Abstract
An important research application of amyloid imaging with positron emission tomography (PET) is detection of the earliest evidence of fibrillar amyloid-beta (Aβ) deposition. Use of amyloid PET for this purpose, requires a reproducible method for defining a cutoff that separates individuals with no significant Aβ deposition from those in which Aβ deposition has begun. We previously reported the iterative outlier approach (IO) for the analysis of Pittsburgh Compound-B (PiB) PET data. Developments in amyloid imaging since the initial report of IO have led us to re-examine the generalizability of this method. IO was developed using full-dynamic atrophy-corrected PiB PET data obtained from a group of control subjects with a fairly distinct separation between PiB-positive [PiB(+)] and PiB-negative [PiB(−)] subjects.
Methods
We tested the performance of IO using late-summed tissue ratio data with atrophy correction or with an automated template method without atrophy correction and tested the robustness of the method when applied to a cohort of older subjects in which separation between PiB(+) and PiB(−) subjects was not so distinct.
Results
The IO method did not perform consistently across analyses and performed particularly poorly when separation was less clear. We found that a sparse k-means (SKM) cluster analysis approach performed significantly better; performing more consistently across methods and subject cohorts. We also compared SKM to a consensus visual read approach and found very good correspondence.
Conclusion
The visual read and SKM methods, applied together, may optimize the identification of early Aβ deposition. These methods have the potential to provide a standard approach to the detection of PiB-positivity that is generalizable across centers.
Keywords: Amyloid, Positron Emission Tomography, Pittsburgh Compound B, Visual Read, Cluster analysis
1. Introduction
Since the initial amyloid imaging studies using PiB (Klunk et al., 2004), it has become widely accepted that this technique provides a quantitative representation of fibrillar Aβ deposition in the brain (Ikonomovic et al., 2008). While the initial focus was on the robust signal seen in symptomatic AD, emphasis in many research studies has moved towards detection of the earliest signs of Aβ deposition in cognitively normal individuals (Klunk & Mathis, 2009). This shift towards initial detection has generated a need for reliable methods that can distinguish brains free of fibrillar Aβ from brains that have early-stage Aβ deposition and that such methods can be standardized and applied across centers.
It should be noted that both fibrillar Aβ deposition and PiB retention are continuous measures and the latter need not be dichotomized into PiB(+) and PiB(−). Many studies have used PiB retention as a continuous variable, correlating PiB retention to a variety of measures (Apostolova et al., 2010; Furst et al., 2010; Mormino et al., 2009; Pike et al., 2007; Rentz et al., 2010; Resnick et al., 2010). While this approach is appealing and, perhaps, preferred for some applications, in other applications it is necessary to dichotomize subjects into those that have no evidence of amyloid deposition and those that are along the continuum of amyloid deposition. This dichotomy is perhaps most important in cognitively normal groups when one attempts to discern effects of normal aging from effects of preclinical AD (Sperling et al., 2011).
Previous studies have presented a variety of approaches to define amyloid-positive cutoffs using PiB PET (e.g. (Jack et al., 2008; Kemppainen et al., 2007; Klunk et al., 2004; Mintun et al., 2006; Mormino et al., 2012; Rowe et al., 2007)). Some have focused on visual reads (Johnson et al., 2007; Ng et al., 2007; Rabinovici et al., 2007; Suotunen et al., 2010; Tolboom et al., 2010), others have used more standard statistical approaches including receiver operating characteristic (ROC) analyses (Devanand et al., 2010; Mormino et al., 2009; Ng et al., 2007; Pike et al., 2007), which requires reference group membership such as AD diagnosis, and cluster analysis (Bourgeat et al., 2010), which does not require reference group membership. We have previously reported an ad hoc method termed the iterative-outlier approach (IO) (Aizenstein et al., 2008). When applied to distribution volume ratio (DVR) data from the first 62 consecutive cognitively normal control subjects studied in our center, this method appeared to provide a good, objective approach to defining PiB(−)/PiB(+) cutoffs.
In the present study, we examine the performance of IO when extended beyond application to DVR, atrophy-corrected data (since these data are rarely available in most centers) and beyond the original set of 62 control subjects. We examined generalization of IO to several analyses including: late-summed data; manual, atrophy-corrected vs. automated-template uncorrected ROIs; global vs. regional approaches; and different cohorts. We paid particular attention to very elderly cohorts in which the frequency of amyloid deposition is likely to be greater than that in our original, 62 controls (Savva et al., 2009).
Here we report the problems of extending IO in these ways. We then explored an alternative method by using SKM cluster analysis to define cutoffs (Witten & Tibshirani, 2010). We refined this method using re-sampling to obtain more robust regional weights and precision in defining cutoffs. We also compared this SKM method to a consensus panel rating of visual reads. When discrepancies were observed between the SKM and visual rating of PiB-positivity on baseline scans, SKM ratings of longitudinal follow-up scans were used to interpret the discrepancies.
2. MATERIALS AND METHODS
2.1 Human Subjects
2.1.1 Original 62 Cognitively Normal Controls
We included the first 62 consecutive cognitively normal controls that were studied with PiB PET in Pittsburgh. Forty-three were community volunteers (Aizenstein et al., 2008) and 19 were volunteers at the University of Pittsburgh Alzheimer’s Disease Research Center (ADRC). Fifty-four were elderly (mean age 72.9±7.2yrs) and 8 were ≤ 55yrs (35–55yrs). The average age of the complete cohort was 69.4±11.5yrs. All subjects were evaluated with standard neuropsychological test battery, designed to assess those areas of cognition known to be impaired in AD and also to be sensitive to Mild Cognitive Impairment (MCI) (Petersen, 2004). All subjects provided informed consent for both the ADRC clinical examination and PET imaging. All studies were approved by the Human Use Subcommittee of the Radioactive Drug Research Committees and the Institutional Review Board of the University of Pittsburgh.
2.1.2 Older 152 Cognitively Normal Controls
We included a group of 152 cognitively normal controls that were studied with PiB PET and were the subject of a separate report (Mathis et al., in press). The average age of this older cohort was 85.4 ± 2.8 yrs, with a range of 82–95 years. As with the original 62 controls, all subjects were evaluated with a standard neuropsychological test battery and were adjudicated as cognitively normal.
2.2 Imaging Studies
[11C]PiB was produced as previously described (Price et al., 2005). Prior to PiB-PET, a 1.5T spoiled-gradient-recalled-MR was obtained for each subject for co-registration and region-of-interest (ROI) definition (Price et al., 2005). PET imaging was conducted using a Siemens/CTI ECAT HR+ (3D mode, 15.2 cm field-of-view, 63 planes, reconstructed image resolution ~6 mm FWHM). The subject’s head was immobilized to minimize head motion. PiB was injected intravenously (12–15 mCi, over 20 sec, specific activity ~1–2 Ci/µmol) and dynamic PET scanning was performed over 90 minutes (34 frames). Analysis of the PiB PET data utilized a Logan graphical analysis of dynamic PiB PET scan with cerebellum as reference (using 35–90 min of data) to produce a DVR (where DVR = VT/VND) (Lopresti et al., 2005) or utilizing standardized uptake value ratio (SUVR) (determined 40–60 and 50–70 min post-injection). To determine SUVR, SUV was determined by normalizing regional tissue radioactivity concentration to injected dose and body mass and each regional SUV was divided by the cerebellar reference SUV that was representative of free and nonspecific radiotracer retention. Thus, the final SUVR is equivalent to a tissue ratio. All subjects received a single PET acquisition that was re-analyzed to generate the various datasets used in each of the comparisons discussed below.
