Abstract
Background and Aims
The gametophyte phase of ferns plays an important role in habitat selection, dispersal, adaptation and evolution. However, ecological studies on fern gametophytes have been impeded due to the difficulty of species identification of free-living gametophytes. DNA barcoding provides an alternative approach to identifying fern gametophytes but is rarely applied to field studies. In this study, an example of field vittarioid gametophyte identification using DNA barcoding, which has not been done before, is given.
Methods
A combination of distance-based and tree-based approaches was performed to evaluate the discriminating power of three candidate barcodes (matK, rbcL and trnL-F) on 16 vittarioid sporophytes. Sequences of the trnL-F region were generated from 15 fern gametophyte populations by tissue-direct PCR and were compared against the sporophyte dataset, using BLAST.
Key Results trnL-F
earns highest primer universality and discriminatory ability scores, whereas PCR success rates were very low for matK and rbcL regions (10·8 % and 41·3 %, respectively). BLAST analyses showed that all the sampled field gametophytes could be successfully identified to species level. Three gametophyte populations were also discovered to be living beyond the known occurrence of their sporophyte counterparts.
Conclusions
This study demonstrates that DNA barcoding (i.e. reference databasing, tissue-direct PCR and molecular analysis), especially the trnL-F region, is an efficient tool to identify field gametophytes, and has considerable potential in exploring the ecology of fern gametophytes.
Keywords: DNA barcoding, field gametophyte, independent gametophyte, vittarioid fern
INTRODUCTION
Fern gametophytes, the free-living haploid stage in the fern life cycle, play a critical role in establishing natural populations. In several fern lineages, gametophytes are perennial via asexual proliferation and production of dispersable gemmae (Bower, 1888; Stokey and Atkinson, 1958; Nayar, 1963; Farrar, 1974; Farrar et al., 2008). Moreover, some ‘species’ have been described as occurring as gametophytes only, lacking a sporophyte phase (Farrar, 1967, 1992; Farrar and Mickel, 1991; Raine et al., 1991). Discovery and identification of such populations of gametophytes living independently of their sporophyte counterparts has brought new insights into previously unsuspected roles of the gametophyte phase in habitat selection and migration (Rumsey and Sheffield, 1996; Dassler and Farrar, 1997; Ebihara et al., 2008; Farrar et al., 2008). However, field identification of fern gametophytes remains difficult due to their morphological simplicity, even though gametophyte morphologies of most fern lineages have been documented (e.g. Stokey, 1950; Atkinson and Stokey, 1964; Nayar and Kaur, 1971). As a result, a full understanding of the role of the gametophyte phase in fern ecology has been impeded.
As an alternative to morphological identification, the comparison of molecular characters offers a new opportunity to overcome the ‘traditional’ identification difficulties. DNA barcoding, using the standardized molecular markers for DNA-based identification, has become an important approach, especially for relatively featureless individuals (Ahrens et al., 2007; Kesanakurti et al., 2011). Recently, such molecular tools and accompanying techniques have been successfully applied in species identification of fern gametophytes of unknown origin (Schneider and Schuettpelz, 2006; Li et al., 2009; de Groot et al., 2011). However, using DNA barcoding for identifying field-collected fern gametophytes has not yet been tested.
Establishing universal DNA barcoding markers for land plants is still in development, especially in ferns. Markers for >20 loci/genes have been applied to DNA barcoding studies (reviewed in CBOL Plant Working Group, 2009; Hollingsworth et al., 2011). Primer universality and sequence variation are two of the most important criteria for selecting DNA barcoding loci. Some of the most frequently suggested regions for ferns include matK, rbcL, trnH-psbA and trnL-F (Nitta, 2008, Ebihara et al., 2010; de Groot et al., 2011; Li et al., 2011). As the most recent study conducted by Li et al. (2011) suggested using rbcL and matK as core DNA barcodes and trnL-F as a back-up locus for ferns, we chose these three loci as candidate markers in our study and tested their performance in field gametophyte identification.
As a case study for the applicability of DNA barcoding to field gametophyte identification, the vittarioid ferns were chosen. Gametophytes of vittarioids differ from most other ferns in their branched ribbon-like growth form (rather than the well-known heart-shaped morphology) and in the production (by most genera, with the exception of Ananthacorus) of dispersable vegetative units called gemmae (Nayar and Kaur, 1971; Farrar, 1974). A number of additional morphological and developmental characters allow for the morphological distinction of vittarioid genera from gametophytes of the few other fern groups that produce gemmae (Sheffield and Farrar, 1988; Raine et al, 1991; Dassler and Farrar, 1997). As a consequence of vegetative reproduction, vittarioid gametophyte populations can persist independently from any sporophyte production (Farrar, 1978; Farrar and Landry, 1987; Farrar and Mickel, 1991). Because of their special morphological characters and the potential for independent gametophyte populations, vittarioid ferns hold considerable promise for studing the ecological parameters of the gametophyte phase.
The vittarioids of Taiwan are a diverse group, representing 16 out of 26 vittarioid taxa that occur in East Asia (Iwatsuki, 1995; Zhang, 1996a, b; Knapp, 2011). These 16 species belong to three genera: Antrophyum, Haplopteris and Vaginularia (sensu Ruhfel et al., 2008). First, we present a step-by-step procedure for field gametophyte identification from (a) DNA barcodes databasing, (b) testing the identification power of the DNA barcode region and (c) molecular identification of field gametophytes. Initial morphological observations were also conducted to test the feasibility of using barcode-identified gametophytes to recognize additional morphological characters that might be diagnostic for species and species groups within the vittarioids.
MATERIALS AND METHODS
DNA barcode databasing of vittarioid ferns
For DNA barcode databasing, 46 mature (i.e. with sori) and well-identified sporophyte samples were used in this study, including seven Haplopteris species, seven Antrophyum species, and two Vaginularia species, representing all the 16 vittarioids species known from Taiwan (Knapp, 2011). For each species, three individuals from different populations were sampled, except for Vaginularia trichoidea because only a single population was found. Detailed information is listed in Table 1 and voucher specimens are deposited in the herbarium (TAIF) of the Taiwan Forestry Research Institute.
Table 1.
