Abstract
Background
Polyploidy is a major component of eukaryote evolution. Estimation of allele copy numbers for molecular markers has long been considered a challenge for polyploid species, while this process is essential for most genetic research. With the increasing availability and whole-genome coverage of single nucleotide polymorphism (SNP) markers, it is essential to implement a versatile SNP genotyping method to assign allelic configuration efficiently in polyploids.
Scope
This work evaluates the usefulness of the KASPar method, based on competitive allele-specific PCR, for the assignment of SNP allelic configuration. Citrus was chosen as a model because of its economic importance, the ongoing worldwide polyploidy manipulation projects for cultivar and rootstock breeding, and the increasing availability of SNP markers.
Conclusions
Fifteen SNP markers were successfully designed that produced clear allele signals that were in agreement with previous genotyping results at the diploid level. The analysis of DNA mixes between two haploid lines (Clementine and pummelo) at 13 different ratios revealed a very high correlation (average = 0·9796; s.d. = 0·0094) between the allele ratio and two parameters [θ angle = tan−1 (y/x) and y′ = y/(x + y)] derived from the two normalized allele signals (x and y) provided by KASPar. Separated cluster analysis and analysis of variance (ANOVA) from mixed DNA simulating triploid and tetraploid hybrids provided 99·71 % correct allelic configuration. Moreover, triploid populations arising from 2n gametes and interploid crosses were easily genotyped and provided useful genetic information. This work demonstrates that the KASPar SNP genotyping technique is an efficient way to assign heterozygous allelic configurations within polyploid populations. This method is accurate, simple and cost-effective. Moreover, it may be useful for quantitative studies, such as relative allele-specific expression analysis and bulk segregant analysis.
Keywords: SNP genotyping, competitive allele-specific PCR, KASPar, allele dosage, Citrus clementina, C. maxima, C. reticulata, polyploid, triploid, tetraploid
INTRODUCTION
Polyploidy is a major component of eukaryote evolution, particularly in angiosperms (Gant, 1981; Soltis and Soltis, 1993; Wendel and Doyle, 2005). Many plant species result from autopolyploidization or allopolyploidization events, and polyploidization is considered the most common sympatric speciation mechanism (Otto and Whitton, 2000). Despite the indisputable importance of polyploid plant species, the genetics of these plant species are less well known than those of their diploid counterparts. Indeed, the estimation of molecular marker allele copy number has long been considered a challenge for polyploid species with polysomic inheritance, while it is essential to assign the allelic configuration for different types of heterozygotes for accurate population genetic studies. In segregating polyploid progeny, the population genetic structure can provide relevant information about the underlying meiosis mechanisms that take place in the formation of these progeny, which also greatly affect character segregation (Hutten et al., 1993; Tai and DeJong, 1997; Douches and Maas, 1998; Barcaccia et al., 2003; Brownfield and Kohler, 2011). Moreover, allelic dosage can affect gene expression and phenotype. Therefore, the determination of allelic dosages is particularly important for marker–trait association studies (De Jong et al., 2003; Sjoling et al., 2005). When parents of a polyploid progeny share one allele, only the dosage allele estimation allows knowledge of the alleles transmitted by each parent to heterozygous progeny. Therefore, knowledge of allelic dosage in polyploids appears to be essential for studies using single nucleotide polymorphism (SNP) markers, most of which are biallelic.
Several techniques have been used to estimate allele dosage in polyploid genotypes or tissues. When analysing microsatellite markers [simple sequence repeats (SSRs)], the microsatellite allele counting–peak ratios method (MAC-PR; Esselink et al., 2004) is especially useful. However, SSR analysis remains relatively costly and time consuming compared with actual SNP genotyping methods. Moreover, with the increasing availability of expressed sequence tag (EST) databases and whole-genome sequences, SNPs have become the most abundant and powerful polymorphic markers that can be selected throughout the entire genome (Edwards and Batley, 2010).
Several SNP genotyping methods have been developed. Some methods are based on electrophoretic separation after PCR amplification, including allele-specific primer extension (Kwok, 2001) and temperature-switch PCR (Tabone et al., 2009). High-throughput genotyping can be obtained using array methodologies (Sapolsky et al., 1999; Ishikawa et al., 2005); other techniques are based on pyrosequencing™ (Ronaghi et al., 1998; Ahmadian et al., 2000). However, the application of SNP markers has been limited primarily to diploid organisms, while the application of these markers to polyploid organisms for allele dosage estimation remains limited. Rickert et al. (2002) reported the use of pyrosequencing™ in polyploid potatoes, with some sequence-specific limitations. The usefulness of SNPlex™ (Berard et al., 2009) and Illumina Golden Gate™ assays (Akhunov et al., 2009) for the genotyping of polyploid wheat has been demonstrated. For genotype calling in tetraploid species with SNP analysis using the Illumina GoldenGate™ array, Voorrips et al. (2011) developed an algorithm using mixture models, but they assumed Hardy–Weinberg equilibrium within the population, which does not occur in all segregating polyploid progeny. Microarray data (Kirov et al., 2006; Meaburn et al., 2006; Steer et al., 2007) have also been used to estimate allelic frequencies in bulk populations or DNA pools, i.e. to perform genome-wide association scans. Array analysis is more suitable for genotyping large numbers of samples over numerous markers than for performing small-scale analysis, as array analysis lacks flexibility in terms of the numbers and panels of SNP loci that can be analysed. Targeted pyrosequencing™ (Gruber et al., 2002; Neve et al., 2002; Wasson et al., 2002; Lavebratt et al., 2004) can be useful for performing allele frequency estimation for a few genes in pooled DNA, but this technique remains relatively costly and time consuming. It is, therefore, important to develop alternative methods that offer a wider spectrum of genotyping possibilities to infer SNP allelic configurations in polyploid plants in small- to larger scale projects.
