Abstract
Background
Genome restructuring is an ongoing process in natural plant populations. The influence of environmental changes on the genome is crucial, especially during periods of extreme climatic fluctuations. Interactions between the environment and the organism manifest to the greatest extent at the limits of the species' ecological niche. Thus, marginal populations are expected to exhibit lower genetic diversity and higher genetic differentiation than central populations, and some models assume that marginal populations play an important role in the maintenance and generation of biological diversity.
Scope
In this review, long-term data on the cytogenetic characteristics of diploid Aegilops speltoides Tauch populations are summarized and discussed. This species is distributed in and around the Fertile Crescent and is proposed to be the wild progenitor of a number of diploid and polyploid wheat species. In marginal populations of Ae. speltoides, numerical chromosomal aberrations, spontaneous aneuploidy, B-chromosomes, rDNA cluster repatterning and reduction in the species-specific and tribe-specific tandem repeats have been detected. Significant changes were observed and occurred in parallel with changes in plant morphology and physiology.
Conclusions
Considerable genomic variation at the chromosomal level was found in the marginal populations of Ae. speltoides. It is likely that a specific combination of gene mutations and chromosomal repatterning has produced the evolutionary trend in each specific case, i.e. for a particular species or group of related species in a given period of time and in a certain habitat. The appearance of a new chromosomal pattern is considered an important factor in promoting the emergence of interbreeding barriers.
Keywords: Aegilops speltoides, wheat, marginal populations, chromosomes, evolution, speciation
INTRODUCTION
The influence of environmental changes on the genome is crucial, especially during periods of extreme climatic fluctuations. It is generally accepted that plant genomes respond to variations in environmental conditions and that the large-scale features of plant systems (such as yield and sustainability) depend on interactions between individual plants and environmental factors (Yin and Struik, 2007; Martienssen, 2008). Interactions between the environment and the organism manifest to the greatest extent at the limits of the species' ecological niche. The central–marginal hypothesis states that two key genetic parameters – the effective population size and the rate of gene flow – should be highest at the range centre and lowest at the range margins (for a review, see Eckert et al., 2008). Thus, marginal populations are expected to exhibit lower genetic diversity and higher genetic differentiation than central populations. At the same time, in marginal populations under the influence of an unusual ecology, intensive processes of raciation and speciation may take place, and some models assume that marginal populations play an important role in the maintenance and generation of biological diversity (Mayr, 1970; Kirkpatrick and Barton, 1997; Grant, 1981; Channell and Lomolino, 2000; Navarro and Barton, 2003). Indeed, the genomic changes in marginal populations that experience strong selective forces could be key for understanding genomic evolution and for predicting the response of a genome to environmental changes.
Here, I summarize our long-term data on the cytogenetic characteristics of natural populations of diploid Aegilops speltoides Tausch (2n = 2x = 14) with special emphasis on small, marginal, stressed populations. This species belong to sect. Sitopsis (Triticeae, Poaceae), is distributed in and around the Fertile Crescent and is proposed to be the wild progenitor of a number of diploid and polyploid wheat species (Sears, 1941; Zohary and Imber, 1963; Kimber and Feldman, 1987). Currently in the Middle East region, we observe recession of a plant range to the north (Tchernov, 1988; Hofreiter and Stewart, 2009; http://www.ipcc.ch) which is a common event when climate changes. This scenario repeatedly occurred during the glacial period and the subsequent Holocene. This is a unique opportunity to observe the reaction of the genome of a model group of species to a changing environment, as Middle Eastern flora might be the first to experience the impact of global warming due to the close proximity to the African–Arabian desert domain. Comparative cytogenetic study of genome evolution in natural plant populations throughout the species' range yields a snapshot of the genome state, and if the cytogenetic findings are combined with data from the fields of molecular genetics and botany, they may shed light on past, current and even future evolutionary events.
MARGINAL POPULATIONS: A SOURCE OF A NEW FORMS AND SPECIES
Defining marginal populations
The importance of clearly defining marginality should be emphasized. The main criteria we used for assigning populations of Ae. speltoides to the marginal category were as follows: (1) the position relative to the centre of the species' range; (2) the population size (the area of small populations is <1000 m2); (3) the degree of population destruction (mainly due to human activity); (4) local ecology (biotic and abiotic components); and (5) elevation (the optimum is from 100 to 1000 m asl). The current centre of the Ae. speltoides range is in the middle of the Fertile Crescent (Zohary et al., 1969; Zohary, 1970; Kimber and Feldman, 1987) and is limited to the approximate geographic coordinates 36–38°N, 37–41°E (a list of the investigated populations and their characteristics is shown in the Appendix). All Israeli populations of Ae. speltoides are peripheral and located at the southern border. The northernmost populations in Turkey could also be regarded as peripheral. Among the ten investigated Israeli populations (Raskina et al., 2011), only two fell under the definition of marginal, specifically a population near the mouth of the Kishon River (Kishon population) and the populations on the northern slope of Carmel Mountain which no longer exist (Technion 2 population). Both populations are extremely small (approx. 100 m2 and 20 m2, respectively). The Kishon population is the only Israeli population that is located at sea level (2 m asl) and is close to the Akko plain terminal of desert plants (Raskina et al., 2004a, b, 2011). Technion 2 and the neighbouring population, Technion 1, represent the remnants of a once large single population; we have observed the continuing, rapid decline of this population over the last decade. Despite the negative impact of anthropogenic factors, the main factor in the rapid decline of the remnants of the original Technion population is climate change, which has led to the displacement of local flora by other species. The northernmost Turkey populations (Appendix) also fall under the definition of marginal.