Sporophyte materials used for DNA databasing in this study
| Taxon | Location | Voucher | GenBank no. |
|---|---|---|---|
| Antrophyum callifolium Blume | Pingtong, Taiwan | Wade1289 | JN869313 |
| Cat Tien, Vietnam | Wade1422 | JN869314 | |
| Tam Dao, Vietnam | Wade1497 | JN869315 | |
| Antrophyum castaneum H. Ito | Hsinchu, Taiwan | Wade1498 | JN869324 |
| Nantou, Taiwan | Wade1638 | JN869326 | |
| Hualian, Taiwan | Wade1536 | JN869325 | |
| Antrophyum formosanum Hieron. | Hualien, Taiwan | Wade1488 | JN869319 |
| Taoyuan, Taiwan | Wade1546 | JN869320 | |
| Taitong, Taiwan | Wade1640 | JN869321 | |
| Antrophyum henryi Hieron. | Hsinchu, Taiwan | Wade379 | JN869322 |
| Hsinchu, Taiwan | Wade1544 | JX040519 | |
| Yunlin, Taiwan | Lu22397 | JN869323 | |
| Antrophyum obovatum Baker | Hualian, Taiwan | Wade1487 | JN869327 |
| Tam Dao, Vietnam | Kuo1792 | JN869328 | |
| Yunnan, China | Kuo1478 | JN869329 | |
| Antrophyum parvulum Blume | Hualian, Taiwan | Wade1537 | JN869332 |
| Hainan, China | Kuo1719 | JN869330 | |
| Banahaw, Philippine | Kuo519 | JN869331 | |
| Antrophyum sessilifolium (Cav.) Spreng. | Taitong, Taiwan | Wade1265 | JN869316 |
| Taitong, Taiwan | Wade1506 | JN869317 | |
| Taitong, Taiwan | Wade1502 | JN869318 | |
| Haplopteris anguste-elongata (Hayata) E. H. Crane | Nantou, Taiwan | Wade1541 | JN869336 |
| Ilan, Taiwan | Wade1479 | JN869335 | |
| Taitong, Taiwan | Wade1641 | JN869337 | |
| Haplopteris elongata (Sw.) E. H. Crane | Hsinchu, Taiwan | Wade1542 | JN869339 |
| Taitong, Taiwan | Wade1642 | JN869340 | |
| Hainan, China | Kuo1724 | JN869338 | |
| Haplopteris ensiformis (Sw.) E. H. Crane | Cat Tien, Vietnam | Wade1395 | JN869343 |
| Pingtong, Taiwan | Wade 1473 | JN869341 | |
| Singapore | Ralf20110214–1 | JN869342 | |
| Haplopteris flexuosa (Fée) E. H. Crane | Ilan, Taiwan | Wade1528 | JN869346 |
| Tam Dao, Vietnam | Kuo1804 | JN869344 | |
| Yakushima, Japan | Kuo1092 | JN869345 | |
| Haplopteris mediosora (Hayata) X. C. Zhang | Nantou, Taiwan | Wade1492 | JN869349 |
| Nantou, Taiwan | Lu22569 | JN869350 | |
| Nantou, Taiwan | Wade2085 | JX040520 | |
| Haplopteris sp. | Taipei, Taiwan | Wade367 | JN869333 |
| Taipei, Taiwan | Wade1711 | JN869334 | |
| Taipei, Taiwan | Wade1700 | JX040518 | |
| Haplopteris taeniophylla (Copel.) E. H. Crane | Nantou, Taiwan | Wade1493 | JN869347 |
| Nantou, Taiwan | Lu22621 | JN869348 | |
| Nantou, Taiwan | Wade2086 | JX040521 | |
| Vaginularia paradoxa Mett. ex Miq. | Nantou, Taiwan | Wade1475 | JN869352 |
| Banahaw, Philippine | Kuo2065 | JN869353 | |
| Taitong, Taiwan | Wade1667 | JN869354 | |
| Vaginularia trichoidea (J. Sm.) Fée | Banahaw, Philippine | Hsu4080 | JN869351 |
GenBank nos. are for trnL-F. All vouchers are deposited in TAIF.
DNA extractions of these 46 samples were carried out either by modified CTAB (Wang et al., 2004) or Plant Genomic DNA Mini Kit (Geneaid, Taipei, Taiwan), following the manufacturer's protocol. Three candidate plastid barcoding regions (i.e. matK, rbcL and trnL-F) that were recommended in previous DNA barcoding studies in ferns were selected (Li et al., 2011). All the primers used in this study are shown in Table 2. The PCR amplifications were performed in 15-μL reaction volumes containing 10–100 ng template DNA, 7·2 µL ddH2O, 1·5 µL 10 × buffer, 1·2 µL of 10 µm dNTPs, 1·5 µL each of 10 µm primers, and 0·5 units of Taq polymerase. Thermal cycles were performed with a 2-min denaturation step at 94 °C, followed by 40 cycles at 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1·5 min, followed by a 10-min final extension at 72 °C. The total volume of PCR products was run on 1 % agarose gels then stained with ethidium bromide, and bands were excised and purified with a gel extraction kit (Geneaid). Sequencing reactions were done by Genomics (Taipei, Taiwan) using the BigDye Terminator V3·1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA, USA). The sequencing products were analysed by an ABI 3730 × l DNA analyser (Applied Biosystems). All the sequences were then deposited in GenBank for field gametophyte identification (Table 1).
Table 2.
Primers used in this study
| Locus | Primer | Sequences (5′–3′) | Reference |
|---|---|---|---|
| trnL-F | FernL 1Ir1 | GGYAATCCTGAGCCAAATC | Li et al., 2009 |
| trnL-F | F | ATTTGAACTGGTGACACGAG | Taberlet et al., 1991 |
| matK | FERN matK fEDR | ATTCATTCRATRTTTTTATTTHTGGARGAYAGATT | Kuo et al., 2011 |
| matK | FERN matK rAGK | CGTRTTGTACTYYTRTGTTTRCVAGC | Kuo et al., 2011 |
| rbcL | F1F | ATGTCACCACAAACAGAAACTAAAGCAAGT | Wolf et al., 1994 |
| rbcL | 1379R | TCACAAGCAGCAGCTAGTTCAGGACTC | Pryer et al., 2001 |
Identification power of DNA barcoding regions in vittarioid ferns
DNA sequences were aligned with the program MUSCLE (Edgar, 2004) using default settings and edited by BioEdit (Hall, 1999) to correct obvious misalignments. Variable and parsimony-informative characters were calculated using DnaSP version 5 (Librado and Rozas, 2009).
To compare and evaluate the identification power of these three DNA barcode regions in vittarioids ferns, both distance and tree-based approaches were performed. For the distance approach, the Kimura 2-parameter (K2P) distance of inter-species and intra-species pairs in each genus were inferred by MEGA5 (Tamura et al., 2011). The genus Vaginularia was excluded from this analysis due to the unavailability of more than two species. Our calculation of species discrimination rate followed Li et al. (2011), which is the percentage of species that could be distinguished among all possible species pairs. A pair of species was scored as successfully distinguished if the interspecific distance was always greater than zero and greater than the intraspecific distance.