The baseline full resolution MR was resliced along the AC-PC line and down-sampled to PET voxel space. After addressing any motion, the PiB PET data was summed to form a static image and co-registered to the down-sampled MR image. The longitudinal, follow-up MR was first co-registered to the baseline MR before co-registering the longitudinal, follow-up PiB PET data. The MR-based partial volume correction corresponded to a two-compartment approach correcting PiB-PET data for diluting effects of expanding cerebrospinal spaces accompanying cerebral atrophy (Meltzer et al., 1999). The definition of both manual and template ROI’s have been previously described in detail (Cohen et al., 2009; Rosario et al., 2011).
2.3 Definition of Cutoffs
2.3.1 Iterative-outlier (IO) Approach
A “mild” outlier was defined as PiB retention that exceeded the third quartile by >1.5 times the interquartile range (Q3+1.5×IQR) or was beneath the first quartile by an amount <1.5 times the interquartile range (Q1-1.5×IQR), that is, any observation outside the “inner fences” of the box-and-whisker plot (Schwertman & de Silva, 2007). Brain regions used for the IO calculations were the seven defined for the original IO method [anterior cingulate (ACG); frontal cortex (FRC); lateral temporal cortex (LTC); lateral occipital cortex (OCC); occipital pole (OCP); parietal cortex (PAR); and precuneus cortex (PRC)] (Aizenstein et al., 2008). After determining which subjects were outliers in any one (or more) of these neocortical brain regions, we removed those subjects and repeated the calculation until there were no outliers remaining. The cutoff values were thus defined as the upper-inner fence of remaining control subjects for each of the seven brain regions. For the regional analyses, any subject who had PiB retention values exceeding the cutoff point for any one (or more) of these seven brain regions was defined as PiB(+). For the global analyses, a composite cortical measure was used by averaging the PiB retention in the seven neocortical regions for each subject and the IO calculation was applied to this single measure.
2.3.2 Clustering Approach- Sparse K-means Clustering (SKM) + Resampling
The method of SKM (Witten & Tibshirani, 2010) was applied to identify PiB(+) and PiB(−) subgroups (k=2). The analysis was performed using statistical computing software R (R development core team, 2011; http://www.r-project.org/) separately on each dataset (e.g. DVR, SUVR40-60, SUVR50-70; manual and template; regional and global ROIs) initially using all ROI’s measured in our PiB analysis [a total of 13 ROIs; see (Cohen et al., 2009; Rosario et al., 2011)]. Unlike the commonly used methods such as k-means (Hartigan & Wong, 1979) hierarchical clustering and model-based clustering, in which all variables are treated equally, the SKM method achieves variable selection (i.e., weighting) while grouping subjects into clusters. This is desirable because brain regions differ in their importance to grouping, for example amyloid deposition levels in anterior cingulate vary more than those in the sensorimotor cortex and should play a bigger role in the classification of subjects into clusters by PiB retention. A weight was calculated for each brain region such that the larger the weight, the more important the region to the clustering. All analysis presented here utilized the six most relevant ROI’s determined in an unbiased fashion by SKM. As described below in results, these included five areas common to the IO method (ACG, FRC, LTC, PAR and PRC) and a sixth ROI unique to the SKM analysis anteroventral striatum (AVS). In general the goal of clustering is to minimize the within-cluster difference while maximizing the between-cluster differences (Hastie et al., 2009). This difference, also called a dissimilarity measure, was defined as squared Euclidean distance multiplied by regional weight in SKM. The final results of SKM are the cluster membership and variable weights. To allow comparison of these weights across different SKM results, we normalized weights by dividing by the sum of weights in each analysis, so weights summed to 1.00 in each clustering.
We used a method combining SKM and a resampling method that is an extension of tight clustering methods (Bi et al., 2011). We resampled each of the eight datasets 1000 times and applied SKM to each sub-sample. For each resampling run, we randomly sampled 70% (i.e., 43/62 subjects) of subjects without replacement, calculated a PiB(+) and PiB(−) cluster and then predicted the cluster membership for the remaining 30% of subjects. This method can obtain average variable weights across resampling runs and a confidence level representing the observed portion of the observation appearing in a cluster among resampling runs. This confidence level is computed as b/B where b is the number of times a subject appears in a specified cluster and B is the total number of resampling runs (1,000). We then constructed tight-clusters including only subjects with a confidence level ≥96% in a specified cluster. This resulted in 9/62 subjects with a ≥96% confidence of being in the PiB(+) cluster and 41/62 subjects with a ≥96% confidence of being in the PiB(−) cluster. We computed tight-cluster centers as centroids of each tight cluster and obtained regional cutoffs as the mid-point between these two tight-cluster centers. Global cortical SKM cutoffs were calculated as weighted averages of regional cutoffs.
2.4 Visual Rater Approach
Five raters (all experienced neuroimaging researchers; ADC, HJA, EM, JMM, WEK) individually rated scans in a group session. Following one training session using PiB-PET images from amyloid-positive and amyloid-negative MCI subjects not included here, raters were asked to rate the 62 control subjects as positive or negative. First, parametric SUVR50-70 images were displayed in a “Hot Metal” scale and readers were instructed to look for high grey matter signal (grey matter signal ≥ signal in white matter). High signal in any grey matter brain area was scored as PiB(+). While readers could view any slice, the following procedure was used to guide viewing. All slices were simultaneously observed for evidence of obvious positive signal. If present, a positive rating could be made. If no obvious positive signal was observed, at minimum, the slices listed below were examined further before a final decision was made. These slices included every fourth axial slice from the first appearance of lateral ventricles to the cerebellar peduncles (with special attention to the anterior and posterior midline) and four sagittal slices to both the left and right of the midline (with special attention to precuneus and inferior frontal regions). Any regions suspect for positive signal could be viewed in orthogonal views for better localization. After raters individually scored cases without discussion, a consensus outcome measure was determined for each subject. In every case, the consensus outcome (after discussion) was identical to the majority outcome of individual ratings before discussion. All 62 baseline scans were rated as either PiB(+) or PiB(−).
2.5 Examination Of Follow-up Data
Fifty-one of the original 62 subjects received follow-up PiB-PET 1–3 years after baseline scans. Follow-up data was analyzed identically to the baseline data using SUVR50-70 data with cerebellum as a reference region generated using the automated template method and was characterized using cutoffs defined with baseline data by SKM.
2.6 Agreement Between Methods
Agreement for the assignment of PiB(+) or PiB(−) status between any two comparable methods was determined by Cohen’s kappa (Cohen, 1960) and also expressed as overall agreement as well as agreement across PiB(+) cases (positive agreement) and agreement across PiB(−) cases (negative agreement (Cicchetti & Feinstein, 1990; Spitzer & Fleiss, 1974).
3. RESULTS
In Table 1, method comparisons are expressed as overall agreement across all 62 subjects in the original cohort (Aizenstein et al., 2008) as well as separate positive agreement for PiB(+) cases and negative agreement for PiB(−) cases. In addition, a Cohen’s kappa value is given for the overall agreement. These methods included both the IO and SKM approaches. For both approaches, cutoffs were derived from the original cohort of 62 controls using three versions of the same PiB PET data: DVR, SUVR40-60 and SUVR50-70. For each version of the PiB PET data, results were calculated using: 1) a partial-volume (i.e., atrophy)-corrected set of manually-drawn “regional” ROIs (MAN-r); 2) an uncorrected set of template-based regional ROIs (Tem-r); and 3) a single, uncorrected, template-based composite global ROI (Tem-g). Finally, for the SUVR50-70 dataset, both IO and SKM cutoffs were calculated using a distinct set of 152 older controls (152OLDER) instead of the original 62 controls using both the Tem-r and Tem-g ROIs. For comparison to the other analyses, these “152OLDER” cutoffs were then used to define the PiB(+) and PiB(−) status of the 62 original controls.
