Abstract
Branchio-oto-renal syndrome is characterized by branchial defects, hearing loss, preauricular pits, and renal anomalies. Mutations in EYA1 are the most common cause of branchio-oto-renal and branchio-otic syndromes. Large chromosomal aberrations of 8q13, including complex rearrangements occur in about 20% of these individuals. However, submicroscopic deletions and the molecular characterization of genomic rearrangements involving the EYA1 gene have rarely been reported. Using the array-comparative genomic hybridization, we identified non-recurrent genomic deletions including the EYA1 gene in three patients with branchio-oto-renal syndrome, short stature, and developmental delay. One of these deletions was mediated by two human endogenous retroviral sequence blocks, analogous to the AZFa microdeletion on Yq11, responsible for male infertility. This report describes the expanded phenotype of individuals, resulting from contiguous gene deletion involving the EYA1 gene and provides a molecular description of the genomic rearrangements involving this gene in branchio-oto-renal syndrome.
Keywords: BOR, EYA1, array-CGH, LTR/ERV1, short stature, developmental delay
INTRODUCTION
Branchio-oto-renal (BOR) syndrome (OMIM #113650) is an autosomal dominant disorder characterized by second branchial arch, otologic, and renal anomalies. Approximately 40% of patients have mutations in the EYA1 gene, the human ortholog of the Drosophila eyes absent gene [Abdelhak et al., 1997]. Mutations in the homeodomain transcription factor Six family, SIX5 are causative in ~5% of individuals with BOR syndrome [Hoskins et al., 2007] and aberrations of the SIX1 gene account for about 4% of patients with the syndrome [Ruf et al., 2004; Kochhar et al., 2008]. Large cytogenetic abnormalities, including complex rearrangements have been estimated to occur in ~20% of individuals with BOR syndrome [Chang et al., 2004]. Such individuals have been identified by G-banded chromosome studies [Haan et al., 1989; Gu et al., 1996] and Southern blot analysis [Abdelhak et al., 1997; Vervoort et al., 2002]. It has been proposed that contiguous gene deletion involving EYA1 can cause oto-facio-cervical (OFC) syndrome (OMIM #166780), characterized by long face, high arched palate, prominent auricles, conductive hearing loss, lateral cervical fistulas, sloping shoulders, winged low, and laterally set scapulae, short stature, and developmental delay [Rickard et al., 2001]. Point mutations in EYA1 have also been reported in rare patients with OFC syndrome [Estefania et al., 2006; Mercer et al., 2006]. Whether BOR and OFC are allelic disorders remains to be established and the question highlights the need for detailed clinical and molecular characterization of additional individuals with contiguous gene deletions involving the EYA1 gene. This report describes the phenotype of three patients with clinical diagnosis of BOR syndrome, who harbor variably sized genomic deletions including the EYA1 gene and lack most of the musculoskeletal characteristics of OFC syndrome. We also demonstrate that LTR/ ERV1-mediated non-allelic homologous recombination is the most likely mechanism of deletion in one of the patients with BOR syndrome.
CLINICAL REPORT
Patient 1
The patient was a product of twin pregnancy, born at 37 weeks gestation by cesarean for breech presentation. His birth weight was 2,040 g (~10th centile), length was 42 cm (<3rd centile), and head circumference was 32 cm (~25th centile). He had feeding difficulties in the first few weeks of life. By 2 weeks of age, he was found to have uremia, with BUN of 63 mg/dl, creatinine of 1.7 mg/dl, and potassium of 6.9 mmol/L. A renal ultrasound showed left renal agenesis and a hypoplastic right kidney, measuring 3 cm in the longest diameter. Ophthalmology exam showed left microphthalmos, left iris and chorioretinal coloboma, sparing the macula. A CT scan of the temporal bones showed bilateral inner ear malformations with incompletely formed cochlea and posterior semicircular canals. Auditory brainstem response (ABR) showed moderate conductive hearing loss in the right ear and mild conductive loss in the left, necessitating the use of hearing aids. He received early childhood intervention for hearing habilitation. He was able to sit unassisted by 8 months of age, pulled up to stand around 12 months of age, and walked independently by 15 months of age. Based on Preschool Language Scale-4, Spanish Edition (SPLS-4) and the Preschool Language Scale-4, English Edition (PLS-4), his comprehension and oral expression were near a 1½ year level at the chronological age of 31 months. His physical examination at 31 months of age showed height (82.8 cm) and head circumference (46.1 cm) 2.5 SD below the mean. His weight was 11.2 kg, ~3rd centile. Dysmorphic features included facial asymmetry with left-sided microphthalmos, left iris coloboma, cup-shaped, simplified asymmetric ears, bilateral preauricular pits, bilateral branchial cleft fistulae, micrognathia, and right clubfoot (Fig. 1A). His renal function was stable with creatinine of 0.5 mg/dl, BUN of 42 mg/ dl, and potassium of 4.8 mmol/L. Karyotype analysis at a resolution of 500–550 bands revealed a 46, XY complement. DNA sequencing of CHD7, SIX1, and PAX2 was normal (data not shown).