In our tree-based approaches, both maximum parsimony (MP) and maximum likelihood (ML) phylogenies were reconstructed. MP phylogeny was reconstructed using PAUP* 4·0 (Swofford, 2003) under the setting of random-taxon-addition, TBR swapping, gaps as missing data and equal weighting. Heuristic bootstrap analysis was performed with 10 000 bootstrap replicates, ten random addition cycles per bootstrap replicate, TBR swapping and equal weighting. ML phylogeny was reconstructed using GARLI 0·96. beta (Zwickl, 2006) with a GTR + I + Γ model of sequence evolution, and the genthreshfortopoterm option was set to 20 000. Branch support was assessed with 5000 bootstrap replicates under the same criteria. Species discrimination was considered successful if a species formed a highly supported monophyletic group with both MP and ML approaches (bootstrap values >70 %).
Collection and molecular identification of field vittarioid gametophytes
In the field, only gametophytes with a ribbon-like growth form and spindle-shaped gemmae (the diagnostic characters of vittarioids) were selected (Farrar, 1974). Based on these criteria, a total of 15 gametophyte samples was collected in Taiwan (Table 3). A duplicate of every gametophyte sample was preserved in 50 % alcohol as a voucher and deposited in the herbarium (TAIF).
Table 3.
Field gametophyte materials used in this study and the results of BLAST
| Voucher | Location | GenBank no. | Result of BLASTn | Max score | MPI (%) |
|---|---|---|---|---|---|
| Wade1696 | Tainan, Taiwan | JX040524 | Antrophyum formosanum | 1563 | 100 |
| Wade2146 | Taipei, Taiwan | KC202411 | Antrophyum formosanum | 1563 | 100 |
| Wade1514 | Taipei, Taiwan | JX040525 | Antrophyum henryi | 1563 | 100 |
| Wade1694 | Nantou, Taiwan | JX040526 | Antrophyum parvulum | 1386 | 96 |
| Wade1519 | Taitong, Taiwan | JX040523 | Antrophyum sessilifolium | 1533 | 100 |
| Wade1511 | Pingtong, Taiwan | JX040530 | Haplopteris anguste-elongata | 1424 | 100 |
| Wade1512 | Nantou, Taiwan | JX040531 | Haplopteris anguste-elongata | 1424 | 100 |
| Wade2145 | Taipei, Taiwan | KC202412 | Haplopteris anguste-elongata | 1424 | 100 |
| Wade1697 | Taitong, Taiwan | JX040532 | Haplopteris elongata | 1424 | 100 |
| Wade1510 | Pingtong, Taiwan | JX040533 | Haplopteris ensiformis | 1447 | 100 |
| Wade1513 | Ilan, Taiwan | JX040535 | Haplopteris flexuosa | 1411 | 100 |
| Wade1681 | Nantou, Taiwan | JX040535 | Haplopteris flexuosa | 1411 | 100 |
| Wade1477 | Taipei, Taiwan | JX040528 | Haplopteris sp. | 1431 | 100 |
| Wade1509 | Taipei, Taiwan | JX040527 | Haplopteris sp. | 1431 | 100 |
| Wade1695 | Nantou, Taiwan | JX040529 | Haplopteris sp. | 1425 | 99 |
GenBank nos. are for trnL-F. All vouchers are deposited in TAIF.
Due to the mat-forming and ribbon-shaped nature of field vittarioid gametophytes (Fig. 1A), DNA contamination from associated bryophytes or gametophytes of other fern species/individuals may easily complicate the traditional DNA extraction process. Thus, tissue-direct PCR methods based on Li et al. (2010) were used to generate DNA barcode sequences that avoided these situations. For each tissue-direct PCR, only a tiny piece (approx. 1 mm2) of tissue sample was taken from a specific gametophyte individual. Mechanical + chemical manipulations (i.e. liquid nitrogen and sonication pretreatment + 1 m betaine and 5 % DMSO in reaction buffer) and secondary PCR were applied in tissue-direct PCRs in this study. Secondary PCRs were conducted using 1 µL of first-round PCR product as the template. Concentration of primers, PCR buffer, dNTP, Taq polymerase and PCR thermal cycles were the same as discribed above. Other details of tissue-direct PCR methods can be found in Li et al. (2010). Eight tissue-direct PCRs were repeated per sample and their success rate was recorded. The results of PCR amplifications were checked by electrophoresis on a 1 % agarose gel in TBE buffer. Sequencing procedures for these amplicons were the same as mentioned above and the sequences are deposited in GenBank (Table 3).
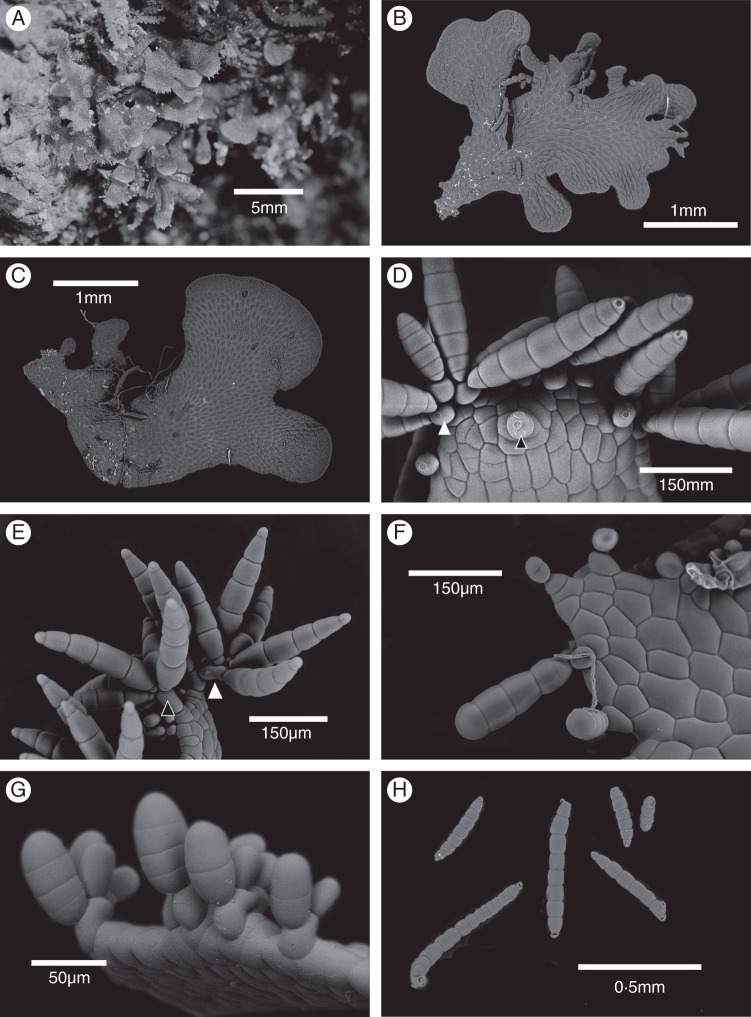
Fig. 1.