The KBiosciences Competitive AlleleSpecific PCR SNP genotyping system (KASPar) is a homogeneous fluorescent endpoint genotyping system (Cuppen, 2007) that utilizes a unique form of competitive, allele-specific PCR and combines the use of a highly specific 5′–3′ exonuclease-deleted Taq DNA polymerase with two competitive, allele-specific, tailed forward primers and one common reverse primer. This system is simple and cost-effective compared with other SNP genotyping assays and is well adapted to low- to medium-throughput genotyping projects (Chen et al., 2010). This technology has been successfully applied to the study of humans, animals and plants (Nijman et al., 2008; Bauer et al., 2009; Cortes et al., 2011; Rosso et al., 2011).
Citrus is mainly diploid. However, many modern breeding projects for the production of seedless mandarins based on the production of triploid hybrids (Ollitrault et al., 2008; Aleza et al., 2010, 2012a, b) and tetraploid rootstocks are promising (Saleh et al., 2008; Dambier et al., 2011; Grosser and Gmitter, 2011). Triploid populations in citrus can arise from unreduced gametes in crosses between diploid parents or from interploid (diploid × tetraploid or tetraploid × diploid) crosses. Sexual polyploidization resulting from 2n mega gametophyte production is routinely exploited for triploid citrus breeding (Aleza et al., 2010). In such crosses, segregation of a marker depends on the parental genetic structure, the relative distance to the centromere and the mode of restitution [first division restitution (FDR) or second division restitution (SDR)]. The MAC-PR method has been successfully applied to demonstrate the SDR origin of the 2n gametes arising from the ‘Fortune’ mandarin cultivar and to locate the centromere in one chromosome (Cuenca et al., 2011). For interploid crosses, marker segregations are almost exclusively dependent on the parental genetic conformation and preferential chromosome pairing. SSR markers were also used to analyse the meiotic behaviour of a tetraploid interspecific somatic hybrid of C. deliciosa + C. lemon (Kamiri et al., 2011), with the authors concluding that there was predominant tetrasomic segregation. However, the low availability of SSR markers displaying a favourable parental allelic structure that can be used to differentiate male and female contributions to the hybrids limits such studies to just a few areas of the genome. Conversely, large SNP resources have become available from extensive sequencing projects (Terol et al., 2007, 2008; Gmitter et al., 2012; Ollitrault et al., 2012b).
The objective of the present work was to evaluate the potential of the KASPar method to assess SNP allelic configurations in polyploid plants. Citrus was chosen as a model system because of its economic importance, the worldwide ongoing polyploidy manipulation projects for cultivar and rootstock breeding, and the increasing availability of SNP markers.
The quantitative value of the KASPar assay was estimated by pooling DNA from two haploid lines at several relative concentrations, simulating, among others, triploid and tetraploid heterozygous progeny. A method was developed for semi-automated polyploid genotype calling and applied for allelic configuration analysis of 170 triploid hybrids from two families arising from both sexual polyploidization and interploid crosses.
MATERIALS AND METHODS
Sample preparation
DNA pool preparation
Genomic DNA from two haploid lines, Citrus maxima (Burm.) Merr. (pummelo) cv Chandler and C. clementina Hort. ex Tan. (clementine; Aleza et al., 2009a), was isolated using a Plant DNAeasy Kit from Qiagen Inc. (Valencia, CA, USA) following the manufacturer's protocol. DNA concentrations were estimated with PicoGreen® and adjusted to 30 ng μL−1. DNA from the two haploid lines was pooled at ratios of 9:1, 5:1, 3:1, 2:1, 3:2, 1:1, 2:3, 1:2, 1:3, 1:5 and 1:9. Five samples (replications) of each haploid line and pool were prepared and used to test the accuracy of the technique.
Simulation of triploid and tetraploid hybrid samples by pooling DNA from haploid lines
Two sub-sets of the haploid DNA pool, one corresponding to 2:1 and 1:2 ratios that simulated heterozygous triploid genotypes and the other corresponding to 3:1, 1:1 and 1:3 ratios that simulated tetraploid heterozygous genotypes, were jointly used with the haploid genotypes to test the capability of the technique to discriminate among different types of heterozygotes within triploid and tetraploid populations.
Natural triploid populations: 2x × 2x crosses
‘Fortune’ (C. clementina × C. tangerina hort. ex Tan.) and ‘Willowleaf’ (C. deliciosa Ten.) diploid mandarins and 39 triploid hybrids segregating from this cross (Aleza et al., 2010) were selected to test the accuracy of the technique by analysing two replicates of each sample. Moreover, 86 triploid hybrids from a ‘Clementine’ (C. clementina) × ‘Nadorcott’ (C. reticulata Blanco) population (Aleza et al., 2010) were also analysed as individual samples to perform genotype calling.
Natural triploid populations: 4x × 2x cross
Tetraploid ‘Clementine’ (C. clementina 4x) and diploid ‘Pink’ pummelo (Citrus maxima 2x) and 88 triploid hybrids segregating from this cross (Aleza et al., 2012b) were also analysed. Tetraploid ‘Clementine’ was obtained by treating buds of the diploid ‘Clementine’ with colchicine (Aleza et al., 2009b); therefore, this genotype should be duplex (aabb) for all heterozygous loci.
SNP selection
SNPs for the analysis of signal–dosage correlation
To validate the quantitative value that was obtained from the KASPar assay using pooled DNA, seven SNPs differentiating the two haploid lines (C. maxima and C. clementina) were selected from previous genotyping data obtained on the Illumina GoldenGate™ platform (Ollitrault et al., 2012a). These SNPs were also used to test the accuracy of the technique in genotyping repetitions of the same sample over the ‘Fortune’ × ‘Willowleaf’ triploid population.