FIG. 1.

Photographs of individuals with EYA1 contiguous gene deletions. A: Patient 1 at the age of 15 months, with facial asymmetry, left-sided microphthalmos with iris coloboma, cup-shaped, simplified asymmetric ears, bilateral preauricular pits, bilateral branchial cleft fistulae, and micrognathia. B: Patient 2 at the age of 34 months, with frontal bossing, small, low set ears with bilateral preauricular pits, midfacial hypoplasia, and micrognathia. C: Patient 3 at the age of 7.5 years, with bilateral preauricular ear pits, small right preauricular tag, and large anteriorly protruding ears with cupped helices.
Patient 2
Patient 2 was born at 36 weeks gestation via elective cesarean, with birth weight of 2,650 g (~50th centile), length of 45.4 cm (25th–50th centile), and head circumference of 33 cm (~60th centile). She was evaluated immediately after birth for feeding difficulties and right-sided neck swelling. The ABR study was consistent with mild to moderate sensorineural hearing loss in the right ear and a severe sensorineural hearing loss in the left ear, requiring hearing aids. The renal ultrasound revealed left renal pelviectasis with no evidence of vesicoureteral reflux by voiding cystourethrogram (VCUG). The renal function studies were normal. She began rolling between 8 and 9 months of age, sat unassisted at 11 months of age and walked at 16 months. Using the PLS-4 Spanish Version at 2 years and 3 months of age, her auditory comprehension was assessed at 70% and expressive communication at 78%, falling below the average range of abilities for comprehension and use of spoken language. She received early childhood intervention therapies for mild gross motor delays and hearing loss. On physical exam, at the age of 34 months, her weight was 11.8 kg (~8th centile), height was 83.4 cm (2.5 SD below the mean), and head circumference was 49 cm (~60th centile). Her dysmorphic features included frontal bossing, small, low set ears with bilateral ear pits, midfacial hypoplasia, and micrognathia (Fig. 1B). There was a palpable soft mass in the right neck region consistent with branchial cleft cyst. Her genetic evaluation revealed normal female chromosome analysis at 575 band resolution. Sequence analysis of EYA1 was normal (data not shown).
Patient 3
Patient 3 was born precipitously after abruptio placenta, requiring neonatal resuscitation at birth. Her birth weight was 3.26 kg (~35th centile) and birth length was 50.8 cm (75th–90th centile). After birth, she had significant respiratory distress, feeding and swallowing problems with post-prandial cyanosis, and repeated episodes of aspiration pneumonia with gastroesophageal reflux. She was found to have bilateral branchial cleft defects. The renal ultrasound was normal. Initial VCUG showed grade III–IV vesicoureteral reflux with blunting of the right upper pole and left mid pole calyces. Subsequent VCUG studies showed improvement, with continued lower grade reflux. An ABR showed mild sensorineural hearing loss in the right ear and mild to moderate sensorineural hearing loss in the left ear. Her brain MRI showed mild bilateral cochlear dysplasia with marked enlargement of the endolymphatic sacs and ducts, right greater than left. The VIII nerves were reported to be smaller than normal bilaterally. At 17 months of age, she was found to have hypoglycemia due to growth hormone (GH) deficiency and was started on GH therapy. She also had seizures associated with hypoglycemia and was treated with oxcarbazepine. She had a Nissen fundoplication and a gastrostomy tube placed for feeding difficulties. She had recurrent para-esophageal hernias that were surgically repaired as well as correction of an umbilical hernia. She also had surgery for correction of strabismus. Her family history was remarkable for a younger sister with bilateral preauricular pits, two paternal cousins with preauricular pits and a maternal uncle with renal aplasia. She started rolling over at 10 months of age, sat unassisted at 18 months, crawled at 27 months, was able to stand at 3 years and 10 months, and walked independently at 5 years of age. On physical examination, at the age of 7.5 years, she was found to be non-verbal, and exhibited many autistic features with repetitive stereotypes and perseverative motor activities. Her height was 120 cm (~25th centile while on growth hormone), weight was 23.15 kg (25th–50th centile), and head circumference was 52.5 cm (50th–75th centile). Her dysmorphic features included bilateral preauricular ear pits, a small right preauricular tag, and large anteriorly protruding ears with cupped helices (Fig. 1C). She had mild hypotonia and pes cavus. She had a normal female chromosome analysis at a resolution of 475 bands and normal fluorescence in situ hybridization (FISH) study for 22q11.2 deletion. Plasma amino acids, urine organic acids, and thyroid studies were also normal.