Morphological and physiological characteristics of plants from peripheral and marginal populations
In marginal populations, changes in plant morphology and physiology have been observed. In the case of the marginal population of Ae. speltoides from Cankiri (Turkey), adult plants grown in a greenhouse were half the size of phenotypically normal plants and had smaller spikes of 3·5–4·5 cm. The time from germination to flowering was up to a year, which is longer than the typical 6–7 months (Appendix). It should be noted that this was the northernmost population of all of those investigated, and the winter-type characteristic of the population explains the delay in development in the absence of vernalization. The exact opposite was observed for the southernmost Kishon population. The time from germination to flowering was found to occur in half the expected time, a maximum of 3–3·5 months. Interestingly, despite the short vegetative phase, the maturation of the seeds in this population occurred only 10–14 d earlier than in the neighbouring populations on Mount Caramel, which exhibit a normal life cycle (Technion, Nahal Mearot and Ramat Hanadiv) (Appendix).
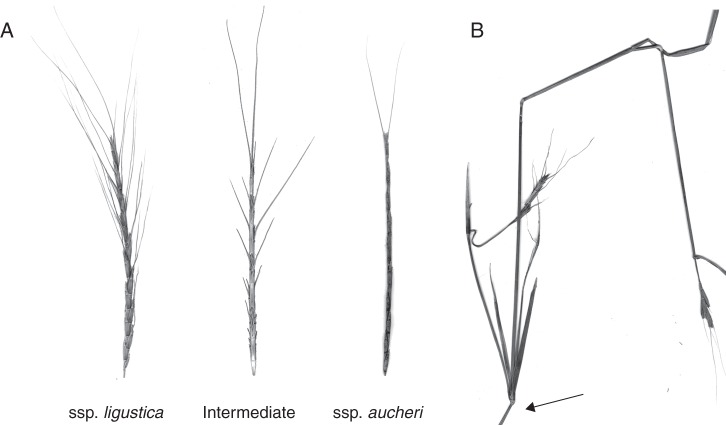
Another important feature of the species is spike morphology. Aegilops speltoides has a unique genetic dimorphism in the fruit types (Fig. 1A). Both morphotypes, dominant ligustica and recessive aucheri, are encoded by tightly linked genes (Sears, 1941; Zohary and Imber, 1963; Kimber and Feldman, 1987). In nature, both subspecies coexist in mixed cross-pollinated populations. The ratio of the ligustica–aucheri morphotypes varies significantly between populations and very probably depends on environmental conditions. Over the past 12 years, we recorded a large shift in the Kishon population towards the ligustica phenotype; however, in the small Technion 1 population, the last few plants with the ligustica morphotype were observed in 2002, and this small population has become homogeneous and currently consists of only plants with the aucheri phenotype. In the last decade, these two populations diverged considerably. I recently discovered a similar small population consisting only of the aucheri morphotype at the foot of the Carmel Mountain in the Nahal Mearot Valley and an En-Efek population in the Akko coastal plain consisting only of the ligustica morphotype (Appendix), which suggests that this phenomenon is not unique. Another morphological alteration was found in populations of Ramat Hanadiv and Givat Koah (Fig. 1A). It is expressed in an intermediate abnormal ligustica–aucheri phenotype, and may be caused by genetic changes within the linked group of genes. These plants have spikes that resemble the aucheri type (i.e. not brittle) but also have pronounced lateral awns similar to the ligustica type. A large number of plants collected in various remote locations from the main population of Ramat Hanadiv indicate that this seemingly neutral mutation has already become fixed in the population. In addition to this abnormality, we also noted the development of secondary tillers from the lateral buds of the main culm and even tertiary tillers, which is atypical for Ae. speltoides (Fig. 1B). The same anomaly was also documented in plants from the Cankiri population (Turkey). Evidently, the identified population-specific alterations in the life cycle and phenotype of Ae. speltoides are caused by as yet unknown gene mutations and/or chromosomal rearrangements, which result in changes in gene arrangement and/or gene expression.
Fig. 1.
Morphological characteristics of Ae. speltoides. (A) Left, normal spike from Ae. speltoides ssp. ligustica; right, normal spike from Ae. speltoides ssp. aucheri; centre, abnormal intermediate phenotype. (B) The atypical development of a secondary tiller from the lateral bud (arrowed) on the main culm.
Chromosomal rearrangements and genomic repatterning in Ae. speltoides
Natural populations are known to be enriched with chromosomal rearrangements that generally occur in the heterozygous state (White, 1978; Rieseberg, 2001; Levin, 2002). The effect of chromosomal rearrangements is suppression of recombination within rearranged regions (inversions), the disruption of existing linkage groups and the creation of new ones (translocations), which may lead to changes in gene expression and in the interactions between genes (Rieseberg, 2001; Strasburg et al., 2009; Brown and O'Neill, 2010). Some chromosomal rearrangements may be neutral without significant effect on the phenotype. Nevertheless, underlying heteromorphism in homologous chromosomes may create intraspecific polymorphisms in the heterochromatin pattern. In contrast, it is thought that the majority of chromosomal rearrangements that involve euchromatin are deleterious and can be maintained in a population only in the heterozygous state because the homozygote is eliminated by natural selection (Levin, 2002; Charlesworth, 2009; Brown and O'Neill, 2010; Faria and Navarro, 2010). However, some chromosomal aberrations may become fixed in the population by positive selection if they are associated with the emergence of an adaptive combination of traits, especially in a changing environment (Kirkpatrick and Barton, 1997; Hoffmann et al., 2004; Coghlan et al., 2005; Orr, 2005; Kirkpatrick and Barton, 2006; Rieseberg and Willis, 2007; Charlesworth, 2009). The frequency and spectrum of chromosomal repatterning, which are determined by external (biotic and abiotic) and internal (such as population size and mating system) factors, are indicators of the population's state in time and space.