Table 1.
Comparison of regional analysis method
| IO overall % agreement |
SKM overall % agreement |
IO Positive agreement |
SKM Positive agreement |
IO Negative agreement |
SKM Negative agreement |
||
|---|---|---|---|---|---|---|---|
|
Regional: Manual vs. Manual |
MAN-rDVR vs. MAN-rSUVR50-70 |
59/62 (95.2%) κ=0.876 |
61/62 (98.4%) κ=0.957 |
30/33* (90.9%) |
30/31* (96.8%) |
88/91* (96.7%) |
92/93* (98.9%) |
|
MAN-rDVR vs. MAN-rSUVR40-60 |
54/62 (87.1%) κ=0.710 |
61/62 (98.4%) κ=0.957 |
32/40 (80.0%) |
30/31 (96.8%) |
76/84 (90.5%) |
92/93 (98.9%) |
|
|
MAN-rSUVR50-70 vs. MAN-rSUVR40-60 |
53/62 (85.5%) κ=0.677 |
62/62 (100%) κ=1.0 |
32/41 (78.0%) |
32/32 100%) |
74/83 (89.2%) |
92/92 (100%) |
|
|
Regional: Manual vs. Template |
MAN-rDVR vs. Tem-rDVR |
53/62 (85.5%) κ=0.668 |
58/62 (93.5%) κ=0.832 |
30/39 (76.9%) |
28/32 (87.5%) |
76/85 (89.4%) |
88/92 (95.7%) |
|
MAN-rSUVR50-70 vs. Tem-rSUVR50-70 |
42/62 (67.7%) κ=0.390 |
52/62 (83.9%) κ=0.597 |
32/52 (61.5%) |
24/34 (70.6%) |
52/72 (72.2%) |
80/90 (88.9%) |
|
|
MAN-rSUVR40-60 vs. Tem-rSUVR40-60 |
57/62 (91.9%) κ=0.826 |
55/62 (88.7%) κ=0.699 |
40/45 (88.9%) |
24/31 (77.4%) |
74/79 (93.7%) |
86/93 (92.5%) |
|
|
Regional: Template vs. Template |
Tem-rDVR vs. Tem-rSUVR50-70 |
48/62 (77.4%) κ=0.563 |
55/62 (88.7%) κ=0.722 |
44/58 (75.9%) |
28/35 (80.0%) |
52/66 (78.8%) |
82/89 (92.1%) |
|
Tem-rDVR vs. Tem-rSUVR40-60 |
57/62 (91.9%) κ=0.822 |
58/62 (93.5%) κ=0.832 |
38/43 (88.4%) |
28/32 (87.5%) |
76/81 (93.8%) |
88/92 (95.7%) |
|
|
Tem-rSUVR50-70 vs. Tem-rSUVR40-60 |
48/62 (77.4%) κ=0.566 |
59/62 (95.2%) κ=0.881 |
42/56 (75.0%) |
32/35 (91.4%) |
54/68 (79.4%) |
86/89 (96.6%) |
|
|
Regional Template vs. Global Template |
Tem-rDVR vs. Tem-gDVR |
50/62 (80.6%) κ=0.536 |
57/62 (91.9%) κ=0.766 |
22/34 (64.7%) |
22/27 (81.5%) |
78/90 (86.7%) |
92/97 (94.8%) |
|
Tem-rSUVR50-70 vs. Tem-gSUVR50-70 |
32/62 (51.6%) κ=0.127 |
54/62 (87.1%) κ=0.656 |
10/40 (25.0%) |
22/30 (73.3%) |
54/84 (64.3%) |
86/94 (91.5%) |
|
|
Tem-rSUVR40-60 vs. Tem-gSUVR40-60 |
47/62 (75.8%) κ=0.346 |
57/62 (91.9%) κ=0.766 |
12/27 (44.4%) |
22/27 (81.5%) |
82/97 (84.5%) |
92/97 (94.8%) |
|
|
Global: Template vs. Template |
Tem-gDVR vs. Tem-gSUVR50-70 |
56/62 (90.3%) κ=0.664 |
62/62 (100%) κ=1.0 |
10/16 (62.5%) |
22/22 (100%) |
102/108 (94.4%) |
102/102 (100%) |
|
Tem-gDVR vs. Tem-gSUVR40-60 |
57/62 (91.9%) κ=0.664 |
62/62 (100%) κ=1.0 |
12/17 (70.6%) |
22/22 (100%) |
102/107 (95.3%) |
102/102 (100%) |
|
|
Tem-gSUVR50-70 vs. Tem-gSUVR40-60 |
61/62 (98.4%) κ=0.900 |
62/62 (100%) κ=1.0 |
10/11 (90.9%) |
22/22 (100%) |
112/113 (99.1%) |
102/102 (100%) |
|
|
62 Younger vs. 152 Older |
62Younger-Tem-rSUVR50-70 vs. 152OLDER-Tem-rSUVR5070 |
27/62 (43.5%) κ=0 |
53/62 (85.5%) κ=0.712 |
0/35 (0.0%) |
20/29 (69.0%) |
54/89 (60.7%) |
86/95 (90.5%) |
|
62Younger-Tem-gSUVR50-70 vs. 152OLDER-Tem-gSUVR5070 |
56/62 (90.3%) κ=0 |
57/62 (91.9%) κ=0.664 |
0/6 (0.0%) |
12/17 (70.6%) |
112/118 (94.9%) |
102/107 (95.3%) |
|
Positive agreement is calculated as follows: if there are 16 positive cases by one method and 17 positive cases by the other and agreement across 15 cases, then 30 (15 + 15) out of a total of 33 positive ratings were in agreement, or 90.9%. The same applies to negative agreement. Conditions with perfect agreement are bolded.