SNPs for triploid population analysis
Three out of seven SNPs used for the previous analysis, and eight other SNP markers, were selected to test the capacity of the technique for differentiating between heterozygous genotypes within two triploid populations, one arising from a 2x × 2x cross, and the other from a 4x × 2x cross. These SNPs are heterozygous for ‘Clementine’ and homozygous or heterozygous with the null allele for ‘Nadorcott’ and ‘Pink’.
SNP genotyping
All samples were genotyped for the SNP markers using KASPar technology by KBioscience® (http://www.kbioscience.co.uk/). The KASPar™ Genotyping System is a competitive, allele-specific dual Förster resonance energy transfer (FRET)-based assay for SNP genotyping. It uses two FRET cassettes where fluorometric dye [FAM (6-carboxy-fluorescein) or VIC®] is conjugated to primer but quenched via resonance energy transfer; ROX dye (6-carboxy-X-rhodamine, succinimidyl ester) is used to normalize the data. Sample DNA is amplified with a thermal cycler using allele-specific primers, leading to the separation of fluorometric dye and quencher when the FRET cassette primer is hybridized with DNA. Primers were designed by KBioscience®, based on the SNP locus-flanking sequence (approx. 50 nucleotides on each side of the SNP). Two 40-mer allele-specific oligonucleotides and one common 20-mer oligonucleotide were defined for each locus. Detailed information for all SNP markers can be found in Supplementary Data Table S1. Additional details about this genotyping method can be found in Cuppen (2007).
Data analysis method
Normalized signals from each SNP allele (x and y) were provided by KBioscience® services, and two-dimensional plot representations were obtained using SNPViewer software (http://www.kbioscience.co.uk/software/SNP%20viewer%20intro.html). From the x and y normalized values, the theta angle [θ = tan−1 (y/x); 0 º ≤ θ ≤ 90 º] and the relative y allele signal [y′ = y/(x + y); 0 ≤ y′ ≤1] of each sample were calculated. Further analyses were carried out that considered the y′ parameter, as this parameter was found to provide better clustering and genotype calling of the samples.
Data from all haploid lines and DNA pools with different allele configurations (9:1, 5:1, 3:1, 2:1, 3:2, 1:1, 2:3, 1:2, 1:3, 1:5 and 1:9) were tested for correlations between doses, and both the theta angle and y′ values that were obtained. Cluster analysis (MacQueen, 1967) using the farthest-neighbour method with standardized squared euclidean distances and analysis of variance (ANOVA) were performed from the normalized allele signals (x, y) jointly and from the y′ parameter data for each SNP.
Data from triploid and tetraploid simulated populations were also analysed separately by cluster analysis and ANOVA. Replications of the same samples were used to test the precision of the technique by genotype calling.
All statistical data were analysed using Statgraphics® Plus v5.1 software (Rockville, MD, USA).
RESULTS
Marker design and data acquisition
Primers for the KASPar assay were successfully designed by KBioscience® for all 15 of the submitted SNP-surrounding sequences. Data acquisition for x and y allele signals allowed successful allelic calling for 2535 out of 2563 marker–genotype combinations (98·91 %). The validity of the genotyping results was verified by comparing the results for 24 diploid varieties with previous data obtained with an Illumina GoldenGate™ array. Complete conformity was observed (data not shown).
Analysis of the correlation between relative allele signals and relative allele frequencies in the DNA pools
To confirm the value of the KASPar assay for producing semi-quantitative data, equimolar DNA extracts from two haploid lines (Clementine and pummelo) were mixed at 13 different relative concentrations, and five replicates were analysed for each of seven SNP markers. The correlations between relative allele signals and relative doses were analysed.
An example of correlation analysis between relative allele dosage and signals is shown in Fig. 1 for the CiC2840-01 SNP marker. From x and y signal values (Fig. 1A), theta angle [θ = tan−1 (y/x); Fig. 1B] and the relative y allele signal [y′ = y/(x + y); Fig. 1C] of each haploid line sample and DNA pool were calculated. High values of correlation coefficients between both parameters and relative allele dosage in the DNA pools were obtained for all analysed SNP markers, with an average of 0·9796 and a standard deviation of 0·0094 for the y′ parameter and an average of 0·9710 and a standard deviation of 0·0176 for the theta angle. Correlation values obtained for the y′ parameter were slightly superior to those obtained by the theta angle for six of the seven SNP markers that were analysed (Table 1).
Fig. 1.