Numerical chromosomal aberrations in Ae. speltoides populations: spontaneous aneuploidy and B-chromosomes
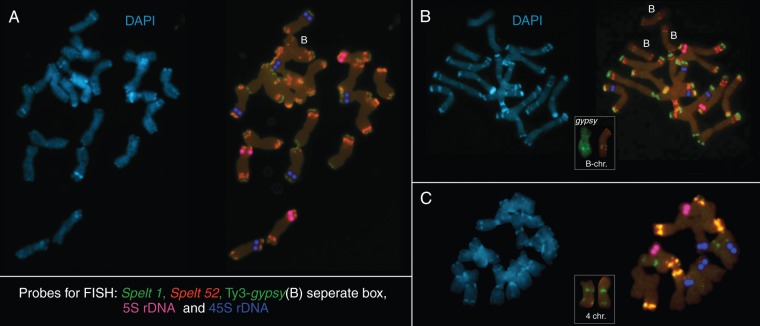
Spontaneous aneuploidy is not a normal and stable state of the diploid genome. Additional chromosomes resulting from meiotic disruption further destabilize the genome through the disruption of normal chromosome pairing and segregation. The result may be lethal or at least cause a decrease in fertility. In populations of Ae. speltoides, aneuploidy may occur for individual chromosomes as a result of gene mutations or/and meiotic disorders, but spontaneous non-disjunction of the entire chromosome complement can also occur. The triploid genotype found in the Ramat Hanadiv population is an example of the latter and is shown in Fig. 2A; curiously, an additional B-chromosome is also present. The emergence of supernumerary B-chromosomes (Bs) is another consequence of genomic aberrations in natural Ae. speltoides populations (Fig. 2B). The presence of Bs, similar to aneuploidy, increases the frequency of recombination, thereby causing new chromosomal abnormalities (Mendelson and Zohary, 1972; Zarchi et al., 1974; Cebria et al., 1994; Camacho et al., 2000, 2002; Puertas, 2002; Jones and Houben, 2003; Jones et al., 2008a, b; Belyayev et al., 2010). We detected up to eight Bs in natural populations of Ae. speltoides (Raskina et al., 2011). Interestingly, the presence of from one to three Bs has a positive effect on the plant, whereas a higher number of Bs reduces fertility and vigour (Mendelson and Zohary, 1972; Belyayev et al., 2010). If there are only two Bs in a genome, they often behave like normal homologues in meiosis by forming a bivalent and then separating normally to opposite poles during anaphase 1. However, increases in the number of Bs further destabilize the genome by promoting heterologous recombination and meiotic aberrations. The ability for heterologous conjugation with the A-chromosomes is due to the structural organization of Bs in Ae. speltoides. A well-known and important feature of Bs in plants and animals is a high proportion of heterochromatin, which is gained through the accumulation of different types of repetitive DNAs, including transposable elements (Puertas, 2002; Jones and Houben, 2003; Jones et al., 2008a, b; Carchilan et al., 2009). In Ae. speltoides, all Bs carry a single intercalary Spelt 1 tandem repeat cluster and a 5S rDNA cluster in both arms (Figs 2A, B and 3A, B) (Raskina et al., 2011). In addition, as shown in Fig. 2B, a large intercalary cluster of Ty3-gypsy elements was found in close proximity to the 5S rDNA and Spelt 1 blocks. Clusters of highly repetitive DNA such as 5S rDNA, tandem repeats, clusters of transposable elements, and telomere and centromeric repeats are the hot spots for homo- and heterologous synapses and recombination between chromosomes of the types A–A and A–B in meiosis. An example of synapses between Bs and A-chromosomes is shown in Fig. 3A, B. The similarity in the B-chromosome structures throughout the species range caused us to hypothesize that they were generated from similar processes, specifically the heterologous recombination of certain A-chromosomes. We suggest that chromosome 4, which only carries the intercalary Spelt 1 cluster, and chromosome 5, which is an exclusive source of 5S rDNA in the genome of Ae. speltoides, may be involved in the heterologous synapses and recombination resulting in the formation of Bs (Raskina et al., 2011). The emergence of Bs in the population usually accompanies outcrossing; however, an important feature of Ae. speltoides is that it is highly self-compatible and reproduces effectively in both cross- and self-pollination mating systems (Zohary and Imber, 1963; Raskina et al., 2004b; Belyayev et al., 2010). It is very likely that the dualism of their reproduction system in combination with dimorphism (the coexistence of the aucheri and ligustica morphotypes in natural panmictic populations) is the basis for the evolutionary lability of the Ae. speltoides genome, determines its direct involvement in the generation of allopolyploid wheats and probably allowed it to become a progenitor for the sect. Sitopsis (Raskina et al., 2004b).
Fig. 2.