3.1 Regional IO Classification of PiB(+)/PiB(−) Status
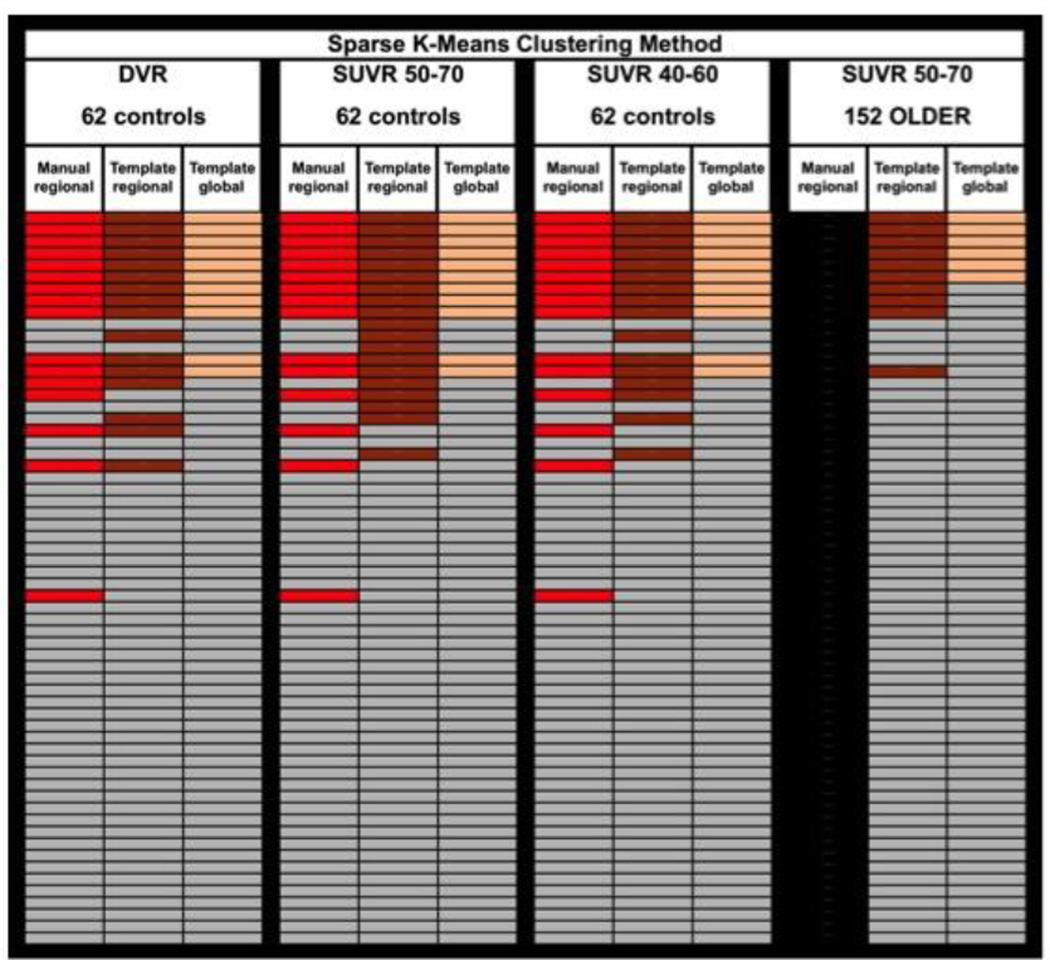
For direct comparability with the original publication of the IO method (Aizenstein et al., 2008), all “regional” IO methods used data from the same seven cortical ROIs defined for the Aizenstein et al. study (ACG, FRC, LTC, OCC, OCP, PAR and PRC). It should be noted that the results did not differ if the six region ROI set selected below by the SKM method was substituted into these analyses. Regional IO methods analyzed the PiB retention data from the seven ROIs in an independent manner, such that positivity in any one region defined a subject as PiB(+) overall. The numbers of iterations and the numbers of high and low outliers removed in each iteration are shown in Supplementary Table A and Fig. 1A–C. All eight subjects ≤55yrs remained within the PiB(−) group after all outliers were removed. This was true of all methods described below. While there were considerable inconsistencies across the various cutoffs derived from the original 62 controls using the IO approach (overall agreement 81.3 ± 15.0%; positive agreement 63.2 ± 29.2%; negative agreement 86.1 ± 11.4%), this was most apparent when comparing the MAN-rSUVR50-70 to Tem-rSUVR50-70 data (overall agreement 67.7%; positive agreement 61.5%; negative agreement 72.2%) (Table 1, Fig. 2). However, the biggest discrepancy was observed when the cutoffs were derived from an independent, older cohort that had a more continuous distribution of PiB retention values (Figure 1E&F). In this case, there were no PiB(+) cases among the 62 original controls using the cutoffs derived from the 152 older controls (i.e., positive agreement = 0%) (Table 1, Fig. 2). This was due to the fact that the more continuous nature of the PiB PET data among the older controls led to fewer outliers and much higher cutoffs (Fig. 1F). For example, the global cutoff determined for SUVR50-70 data from the 152 older controls was 2.80, compared to 1.73 from the 62 original controls (Supplementary Table B). Note that manual ROI data was not available for the older cohort, so no manual-regional cutoffs were determined or applied here. All IO cutoff values are listed in Supplementary Table B.
Figure 1.
(A–C) Depiction of IO for the MAN-rDVR dataset. Subjects with PiB retention above the upper-inner fence or below the lower-inner fence [circles outside the whiskers of the plot in (A)] were excluded and box plots were recalculated (B). After 2 iterations, no more high outliers remained and the upper-inner fence was taken as the IO-MAN-rDVR cutoff (C). (D) Box and whisker plots of 62 subject group for IO-Tem-rSUVR50-70 dataset (red) compared to IO-Tem-rSUVR40-60 dataset (black). (E) Box and whisker plots of 62 subject control group for IO-Tem-rSUVR50-70 dataset (red) compared to IO-152OLDER-Tem-rSUVR50-70 dataset (black). (F) Histograms showing the more bimodal 62 control dataset (MAN-rSUVR50-70; yellow) and the more continuously distributed 152 older control dataset (Tem-rSUVR50-70; blue). The IO cutoff determined with the MAN-rSUVR50-70 dataset from the 62 younger controls is shown with a solid red line and the cutoff determined with the 152OLDER-Tem-rSUVR50-70 dataset is shown with a dashed red line.
Figure 2.
Schematic demonstrating agreement of PiB(+)/PiB(−) status using IO cutoffs derived from three versions of the PiB PET data (DVR, SUVR50-70 and SUVR40-60; all determined from a single acquisition) and from SUVR50-70 cutoffs derived from a distinct group of 152 older controls. All cases are arranged in the same order of increasing SUVR50-70 global cortical values with highest at the top. Manual-regional analyses (with partial-volume correction) are always shown in the left-most of the three columns with PiB(+) cases in red, template-based-regional analyses (without partial-volume correction) are always shown in the middle column with PiB(+) cases shown in brown and template-based-global analyses (without partial-volume correction) are always shown in the right-most of the three columns with PiB(+) cases shown in tan. For all analyses, PiB(−) cases are shown in gray. Manual ROI data was not available for the older control group. Note that, even when the cutoffs from the 152 older controls were used, it is the original 62 younger controls that are shown rated in this figure.