Morphology of vittarioid fern gametophytes: (A) field gametophyte (Haplopteris flexuosa, Wade1681); (B) ribbon-like morphology of Haplopteris auguste-elongata gametophyte (Wade2145); (C) ribbon-like morphology of Antrophyum formosanum gametophyte (Wade2146); (D) gametophyte with gemmae, showing position of gemmifers is variable (A. formosanum, Wade2146) – white and black arrowheads indicate gemmifers produced from marginal and inner cells of gametophyte, respectively; (E) gametophyte with gemmae, showing basal gemmifer produced several gemmifers (white arrowhead) or gemmae only (black arrowhead) (Haplopteris sp., Wade1477); (F) extended (A. formosanum, Wade2146) gemmae development; (G) reflexed gemmae development (A. formosanum, Wade2146); (H) mature gemmae showing variation in body cell numbers (Haplopteris anguste-elongata, Wade2145).
Using a complete DNA barcode database available for vittarioid species in Taiwan, both tree-based and BLAST methods were used for the molecular identification of our gametophyte samples. For the tree-based method, the sequences generated from gametophyte samples were first aligned with database sequences generated from sporophytes using MUSCLE (Edgar, 2004). Based on this matrix, MP and ML phylogenetic analyses were performed using the same criteria as mentioned above. For the BLAST method, NCBI BLASTn searches were applied to all sequences generated from field gametophytes. Identifications were considered to be successful only when the highest maximal percentage identity (MPI) included a single species and scored above 95 %.
Morphological observation of field vittarioids gametophytes
To document the morphology of gametophytes, cryo SEM was employed using the tabletop scanning electron microscope (TM-3000, Hitachi). Cryo SEM has been shown to be particularly effective for fragile and hydrated materials such as fern gametophytes (Sheffield and Farrar, 1988). Fresh materials were mounted on aluminium stubs with carbon tape. The stubs were placed on a cold stage that was pre-frozen by liquid nitrogen, and then moved into the chamber for observation. We paid detailed attention to the formation and construction of gemmae, features suggested as possibly diagnostic for identification of vittarioid species (Farrar, 1974, 1978; Crane, 1997).
RESULTS
Constructing the DNA barcode database
PCR amplifications of trnL-F succeeded in all 46 sporophyte samples (Table 1). By comparison, the success rates were very low for the matK and rbcL regions (10·8 % and 41·3 %, respectively); therefore, they were excluded from the following analyses. The sequences of trnL-F were mostly generated from one-direction reads. Only the trnL-F sequences of Antrophyum parvulum (Wade1537, Wade1719 and Kuo519) were assembled from both direction reads, since they were truncated by polyA/T in an intergenic spacer region (IGS). trnL-F sequences generated from these samples ranged from 772 to 860 bp. The shortest was in Haplopteris taeniophylla (Wade1493, Wade2086 and Lu22621), and the longest was in the two samples of Antrophyum callifolium (Wade1289 and Wade1422).
Species discrimination power of trnL-F: distance and tree-based approaches
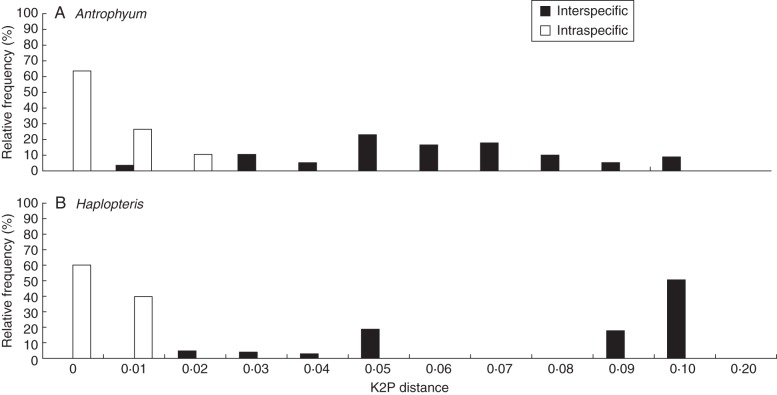
The alignment matrix had a total of 1002 characters, with 471 (47 %) and 441 (44 %) variable and parsimony-informative characters, respectively. The intra- versus interspecific K2P distances for Antrophyum and Haplopteris are shown in Fig. 2. The genus Vaginularia was excluded because it was a single sample. The intra- and interspecific K2P distances do not overlap in Haplopteris, but do overlap a little in Antrophyum. In Antrophyum, the minimal interspecific distance (between A. formosanum and A. henryi, K2P distance = 0·002) is smaller than the maximal intraspecific distance of A. callifolium (K2P distance = 0·017). The species discrimination rates for Antrophyum and Haplopteris are 95 % and 100 %, respectively (Table 4).
Fig. 2.
Comparison of inter- and intraspecific distance across sporophytes of all species pairs for two genera: (A) Antrophyum, and (B) Haplopteris. The x-axis is the K2P variation with bars corresponding to 0·01 intervals; the y-axis is relative frequency within the dataset.
Table 4.
