes.
Table S3 Tissue distribution of famitinib in rats after a single 5 mg kg−1 oral dose.
References
- Ayoub WS, Geller SA, Tran T, Martin P, Vierling JM, Poordad FF. Imatinib (Gleevec)-induced hepatotoxicity. J Clin Gastroenterol. 2005;39:75–77. [PubMed] [Google Scholar]
- Baillie TA, Cayen MN, Fouda H, Gerson RJ, Green JD, Grossman SJ, et al. Drug metabolites in safety testing. Toxicol Appl Pharmacol. 2002;182:188–196. doi: 10.1006/taap.2002.9440. [DOI] [PubMed] [Google Scholar]
- Bello C, Houk B, Sherman L, Misbah S, Sarapa N, Smeraglia J, et al. Effect of rifampin on the pharmacokinetics of SU11248 in healthy volunteers [abstract] J Clin Oncol. 2005;23:3078. [Google Scholar]
- Bonvin A, Mesnil A, Nicolini FE, Cotte L, Michallet M, Descotes J, et al. Dasatinib-induced acute hepatitis. Leuk Lymphoma. 2008;49:1630–1632. doi: 10.1080/10428190802136384. [DOI] [PubMed] [Google Scholar]
- Bort R, Ponsoda X, Jover R, Gomez-Lechon MJ, Castell JV. Diclofenac toxicity to hepatocytes: a role for drug metabolism in cell toxicity. J Pharmacol Exp Ther. 1999;288:65–72. [PubMed] [Google Scholar]
- Cabebe E, Wakelee H. Sunitinib: a newly approved small-molecule inhibitor of angiogenesis. Drugs Today (Barc) 2006;42:387–398. doi: 10.1358/dot.2006.42.6.985633. [DOI] [PubMed] [Google Scholar]
- Dahlin DC, Miwa GT, Lu AY, Nelson SD. N-acetyl-p-benzoquinone imine: a cytochrome P-450-mediated oxidation product of acetaminophen. Proc Natl Acad Sci U S A. 1984;81:1327–1331. doi: 10.1073/pnas.81.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen T, Sternberg CN, Robert C, Mulders P, Pyle L, Zbinden S, et al. Targeted therapies for renal cell carcinoma: review of adverse event management strategies. J Natl Cancer Inst. 2012;104:93–113. doi: 10.1093/jnci/djr511. [DOI] [PubMed] [Google Scholar]
- Goodman VL, Rock EP, Dagher R, Ramchandani RP, Abraham S, Gobburu JV, et al. Approval summary: sunitinib for the treatment of imatinib refractory or intolerant gastrointestinal stromal tumors and advanced renal cell carcinoma. Clin Cancer Res. 2007;13:1367–1373. doi: 10.1158/1078-0432.CCR-06-2328. [DOI] [PubMed] [Google Scholar]
- Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- Hartmann JT, Haap M, Kopp HG, Lipp HP. Tyrosine kinase inhibitors – a review on pharmacology, metabolism and side effects. Curr Drug Metab. 2009;10:470–481. doi: 10.2174/138920009788897975. [DOI] [PubMed] [Google Scholar]
- Ho C, Davis J, Anderson F, Bebb G, Murray N. Side effects related to cancer treatment: CASE 1. Hepatitis following treatment with gefitinib. J Clin Oncol. 2005;23:8531–8533. doi: 10.1200/JCO.2004.00.6650. [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Pelkonen O, Hakkola J, Raunio H. Expression and regulation of xenobiotic-metabolizing cytochrome P450 (CYP) enzymes in human lung. Crit Rev Toxicol. 2002;32:391–411. doi: 10.1080/20024091064273. [DOI] [PubMed] [Google Scholar]
- Kang P, Dalvie D, Smith E, Renner M. Bioactivation of lumiracoxib by peroxidases and human liver microsomes: identification of multiple quinone imine intermediates and GSH adducts. Chem Res Toxicol. 2008;22:106–117. doi: 10.1021/tx8002356. [DOI] [PubMed] [Google Scholar]
- Kim A, Balis FM, Widemann BC. Sorafenib and sunitinib. Oncologist. 2009;14:800–805. doi: 10.1634/theoncologist.2009-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammert C, Einarsson S, Saha C, Niklasson A, Bjornsson E, Chalasani N. Relationship between daily dose of oral medications and idiosyncratic drug-induced liver injury: search for signals. Hepatology. 2008;47:2003–2009. doi: 10.1002/hep.22272. [DOI] [PubMed] [Google Scholar]
- Li X, He Y, Ruiz CH, Koenig M, Cameron MD, Vojkovsky T. Characterization of dasatinib and its structural analogs as CYP3A4 mechanism-based inactivators and the proposed bioactivation pathways. Drug Metab Dispos. 2009a;37:1242–1250. doi: 10.1124/dmd.108.025932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kamenecka TM, Cameron MD. Bioactivation of the epidermal growth factor receptor inhibitor gefitinib: implications for pulmonary and hepatic toxicities. Chem Res Toxicol. 2009b;22:1736–1742. doi: 10.1021/tx900256y. [DOI] [PubMed] [Google Scholar]
- Li X, Kamenecka TM, Cameron MD. Cytochrome P450-mediated bioactivation of the epidermal growth factor receptor inhibitor erlotinib to a reactive electrophile. Drug Metab Dispos. 2010;38:1238–1245. doi: 10.1124/dmd.109.030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Makrauer FL, Qamar AA, Janne PA, Odze RD. Fulminant hepatic failure secondary to erlotinib. Clin Gastroenterol Hepatol. 2007;5:917–920. doi: 10.1016/j.cgh.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Loriot Y, Perlemuter G, Malka D, Penault-Llorca F, Boige V, Deutsch E, et al. Drug insight: gastrointestinal and hepatic adverse effects of molecular-targeted agents in cancer therapy. Nat Clin Pract Oncol. 2008;5:268–278. doi: 10.1038/ncponc1087. [DOI] [PubMed] [Google Scholar]