3.2 Sparse K-means + resampling Weights and Cutoffs
3.2.1 Determination of regional weights
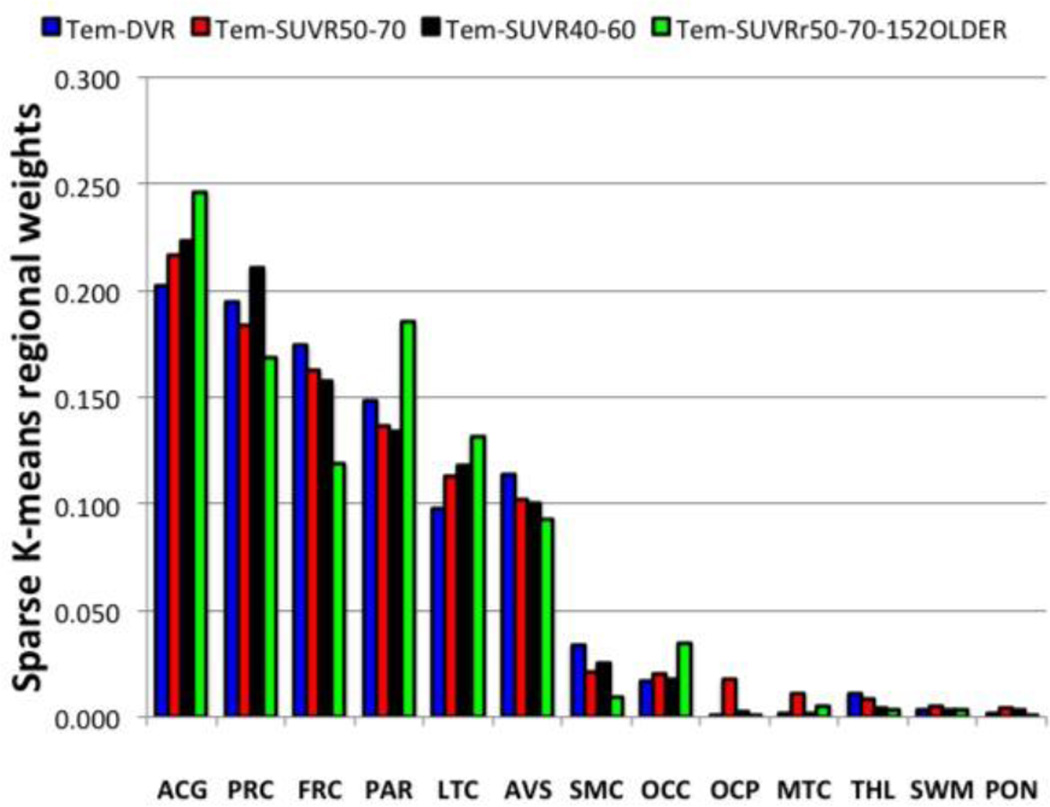
SKM analysis with resampling identified 6 of 13 ROI’s with markedly higher average weights than other ROI’s. Five regions (ACG, FRC, LTC, PAR and PRC) corresponded to those previously chosen for the original IO analysis method. One new subcortical region was added (AVS) to the analyses and the two occipital regions (OCC and OCP) were not weighted highly and were not used in the SKM analyses. ACG and PRC were generally found to have the largest weights, followed by FRC, PAR, and LTC and AVS (Fig. 3).
Figure 3.
Regional weights determined by the SKM method for the all template-based datasets are shown in order of decreasing weights for the SUVR50-70 dataset.
3.2.2 Regional Tight cluster Cutoffs
After 1,000 resampling analyses on each dataset from the original 62 subjects, 9/62 were classified PiB(+) by ≥96% of samplings (for each dataset) and 41 subjects were classified PiB(−) by ≥96% of samplings (for each dataset). For older 152 control dataset, 54/152 observations were classified PiB(+) by ≥99% of the samplings and 92 were classified PiB(−) by ≥99% of the samplings. In all datasets, the remaining subjects were much more variable in cluster membership and were not used for determination of tight-cluster centers or cutoffs. We defined the tight-cluster centers using only 9 PiB(+) and 41 PiB(−) [or 54 PiB(+) and 92 PiB(−) for older 152]. The tight-cluster centers defined regional SKM cutoffs and global cutoffs were created using the weighted average of the six regions (Supplemental Table C).
3.3 Regional SKM Classification of PiB(+)/PiB(−) Status
Using the six cortical regions identified above by SKM in a regionally independent manner, the SKM method performed far more consistently across data versions and ROI sampling methods than did the IO method (Table 1) (overall agreement 96.6 ± 4.3%; positive agreement 79.9 ± 10.1%; negative agreement 94.6 ± 2.9%). In fact, agreement across all three PiB PET data versions (DVR, SUVR40-60 and SUVR50-70) was perfect using the template-based global ROI and nearly perfect when using manual regional ROI data (Fig. 4). Most notable however, was the superior performance of the SKM over the IO approach when SUVR50-70 cutoffs were derived from the independent, older cohort. In comparison to the failure of the IO approach to identify any PiB(+) cases among the 62 original controls using cutoffs derived from the 152 older controls (Fig. 2), the SKM approach identified more than half of the corresponding PiB(+) cases (i.e., positive agreement = 69–71%) (Table 1, Fig. 4). Despite the more continuous nature of the PiB PET data among the older controls, the SKM cutoffs were much more similar whether derived from the 62 original or the 152 older controls (Supplementary Table C). For example, the global cutoff determined for SUVR50-70 data from the 152 older controls was 1.84, compared to 1.64 from the 62 original controls. This represents only a 12% increase, compared to the 62% increase described above for the IO approach. All SKM cutoff values are listed in Supplementary Table C.
Figure 4.
Depiction of PiB(−) (gray) and PiB(+) (colored) subjects across datasets and analysis methods for SKM. Cases have the same arrangement and color-coding as Figure 2.
3.4 Effect of Global vs. Regional Definition of PiB(+)/PiB(−) Status
As expected, requiring positivity in a global ROI (composed of a combination of 7 cortical ROIs for the IO approach and 6 ROIs for the SKM approach) resulted in fewer PiB(+) cases than with a regional approach (Table 1, Figs. 2 & 4). Similar to the comparison across analysis methods described above the SKM approach proved more consistent than the IO approach (Table 1). With all of methods, there was no subject identified as PiB(+) globally that was not also identified as PiB(+) by the corresponding regional approach. The global cutoffs for each method are listed in Supplemental Tables B and C and the iterations required for the IO approach are listed in Supplemental Table D.
3.5 Visual Read Classification of PiB(+)/PiB(−) Status
By its nature, a visual read is not corrected for partial-volume (atrophy) effects, therefore, the visual read method used is more analogous to the regional-template approaches discussed above. Since visual reads were performed on SUVR50-70 parametric images, we compared them to the template-based ROI, regional SUVR50-70 cutoffs derived from the more consistent SKM approach (SKM-Tem-rSUVR50-70) (Figure 5). The consensus visual read measure rated 21 subjects as PiB(+) [1 rater 21/21 (100%); 2 raters 20/21 (95.2%); 2 raters 19/21(90.5%)]. These 21 subjects included 15/19 of subjects identified as PiB(+) by SKM-Tem-rSUVR50-70 and 3 subjects just below the SKM cutoffs and 3 others substantially below the cutoffs (Fig. 5). Comparing the SKM-Tem-rSUVR50-70 method and the visual read method, there was an overall agreement of 83.9%, a positive agreement of 75.0% and a negative agreement of 88.1%. The Cohen’s kappa was 0.63, a value commonly defined as good (Landis & Koch, 1977). Among the readers, there was a 94.5% overall agreement between readers with a kappa of 0.89. Additionally, 2 of the 5 readers were asked to perform the visual reads twice; comparison of the two reading sessions revealed an overall agreement of 93.5% between the first and second sessions for both readers.
Figure 5.
Schematic demonstrating agreement among baseline SKM Tem-rSUVR50-70 quantitative analysis [left; PiB(+) in brown], baseline visual reads [middle; PiB(+) in green] and longitudinal SKM TEM-rSUVR50-70 quantitative analysis [right; PiB(+) in brown]. Gray cells always indicate subjects rated as PiB(−) by the indicated method. Black cells indicate that there was not longitudinal follow-up available. All cases have the same arrangement as in figures 2 and 4.