MATERIALS AND METHODS
The clinical diagnosis of BOR was made in all three patients based on the established criteria [Chang et al., 2004]. They had three or more of the major criteria (second branchial arch anomalies, deafness, preauricular pits, auricular deformity, and renal anomalies) or two major and two minor criteria (external auditory canal anomalies, middle ear anomalies, inner ear anomalies, pre-auricular tags, facial asymmetry, or palate abnormalities), required for the diagnosis. Written informed consents were obtained from the guardian of each subject. The study was performed in accordance with the Institutional Guidelines for Human Research with approval by the Institutional Review Board at Baylor College of Medicine.
Clinical Chromosome Microarray Analysis (CMA) and Fluorescence In Situ Hybridization (FISH) Studies
The array-comparative genomic hybridizations (array-CGH), consisting of 42,640 oligonucleotides were performed using CMA V6.3 OLIGO, designed by Baylor Medical Genetics Laboratories and manufactured by Agilent Technology (Santa Clara, CA) [http://www.bcm.edu/geneticlabs/cma/tables.html] [Ou et al., 2008].
FISH experiments on metaphase spreads were performed to confirm the deletion of EYA1 in this region, using BAC clones, RP11 -326E22 in Patient 1, and RP11-403D15 in Patients 2 and 3. Miniprep BAC DNA (100 ng) was labeled with Spectrum Orange -dUTP or Spectrum Green-dUTP (Vysis, Downers Grove, IL), according to the manufacturer’s instructions, and used as probes for FISH analysis using standard protocols [Trask, 1991].
Genome-Wide Oligonucleotide Array-CGH
To precisely map the deletion breakpoints, genome-wide analysis for DNA copy-number alterations was performed in the three patients using the Agilent Human Genome Microarray Kit 244A (Agilent Technology) according to the manufacturer’s specifications, as described [Fan et al., 2007]. DNA samples (~2 μg) from patients and gender-matched normal individuals as reference were digested with AluI and RsaI and labeled with Cy5 or Cy3 using a BioPrime array-CGH genomic labeling module (Invitrogen, Carlsbad, CA).
After hybridization, slides were scanned and analyzed for relative gain or loss of fluorescent signals from hybridization of the patient and reference DNAs. Genomic region analyses were performed according to the human reference sequence Build 36.1 (UCSC genome browser: http://genome.ucsc.edu) with the software provided by Agilent Technology. Additionally, in Patients 1 and 3, analyses were performed using NimbleGen array HG18_ WG_CGH_v1 with 2.1 M oligonucleotides (NimbleGen Systems, Madison, WI) according to the manufacturer’s instructions. The array contained 2.1 million probes of variable length oligonucleotides designed for unique sequence regions. Total genomic patient DNA was directly labeled with 9-prime-Cy5 mers, and the reference DNA was labeled with 9-prime-Cy3. Probes were co-hybridized to the slide using MAUI Hybridization System (BioMicro Systems, Salt Lake City, UT). After post-hybridization washing, the arrays were scanned using a GenePix 4000B Scanner (Molecular Devices Corporation, Sunnyvale, CA). Data were analyzed using NimbleS-can v2.4 software and visualized with SignalMap (Nimblegen Systems). The relative intensity of the test sample versus the reference DNA was indicated on a log 2 scale. A positive result was determined when a segment of oligo probes showed a 0.2-fold average difference from the reference normal DNA.