The species discrimination rate of trnL-F and the proportions of monophyletic species in phylogenetic analysis
| Genus | Species discrimination rate (successfully distinguished pairs/total pairs) | Proportion of monophyletic species (monophyletic/total species) |
|---|---|---|
| Antrophyum | 95 % (20/21) | 85·7 % (6/7) |
| Haplopteris | 100 % (21/21) | 100 % (7/7) |
| Vaginularia | 100 % (1/1) | 100 % (2/2) |
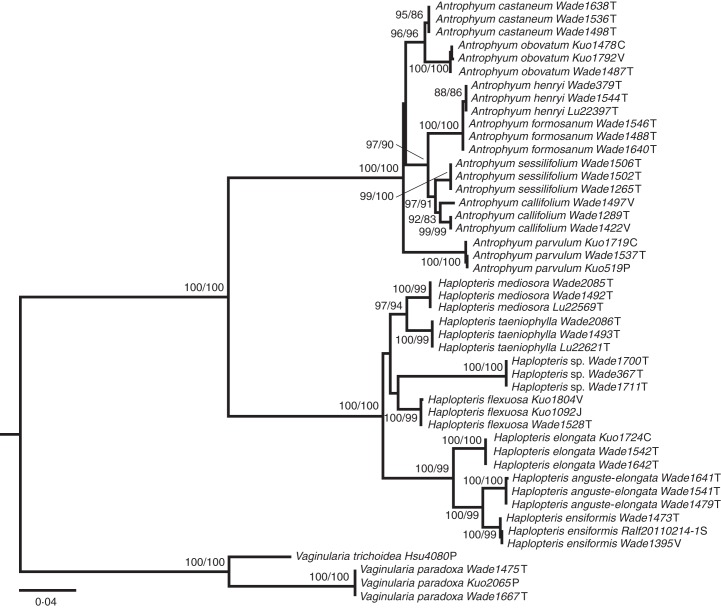
Four of the most parsimonious trees, all with 732 steps, were found in MP analysis and the ln value of the most likely phylogeny yielded from ML analyses was –4736·9411. As there is no conflict between the MP and ML topologies, only the ML phylogram is shown (Fig. 3). These results are consistent with previous phylogenies of vittarioids, in which three genera (i.e. Antrophyum, Haplopteris and Vaginularia) each forms a monophyletic clade (Crane, 1997; Ruhfel et al., 2008). Except for Antrophyum formosanum, all the species are monophyletic (bootstrap value over 80 %). Proportions of monophyletic species in each genus are summarized in Table 4.
Fig. 3.
Phylogeny of vittarioids including only sporophytes from ML analysis of trnL-F region. Numbers above nodes indicate bootstrap values. Abbreviations: C, China; J, Japan; P: Philippines; S, Singapore; T, Taiwan; V, Vietnam.
Molecular identification of gametophyte samples
trnL-F for all 15 gametophyte populations were successfully amplified by tissue-direct PCR with a success rate of 89 % (i.e. 107 from 15 populations × 8 reactions). All trnL-F sequences of the gametophyte samples were generated by one-direction reads, except for Wade1694, which had polyA/T in IGS region and was sequenced in both directions.
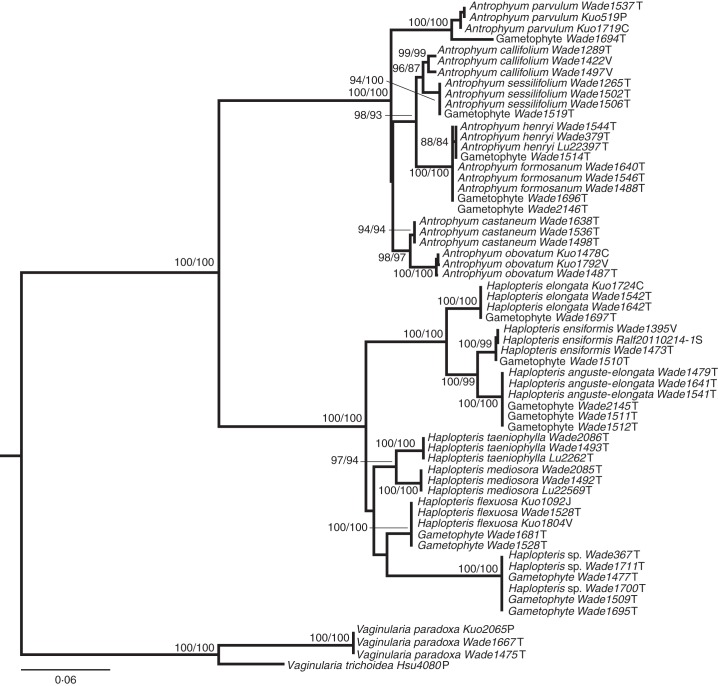
For the MP analysis, four most parsimony trees, all with 757 steps, were found by using the combined dataset of both sporophyte and gametophyte sequences. For the ML analysis, the ln value of the most likely tree found by using the combined dataset was –4896·5286. Only the ML phylogram (Fig. 4) is shown because there is no conflict between the MP and ML analyses. Each of the 15 gametophyte sequences formed a highly supported monophyletic group with a single sporophyte species. Through online NCBI BLASTn searches, each of 15 gametophyte sequences highly matched (MPI over 95 %) a single sequence generated from sporophytes (Table 3). As a result, 15 field gametophyte populations could easily be assigned to nine taxa belonging to two genera (i.e. Antrophyum and Haplopteris) by using trnL-F DNA barcoding.
Fig. 4.
Phylogeny of vittarioids including both sporophytes and gametophytes derived from ML analysis of trnL-F region. Numbers above nodes indicate bootstrap values under ML (left) and MP (right) analysis. Abbreviations: C, China; J, Japan; P: Philippines; S, Singapore; T, Taiwan; V, Vietnam.
In most cases, gametophyte populations were found in locations where their counterpart sporophytes were also found nearby. However, three gametophyte populations were located >20 km away from the nearest known sporophyte population of the species: Antrophyum henryi (Wade1514), A. parvulum (Wade1694), and Haplopteris sp. (Wade1695).
Morphological observations of gametophyte samples
All of the mature gametophytes of these 15 populations were ribbon-shaped with amorphous branching (Fig. 1B, C) and were producing gemmae. Gemmae development varied both within and among species (Table 5). For example, the gemmifer (supporting gemmae) could be produced from marginal cells (in most cases) and also from the inner cells of the gametophyte thallus (Fig. 1D); a gemmifer tended to produce only gemma(e), but sometimes it also produced more gemmifers (Fig. 1E); young gemmae in both extended (horizontal to the gametophyte margin, Fig. 1F) and reflexed (reflexed to the concave side of the gametophyte, Fig. 1G) positions were observed in different individuals of a population; the cell number of mature gemmae varied from four to 12 (Fig. 1H). Additional variation in gemma shape and development was noted but detailed analysis and correlation of variables with species identification has not yet been completed.
Table 5.
Comparison of gemmae formation in species of vittarioids
| Taxon | Voucher | Position of young gemmae | No. of gemmae body cells | No. of gemmae/terminal gemmifer | Basal gemmifer producing more gemmifers |
|---|---|---|---|---|---|
| A. formosanum | Wade1696 | Extended or reflexed | 5–7 | 1–3 | Yes |
| A. henryi | Wade1514 | Extended | 7–8 | 1–2 | – |
| A. parvulum (or A. sp.)* | Wade1694 | – | 5–7 | 1–3 | Yes |
| A. sessilifolium | Wade1519 | – | 5–8 | 1–3 | – |
| H. anguste-elongata | Wade1511 | Extended or reflexed | 4–12 | 1–3 | Yes |
| H. elongata | Wade1697 | Extended | 4–9 | 1–3 | Yes |
| H. ensiformis | Wade1510 | Extended | 6–12 | 1–3 | – |
| H. flexuosa | Wade1681 | Extended | 5–10 | – | – |
| H. sp. | Wade1509 | – | 3–4 | 1–4 | Yes |
‘–’ indicates no observation.