Fluorescence in situ hybridization (FISH) on somatic chromosomes of Ae. speltoides with Spelt 1 (green), Spelt 52 (red), As5SDNAE (5S rDNA, pink pseudocolour) and pTa71 (45S rDNA, blue pseudocolour) DNA probes, and differential staining with 4′,6-diamidino-2-phenylindole (DAPI). (A) Original triploid genotype from the Ramat Hanadiv population; one B-chromosome also appears. (B) Diploid genotype with three Bs from the Ramat Hanadiv population. Inset B-chromosomes: left, large intercalary Ty3-gypsy cluster (Belyayev et al., 2001) in the long arm; right, intercalary Spelt 1 and 5S rDNA clusters in the long arm and a distal 5S rDNA cluster in the short arm. (C) Metaphase plate of the plant from the marginal Cankiri population: loss of almost all terminal Spelt 1 and a reduced number of Spelt 52 clusters in comparison with the Ramat Hanadiv population. Inset: chromosome 4 contains intercalary and near-centromeric Spelt 1 clusters.
Fig. 3.
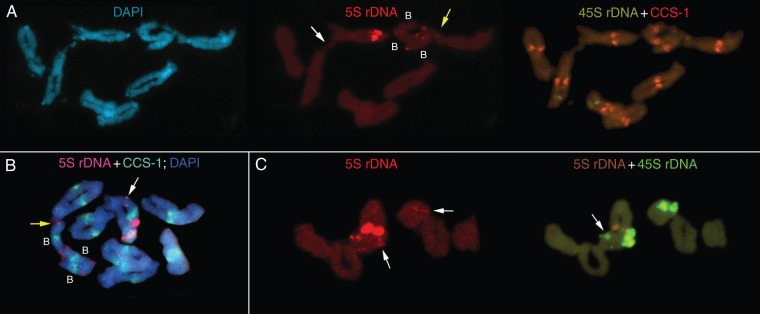
Fluorescence in situ hybridization (FISH) on meiotic chromosomes of Ae. speltoides from the Kishon population with As5SDNAE (5S rDNA, red), pTa71 (45S rDNA, green), CCS-1 [cereal centromere sequence, green (Aragon-Alcaide et al., 1996)]. (A) Heterologous synapses between the long arms of chromosomes 5 and 1 (white arrow) and between B- and A-chromosomes (yellow arrow). (B) A synapse between A- and B-chromosomes is shown with a yellow arrow; chromosome 5 is heterozygous for pericentric inversion and carries an intercalary additional 5S rDNA cluster (white arrow). (C) Left, two additional 5S rDNA clusters are marked with arrows; right, a cell-specific additional 45S rDNA intercalary cluster is marked with an arrow.
Structural chromosome aberrations: reduction in the species-specific Spelt 1 and tribe-specific Spelt 52 tandem repeats in peripheral and marginal populations of Ae. speltoides
Significant intraspecific polymorphisms in the distribution of Spelt 1and Spelt 52 tandem repeats that are integral parts of heterochromatin were found in Ae. speltoides (Raskina et al., 2011). The heterochromatin chromosomal pattern is one of the most important characteristics of a species. Nevertheless, there is a significantly high level of intraspecific C-banding polymorphisms in the Aegilops/Triticum complex (Friebe and Gill, 1996; Maestra and Naranjo, 1999; Maestra and Naranjo, 2000; Badaeva et al., 2002, 2004, 2007). In the 1970s, research on the inheritance of Giemsa C-bands showed Mendelian segregation in successive generations under self-pollination, which provided direct evidence that meiotic crossing-over caused the variation in the heterochromatic patterning of rye (Singh, 1977), barley (Linde-Laursen, 1979) and maize (Handlaczky and Kalman, 1975). Tandem repeats comprise a significant portion of the distal and terminal heterochromatin of the Aegilops and Triticum genomes (Anamthawat-Jonsson and Heslop-Harrison, 1993; Salina et al., 2006; Zoshchuk et al., 2007). The intraspecific chromosome patterns of the species-specific Spelt 1 and tribe-specific Spelt 52 tandem repeats in Ae. speltoides are highly variable. We observed an almost complete loss of the terminal Spelt 1 repeats in marginal populations (Fig. 2C), in which the number of blocks was 12–14 times lower than in central populations (Raskina et al., 2011). The number of Spelt 52 blocks was also 1·5–2 times lower in intermediate and marginal populations than in central populations (Fig. 2B). Intraspecific polymorphisms of the Spelt 1 and Spelt 52 tandem repeat chromosomal patterns are a special case of heterochromatin pattern polymorphism and a result of complex chromosomal rearrangements in the panmictic populations of outcrossing Ae. speltoides, as these polymorphisms reflect random chromosomal recombination under random mating (Raskina et al., 2004a; Belyayev et al., 2010). The number of Spelt 52 clusters in successive generations under self-pollination follows chromosomal segregation, while the copy number abundance of this tandem repeat in each successive genome is subject to amplification or reduction as a consequence of homologous and/or heterologous recombination in distal/terminal chromosomal regions. We propose that the depletion of tandem repeats in the marginal populations of Ae. speltoides could be a result of either elimination of the repeats under stressful environmental conditions in the peripheral populations or amplification of the repeats in conducive climatic and/or edaphic environments in the centre of the species' geographical distribution. It is likely that both scenarios have occurred simultaneously and that we observed a bidirectional shift in the repetitive DNA genomic patterns that led to interpopulation diversification. In the central populations with optimal environmental conditions, the current chromosomal rearrangements, such as duplications and insertions caused by unequal crossing-over and/or reciprocal balanced translocations, contribute to the accumulation of tandem repeats in the population. In peripheral and marginal populations, an increase in the recombination frequency under stressful conditions (Grant, 1981; Levin, 2002; Belyayev et al., 2010; Raskina et al., 2011) leads to unbalanced translocations and multiple deletions that involve more breakpoints, which dominate over other types of structural chromosomal mutations. Consequently, we witnessed the loss of a large number of Spelt 1 and Spelt 52 clusters. The appearance of a new chromosomal pattern is considered an important factor in the diversification of populations and the prevention of cross-breeding.