Long-Range Polymerase Chain Reaction (PCR) and Sequencing
Long-range (LR) PCR reaction was performed to amplify the predicted junction fragments in the breakpoints regions according to the manufacturer’s specifications (Takara Bio, Shiga, Japan). The products of PCR were purified with the PCR Purification Kit (Qiagen, Valencia, CA) and sequenced (Lone Star, Houston, TX).
Bioinformatics and In Silico Sequence Analysis
Genomic sequence based on the oligonucleotide coordinates from the array-CGH experiment was downloaded from the UCSC genome browser (Build 36, UCSC genome browser, March 2006) and assembled using the Sequencher software (Gene Codes, Ann Arbor, MI). Interspersed repeat sequences were analyzed by RepeatMasker [http://www.repeatmasker.org].
RESULTS
Patient 1
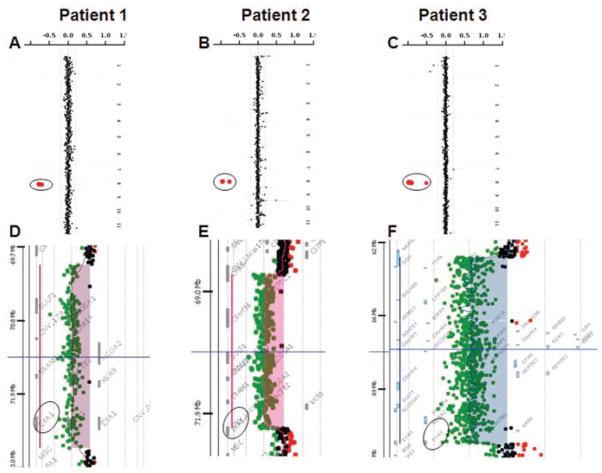
The clinical oligonucleotide-based array-CGH (Fig. 2A) showed an interstitial loss in copy number in 8q13.3 region, involving the EYA1 gene, encompassing at least 0.5 Mb and confirmed by FISH analysis. The genome-wide oligonucleotide array-CGH mapped the distal breakpoint between 72748049 and 72756201 and the proximal breakpoint between 70047512 and 70053688, consistent with ~2.7 Mb deletion (Fig. 2D). The 6.5 kb junction fragment of the deletion was amplified using LR PCR with primers F: CCGTAGCCCTGAATGTTGACTACTATACCC and R: AACAATACGGTGACTGAAAGATGTGTGTTG and sequenced. The distal breakpoint mapped between 72752316 and 72752423 and the proximal breakpoint mapped between 70050577 and 70050684. The DNA sequence analysis of the breakpoint regions revealed that the distal and proximal deletion breakpoints map within the perfect homology portions (107 bp) within the LTR/ERV1 elements on both sides, suggesting non-allelic homologous recombination as the most likely mechanism of deletion. This 2.7 Mb deletion includes the entire EYA1 and six other contiguous genes (Table I). The parental FISH studies were normal, suggesting that the deletion was apparently de novo in this patient.
FIG. 2.
Summary of the results using customized array-CGH and genome-wide array-CGH analyses in Patients 1, 2, and 3. Note that the genomic loss of copy number of EYA1 in the three patients on 8q13.3 identified on the clinical array-CGH (A, B, C circled). Fine mapping of the breakpoints using 244k Human Genome Agilent microarray, showing a 2.7 Mb deletion involving EYA1 and six other contiguous genes in Patient 1 (D), 3.6 Mb deletion in Patient 2 (E), and 8.7 Mb deletion in Patient 3 (F).
TABLE I.