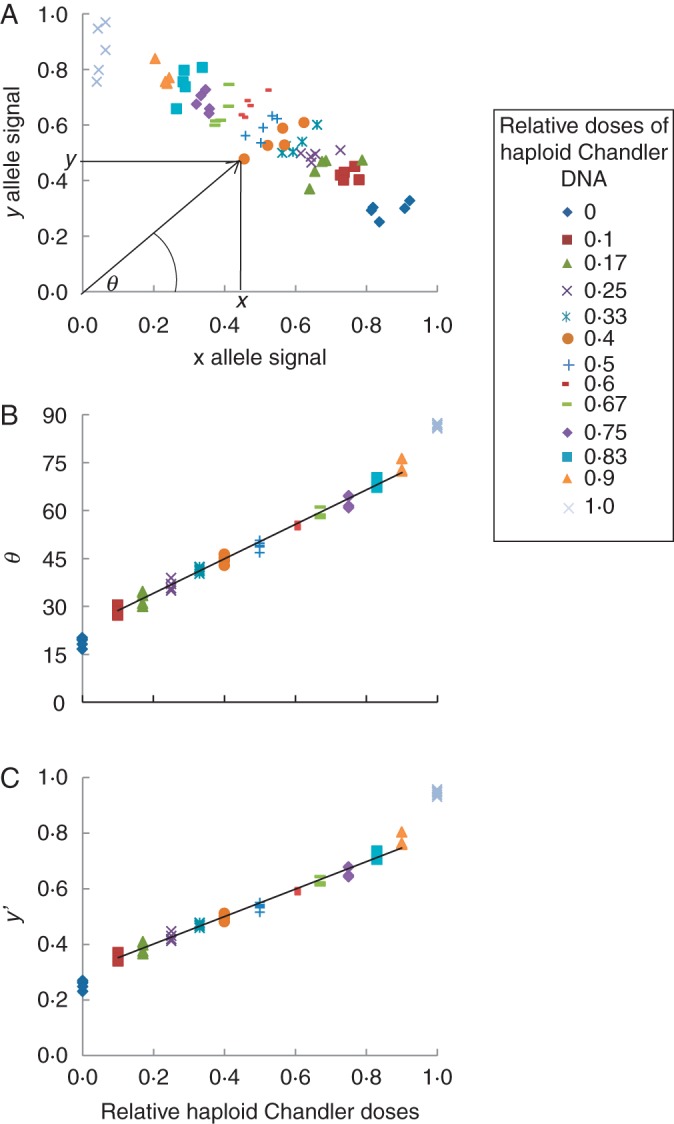
Correlation study of allele doses and x, y signals for the CiC2840-01 SNP marker. (A) Plot of normalized x, y allele signals. (B) Correlation between relative haploid Chandler doses and theta angle [θ = tan−1 (y/x)]. (C) Correlation between relative haploid Chandler doses and the y′ parameter [y′ = y(x + y)].
Table 1.
Correlation coefficients between relative allele dosage and allele signals from DNA pools at intermediate proportions for the theta angle and y′ parameter for the seven SNP markers analysed
| SNP marker | Correlation coefficient for angle θ | Correlation coefficient for y′ parameter |
|---|---|---|
| CiC2840-01 | 0·9941 | 0·9919 |
| CiC5089-06 | 0·9753 | 0·9779 |
| CiC5785-01 | 0·9580 | 0·9747 |
| DXS-M618 | 0·9881 | 0·9923 |
| F3H-M309 | 0·9788 | 0·9803 |
| FLS-M400 | 0·9535 | 0·9717 |
| TRPA-M593 | 0·9492 | 0·9684 |
| Average | 0·9710 | 0·9796 |
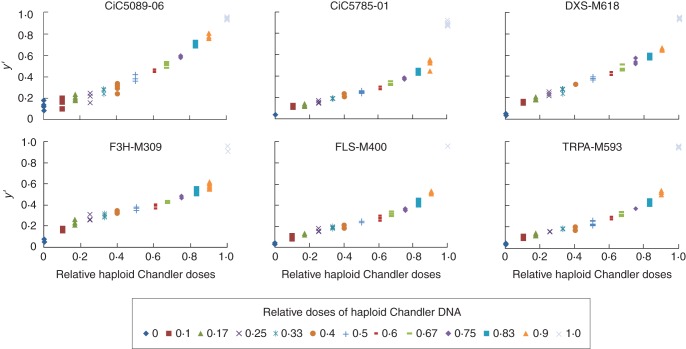
For some markers (CiC5785-01, F3H-M309, FLS-M400 and TRPA-M593; Fig. 2), the linear regression established from the mixed sample did not fit with the signals from the pure sample. For all these markers, the relative signals corresponding to the haploid ‘Chandler’ allele in the DNA mixes appear to be lower than expected in relation to the relative DNA dosages. This can probably be attributed to PCR allele competition between the ‘Clementine’ and ‘Chandler’ alleles in the DNA mixes.
Fig. 2.
Correlation study between the relative haploid Chandler doses and the y′ parameter for six SNP markers.
However, in the DNA mixes, the correlations between allele signals and allele doses remained high for these markers (between 0·9684 and 0·9803), testifying to a very good linear regression between relative signals of the two alleles and relative allele dosages.
These data indicate that the KASPar technique, using either the y′ parameter or the theta angle, can be useful for a quantitative analysis of the relative allele frequency in a genotype or DNA pool. Because y′ produced a slightly higher correlation coefficient, this parameter was employed in further analyses.