Among structural chromosomal rearrangements, inversions are especially important for the creation of interbreeding barriers. In the genome of Ae. speltoides, the intercalary position of Spelt 1 cluster(s) indicates the probable presence of an inversion(s) in chromosome 4 (Raskina et al., 2011) (Fig. 2C inset). An important feature of inversions is that they appear to be a source of particular adaptive combinations of genes (Grant, 1981; Rieseberg and Willis, 2007), and could capture and spread locally adapted alleles in a population by suppressing recombination between the loci (Hoffmann et al., 2004; Kirkpatrick and Barton, 2006). Survival and reproduction of self-compatible genotypes may give rise to an endemic form of the species that is adapted to the new environment, while the parental species recedes or disappears (Lewis and Raven, 1958; Lewis, 1962; Grant, 1981; Tchernov, 1988). We propose that the scenario of sympatric speciation has occurred in the sect. Sitopsis in the periphery of the Ae. speltoides range subjected to a changing environment (Raskina et al., 2004b).
Rearrangements of ribosomal DNA sites
In addition to the major chromosomal rearrangements that I described above, it is also possible to estimate the level of microevolutionary genomic change indirectly by evaluating the repatterning of well-defined chromosomal markers and, primarily, by the mobility of rDNA clusters. Both the location and number of rDNA sites vary intraspecifically (Eickbush and Eickbush, 2007). The variation may involve major loci, or fragments of the unit (Fig. 3C), and these are often not known to be transcribed (Heslop-Harrison, 2000). The mechanism for rDNA cluster repatterning could be unequal crossing-over (Eickbush and Eickbush, 2007) or the activity of adjacent transposable elements (Raskina et al., 2004a, b, 2008). It is obvious that chromosomal repatterning further increases/decreases the number of rDNA sites or their repositioning, but the dynamics of rDNA clusters may be regarded as a strong indicator for ongoing significant microevolutionary processes (Jiang and Gill, 1994; Raskina et al., 2004b).
In the marginal Kishon population, we discovered modified genotypes of Ae. speltoides with rDNA patterns similar to those of the closely related species Ae. sharonensis. Likewise, Ae. sharonensis plants from the bordering population in the Kishon area possessed Ae. speltoides-like features, including additional 5S and 45S ribosomal sites in both species on chromosomes 1, 5 and 6, which differed from the usual rDNA patterning for this species (Fig. 4). We speculate that the increased ratio of self-pollination and inbreeding in a stressful environment induced rDNA repatterning in this small marginal population of Ae. speltoides (Raskina et al., 2004b). We found further evidence to support this hypothesis. In the third successive generation of self-pollinated plants of Ae. speltoides, we found the de novo appearance of additional 5S rDNA clusters in regions of secondary constriction in chromosomes 1 and 6, but the maternal plant had normal rDNA patterning (Belyayev et al., 2010). The process of rDNA repatterning is permanent in the Kishon population, and the emerging variants in most cases resemble the chromosomal rDNA pattern found in closely related species of the Sitopsis group. Thus, we propose that canalized repatterning of rDNA sites may eventually lead to sympatric speciation in marginal populations of Ae. speltoides (Raskina et al., 2004b).
Fig. 4.
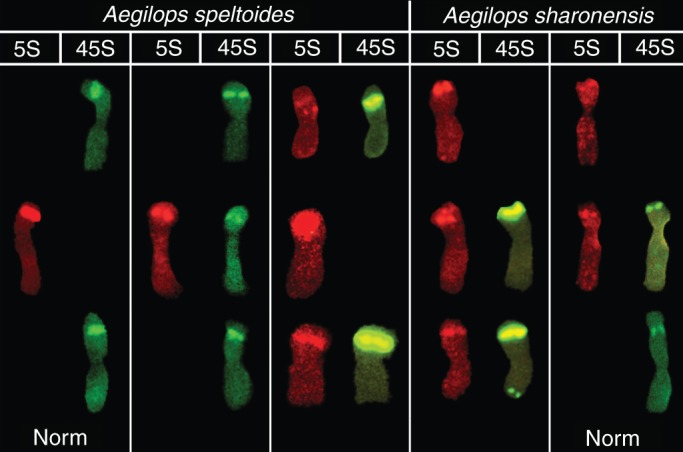
Chromosomal pattern of 5S rDNA (red) and 45S rDNA (green) of Ae. speltoides and Ae. sharonensis from the Kishon populations (Raskina et al., 2004b). Modified genotypes of both species carry additional rDNA clusters on the chromosomes 1, 5 and 6.
CONCLUDING REMARKS
In combination with gene mutations, the role of chromosomal rearrangements in the evolution of the eukaryotic genome has been debated for a long time (Dobzhansky, 1935; Mayr, 1970; White, 1978; Grant, 1981; Rieseberg and Willis, 2007; Brown and O'Neill, 2010). It is likely that a specific combination of gene mutations and chromosomal repatterning has produced the evolutionary trend in each specific case, i.e. for a particular species or group of related species in a given period of time and in a certain habitat (Dobzhansky, 1935). Changes in genomic structure are an ongoing process that occurs in natural populations. I have no uncontested evidence that there is a direct link between chromosomal rebuilding and changes in plant morphology and physiology, but I can at least say that these changes occur simultaneously. I propose that chromosomal repatterning in these cases might play a role in the development of new traits by breaking apart linkage groups, altering the interaction between genes and/or affecting gene expression.