Genes and Related Disorders Within the Deleted Region of 8q13
| Genes | OMIM No. | Name | Disorder | Deleted in patient |
|---|---|---|---|---|
| EYA1 | 601653 | Eyes absent, Drosophila, homolog of, 1 | BOR syndrome (AD) | 1, 2, 3 |
| LACTB2 | Lactamase, beta 2 | 1, 2, 3 | ||
| TRAM1 | 605190 | Translocation-associating membrane protein 1 | 1, 2, 3 | |
| NCOA2 | 601993 | Nuclear receptor coactivator 2 | 1, 2, 3 | |
| PRDM14 | 611781 | PR domain-containing protein 14 | 1,2,3 | |
| SLCO5A1 | Solute carrier organic anion transporter family, member 5A1 | 1, 2, 3 | ||
| SULF1 | 610012 | Sulfatase 1 | 1, 2, 3 | |
| PREX2 | 612139 | DEP domain-containing protein 2 | 2, 3 | |
| CPA6 | 609562 | Carboxypeptidase A6 | 3 | |
| ARFGEF1 | 604141 | ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) | 3 | |
| CSPP1 | 611654 | Centrosome spindle pole-associated protein 1 | 3 | |
| COPS5 | 604850 | COP9, subunit 5 | 3 | |
| C8orf45 | Homo sapiens chromosome 8 open reading frame 45 | 3 | ||
| SGK3 | 607591 | Serum/glucocorticoid-regulated kinase-like protein | 3 | |
| VCPIP1 | 611745 | VCP/p47 complex-interacting protein 1 | ||
| MYBL1 | 159405 | V-Myb Avian Myeloblastosis viral oncogene homolog-like 1 | 3 | |
| C8orf46 | Homo sapiens chromosome 8 open reading frame 46 | 3 | ||
| ADHFE1 | 611083 | Alcohol dehydrogenase, iron-containing, 1 | 3 | |
| RRS1 | RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) | 3 | ||
| CRH | 122560 | Corticotropin-releasing hormone | 3 | |
| TRIM55 | Tripartite motif-containing 55 | 3 | ||
| DNAJC5B | DnaJ (Hsp40) homolog, subfamily C, member 5 beta | 3 | ||
| PDE7A | 171885 | Phosphodiesterase 7A | 3 | |
| MTFR1 | Mitochondrial fission regulator 1, nuclear gene encoding mitochondrial protein | 3 | ||
| CYP7B1 | 603711 | Cytochrome p450, family 7, subfamily B, polypeptide 1 | Congenital bile acid synthesis defect; Spastic Paraplegia 5A (AR) | 3 |
| BHLHE22 | Basic helix-loop-helix family, member e22 | 3 | ||
| YTHDF3 | YTH domain family, member 3 | 3 | ||
| TTPA | 600415 | Tocopherol transfer protein, alpha | Ataxia, friedreich-like with vitamin E deficiency (AVED) (AR) | 3 |
| GGH | 601509 | Gamma-glutamyl hydrolase | 3 |
AR, autosomal recessive; AD, autosomal dominant.
Patient 2
The clinical oligonucleotide-based array-CGH showed an interstitial loss in copy number spanning a minimum of 3.2 Mb in the 8q13.3 region, involving the 3′ end of the EYA1 gene (Fig. 2B). The genome-wide oligonucleotide array-CGH mapped the distal breakpoint between 72285931 and 72291783 and the proximal breakpoint between 68678326 and 68683974 (Fig. 2E), encompassing approximately 3.6 Mb. The 4 kb junction fragment of the deletion was amplified using PCR with primers F: CCGTTCTCTGGTTTTCTCTAGGAGGTTCTC and R: GGGTCGTTCACAGCTGTTAGAGGATAACTG and sequenced. The distal deletion breakpoint maps within a unique sequence and the proximal breakpoint maps within LTR/ERVL-MalR element. The distal breakpoint mapped between 72290630 and 72290633 and the proximal breakpoint mapped between 68682573 and 68682576. This 3.6 Mb deletion removed exons 15–18 in the isoform b and 13–16 in the isoform a of EYA1, in addition to seven other proximal genes (Table I). This deletion was apparently de novo in the patient after determining normal parental FISH analyses.
Patient 3
The clinical array-CGH analysis showed a loss in copy number in the 8q12.3–q13.3 region, encompassing a segment of at least 6.4 Mb in size and confirmed by FISH analysis (Fig. 2C). The oligonucleotide array-CGH mapped the distal breakpoint between 72390314 and 72391812 and the proximal breakpoint between 63665198 and 63666882, consistent with ~8.7 Mb deletion (Fig. 2F). The 2 kb junction fragment of the deletion was amplified using PCR with primers F: CTATCTAGTATACAGTGTGGACTGCCCTCTGG and R: AGTCAGCCACGTAGGAGGTTGTGATATG and sequenced. The distal deletion breakpoint was mapped within a unique sequence and the proximal breakpoint within short interspersed repeat element, SINE/MIR. The distal breakpoint mapped between 72391528 and 72391530 and the proximal breakpoint mapped between 63666394 and 63666396. This 8.7 Mb deletion removed exons 8–18 in the isoform b and exons 6–16 in isoform a. The parental samples were unavailable for further studies.