3.6 Examination of Follow-up data
Of the 62 subjects with baseline scans, 51 returned for follow-up. Looking at these 51 “follow-up” subjects at baseline, the SKM-Tem-rSUVR50-70 method and visual read method had an overall agreement of 82.4%, a positive agreement of 75.7% and a negative agreement of 86.2% - numbers very representative of the full cohort of 62 at baseline (see above). Re-evaluating the agreement at follow-up showed the SKM-Tem-rSUVR50-70 method and visual read method to have an overall agreement of 88.2%, a positive agreement of 85.7% and a negative agreement of 90.0% - numbers clearly improved from baseline. All cases defined as PiB(+) by SKM-Tem-rSUVR50-70 remained PiB(+) at follow-up. The improvement was because four of the five follow-up cases called PiB(+) by visual read and PiB(−) by SKM-Tem-rSUVR50-70 at baseline became PiB(+) by SKM-Tem-rSUVR50-70 at follow-up (using the same cutoffs defined with the baseline data). In addition, one case which both the visual read and SKM-Tem-rSUVR50-70 methods called PiB(−), became PiB(+) by SKM-Tem-rSUVR50-70 at follow-up. Visual reads were not available on follow-up scans.
4. Discussion
Identifying objective approaches for establishing amyloid-positivity is critical to standardization of amyloid-PET data analysis. Previous studies have used a variety of approaches, most of which involve subjective choices, such as the number of standard deviations above control means, the exact location of natural breakpoints and interpretation of visual reads (e.g. (Jack et al., 2008; Kemppainen et al., 2007; Klunk et al., 2004; Mintun et al., 2006; Mormino et al., 2012; Rowe et al., 2007) (Johnson et al., 2007; Ng et al., 2007; Rabinovici et al., 2007; Suotunen et al., 2010; Tolboom et al., 2010) (Bourgeat et al., 2010; Devanand et al., 2010; Mormino et al., 2009; Ng et al., 2007; Pike et al., 2007). Others, such as ROC analyses, rely on the particular composition of groups other than the cognitively normal controls (such as AD groups), which can vary widely depending on the population utilized (Devanand et al., 2010; Mormino et al., 2009; Ng et al., 2007; Pike et al., 2007). Here, we sought to minimize these limitations with approaches that do not rely on AD subjects and involve no subjective choices. We compared two objective approaches to define PiB(+)/PiB(−) cutoffs: IO and SKM. Since visual read methods can be universally employed and are likely to be preferred clinically, we also compared objective approaches to consensus visual reads.
IO was initially thought to be particularly well-suited to defining the PiB(−)/PiB(+) cutoffs in a consistent fashion (Aizenstein et al., 2008). However, when expanding this approach, IO did not produce consistent results (Table 1, Fig. 2). IO was sensitive to small variations in data, as seen in the relatively poor 77.4% (manual) to 85.5% (template) overall agreement between rSUVR50-70 and rSUVR40-60 (Table 1, Fig. 2). This was surprising given the similar distribution of data in these two datasets (Fig. 1D). The idiosyncrasies of IO required more iterations for rSUVR40-60 (manual) or rSUVR50-70 (template), resulting in more high outliers, lower cutoffs and more PiB(+) cases (Supplemental Table A).
The most troublesome problem with IO was the sensitivity to variations in cohort composition, as was observed with the 152OLDER dataset (Fig. 1F). When outliers were more evident among the 62 younger controls, IO performed well, but with a more continuous distribution of data from the older 152 dataset (Fig. 1E&F), IO performed very poorly – identifying no PiB(+) cases (Table 1, Fig. 2). Given variations in data collection and subject populations across imaging centers, IO did not show sufficient consistency to be widely applicable.
SKM with re-sampling was robust and consistent, producing similar results across methods. The differences among SKM methods were generally seen in a small zone near the cutoff (Fig. 4), whereas differences among IO methods could be seen far from the cutoff zones in some cases (Fig. 2). Further, the primary differences observed using SKM were between the manual- and template-based ROI approaches, which can likely be explained by differences in cortical and reference ROI placements. SKM performed very well when the ROIs were consistent. For example, there was near-perfect (61/62 or 62/62) agreement across all three regional-manual SKM methods and perfect agreement across all three global-template SKM methods (Table 1, Fig. 4). This latter fact should be emphasized because, currently, the global-template approach is the approach most commonly used in the expression of quantitative amyloid imaging results. The SKM approach also performed superiorly when the older cohort of 152 controls was used to define cutoffs. Because of the consistency across methods and diverse cohorts, we believe SKM could be widely applicable and conclude SKM is the superior objective approach. SKM also has the advantage that different numbers of clusters can be explored, in these studies only k=2 were utilized but there is the possibility of using the SKM method to identify three groups of subjects; PiB(+), PiB(−) and a PiB-intermediate group, ongoing work is focusing on the stability of k=2 vs. k=3.
Our data indicate that there can be good agreement between visual reads and SKM. The visual readers did rate six scans PiB(+) that were rated PiB(−) at baseline by SKM-Tem-rSUVR50-70. When the follow-up SKM-tem-rSUVR50-70 data were analyzed, the baseline visual reads more closely matched the follow-up SKM assignments, suggesting that the visual reads may be detecting PiB-positivity before the quantitative method. Both SKM and visual reads have strengths and weaknesses. Although it cannot be determined with certainty in the absence of pathological confirmation, there were certain types of cases in which SKM appeared to perform better than visual reads, but more frequent cases where visual reads appeared to be more sensitive. Visual reads were more sensitive to focal and asymmetric increases in PiB, frequently “missed” by SKM, likely due to the ROI approach used, averaged across hemispheres. However, this same sensitivity to asymmetries could produce false positives. The visual approach is probably more comparable to voxel-based approaches and work is ongoing to develop quantitative cutoffs for voxel-based data (Bi et al., 2011). Considered together, optimal approaches for accurate detection of early stages of fibrillar Aβ deposition may be a combination of quantitative SKM with visual reads.
Before broadly applying the results of this paper, three caveats should be kept in mind. First, the discussion above is targeted at detecting the first signs of Aβ deposition in preclinical AD. A definition of amyloid-positivity that is more specific for dementia due to AD, may require widespread Aβ deposition at levels higher than the cutoff values reported here, i.e., an AD-like cutoff. Second, consideration should be given to how the field interprets focal and near-threshold levels of PiB that could be observed in subjects with clear clinical dementia, since these same levels can be found in 20–50% of cognitively normal subjects depending on age, apolipoprotein-E genotype and other factors (Aizenstein et al., 2008; Mintun et al., 2006; Morris et al., 2010; Rowe et al., 2010). Such subjects could be cognitively impaired for reasons other than AD. That is, they may be in a presymptomatic stage of AD, but are in symptomatic stages of a non-AD dementia. Third, it is important to note that the exact values reported here are not generalizable to all other analysis methods or even similar analysis methods at other centers – particularly on data acquired on different scanners. This suggests a more standardized approach to the expression of amyloid imaging results is necessary. However, application of SKM to any large PiB dataset in cognitively normal controls should result in equivalent, center-specific definitions of PiB-positivity. While standardization remains a challenge for any quantitative method, it is not yet clear how standardized visual reads can be made. If a high degree of consistency can be achieved across centers with visual read outcomes, this would be a significant advantage to visual read approaches.
5. CONCLUSION
This report describes two objective approaches (IO and SKM) to defining a cutoff for amyloid-positivity with PiB-PET. Other factors were explored including: 1) regional vs. global approaches; 2) DVR vs. SUVR; 3) manual- vs. template-based ROIs; and 4) different distributions of PiB retention within populations. Discordant results across visual reads and one SKM method were compared to SKM outcomes over 1–3yrs of longitudinal follow-up. While IO appears to be challenged by differing datasets and analysis methods, SKM produced much more similar results across methods. Regional approaches proved to be predictably more sensitive than global approaches. DVR and SUVR data appeared to yield essentially equivalent results with SKM, the differences being mainly due to differences in cortical and reference ROI placement. The cutoffs described for regional data appear very useful for the identification of early stages of amyloid deposition but may not be optimal for specific identification of dementia due to AD. While this process should be generalizable across centers, the exact cutoff numbers may not be generalizable. The high concordance of these objective methods with consensus visual reads is encouraging; given the likelihood that clinical application of amyloid-PET is likely to be highly dependent on visual analysis alone.