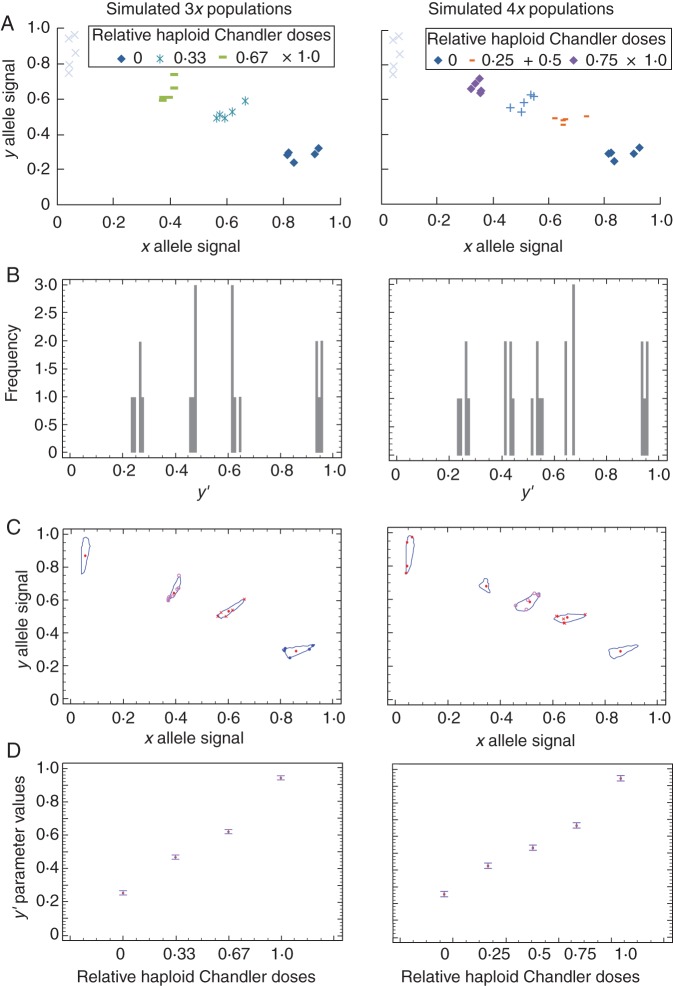
Cluster analysis and ANOVA for simulated triploid and tetraploid allele dosage
Separated cluster analyses and ANOVA from relative y allele signals (y′ parameter) in triploid and tetraploid simulated populations were performed. With diallelic markers, for a triploid heterozygous genotype, there are only two allelic configurations to distinguish: aab and abb (duplex and simplex of a-allele). For a heterozygous tetraploid genotype, three allelic configurations may be differentiated: aaab, aabb and abbb (triplex, duplex and simplex of a-allele). With higher ploidy levels, the number of possible allelic configurations becomes even larger (n–1 configurations for n ploidy).
The ANOVA (Table 2) revealed a complete and correct classification of the average value of the different configurations that were simulated. An example of the x and y allele signals, the frequency histogram for the y′ parameter and the l.s.d. intervals for the mean from ANOVA for simulating triploid and tetraploid populations is provided for the CiC2840-01 SNP marker in Fig. 3.
Table 2.
Homogeneous groups formed, and F-values from ANOVA of SNPs from DNA pools simulating triploid and tetraploid populations showing the percentage of correctly classified replications by cluster analysis based on the y′ parameter
| SNP marker | Simulating 3n populations |
Simulating 4n populations |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1:0 | 2:1 | 1:2 | 0:1 | F-value from ANOVA | % 3n correctly classified | 1:0 | 3:1 | 1:1 | 1:3 | 0:1 | F-value from ANOVA | % 4n correctly classified | |
| CiC2840-01 | a | b | c | d | 2975·34 | 100 | a | b | c | d | e | 1644·48 | 100 |
| CiC5089-06 | a | b | c | d | 1045·50 | 100 | a | b | c | d | e | 673·68 | 96 |
| CiC5785-01 | a | b | c | d | 5110·86 | 100 | a | b | c | d | e | 6084·71 | 100 |
| DXS-M618 | a | b | c | d | 2938·98 | 100 | a | b | c | d | e | 3013·55 | 100 |
| F3H-M309 | a | b | c | d | 2063·52 | 100 | a | b | c | d | e | 1445·66 | 100 |
| FLS-M400 | a | b | c | d | 10046·43 | 100 | a | b | c | d | e | 10459·43 | 100 |
| TRPA-M593 | a | b | c | d | 11140·59 | 100 | a | b | c | d | e | 6666·05 | 100 |
| Average | 100 | 99·43 | |||||||||||
Fig. 3.
Study of simulating triploid and tetraploid populations for the CiC2840-01 SNP marker. (A) Plot of normalized x, y allele signals. (B) Frequency histogram for the y′ parameter. (C) Cluster analysis. (D) Least significant difference intervals for the mean obtained from ANOVA.
Moreover, all expected homogeneous groups were formed by cluster analysis using the farthest-neighbour method with standardized squared euclidean distances. All of the triploid sample replications and 99·43 % of the tetraploid ones were correctly classified; only one replication for the CiC5089-06 SNP marker was classified into an incorrect cluster (Table 2).
Allelic configuration of triploid populations
Accuracy of genotype calling for duplicated triploid samples
Thirty-nine triploid hybrids arising from ‘Fortune’ 2n gametes in the ‘Fortune × Willowleaf’ hybridization were analysed for seven SNPs, with two technical replications. All SNPs had the following allelic configuration: ‘Fortune’, ab; and ‘Willowleaf’ mandarin, aa. Therefore, depending on the origin of the diploid gamete, three genotypic clusters were expected: aaa, aab and abb. Samples with replications that were classified in the same cluster and thus genotyped with the same allelic conformation reached 97·44 %. Errors in classification were observed in five of the seven SNP markers analysed. Considering a replicate for the same DNA sample (with different allele calling between replicates) to be classified correctly, the average error rate for further routinely genotyping without replicates was estimated to be 1·28 %.
To perform the genotype calling of triploid progeny, cluster analyses were performed according to the expected genotypes for each population and the parental-specific allelic configuration of each marker.
2x × 2x triploid progeny
When crossing a heterozygous female parent (ab) with homozygous parents (aa), maternal heterozygosity restitution (HR) is reflected in the duplex (aab) triploid hybrids. Under the SDR mechanism, HR is directly linked to the distance from the locus under consideration to the centromere and, therefore, the frequency of HR can be estimated from this distance, as proposed by Cuenca et al. (2011). To validate genotype calling for triploids resulting from a 2x × 2x cross, three SNP markers (CiC3440-07, CiC5785-01 and CiC6278-01) mapped in chromosome II were selected. Indeed, Cuenca et al. (2011) located the centromere position for the corresponding linkage group at 59·6 cM of the current reference ‘Clementine’ genetic map (Ollitrault et al., 2012a) using the Cx(Co)4 partial interference model. The expected HR for the three considered markers (also mapped in the ‘Clementine’ map) was estimated using the same partial interference model.