DISCUSSION
This report describes the molecular characterization of genomic rearrangements involving the EYA1 gene in three individuals with BOR syndrome. Large, cytogenetically visible complex rearrangements, involving EYA1, result in variable phenotype including BOR/BO with developmental delay and other congenital anomalies [Haan et al., 1989; Gu et al., 1996]. Vincent et al. [1994] described complex phenotype comprised of BOR syndrome, Duane syndrome, hydrocephalus, and trapezius aplasia resulting from large deletion of 8q12.2–q21.2. Contiguous gene deletion including the EYA1 gene has been reported in an individual described with OFC syndrome [Rickard et al., 2001], with moderate hearing loss, bilateral preauricular pits, a cupped auricle, sloping shoulders, laterally displaced scapulae, small left kidney with a bifid right pelvis, short stature, and mild developmental delay. While all three of the patients reported here with deletion of EYA1 and contiguous genes had a clinical diagnosis of BOR syndrome, they lacked the typical facies and most of the skeletal features characteristic of OFC syndrome. The BOR syndrome typically manifests variable expressivity among and within families [Heimler and Lieber, 1986]. Based on the reports of single-nucleotide substitutions in EYA1 in OFC syndrome [Estefania et al., 2006], and in an individual with features of both OFC and BOR syndromes [Mercer et al., 2006], it is plausible that OFC syndrome is a manifestation of the severe end of the spectrum of the musculoskeletal abnormalities due to EYA1 aberrations. Indeed, long narrow facies and palatal abnormalities were described in some of the earliest reports of large dominant pedigrees, affected with BOR syndrome [Melnick et al., 1976; Heimler and Lieber, 1986]. Short stature, a well-defined feature of OFC, was observed in all three of the patients reported here, presenting as intrauterine growth retardation in Patient 1 and necessitating growth hormone treatment in Patient 3. Mild developmental delay is a consistent feature of OFC syndrome, although speech delay due to hearing loss is not unusual in BOR syndrome. Mild speech delay was observed in Patients 1 and 2, while Patient 3 with a larger 8.7 Mb deletion had moderate-severe developmental delay, in part, due to hypoxic ischemic injury at birth. Our data suggest that in the absence of a well-defined molecular diagnosis of OFC syndrome, multiple overlapping features of this syndrome with BOR, and the apparent variable expressivity due to EYA1 mutations, OFC is likely a representation of furthest expression of skeletal phenotype due to EYA1 disruption.
Despite the fact that EYA1 plays an important role in eye development [Bonini et al., 1993], eye defects are rarely reported in humans with EYA1 haploinsufficiency. Mice homozygous for Eya1 deletion die perinatally because of severe craniofacial defects and often exhibit open eyelids [Xu et al., 1999]. In this series, Patient 1 had unilateral microphthalmos, and iris and chorioretinal colobomas. To exclude a concurrent mutation in the SIX1 gene and to exclude other co-morbid conditions responsible for coloboma in this individual, particularly CHARGE (OMIM #214800) and Renal-Coloboma syndrome (OMIM #120330), we additionally sequenced the coding region and the intron–exon boundaries of SIX1, CHD7, and PAX2, and did not detect any mutations in these genes (data not shown). It is possible that another dosage sensitive gene within the deletion interval in this patient is important for the patterning of the human eye (Table I). Alternatively, a functional polymorphism of the EYA1 hemizygous allele could potentially be responsible for this phenotype.