Supplementary Material
Acknowledgements
Supported by: The National Institutes of Health grants: K01 AG037562, R01 AG033042, P50 AG005133, R37 AG025516, P01 AG025204.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: GE Healthcare holds a license agreement with the University of Pittsburgh. Drs. Klunk and Mathis are co-inventors of PiB and, as such, have a financial interest in this license agreement. GE Healthcare provided no grant support for this study and had no role in the design or interpretation of results or preparation of this manuscript.
All other authors have no conflicts of interest with this work.
References
- Aizenstein HJ, Nebes RD, Saxton JA, Price JC, Mathis CA, Tsopelas ND, Ziolko SK, James JA, Snitz BE, Houck PR, Bi W, Cohen AD, Lopresti BJ, DeKosky ST, Halligan EM, Klunk WE. Frequent amyloid deposition without significant cognitive impairment among the elderly. Arch Neurol. 2008;65(11):1509–1517. doi: 10.1001/archneur.65.11.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Hwang KS, Andrawis JP, Green AE, Babakchanian S, Morra JH, Cummings JL, Toga AW, Trojanowski JQ, Shaw LM, Jack CR, Jr, Petersen RC, Aisen PS, Jagust WJ, Koeppe RA, Mathis CA, Weiner MW, Thompson PM. 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiol Aging. 2010;31(8):1284–1303. doi: 10.1016/j.neurobiolaging.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Tseng GC, Weissfeld LA, Price JC. Sparse Clustering with Resampling for Subject Classificaiton in PET Amyloid Imaging Studies; IEEE Nuclear Science Symposium Conference Record- Valencia; 2011. MIC12.M-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeat P, Chetelat G, Villemagne VL, Fripp J, Raniga P, Pike K, Acosta O, Szoeke C, Ourselin S, Ames D, Ellis KA, Martins RN, Masters CL, Rowe CC, Salvado O. Beta-amyloid burden in the temporal neocortex is related to hippocampal atrophy in elderly subjects without dementia. Neurology. 2010;74(2):121–127. doi: 10.1212/WNL.0b013e3181c918b5. [DOI] [PubMed] [Google Scholar]
- Cicchetti DV, Feinstein AR. High agreement but low kappa: II. Resolving the paradoxes. Journal of clinical epidemiology. 1990;43(6):551–558. doi: 10.1016/0895-4356(90)90159-m. [DOI] [PubMed] [Google Scholar]
- Cohen AD, Price JC, Weissfeld LA, James J, Rosario BL, Bi W, Nebes RD, Saxton JA, Snitz BE, Aizenstein HA, Wolk DA, DeKosky ST, Mathis CA, Klunk WE. Basal Cerebral Metabolism May Modulate the Cognitive Effects of A{beta} in Mild Cognitive Impairment: An Example of Brain Reserve. J. Neurosci. 2009;29(47):14770–14778. doi: 10.1523/JNEUROSCI.3669-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JA. A coefficient of agreement for nominal scales. Educational and Psychological Measurement. 1960;20:37–46. [Google Scholar]
- Devanand DP, Mikhno A, Pelton GH, Cuasay K, Pradhaban G, Kumar JS, Upton N, Lai R, Gunn RN, Libri V, Liu X, Heertum RV, Mann JJ, Parsey RV. Pittsburgh compound B (11C-PIB) and fluorodeoxyglucose (18F-FDG) PET in patients with Alzheimer Disease, Mild Cognitive Impairment, and healthy controls. J Geriatr Psychiatry Neurol. 2010;23(3):185–198. doi: 10.1177/0891988710363715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst AJ, Rabinovici GD, Rostomian AH, Steed T, Alkalay A, Racine C, Miller BL, Jagust WJ. Cognition, glucose metabolism and amyloid burden in Alzheimer's disease. Neurobiol Aging. 2010;33(2):215–225. doi: 10.1016/j.neurobiolaging.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan JA, Wong MA. A K-means clustering algorithm. Applied Statistics. 1979;28:100–108. [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Vol. 2. Stanford: Springer; 2009. [Google Scholar]
- Ikonomovic MD, Klunk WE, Abrahamson EE, Mathis CA, Price JC, Tsopelas ND, Lopresti BJ, Ziolko S, Bi W, Paljug WR, Debnath ML, Hope CE, Isanski BA, Hamilton RL, DeKosky ST. Post-mortem correlates of in vivo PiB-PET amyloid imaging in a typical case of Alzheimer's disease. Brain. 2008;131(Pt 6):1630–1645. doi: 10.1093/brain/awn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Lowe VJ, Senjem ML, Weigand SD, Kemp BJ, Shiung MM, Knopman DS, Boeve BF, Klunk WE, Mathis CA, Petersen RC. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer's disease and amnestic mild cognitive impairment. Brain. 2008;131(Pt 3):665–680. doi: 10.1093/brain/awm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Gregas M, Becker JA, Kinnecom C, Salat DH, Moran EK, Smith EE, Rosand J, Rentz DM, Klunk WE, Mathis CA, Price JC, Dekosky ST, Fischman AJ, Greenberg SM. Imaging of amyloid burden and distribution in cerebral amyloid angiopathy. Ann Neurol. 2007;62(3):229–234. doi: 10.1002/ana.21164. [DOI] [PubMed] [Google Scholar]
- Kemppainen NM, Aalto S, Wilson IA, Nagren K, Helin S, Bruck A, Oikonen V, Kailajarvi M, Scheinin M, Viitanen M, Parkkola R, Rinne JO. PET amyloid ligand [11C]PIB uptake is increased in mild cognitive impairment. Neurology. 2007;68(19):1603–1606. doi: 10.1212/01.wnl.0000260969.94695.56. [DOI] [PubMed] [Google Scholar]
- Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Bergström M, Savitcheva I, Huang GF, Estrada S, Ausén B, Debnath ML, Barletta J, Price JC, Sandell J, Lopresti BJ, Wall A, Koivisto P, Antoni G, Mathis CA, Långström B. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol. 2004;55(3):306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- Klunk WE, Mathis CA. Amyloid Imaging and (What is “Normal”) Aging. In: Jagust W, D'Esposito M, editors. Imaging the Aging Brain. Vol. 1. New York: Oxford University Press; 2009. pp. 191–245. [Google Scholar]
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- Lopresti BJ, Klunk WE, Mathis CA, Hoge JA, Ziolko SK, Lu X, Meltzer CC, Schimmel K, Tsopelas ND, Dekosky ST, Price JC. Simplified Quantification of Pittsburgh Compound B Amyloid Imaging PET Studies: A Comparative Analysis. J Nucl Med. 2005;46(12):1959–1972. [PubMed] [Google Scholar]
- Mathis C, Kuller LWK, Snitz B, Price J, Weissfeld L, Rosario B, Lopresti B, Saxton J, Aizenstein H, McDade E, Kamboh M, DeKosky S, Lopez O. In vivo assessment of amyloid-β deposition in nondemented very elderly subjects. Annals of Neurology. doi: 10.1002/ana.23797. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer C, Kinahan P, Greer P, et al. Comparative evaluation of MR-based partial volume correction schemes for PET. J. Nucl. Med. 1999;40:2053–2065. [PubMed] [Google Scholar]