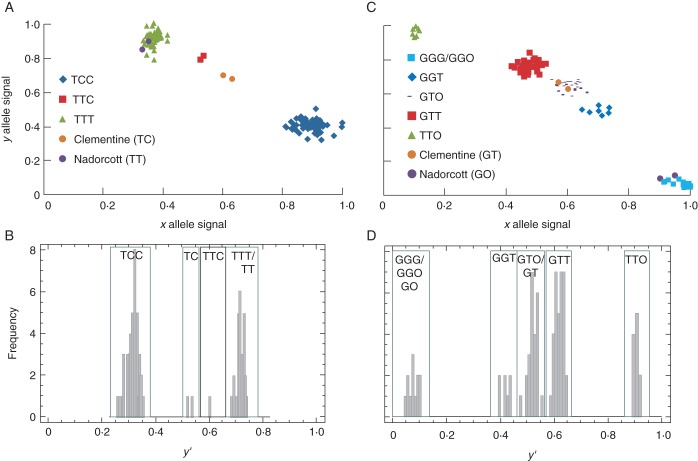
Cluster analyses were performed from y′ parameter values of each hybrid over 11 analysed SNPs (including the three markers on chromosome II) within the ‘Clementine × Nadorcott’ population to carry out genotype calling. Figure 4A,B shows an example of cluster analysis for the CiC2840-01 SNP marker.
Fig. 4.
Plot of normalized x, y allele signals and histogram representing genotype calling from cluster analysis over 85 triploids from the ‘Clementine × Nadorcott’ cross. (A, B) CiC2840-01 (ab × aa); (C, D) CiC1749-05 (ab × a0).
The cluster analysis allowed the detection of null alleles in the male parent for two markers. Indeed, if the supposed homozygous parent in fact had heterozygosity for a null allele, five clusters should be obtained (ab × a0: aab, ab0, aaa/aa0, abb and bb0), where one cluster contains both ab0 triploids and ab diploid genotypes. Such a cluster configuration was observed for the CiC0610-01 and CiC1749-05 SNP markers (Fig. 4C,D).
On average, over all of the markers, 99·37 % of the samples were assigned to a cluster and, therefore, could be accurately called.
For the three markers of LGII, the observed HR values were not significantly different from those estimated from the espective markers and centromere locations in the ‘Clementine’ map (Table 3). This provides additional validation of the accuracy of polyploid genotype calling using the method presented in this study. For the eight remaining markers, the observed HR allowed us to estimate the relative distances of these markers from the centromere (Table 4), revealing markers with centromeric locations (<5 cM distance from the centromere: CiC1380-05, CiC2840-01 and CiC4581-01).
Table 3.
Results of genotype calling for three SNP loci in the ‘Clementine × Nadorcott’ triploid population from 2n gametes for markers heterozygous for ‘Clementine’ and homozygous for ‘Nadorcott’, including the conformity (χ2 test) of the observed %HR with the theoretical one calculated from the distance from each locus to the centromere on chromosome II
| SNP marker | Map position (cM) | NI | aaa | aab | abb | %HR (aab) observed | %HR estimated from map position (centromere at 59·6 cM) | χ2 (P-value) |
|---|---|---|---|---|---|---|---|---|
| CiC3440-07 | 67·22 | 86 | 37 | 10 | 39 | 11·63 | 13·59 | 0·282; NS (P = 0·5954) |
| CiC5785-01 | 44·73 | 84 | 32 | 17 | 35 | 20·24 | 28·23 | 2·648; NS (P = 0·1037) |
| CiC6278-01 | 57·01 | 86 | 46 | 0 | 40 | 0·00 | 4·21 | 3·780; NS (P = 0·0519) |
NI, number of individuals genotyped; aaa, aab and abb: number of individuals of each genotype; NS, non-significantly different at α = 0·05).
Table 4.
Results of genotype calling for eight SNP loci in the ‘Clementine × Nadorcott’ triploid population from 2n gametes for markers heterozygous for ‘Clementine’ and homozygous or heterozygous with a null allele for ‘Nadorcott’, showing the estimated marker–centromere distance
| SNP marker | NI | aaa | Aab | abb | ab0 | bb0 | % HR (aab + ab0) observed | Estimated distance to centromere (cM) |
|---|---|---|---|---|---|---|---|---|
| CiC0610-01 | 86 | 22 | 22 | 14 | 22 | 6 | 51·16 | 27·42 |
| CiC0868-01 | 86 | 39 | 14 | 33 | – | – | 16·8 | 9·23 |
| CiC1380-05 | 86 | 43 | 1 | 42 | – | – | 1·16 | 0·75 |
| CiC1749-05 | 85 | 11 | 33 | 7 | 24 | 10 | 67·06 | 40·61 |
| CiC1757-02 | 86 | 42 | 12 | 32 | – | – | 13·95 | 7·80 |
| CiC2840-01 | 85 | 46 | 2 | 37 | – | – | 2·35 | 1·49 |
| CiC4581-01 | 86 | 37 | 6 | 43 | – | – | 6·98 | 4·15 |
| CiC5089-06 | 84 | 27 | 40 | 17 | – | – | 47·62 | 25·23 |
NI, number of individuals genotyped; aaa, aab, abb, ab0 and bb0, number of individuals of each genotype.