ACKNOWLEDGEMENTS
The author is most grateful to Olga Raskina and anonymous reviewers for helpful comments. This work was supported by the Israel Science Foundation under grant number 723/07.
APPENDIX
Characteristics of the Ae. speltoides populations studied.
| Populations, origin, source | Geographical zone, elevation, co-ordinates | Population size; location | Morphotype |
|---|---|---|---|
| Kirklareli, Turkey*, PI 170203 | Euro-Siberian, 64 m, 41°20′N, 27°29′E | N/A; cultivated field¶ | ssp. ligustica |
| Cankiri, Turkey†, PI 573448 | Euro-Siberian, 680 m, 40°31′N, 33°38′E | N/A; cultivated field¶ | ssp. ligustica |
| Ankara, Turkey*, PI 573452 | Irano-Turanian, 575 m, 36°59′N, 32°56′E | N/A; cultivated field¶ | ssp. ligustica |
| Eregli, Turkey‡, TS-24, G-1038 | Irano-Turanian, 1200 m, 37°25′N, 34°15′E¶ | N/A; natural habitat¶ | ssp. ligustica |
| Gaziantep, Turkey†, TR 50279 | Mediterranean, 940 m, 37°05′N, 37°24′E¶ | N/A; urbanistic area¶ | ssp. aucheri |
| Urfa, Turkey*, PI 542262 | Mediterranean, 700 m, 37°17′N, 38°46′E | N/A; cultivated field¶ | ssp. ligustica |
| Arbil, Iraq*, PI 219867 | Irano-Turanian, 570 m, 36°24′N, 44°08′E | N/A; uncultivated area¶ | ssp. ligustica |
| Latakia, Syria†,§, PI 487235, TS-84 | Mediterranean, 200 m, 35°38′N, 35°59′E | N/A; uncultivated area¶ | ssp. aucheri |
| Tartus, Syria*, PI 487238 | Mediterranean, 600 m, 35°07′N, 36°07′E | N/A; cultivated field¶ | ssp. aucheri |
| Achihood, Israel§, 2·16 | Mediterranean, 45–75 m, 32°55′N, 35°10′E | Large; cultivated field and natural habitat | ssp. ligustica, ssp. aucheri |
| En-Efek, Israel§, 2·37 | Mediterranean, 16 m, 32°50′N, 35°06′E | Small; uncultivated area; endangered | ssp. ligustica |
| Kishon, Israel§, 2·22 | Mediterranean, 2 m, 32°48′N, 35°02′E | Small; natural habitat; endangered | ssp. ligustica, ssp. aucheri, |
| Technion-1, Israel§, 2·36 | Mediterranean, 224 m, 32°46′N, 35°00′E | Small; urbanistic area; endangered | ssp. aucheri |
| Technion-2, Israel§, 2·36 | Mediterranean, 265 m, 32°46′N, 35°00′E | Small; natural abitat; extinct | ssp. ligustica,, ssp. aucheri |
| Nahal Mearot, Israel§, 2·48 | Mediterranean, 52 m, 32°40′N, 34°58′E | Small; natural habitat | ssp. aucheri |
| Ramat Hanadiv, Israel§, 2·46 | Mediterranean, 100–125 m, 32°33′N, 34°56′E | Large; natural habitat; interrupted area | ssp. ligustica, ssp. aucheri, intermediate |
| Katzir, Israel†,§, TS 89 | Mediterranean, 233–250 m, 32°29′N, 35°05′E | Large; natural habitat | ssp. aucheri |
| Givat Koah, Israel§, TS 43 | Mediterranean, 75 m, 32°02′N, 34°58′E¶ | Small; uncultivated area; extinct | Intermediate |
| Ashdod, Israel‡, TS 93 | Mediterranean, 35 m, 31°51′N, 34°45′E¶ | N/A; uncultivated area; extinct | ssp. ligustica |
| Ashkelon, Israel‡, TS 01 | Mediterranean, 45 m, 31°40′N, 34°38′E¶ | N/A; uncultivated area; extinct | ssp. aucheri |
N/A, not available.
Source: *USDA, United States Department of Agriculture; †AARI, Aegean Agricultural Research Institute, Turkey; ‡kindly provided by Professor M. Feldman, Weizmann Institute collection, Rehovot, Israel; § IE, Institute of Evolution collection, Haifa, Israel.
¶ Data obtained from Google Earth.