The vertebrate Eya1 is essential for the formation of multiple organs during embryonic development. Eya1-deficient mice show anomalies in the formation of the inner ear, kidney, cranial ganglia, and branchial arch derivatives [Xu et al., 1999; Zou et al., 2006]. EYA1 has 16 coding exons that extend over 156 kb. It has at least four alternatively spliced transcripts. The 8q13 deletion in Patient 2 removes exons 15–18 in isoform b and exons 8–18 in Patient 3. In both of these patients, the distal breakpoints map within unique sequences within the EYA1 gene, and the proximal breakpoint maps within repetitive elements, LTR/ERVL-MalR and SINE/MIR, respectively. In Patient 1, the EYA1 deletion is mediated by the adjacent ERV1 blocks. The breakpoints map within the perfect homology portions (107 bp) within the LTR/ERV1 elements, suggesting non-allelic homologous recombination (NAHR) as the most likely mechanism of deletion. Human Endogenous Retroviral sequences (HERVs) which are remnants of past retroviral infection, make up about 8% of our genome [Bock and Stoye, 2000]. Homologous recombination between distant HERVs has been shown to cause genomic rearrangements, as in the case of inherited male infertility due to the loss of the 792-kb fragment encompassing the Azoospermia factor, AZFa by intrachromosomal recombination on Yq11 [Kamp et al., 2000]. In addition, HERV loci have been implicated in the diversity and stability of the human MHC class II and in insertion/deletion and duplication events in class I region of the human MHC [Andersson et al., 1998; Kulski et al., 2005]. The deletions observed in our series are non-recurrent. Although identified to be responsible for genomic rearrangement in Patient 1, HERV-mediated NAHR does not appear to be a common deletion mechanism in individuals with EYA1 deletions.
In summary, we have demonstrated the clinical spectrum associated with contiguous gene deletions involving EYA1, and molecularly characterized these novel deletions in three patients with BOR syndrome. Our data suggest that contiguous gene deletions of EYA1 cause BOR phenotype with variable musculoskeletal manifestations and developmental delay.
Acknowledgments
We thank the patients and their families who participated in this study. This work was supported by the Doris Duke Charitable Foundation and the Gillson Longenbaugh Foundation (SRL).
Grant sponsor: Doris Duke Charitable Foundation; Grant sponsor: Gillson Longenbaugh Foundation (SRL).
References
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal [BOR] syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- Andersson G, Svensson AC, Setterblad N, Rask L. Retroelements in the human MHC class II region. Trends Genet. 1998;14:109–114. doi: 10.1016/s0168-9525(97)01359-0. [DOI] [PubMed] [Google Scholar]
- Bock M, Stoye JP. Endogenous retroviruses and the human germline. Curr Opin Genet Dev. 2000;10:651–655. doi: 10.1016/s0959-437x(00)00138-6. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: Genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Chang EH, Menezes M, Meyer NC, Cucci RA, Vervoort VS, Schwartz CE, Smith RJ. Branchio-oto-renal syndrome: The mutation spectrum in EYA1 and its phenotypic consequences. Hum Mutat. 2004;23:582–589. doi: 10.1002/humu.20048. [DOI] [PubMed] [Google Scholar]
- Estefania E, Ramirez-Camacho R, Gomar M, Trinidad A, Arellano B, Garcia-Berrocal JR, Verdaguer JM, Vilches C. Point mutation of an EYA1-gene splice site in a patient with oto-facio-cervical syndrome. Ann Hum Genet. 2006;70:140–144. doi: 10.1111/j.1529-8817.2005.00204.x. [DOI] [PubMed] [Google Scholar]
- Fan YS, Jayakar P, Zhu H, Barbouth D, Sacharow S, Morales A, Carver V, Benke P, Mundy P, Elsas LJ. Detection of pathogenic gene copy number variations in patients with mental retardation by genomewide oligonucleotide array comparative genomic hybridization. Hum Mutat. 2007;28:1124–1132. doi: 10.1002/humu.20581. [DOI] [PubMed] [Google Scholar]