* Wade1694 was identified as A. parvulum because of its 96 % identity to trnL-F sequence of Antrophyum parvulum, but it could be an independent gametophyte of an unknown Antrophyum species.
DISCUSSION
Using trnL-F as a DNA barcode for the identification of fern gametophytes
Species identification of fern gametophytes by using DNA sequences has been applied in several studies. Most of these focus either on single species identification (Schneider and Schuettpelz, 2006; Li et al., 2009) or broad surveys (de Groot et al., 2011). This study is the first to test DNA-based fern gametophyte identification focusing on a specific taxonomic group in a defined geographical region. Most importantly, we demonstrate that with a complete reference database, all the sampled field gametophytes could be easily identified to species level.
DNA barcoding has been broadly applied in ecological studies (Pfenninger et al., 2007; Wilson et al., 2010; Kesanakurti et al., 2011; Pompanon et al., 2012). According to the suggestion of the Consortium for the Barcode of Life (CBOL), an ideal DNA barcode should satisfy three general criteria: primer universality, sequence quality and species discrimination. However, among these three criteria, primer universality earns the first priority in ecological studies where researchers do not have a priori knowledge of the study samples (reviewed in Valentini et al., 2008; Chariton, 2012). Among the three candidate regions tested in this study, only trnL-F showed a high PCR success rate (100 % in both in sporophytes and gametophytes). Moreover, primer universality was even more critical to our study because, with tissue-direct PCR, if primer universality is low then PCR success rate might be greatly reduced due to higher amounts of secondary metabolites coming from the tissue present in the PCR reaction.
In addition to primer universality, our results show that trnL-F also has both high sequence quality and species discrimination. For sequence quality, among the 16 taxa included in this study, two-strand sequencing was necessary for only one species (i.e. Antrophyum parvulum) due to a repeating polymer in its IGS region. For species discrimination, two complementary methods were adopted: an obvious gap between inter- and intraspecies distances was observed in most comparisons in our distance-based analysis (Fig. 2); most of the species also formed their own well-supported monophyletic clade in both MP and ML analyses (Fig. 3).
Within these analyses, Antrophyum formosanum and A. henryi were the only two species that could not be discriminated clearly. Morphologically, these two species share the character state of band-shaped paraphyses and are thought to be closely related (Zhang, 1996a). The size and shape of leaves (the former with a larger plant size and broader leaves, the latter with a smaller plant size and linear leaves) are the only characters used to identify these two species (Zhang, 1996a). The taxonomic status of these two species needs further study.
In conclusion, although rbcL and matK are the two recommended markers for DNA barcoding by CBOL, in our study only trnL-F satisfied the three most important criteria: primer universality, sequence quality and species discrimination. Thus we recommend using trnL-F as a DNA barcode for field gametophyte identification in future studies, at least for vittarioid ferns.
Identification and ecology of vittarioid gametophytes
In this study, we used DNA barcoding to identify field fern gametophytes and the morphology of these gametophytes was observed for comparative purposes. For DNA-based identifications, the trnL-F sequences for each of the 13 populations were 100 % identical to sequences generated from single sporophytes; therefore, identification to species was straightforward. For the other two populations, Wade1695 was 99 % identical (with only 1 bp difference) to an undetermined Haplopteris and there can be little doubt as to the correctness of these identifications because of the high degree of sequence congruence. For Wade1694, although BLASTn analysis showed a 96 % identity (28 bp difference) to Antrophyum parvulum, we cannot exclude the possibility that this population is an independent gametophyte whose sporophyte counterpart may not occur in Taiwan. In summary, our DNA barcoding study successfully classified 15 populations of field gametophytes into nine taxa.
Farrar (1974, 1978) suggested that the development and form of gemmae are useful for identifying vittarioid gametophytes. For example, at the generic level, all gemmae differentiating directly from gemmifer cells separate species of Antrophyum and Radiovittaria from species of Polytaenium and Vittaria, in which the gemmae can differentiate from other gemmae (Crane et al., 1995). At the species level, ongoing studies (D. R. Farrar, unpubl. res.) have shown that gametophytes of Polytaenium lanceolatum can be separated from those of P. lineatum on the basis of protrusions on the gemmifer apex of the former that may function in gemma abscission. Also, the gametophytes of Radiovittaria remota can be separated from the very similar gametophytes of R. minima based on their filiform versus plate-like initial stages of gemma formation. Farrar (1974) also found that mature gemmae of V. graminifolia were regularly composed of four body cells plus two rhizoid primordia, whereas those of V. lineate were composed of up to 12 body cells plus three or more rhizoid primordia.
The initial morphological characteristics of gametophytes we observed in our study were highly variable both within and between samples (Table 5). For example, Wade2146, the gametophyte identified as A. formosanum by DNA barcoding, showed that young gemmae development can be either extended or reflexed and the gemmifer can be produced from either marginal or inner cells of a gametophyte. In another case, Wade2145, the gametophyte identified as H. anguste-elongata by DNA barcoding, showed that the cell number of mature gemmae can vary from four to 12. As a result, the range of morphological variation within species may be greater than the difference between species, or even genera, thus making identifications based purely on morphology unreliable. We conclude that species identification is reliable only with DNA barcoding and that these results will now allow us to intensively study the morphological variability to determine which characters are taxon-diagnostic. If we are successful in this endeavour, the concept of initially using DNA barcoding to recognize taxon-specific morphological markers may be applicable to gametophytes of other fern groups.
Farrar (1985) defined gametophytic independence to be the occurrence of gametophyte populations beyond the distribution and habitat preference of the sporophyte phase. Rumsey and Sheffield (1996) recognized two categories: obligate and facultative independence, i.e. those that could not and those that could, on occasion, also produce sporophytes. The phenomenon of gametophyte independence has been reported in four fern familes: Hymenophyllaceae (Hymenophyllum and Trichomanes), Lomariopsidaceae (Lomariopsis), Polypodiaceae (Grammitis) and Pteridaceae (vittarioids). For vittarioid ferns, despite their wide distribution in tropical Asia, gametophyte independence has only been previously reported in temperate America (Farrar, 1967, 1978; Farrar and Mickel, 1991). In this study, facultative independent gametophytes of vittarioids are reported in Asia for the first time. In Taiwan, sporophytes of Antrophyum spp., including A. henryi, usually grow as lithophytes along humid rock crevices. The population of gametophytes identified as A. henryi in this study was found on the surface of a rock along a dry trail. Similarly, an undetermined Haplopteris species has sporophytes found only at low elevations in northern Taiwan, but gametophytes extend to mid-elevations in central Taiwan. In another case, although the gametophyte Wade1694 was identified as A. parvulum, it could be another case of an independent gametophyte of an unknown Antrophyum species due to the extent of its sequence variation. In congruence with previous studies, these observations imply that gametophytes have a greater tolerance for environmental stresses such as desiccation and low temperatures than do their counterpart sporophytes (Martin et al., 1995; Watkins et al., 2007; Farrar et al., 2008; Watkins and Cardelús, 2012).