LITERATURE CITED
- Anamthawat-Jonsson K, Heslop-Harrison JS. Isolation and characterization of genome-specific DNA sequences in Triticeae species. Molecular and General Genetics. 1993;240:151–158. doi: 10.1007/BF00277052. [DOI] [PubMed] [Google Scholar]
- Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G. A cereal centromere sequence. Chromosoma. 1996;105:261–268. doi: 10.1007/BF02524643. [DOI] [PubMed] [Google Scholar]
- Badaeva ED, Amosova AV, Muravenko OV, et al. Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Plant Systematics and Evolution. 2002;231:163–190. [Google Scholar]
- Badaeva ED, Amosova AV, Samatadze TE, et al. Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Systematics and Evolution. 2004;246:45–76. [Google Scholar]
- Badaeva ED, Dedkova OS, Gay G, et al. Chromosomal rearrangements in wheat: their types and distribution. Genome. 2007;50:907–926. doi: 10.1139/g07-072. [DOI] [PubMed] [Google Scholar]
- Belyayev A, Raskina O, Nevo E. Chromosomal distribution of reverse transcriptase-containing retroelements in two Triticeae species. Chromosome Research. 2001;9:129–136. doi: 10.1023/a:1009231019833. [DOI] [PubMed] [Google Scholar]
- Belyayev A, Kalendar R, Brodsky L, Nevo E, Schulman AH, Raskina O. Transposable elements in a marginal plant population: temporal fluctuations provide new insights into genome evolution of wild diploid wheat. Mobile DNA. 2010;1(6) doi: 10.1186/1759-8753-1-6. http://dx.doi.or/10.1186/1759-8753-1-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, O'Neill R. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annual Review of Genomics and Human Genetics. 2010;11:291–316. doi: 10.1146/annurev-genom-082509-141554. [DOI] [PubMed] [Google Scholar]
- Camacho JP, Sharbel TF, Beukeboom LV. B-chromosome evolution. Philosophical Transactions of the Royal Society B: Biological Sciences. 2000;355:163–178. doi: 10.1098/rstb.2000.0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho JP, Bakkali M, Corral JM, et al. Host recombination is dependent on the degree of parasitism. Proceedings of the Royal Society B: Biological Sciences. 2002;269:2173–2177. doi: 10.1098/rspb.2002.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carchilan M, Kumke K, Mikolajewski S, Houben A. Rye B chromosomes are weakly transcribed and might alter the transcriptional activity of A chromosome sequences. Chromosoma. 2009;118:607–616. doi: 10.1007/s00412-009-0222-8. [DOI] [PubMed] [Google Scholar]
- Cebria A, Navarro ML, Puertas MJ. Genetic control of B chromosome transmission in Aegilops speltoides (Poaceae) American Journal of Botany. 1994;81:1502–1507. [Google Scholar]
- Channell R, Lomolino MV. Dynamic biogeography and conservation of endangered species. Nature. 2000;403:84–86. doi: 10.1038/47487. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Effective population size and patterns of molecular evolution and variation. Nature Reviews Genetics. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- Coghlan A, Eichler EE, Oliver SG, Paterson AH, Stein L. Chromosome evolution in eukaryotes: a multi-kingdom perspective. Trends in Genetics. 2005;21:673–682. doi: 10.1016/j.tig.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. The Y chromosome of Drosophila pseudoobscura. Genetics. 1935;20:366–376. doi: 10.1093/genetics/20.4.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC. Genetic variation across species geographical ranges: the central–marginal hypothesis and beyond. Molecular Ecology. 2008;17:1170–1188. doi: 10.1111/j.1365-294X.2007.03659.x. [DOI] [PubMed] [Google Scholar]
- Eickbush TH, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria R, Navarro A. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends in Ecology and Evolution. 2010;25:660–668. doi: 10.1016/j.tree.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Friebe B, Gill BS. Methods of genome analysis in plants. Boca Raton, FL: CRC Press; 1996. Chromosome banding and genome analysis in diploid and cultivated polyploid wheats. In: Jauhar PP; pp. 39–60. [Google Scholar]
- Grant V. Plant speciation. 2nd edn. New York: Columbia University Press; 1981. [Google Scholar]
- Handlaczky G, Kalman L. Discrimination of homologous chromosomes of maize with Giemsa staining. Heredity. 1975;35:371–374. [Google Scholar]
- Heslop-Harrison JS. RNA, genes, genomes and chromosomes: repetitive DNA sequences in plants. Chromosomes Today. 2000;13:45–56. [Google Scholar]
- Hoffmann AA, Sgrò CM, Weeks AR. Chromosomal inversion polymorphisms and adaptation. Trends in Ecology and Evolution. 2004;19:482–488. doi: 10.1016/j.tree.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Stewart J. Ecological change, range fluctuations and population dynamics during the Pleistocene. Current Biology. 2009;19:584–594. doi: 10.1016/j.cub.2009.06.030. [DOI] [PubMed] [Google Scholar]
- Jiang J, Gill BS. New 18S.26S ribosomal RNA gene loci: chromosomal landmarks for the evolution of polyploid wheats. Chromosoma. 1994;103:179–185. doi: 10.1007/BF00368010. [DOI] [PubMed] [Google Scholar]
- Jones N, Houben A. B chromosomes in plants: escapees from the A chromosome genome? Trends in Plant Sciences. 2003;8:417–423. doi: 10.1016/S1360-1385(03)00187-0. [DOI] [PubMed] [Google Scholar]
- Jones RN, González-Sánchez M, González-García M, Vega JM, Puertas MJ. Chromosomes with a life of their own. Cytogenetics and Genome Research. 2008a;120:265–280. doi: 10.1159/000121076. [DOI] [PubMed] [Google Scholar]