- Mueller EW, Rockey ML, Rashkin MC. Sunitinib-related fulminant hepatic failure: case report and review of the literature. Pharmacotherapy. 2008;28:1066–1070. doi: 10.1592/phco.28.8.1066. [DOI] [PubMed] [Google Scholar]
- Park BK, Kitteringham NR, O'Neill PM. Metabolism of fluorine-containing drugs. Annu Rev Pharmacol Toxicol. 2001;41:443–470. doi: 10.1146/annurev.pharmtox.41.1.443. [DOI] [PubMed] [Google Scholar]
- Smolarek TA, Higgins CV, Amacher DE. Metabolism and cytotoxicity of acetaminophen in hepatocyte cultures from rat, rabbit, dog, and monkey. Drug Metab Dispos. 1990;18:659–663. [PubMed] [Google Scholar]
- Socinski MA, Novello S, Brahmer JR, Rosell R, Sanchez JM, Belani CP, et al. Multicenter, phase II trial of sunitinib in previously treated, advanced non-small-cell lung cancer. J Clin Oncol. 2008;26:650–656. doi: 10.1200/JCO.2007.13.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed B, Bu HZ, Pool WF, Peng GW, Wu EY, Patyna S, et al. Pharmacokinetics, distribution, and metabolism of [14C]sunitinib in rats, monkeys, and humans. Drug Metab Dispos. 2012;40:539–555. doi: 10.1124/dmd.111.042853. [DOI] [PubMed] [Google Scholar]
- Speed W, Bello C, Peng G, Patyna S, Wu E. 2008. In vitro and in vivo metabolism of sunitinib in nonclinical species and humans. AACR Meeting Abstracts 2008: 1285.
- Stepan AF, Walker DP, Bauman J, Price DA, Baillie TA, Kalgutkar AS, et al. Structural alert/reactive metabolite concept as applied in medicinal chemistry to mitigate the risk of idiosyncratic drug toxicity: a perspective based on the critical examination of trends in the top 200 drugs marketed in the United States. Chem Res Toxicol. 2011;24:1345–1410. doi: 10.1021/tx200168d. [DOI] [PubMed] [Google Scholar]
- Tafazoli S, O'Brien PJ. Amodiaquine-induced oxidative stress in a hepatocyte inflammation model. Toxicology. 2009;256:101–109. doi: 10.1016/j.tox.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Tamai N, Miyasaka H. Ultrafast dynamics of photochromic systems. Chem Rev. 2000;100:1875–1890. doi: 10.1021/cr9800816. [DOI] [PubMed] [Google Scholar]
- Teng WC, Oh JW, New LS, Wahlin MD, Nelson SD, Ho HK, et al. Mechanism-based inactivation of cytochrome P450 3A4 by lapatinib. Mol Pharmacol. 2010;78:693–703. doi: 10.1124/mol.110.065839. [DOI] [PubMed] [Google Scholar]
- Teo YL, Saetaew M, Chanthawong S, Yap YS, Chan EC, Ho HK, et al. Effect of CYP3A4 inducer dexamethasone on hepatotoxicity of lapatinib: clinical and in vitro evidence. Breast Cancer Res Treat. 2012;133:703–711. doi: 10.1007/s10549-012-1995-7. [DOI] [PubMed] [Google Scholar]
- Villard PH, Herber R, Seree EM, Attolini L, Magdalou J, Lacarelle B. Effect of cigarette smoke on UDP-glucuronosyltransferase activity and cytochrome P450 content in liver, lung and kidney microsomes in mice. Pharmacol Toxicol. 1998;82:74–79. doi: 10.1111/j.1600-0773.1998.tb01401.x. [DOI] [PubMed] [Google Scholar]
- Washington C, Eli M, Bello C. The effect of ketoconazole (KETO), a potent CYP3A4 inhibitor, on SU011249 pharmacokinetics (PK) in Caucasian and Asian healthy subjects [abstract 553] J Clin Oncol. 2003;22:553. [Google Scholar]
- Wen B, Moore DJ. Bioactivation of glafenine by human liver microsomes and peroxidases: identification of electrophilic iminoquinone species and GSH conjugates. Drug Metab Dispos. 2011;39:1511–1521. doi: 10.1124/dmd.111.039396. [DOI] [PubMed] [Google Scholar]
- Xie C, Yu K, Zhong D, Yuan T, Ye F, Jarrell JA, et al. Investigation of isomeric transformations of chlorogenic acid in buffers and biological matrixes by ultraperformance liquid chromatography coupled with hybrid quadrupole/ion mobility/orthogonal acceleration time-of-flight mass spectrometry. J Agric Food Chem. 2011;59:11078–11087. doi: 10.1021/jf203104k. [DOI] [PubMed] [Google Scholar]
- Xie C, Zhong D, Chen X. Identification of the ortho-benzoquinone intermediate of 5-O-caffeoylquinic acid in vitro and in vivo: comparison of bioactivation under normal and pathological situations. Drug Metab Dispos. 2012;40:1628–1640. doi: 10.1124/dmd.112.045641. [DOI] [PubMed] [Google Scholar]
- Zhou A, Zhang W, Chang C, Chi Y, Wang J. Tolerability of famitinib malate: the preliminary results from phase I clinical trial. Chin J New Drugs. 2011;20:1678–1690. [Google Scholar]
- Zwick E, Bange J, Ullrich A. Receptor tyrosine kinase signalling as a target for cancer intervention strategies. Endocr Relat Cancer. 2001;8:161–173. doi: 10.1677/erc.0.0080161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.