New information on the habitat preferences and distribution of the gametophyte phase is shedding new light on a more complete understanding of fern biology (Farrar, 1967, 1971; Farrar et al., 1983, 2008; Rumsey et al., 1991; Dassler and Farrar, 1997; Ebihara et al., 2008, 2010). Only with more comprehensive investigations that include the entire fern life cycle and with reliable identifications of the gametophyte phase will we be in a position to answer questions critical to fern ecology, distribution and evolution, such as ‘What is the limiting stage in the establishment of new local and distant sporophyte populations?’ and ‘What is the role of the gametophyte phase in adaptation to changing habitats?’ The use of DNA barcoding, as demonstrated in this study, can enhance our capability to answer these and similar questions.
ACKNOWLEDGEMENTS
The authors are indebted to the many people who contributed to this study: Dr Fay-Wei Li for helping with data analysis; Dr Yi-Shan Chao for providing helpful comments; Mr Ralf Knapp and Mrs Pi-Fong Lu for providing the materials; and Miss De-Yen Tang for assisting with laboratory experiments. The study was funded by National Science Council: NSC99-2313-B-054-004-MY3 for Y.M.H. and NSC 99-2621-B-001-MY3 for W.L.C.
LITERATURE CITED
- Ahrens DM, Monaghan T, Vogler AP. DNA-based taxonomy for associating adults and larvae in multi-species assemblages of chafers (Coleoptera: Scarabaeidae) Molecular Phylogenetics and Evolution. 2007;44:436–449. doi: 10.1016/j.ympev.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Atkinson LR, Stokey AG. Comparative morphology of the gametophyte of homosporous ferns. Phytomorphology. 1964;14:51–70. [Google Scholar]
- Bower FO. On some normal and abnormal developments of the oophyte in Trichomanes. Annals of Botany. 1888;1:269–305. [Google Scholar]
- CBOL Plant Working Group. A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chariton A. Short and informative DNA products to indirectly measure vascular plant biodiversity. Molecular Ecology. 2012;21:3637–3639. doi: 10.1111/j.1365-294X.2012.05633.x. [DOI] [PubMed] [Google Scholar]
- Crane EH. A revised circumscription of the genera of the fern family Vittariaceae. Systematic Botany. 1997;22:509–517. [Google Scholar]
- Crane EH, Farrar DR, Wendel JF. Phylogeny of the Vittariaceae: convergent simplification leads to a polyphyletic Vittaria. American Fern Journal. 1995;85:283–305. [Google Scholar]
- Dassler CL, Farrar DR. Significance of form in fern gametophytes: clonal, gemmiferous gametophyes of Callistopteris baueriana (Hymenophyllaceae) International Journal of Plant Sciences. 1997;158:622–639. [Google Scholar]
- Ebihara A, Farrar DR, Ito M. The sporophyte-less filmy fern of eastern North America Trichomanes intricatum (Hymenophyllaceae) has the chloroplast genome of an Asian species. American Journal of Botany. 2008;95:1645–1651. doi: 10.3732/ajb.0800122. [DOI] [PubMed] [Google Scholar]
- Ebihara A, Nitta LH, Ito M. Molecular species identification with rich floristic sampling: DNA barcoding the Pteridophyte flora of Japan. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0015136. e15136. http://dx.doi.org/10.1371/journal.pone.0015136 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar DR. Gametophytes of four tropical fern genera reproducing independently of their sporophytes in the southern Appalachians. Science. 1967;155:1266–1267. doi: 10.1126/science.155.3767.1266. [DOI] [PubMed] [Google Scholar]
- Farrar DR. Ann Arbor, MI, USA: PhD Thesis, University of Michigan; 1971. The biology of ferns with asexually reproducing gametophytes in the Eastern United States; p. 239. [Google Scholar]
- Farrar DR. Gemmiferous fern gametophytes: Vittariaceae. American Journal of Botany. 1974;61:146–155. [Google Scholar]
- Farrar DR. Problems in identity and origin of the Appalachian Vittaria gametophyte, a sporophyteless fern of the Eastern United States. American Journal of Botany. 1978;65:1–12. [Google Scholar]
- Farrar DR. Independent fern gametophytes in the wild. Proceedings of the Royal Society of Edinburgh Section B: Biological Sciences. 1985;86:68–74.. [Google Scholar]
- Farrar DR. Trichomanes intricatum: the independent Trichomanes gametophyte in the Eastern United States. American Fern Journal. 1992;82:68–74. [Google Scholar]
- Farrar DR, Landry GP. Vittaria graminifolia in the United States, again. American Journal of Botany. 1987;74:709–710. [Google Scholar]
- Farrar DR, Mickel JT. Vittaria appalachiana: a name for the ‘Appalachian gametophyte’. American Fern Journal. 1991;81:69–75. [Google Scholar]
- Farrar DR, Parks JC, McAlpin BW. The fern genera Vittaria and Trichomanes in the northeastern United States. Rhodora. 1983;85:83–92. [Google Scholar]
- Farrar DR, Dassler C, Watkins JE, Skelton C. Gametophyte ecology. In: Ranker TA, Haufler CH, editors. Biology and evolution of ferns and lycophytes. Cambridge: Cambridge University Press; 2008. pp. 222–256. [Google Scholar]
- de Groot GA, During HJ, Maas JW, Schneider H, Vogel JC. Use of rbcL and trnL-F as a two-locus DNA barcode for identification of NW European ferns: an ecological perspective. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0016371. e16371. http://dx.doi.org/10.1371/journal.pone.0016371 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0019254. e19254. http://dx.doi.org/10.1371/journal.pone.0019254 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki K. Vittariaceae. In: Iwatsuki K, Yamazaki T, Boufford DE, Ohba H, editors. Flora of Japan. Vol. 1. Tokyo: Kodansha; 1995. pp. 86–88. [Google Scholar]
- Kesanakurti PR, Fazekas AJ, Burgess KS, et al. Spatial patterns of plant diversity below-ground as revealed by DNA barcoding. Molecular Ecology. 2011;20:1289–1302. doi: 10.1111/j.1365-294X.2010.04989.x. [DOI] [PubMed] [Google Scholar]