- Jones RN, Viegas W, Houben A. A century of B chromosomes in plants: so what? Annals of Botany. 2008b;101:767–775. doi: 10.1093/aob/mcm167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimber G, Feldman M. Wild wheat, an introduction. Columbia: University of Missouri; 1987. College of Agriculture. [Google Scholar]
- Kirkpatrick M, Barton NH. Evolution of a species' range. American Naturalist. 1997;150:1–23. doi: 10.1086/286054. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. New York: Oxford University Press; 2002. [Google Scholar]
- Lewis H. Catastrophic selection as a factor in speciation. Evolution. 1962;16:57–271. [Google Scholar]
- Lewis H, Raven PH. Rapid evolution in Clarkia. Evolution. 1958;12:319–336. [Google Scholar]
- Linde-Laursen IB. Giemsa C-banding of barley chromosomes. 111. Segregation and linkage of C-bands on chromosomes 3, 6, and 7. Hereditas. 1979;91:73–77. [Google Scholar]
- Maestra B, Naranjo T. Structural chromosome differentiation between Triticum timopheevii and T. turgidum and T. aestivum. Theoretical and Applied Genetics. 1999;98:744–750. [Google Scholar]
- Maestra B, Naranjo T. Genome evolution in Triticeae. Chromosomes Today. 2000;13:155–167. [Google Scholar]
- Martienssen R. Great leap forward? Transposable elements, small interfering RNA and adaptive Lamarckian evolution. New Phytologist. 2008;179:572–574. doi: 10.1111/j.1469-8137.2008.02567.x. [DOI] [PubMed] [Google Scholar]
- Mayr E. Populations species and evolution. An abridgment of animal species and evolution. Cambridge: Belknap Press; 1970. [Google Scholar]
- Mendelson D, Zohary D. Behavior and transmission of supernumerary chromosomes in Aegilops speltoides. Heredity. 1972;29:329–339. [Google Scholar]
- Navarro A, Barton NH. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution. 2003;57:447–459. doi: 10.1111/j.0014-3820.2003.tb01537.x. [DOI] [PubMed] [Google Scholar]
- Orr HA. The genetic theory of adaptation: a brief history. Nature Reviews Genetics. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Puertas MJ. Nature and evolution of B chromosomes in plants: a non-coding but information-rich part of plant genomes. Cytogenetics and Genome Research. 2002;96:198–205. doi: 10.1159/000063047. [DOI] [PubMed] [Google Scholar]
- Raskina O, Belyayev A, Nevo E. Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosome Research. 2004a;12:153–161. doi: 10.1023/b:chro.0000013168.61359.43. [DOI] [PubMed] [Google Scholar]
- Raskina O, Belyayev A, Nevo E. Quantum speciation in Aegilops: molecular cytogenetic evidence from rDNA cluster variability in natural populations. Proceedings of the National Academy of Sciences, USA. 2004b;101:14818–14823. doi: 10.1073/pnas.0405817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskina O, Barber J, Nevo E, Belyayev A. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genome. Cytogenetic and Genome Research. 2008;120:351–357. doi: 10.1159/000121084. [DOI] [PubMed] [Google Scholar]
- Raskina O, Brodsky L, Belyayev A. Tandem repeats on an eco-geographical scale: outcomes from the genome of Aegilops speltoides. Chromosome Research. 2011;19:607–623. doi: 10.1007/s10577-011-9220-9. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends in Ecology and Evolution. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Rieseberg L, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina EA, Lim YL, Badaeva ED, et al. Phylogenetic reconstruction of Aegilops section Sitopsis and the evolution of tandem repeats in the diploids and derived wheat polyploids. Genome. 2006;49:1023–1035. doi: 10.1139/g06-050. [DOI] [PubMed] [Google Scholar]
- Sears E. Amphidiploids in the seven-chromosome Triticina. University of Missouri Agricultural Experimental Station Research Bulletin. 1941;336:1–46. [Google Scholar]
- Singh RJ. Evidence of heterochromatin polymorphism through crossing-over. Wheat Information Service. 1977;44:9–21. [Google Scholar]
- Strasburg JL, Scotti-Saintagne C, Scotti I, Lai Z, Rieseberg LH. Genomic patterns of adaptive divergence between chromosomally differentiated sunflower species. Molecular Biology and Evolution. 2009;26:1341–1355. doi: 10.1093/molbev/msp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchernov E. The zoogeography of Israel. The distribution and abundance at a zoogeographical crossroad. Dordrecht: Kluwer: Yom-Tov Y, Tchernov E; 1988. The biogeographical history of southern Levant; pp. 159–251. [Google Scholar]
- White MJD. Modes of speciation. W. H. Freeman; 1978. CTSan Francisco. [Google Scholar]
- Yin X, Struik PC. Crop systems biology. In: Spiertz JHJ, Struik PC, van Laar HH, editors. Scale and complexity in plant systems research: gene–plant–crop relations. Dordrecht: Springer; 2007. pp. 63–73. [Google Scholar]
- Zarchi Y, Hillel J, Simchen G. Supernumerary chromosomes and chiasma distribution in Triticum speltoides. Heredity. 1974;33:173–180. [Google Scholar]
- Zohary D, Imber D. Genetic dimorphism in fruit types in Aegilops speltoides. Heredity. 1963;18:223–231. [Google Scholar]
- Zohary D, Harlan JR, Vardi A. The wild diploid progenitors of wheat and their breeding value. Euphytica. 1969;18:58–65. [Google Scholar]
- Zohary M. Botany. In: Amiran DHK, Elster J, Gilead M, Rosenan N, Kadmon N, Parah U, editors. Atlas of Israel. Amsterdam: Elsevier; 1970. [Google Scholar]
- Zoshchuk SA, Badaeva ED, Zoshchuk NV, Adonina IG, Shcherban AB, Salina EA. Intraspecific divergence in wheats of the Timopheevi group as revealed by in situ hybridization with tandem repeats of the Spelt1 and Spelt52 families. Russian Journal of Genetics. 2007;43:771–788. [PubMed] [Google Scholar]