- Mintun MA, Larossa GN, Sheline YI, Dence CS, Lee SY, Mach RH, Klunk WE, Mathis CA, DeKosky ST, Morris JC. [11C]PIB in a nondemented population: potential antecedent marker of Alzheimer disease. Neurology. 2006;67(3):446–452. doi: 10.1212/01.wnl.0000228230.26044.a4. [DOI] [PubMed] [Google Scholar]
- Mormino EC, Brandel MG, Madison CM, Rabinovici GD, Marks S, Baker SL, Jagust WJ. Not quite PIB-positive, not quite PIB-negative: Slight PIB elevations in elderly normal control subjects are biologically relevant. Neuroimage. 2012;59(2):1152–1160. doi: 10.1016/j.neuroimage.2011.07.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mormino EC, Kluth JT, Madison CM, Rabinovici GD, Baker SL, Miller BL, Koeppe RA, Mathis CA, Weiner MW, Jagust WJ. Episodic memory loss is related to hippocampal-mediated beta-amyloid deposition in elderly subjects. Brain. 2009;132(Pt 5):1310–1323. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, Mintun MA. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol. 2010;67(1):122–131. doi: 10.1002/ana.21843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Villemagne VL, Berlangieri S, Lee ST, Cherk M, Gong SJ, Ackermann U, Saunder T, Tochon-Danguy H, Jones G, Smith C, O'Keefe G, Masters CL, Rowe CC. Visual assessment versus quantitative assessment of 11C-PIB PET and 18F-FDG PET for detection of Alzheimer's disease. J Nucl Med. 2007;48(4):547–552. doi: 10.2967/jnumed.106.037762. [DOI] [PubMed] [Google Scholar]
- Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256(3):183–194. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- Pike KE, Savage G, Villemagne VL, Ng S, Moss SA, Maruff P, Mathis CA, Klunk WE, Masters CL, Rowe CC. β-Amyloid imaging and memory in non-demented individuals: evidence for preclinical Alzheimer's disease. Brain. 2007;130(Pt 11):2837–2844. doi: 10.1093/brain/awm238. [DOI] [PubMed] [Google Scholar]
- Price JC, Klunk WE, Lopresti BJ, Lu X, Hoge JA, Ziolko SK, Holt DP, Meltzer CC, Dekosky ST, Mathis CA. Kinetic modeling of amyloid binding in humans using PET imaging and Pittsburgh Compound-B. J Cereb Blood Flow Metab. 2005;25:1528–1547. doi: 10.1038/sj.jcbfm.9600146. [DOI] [PubMed] [Google Scholar]
- Rabinovici GD, Furst AJ, O'Neil JP, Racine CA, Mormino EC, Baker SL, Chetty S, Patel P, Pagliaro TA, Klunk WE, Mathis CA, Rosen HJ, Miller BL, Jagust WJ. 11C-PIB PET imaging in Alzheimer disease and frontotemporal lobar degeneration. Neurology. 2007;68(15):1205–1212. doi: 10.1212/01.wnl.0000259035.98480.ed. [DOI] [PubMed] [Google Scholar]
- Rentz DM, Locascio JJ, Becker JA, Moran EK, Eng E, Buckner RL, Sperling RA, Johnson KA. Cognition, reserve, and amyloid deposition in normal aging. Ann Neurol. 2010;67(3):353–364. doi: 10.1002/ana.21904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick SM, Sojkova J, Zhou Y, An Y, Ye W, Holt DP, Dannals RF, Mathis CA, Klunk WE, Ferrucci L, Kraut MA, Wong DF. Longitudinal cognitive decline is associated with fibrillar amyloid-beta measured by [11C]PiB. Neurology. 2010;74(10):807–815. doi: 10.1212/WNL.0b013e3181d3e3e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario BL, Weissfeld LA, Laymon CM, Mathis CA, Klunk WE, Berginc MD, James JA, Hoge JA, Price JC. Inter-rater reliability of manual and automated region-of-interest delineation for PiB PET. Neuroimage. 2011;55(3):933–941. doi: 10.1016/j.neuroimage.2010.12.070. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't Research Support, U.S. Gov't, Non-P.H.S.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe CC, Ellis KA, Rimajova M, Bourgeat P, Pike KE, Jones G, Fripp J, Tochon-Danguy H, Morandeau L, O'Keefe G, Price R, Raniga P, Robins P, Acosta O, Lenzo N, Szoeke C, Salvado O, Head R, Martins R, Masters CL, Ames D, Villemagne VL. Amyloid imaging results from the Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging. Neurobiol Aging. 2010;31(8):1275–1283. doi: 10.1016/j.neurobiolaging.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Rowe CC, Ng S, Ackermann U, Gong SJ, Pike K, Savage G, Cowie TF, Dickinson KL, Maruff P, Darby D, Smith C, Woodward M, Merory J, Tochon-Danguy H, O'Keefe G, Klunk WE, Mathis CA, Price JC, Masters CL, Villemagne VL. Imaging beta-amyloid burden in aging and dementia. Neurology. 2007;68(20):1718–1725. doi: 10.1212/01.wnl.0000261919.22630.ea. [DOI] [PubMed] [Google Scholar]
- Savva GM, Wharton SB, Ince PG, Forster G, Matthews FE, Brayne C. Age, neuropathology, and dementia. N Engl J Med. 2009;360(22):2302–2309. doi: 10.1056/NEJMoa0806142. [DOI] [PubMed] [Google Scholar]
- Schwertman NC, de Silva R. Identifying outliers with sequential fences. Computational Statistics & Data Analysis. 2007;51(8):3800–3810. [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Jr, Kaye J, Montine TJ, Park DC, Reiman EM, Rowe CC, Siemers E, Stern Y, Yaffe K, Carrillo MC, Thies B, Morrison-Bogorad M, Wagster MV, Phelps CH. Toward defining the preclinical stages of Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer RL, Fleiss JL. A re-analysis of the reliability of psychiatric diagnosis. British Journal of Psychiatry. 1974;125:341–372. doi: 10.1192/bjp.125.4.341. [DOI] [PubMed] [Google Scholar]
- Suotunen T, Hirvonen J, Immonen-Raiha P, Aalto S, Lisinen I, Arponen E, Teras M, Koski K, Sulkava R, Seppanen M, Rinne JO. Visual assessment of [(11)C]PIB PET in patients with cognitive impairment. Eur J Nucl Med Mol Imaging. 2010;37(6):1141–1147. doi: 10.1007/s00259-010-1382-8. [DOI] [PubMed] [Google Scholar]
- Tolboom N, van der Flier WM, Boverhoff J, Yaqub M, Wattjes MP, Raijmakers PG, Barkhof F, Scheltens P, Herholz K, Lammertsma AA, van Berckel BN. Molecular imaging in the diagnosis of Alzheimer's disease: visual assessment of [11C]PIB and [18F]FDDNP PET images. J Neurol Neurosurg Psychiatry. 2010;81(8):882–884. doi: 10.1136/jnnp.2009.194779. [DOI] [PubMed] [Google Scholar]
- Witten DM, Tibshirani R. A framework for feature selection in clustering. J Am Stat Assoc. 2010;105(490):713–726. doi: 10.1198/jasa.2010.tm09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.