- Knapp R. Taipei: KBCC Press and Yuan-Liou Publishing; 2011. Ferns and fern allies of Taiwan. [Google Scholar]
- Kuo LY, Li FW, Chiou WL, Wang CN. First insights into fern matK phylogeny. Molecular Phylogenetics and Evolution. 2011;59:556–566. doi: 10.1016/j.ympev.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Li FW, Tan BC, Buchbender V, et al. Identifying a mysterious aquatic fern gametophyte. Plant Systematics and Evolution. 2009;281:77–86. [Google Scholar]
- Li FW, Kuo LY, Huang YM, Chiou WL, Wang CN. Tissue-direct PCR, a rapid and extraction-free method for barcoding of ferns. Molecular Ecology Resources. 2010;10:92–95. doi: 10.1111/j.1755-0998.2009.02745.x. [DOI] [PubMed] [Google Scholar]
- Li FW, Kuo LY, Rothfels CJ, et al. rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0026597. e26597. http://dx.doi.org/10.1371/journal.pone.0026597 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: aA software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Martin CE, Allen MT, Haufler CH. C3 photosynthesis in the gametophyte of the epiphytic CAM fern Pyrrosia longifolia (Polypodiaceae) American Journal of Botany. 1995;82:441–444. [Google Scholar]
- Nitta JH. Exploring the utility of three plastid loci for biocoding the filmy ferns (Hymenophyllaceae) of Moorea. Taxon. 2008;57:725–736. [Google Scholar]
- Nayar BK. Contributions to the morphology of some species of Microsorium. Annals of Botany. 1963;27:89–100. [Google Scholar]
- Nayar BK, Kaur S. Gametophytes of homosporous ferns. Botanical Review. 1971;37:295–396. [Google Scholar]
- Pfenninger M, Nowak C, Kley C, Steinke D, Streit B. Utility of DNA taxonomy and barcoding for the inference of larval community structure in morphologically cryptic Chironomus (Diptera) species. Molecular Ecology. 2007;16:1957–1968. doi: 10.1111/j.1365-294X.2006.03136.x. [DOI] [PubMed] [Google Scholar]
- Pompanon F, Deagle BE, Symondson WOC, Brown DS, Jarman SN, Taberlet P. Who is eating what: diet assessment using next generation sequencing. Molecular Ecology. 2012;21:1931–1950. doi: 10.1111/j.1365-294X.2011.05403.x. [DOI] [PubMed] [Google Scholar]
- Pryer KM, Smith AR, Hunt JS, Dubuisson JY. rbcL data reveal two monophyletic groups of filmy ferns (Filicopsida: Hymenophyllaceae) American Journal of Botany. 2001;88:1118–1130. [PubMed] [Google Scholar]
- Raine CA, Farrar DR, Sheffield E. A new Hymenopyllum species in the Appalachians represented by independent gametophyte colonies. American Fern Journal. 1991;81:109–118. [Google Scholar]
- Ruhfel B, Lindsay S, Davis CC. Phylogenetic placement of Rheopteris and the polyphyly of Monogramma (Pteridaceae s.l.): evidence from rbcL sequence data. Systematic Botany. 2008;33:37–43. [Google Scholar]
- Rumsey FJ, Sheffield E. Inter-generational ecological niche separation and the ‘independent gametophyte’ phenomenon. In: Camus JM, Gibby M, Johns RJ, editors. Pteridology in perspective. Richmond, Surrey: Royal Botanic Gardens, Kew; 1996. pp. 563–570. [Google Scholar]
- Rumsey FJ, Headley AD, Farrar DR, Sheffield E. The killarney fern (Trichomanes speciosum) in Yorkshire. Naturalist. 1991;116:41–43. [Google Scholar]
- Schneider H, Schuettpelz E. Identifying fern gametophytes using DNA sequences. Molecular Ecology Notes. 2006;6:989–991. [Google Scholar]
- Sheffield E, Farrar DR. Cryo SEM examination of gemma formation in Vittaria graminifolia. American Journal of Botany. 1988;75:894–899. [Google Scholar]
- Stokey AG. The contribution by the gametophyte to classification of the homosporous ferns. Phytomorphology. 1950;1:1–20. [Google Scholar]
- Stokey AG, Atkinson LR. The gametophytes of the Grammitidaceae. Phytomorphology. 1958;8:391–403. [Google Scholar]
- Swofford DL. Sunderland, MA: Sinauer Associates; 2003. PAUP*. Phylogenetic analysis using Parsimony (* and Other Methods). Version 4·0b10. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using Maximum Likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini A, Pompanon F, Taberlet P. DNA barcoding for ecologists. Trends in Ecology and Evolution. 2008;24:110–117. doi: 10.1016/j.tree.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Wang CN, Moller M, Cronk QC. Phylogenetic position of Titanotrichum oldhamii (Gesneriaceae) inferred from four different gene regions. Systematic Botany. 2004;29:407–418. [Google Scholar]
- Watkins JE, Jr, Cardelús CL. Ferns in an angiosperm world: cretaceous radiation into the epiphytic niche and diversification on the forest floor. International Journal of Plant Sciences. 2012;173:695–710. [Google Scholar]
- Watkins JE, Jr, Mack MK, Mulkey SS. Gametophyte ecology and demography of epiphytic and terrestrial tropical ferns. American Journal of Botany. 2007;94:701–708. doi: 10.3732/ajb.94.4.701. [DOI] [PubMed] [Google Scholar]
- Wilson EE, Siduh CS, LeVan KE, Holway DA. Pollen foraging behaviour of solitary Hawaiian bees revealed through molecular pollen analysis. Molecular Ecology. 2010;19:4823–4829. doi: 10.1111/j.1365-294X.2010.04849.x. [DOI] [PubMed] [Google Scholar]
- Wolf PG, Soltis PS, Soltis DE. Phylogenetic relationships of dennstaedtioid ferns: evidence from rbcL sequences. Molecular Phylogenetics and Evolution. 1994;3:383–392. doi: 10.1006/mpev.1994.1044. [DOI] [PubMed] [Google Scholar]
- Zhang XC. Antrophyaceae. In: Chu WM, editor. Flora Republicae Popularis Sinicae. Vol. 3. Beijing: Science Press; 1996a. pp. 1–12. [Google Scholar]
- Zhang XC. Vittariaceae. In: Chu WM, editor. Flora Reipublicae Popularis Sinicae. Vol. 3. Beijing: Science Press; 1996b. pp. 12–31. [Google Scholar]
- Zwickl DJ. Austin, TX, USA: Ph.D. dissertation, The University of Texas; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. [Google Scholar]