4x × 2x triploid progeny
Triploid genotyping was also performed for progeny arising from a cross between doubled diploid ‘Clementine’ (Aleza et al., 2009b) and ‘Pink’ pummelo. When crossing a duplex tetraploid parent (aabb) with a homozygous diploid parent (aa), three clusters can be expected (triplex-aaa, duplex-aab and simplex-abb), where maternal HR is reflected into the duplex (aab) triploid hybrids. On average, over all of the markers, 97·08 % of the samples were assigned to a cluster and, therefore, could be accurately called.
The genetic structure of the triploid progeny arising from ‘Clementine 4x’ × ‘Pink’ is shown in Table 5, which indicates the percentage of HR. For the other loci, HR values varied, ranging from 52·94 % for CiC4581-01 to 69·41 % for CiC5089-06. The average HR value over all loci was 59·64 %.
Table 5.
Results of genotype calling for seven SNP loci in the ‘Clementine 4x × Pink’ triploid population, indicating the heterozygosity restitution in Clementine 4x (%HR) at each locus
| SNP marker | NI | aaa | aab | abb | %HR (aab) observed |
|---|---|---|---|---|---|
| CiC3440-07 | 88 | 17 | 50 | 21 | 56·82 |
| CiC0868-01 | 87 | 11 | 58 | 18 | 66·67 |
| CiC1380-05 | 87 | 19 | 53 | 15 | 60·92 |
| CiC4581-01 | 85 | 19 | 45 | 21 | 52·94 |
| CiC5089-06 | 85 | 15 | 59 | 11 | 69·41 |
| CiC5785-01 | 80 | 22 | 43 | 15 | 53·75 |
| CiC6278-01 | 86 | 22 | 49 | 15 | 56·98 |
NI, number of individuals genotyped; aaa, aab and abb, number of individuals of each genotype.
DISCUSSION
The KASPar method is a powerful technique for assigning SNP allelic configurations in polyploid progeny
Several techniques have been used to estimate allele dosage in polyploids, such as the MAC-PR method (Esselink et al., 2004) for SSR markers, and techniques for SNP genotyping, including allele-specific primer extension (Kwok, 2001), temperature-switch PCR (Tabone et al., 2009), array methodologies (Ishikawa et al., 2005) and targeted pyrosequencing™ (Ahmadian et al., 2000). Our study demonstrates that the KASPar technique (Cuppen, 2007) is an alternative method to infer SNP allelic configurations in polyploid plants that offers a wider spectrum of genotyping possibilities. The KASPar method is simple and cost-effective compared with other SNP genotyping assays and is well adapted to low- to medium-throughput genotyping projects. In addition to the markers published herein, 51 KASPar markers were successfully developed to analyse triploid and tetraploid citrus populations (Aleza et al., 2012c, d; Cuenca et al., 2012). KASPar markers were also successfully developed (41 over 42 tested) and transferred in the true citrus group (Citrus, Fortunella, Poncirus, Microcitrus and Eremocitrus genera) from SNP mining by sequencing within a Citrus collection (Garcia-Lor et al., 2013). When SNPs are mined in a large discovery panel, this offers the opportunity to select markers without additional variation in the flanking DNA sequence used as template for the competitive PCR of the KASPar assay and therefore to have a high degree of success in marker development. KASPar markers were successfully developed in a large range of plant (Cortes et al., 2011; Rosso et al., 2011; Byers et al., 2012) and animal (Nijman et al., 2008; Murad et al., 2009; Luciano et al., 2010) species, demonstrating its universal applicability.
The SNP genotyping and data analysis method presented in this study is simple and effective for genotyping triploid and tetraploid progeny and can also be used in the quantitative analysis of allele-specific expression. Allele signals (x, y) obtained from KBioscience® can easily be transformed into y′ [y′ = y/(x + y); 0 ≤ y′ ≤ 1], which is a very useful parameter to cluster analysed samples. Theta angles [θ = tan−1 (y/x); 0 º ≤ θ ≤ 90 º] can also be used to analyse data, but the y′ parameter offers better clustering results. Quantitative analyses for correlation of the allele signals, and the allele doses and sample clustering carried out in this work, were powerful techniques for assigning allelic configurations in simulated triploid and tetraploid citrus genotypes for all SNP markers that were analysed (100 % of the triploids were correctly classified as well as 99·43 % of the tetraploids). The analysis of concrete triploid hybrids with technical replications confirmed the high degree of accuracy of the technique (error <1·5 %). This SNP genotyping and data analysis method allowed us to distinguish among very close allele ratios, and it can also be efficiently employed for the analysis of higher ploidy levels. Moreover, the segregations observed with this technique have allowed us to identify heterozygous null alleles in one parent for some of the markers. Diploid progeny genotyping confirmed these conclusions for null alleles in Pink pummelo (Ollitrault et al., 2012a).
PCR drift can affect allelic configuration inference in natural polyploid germplasm
Interpretation of relative allele dosage for markers based on relative PCR product intensities has been reported for various plants (Buteler et al., 1999; Julier et al., 2003; Landergott et al., 2006; Martins et al., 2009) and animals (McQuown et al., 2002). The limits of evaluation of such direct allele doses are associated with PCR selection caused by differential primer affinity and PCR drift resulting from random events during early cycles of PCR (Wagner et al., 1994).
In this study, such PCR drift has been observed for some markers, displaying incongruence between the linear regressions established from the mixed DNA pools and the pure sample. However, the correlations between allele signals and allele doses in the DNA pools remained high for these markers. Therefore, as linear regression appears to offer a good approximation of the doses/relative signal relationship, a control with two dosage points should be sufficient to establish a function that correlates both parameters.
Heterozygous diploid genotypes are suitable for determining the 1:1 ratios that are used as a baseline for calculations of allele quantification in the other heterozygous genotype. In the analysis of citrus triploid progeny, the location of different clusters relative to the heterozygous diploid parent allowed us to assign the alternative theoretical triploid heterozygous allelic configuration.