- Gu JZ, Wagner MJ, Haan EA, Wells DE. Detection of a megabase deletion in a patient with branchio-oto-renal syndrome [BOR] and tricho-rhino-phalangeal syndrome [TRPS]: Implications for mapping and cloning the BOR gene. Genomics. 1996;31:201–206. doi: 10.1006/geno.1996.0032. [DOI] [PubMed] [Google Scholar]
- Haan EA, Hull YJ, White S, Cockington R, Charlton P, Callen DF. Tricho-rhino-phalangeal and branchio-oto syndromes in a family with an inherited rearrangement of chromosome 8q. Am J Med Genet. 1989;32:490–494. doi: 10.1002/ajmg.1320320412. [DOI] [PubMed] [Google Scholar]
- Heimler A, Lieber E. Branchio-oto-renal syndrome: Reduced penetrance and variable expressivity in four generations of a large kindred. Am J Med Genet. 1986;25:15–27. doi: 10.1002/ajmg.1320250104. [DOI] [PubMed] [Google Scholar]
- Hoskins BE, Cramer CH, Silvius D, Zou D, Raymond RM, Orten DJ, Kimberling WJ, Smith RJ, Weil D, Petit C, Otto EA, Xu PX, Hildebrandt F. Transcription factor SIX5 is mutated in patients with branchio-oto-renal syndrome. Am J Hum Genet. 2007;80:800–804. doi: 10.1086/513322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- Kochhar A, Orten DJ, Sorensen JL, Fischer SM, Cremers CW, Kimberling WJ, Smith RJ. SIX1 mutation screening in 247 branchio-oto-renal syndrome families: A recurrent missense mutation associated with BOR. Hum Mutat. 2008;29:565. doi: 10.1002/humu.20714. [DOI] [PubMed] [Google Scholar]
- Kulski JK, Anzai T, Inoko H. ERVK9, transposons and the evolution of MHC class I duplicons within the alpha-block of the human and chimpanzee. Cytogenet Genome Res. 2005;110:181–192. doi: 10.1159/000084951. [DOI] [PubMed] [Google Scholar]
- Melnick M, Bixler D, Nance WE, Silk K, Yune H. Familial branchio-oto-renal dysplasia: A new addition to the branchial arch syndromes. Clin Genet. 1976;9:25–34. doi: 10.1111/j.1399-0004.1976.tb01546.x. [DOI] [PubMed] [Google Scholar]
- Mercer C, Gilbert R, Loughlin S, Foulds N. Patient with an EYA1 mutation with features of branchio-oto-renal and oto-facio-cervical syndrome. Clin Dysmorphol. 2006;15:211–212. doi: 10.1097/01.mcd.0000204986.54366.7c. [DOI] [PubMed] [Google Scholar]
- Ou Z, Kang SH, Shaw CA, Carmack CE, White LD, Patel A, Beaudet AL, Cheung SW, Chinault AC. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard S, Parker M, van’t Hoff W, Barnicoat A, Russell-Eggitt I, Winter RM, Bitner-Glindzicz M. Oto-facio-cervical [OFC] syndrome is a contiguous gene deletion syndrome involving EY A1: Molecular analysis confirms allelism with BOR syndrome and further narrows the Duane syndrome critical region to 1 cM. Hum Genet. 2001;108:398–403. doi: 10.1007/s004390100495. [DOI] [PubMed] [Google Scholar]
- Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM, Jr, Brophy PD, Berkman J, Gattas M, Hyland V, Ruf EM, Schwartz C, Chang EH, Smith RJ, Stratakis CA, Weil D, Petit C, Hildebrandt F. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1–SIX1–DNA complexes. Proc Natl Acad Sci USA. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask BJ. Fluorescence in situ hybridization: Applications in cytogenetics and gene mapping. Trends Genet. 1991;7:149–154. doi: 10.1016/0168-9525(91)90378-4. [DOI] [PubMed] [Google Scholar]
- Vervoort VS, Smith RJ, O’Brien J, Schroer R, Abbott A, Stevenson RE, Schwartz CE. Genomic rearrangements of EYA1 account for a large fraction of families with BOR syndrome. Eur J Hum Genet. 2002;10:757–766. doi: 10.1038/sj.ejhg.5200877. [DOI] [PubMed] [Google Scholar]
- Vincent C, Kalatzis V, Compain S, Levilliers J, Slim R, Graia F, Pereira ML, Nivelon A, Croquette MF, Lacombe D. A proposed new contiguous gene syndrome on 8q consists of Branchio-Oto-Renal (BOR) syndrome, Duane syndrome, a dominant form of hydrocephalus and trapeze aplasia; implications for the mapping of the BOR gene. Hum Mol Genet. 1994;3:1859–1866. doi: 10.1093/hmg/3.10.1859. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Zou D, Silvius D, Rodrigo-Blomqvist S, Enerback S, Xu PX. Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas in the inner ear. Dev Biol. 2006;298:430–441. doi: 10.1016/j.ydbio.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]