The situation is much more complicated when analysing polyploid germplasm of unknown origin. Indeed, the variability in the flanking regions of the SNPs that were studied (where the primers were defined) should result in different levels of relative PCR competition and, therefore, should avoid proper allele dose identification from relative x/y signals. This is inherent in all PCR genotyping methods. Perhaps, as suggested by Landergott et al. (2006) for the MAC-PR method, the KASPar assay may be very useful for determining the allelic configuration within crossing families, but it would not be generally applicable for estimating allelic dosage in polyploid germplasm without previous verification of the stability of relative allele amplification. An approach to limit the PCR drift associated with variations in the flanking area of the studied SNPs should be to select SNPs flanked by conserved sequences. Such information is available in SNP mining studies where large discovery panels are used, while there is generally no information on flanking sequences of microsatellite markers. This should be an important advantage of using SNPs rather than microsatellite markers for assignment of allelic configuration in polyploids.
Potential of KASPar for semi-quantitative estimation of allele-specific expression analysis or allelic frequency estimation in DNA extracted from pools
Many genetic variants resulting in phenotypic differences are mediated through changes in gene expression. Variation in gene expression can be due to polymorphisms either at the gene locus (cis) or in other genes that influence gene expression (trans) or cis/trans interactions (Rockman and Kruglyak, 2006). Allele-specific expression (ASE) studies have introduced a creative method to uncover the respective contributions of cis- and trans-regulatory variation (Ronald et al., 2005; Main et al., 2009). Allelic imbalance in non-imprinted genes has been shown to be common in humans, maize and arabidopsis (Lo et al., 2003; Guo et al., 2004; Zhang and Borevitz, 2009). Moreover, ASE analysis should enable the integration of potentially differential allelic functionality in association models between gene expression and phenotype. Therefore, gene expression analysis is a critical step for better understanding of genotype–phenotype relationships.
Analysis of allele-specific expression in relation to genomic structure requires the assessment of DNA and RNA allele dosage. This can be done using different methods: northern (Guo et al., 1996), RNA-FISH (Herzing et al., 2002), SNP-specific array-based (Bjornsson et al., 2008), Solexa (Main et al., 2009) or RNA-seq (Rozowsky et al., 2011).
Furthermore, the estimation of allelic frequencies on pooled DNA is of great interest both in ecological studies of plants (Ritland, 2002), animals (Shaw et al., 1998; Coop et al., 2010; Grant, 2010) or micro-organisms (Brauer et al., 2006; Wenger et al., 2010), and in bulk segregant analysis to locate genes involved in phenotypic variation (Quarrie et al., 1999; Tabor et al., 2000; Yang and Fann, 2007).
The high correlation coefficient values between relative allele dosage and SNP allele signals obtained with the KASPar technique, and the ability of this technique to distinguish between close relative allele dosages at the DNA level, has been demonstrated in this study. Moreover, we were able to detect a 0·1 allele frequency within DNA pools. This technique is therefore a promising method for performing semi-quantitative analysis of relative allele-specific expression by analysing cDNA compared with genomic DNA, to complement global gene expression studies performed by real-time PCR. The KASPar technique may also be useful for allele frequency estimation in populations from DNA pools as mentioned before. For such studies, it should be interesting to extend the range of relative allele dosages to estimate the lowest differences distinguishable with this technique.
Application for citrus genetics and breeding
Triploid citrus breeding is one of the most efficient techniques for the production of seedless mandarins (Ollitrault et al., 2008; Aleza et al., 2010, 2012c, d), and tetraploid rootstocks are promising tools that enable plants to adapt to various abiotic stresses (Saleh et al., 2008; Dambier et al., 2011). Triploid populations in citrus can arise from 2x × 2x crosses or from interploid crosses. Discriminating between different types of heterozygotes within triploid progeny is especially useful for population genetic structure studies and marker–trait association analysis.
Knowing the allelic configuration in triploid and tetraploid progeny is also necessary to identify the mechanism of 2n gamete formation. The maternal HR values of <50 % obtained in this study, which were estimated from a ‘Clementine × Nadorcott’ progeny for nine markers (CiC0868-01, CiC1380-05, CiC1757-02, CiC2840-01, CiC3440-07, CiC4581-01, CiC5089-06, CiC5785-01 and CiC6278-01), confirm the conclusion of Luro et al. (2001) that the 2n gamete in Clementine arose from SDR, as in the Fortune mandarin (Cuenca et al., 2011), while Chen et al. (2008) proposed FDR for sweet orange. Moreover, this study allowed us to identify several centromeric markers that should be very useful for further analyses of the origin of 2n gametes in different cultivars and genotypes, as was done for potatoes (Douches and Quiros, 1988; Werner et al., 1992).
Most tetraploid citrus germplasm arose from chromosome duplication of nucellar cells (Aleza et al., 2011) or were obtained by bud chemical treatment (Aleza et al., 2009b) of diploid genotypes. These tetraploids are, therefore, doubled diploids with the same aabb genomic structure at each heterozygous locus (ab) of the parental diploid line. For such tetraploids, the parental restitution (PR) of the heterozygosity to the diploid gamete depends on preferential pairing between chromosomes. In the case of total preferential pairing (disomic segregation), parental heterozygosity is transferred to all gametes (PR = 100 %). In the case of total random pairing (tetrasomic segregation), the PR ranged from 55 to 66 %, depending on the double reduction frequency (Marsden et al., 1987). In this study, the PR results for the tetraploid (doubled diploid) Clementine ranged from 52·94 % for the CiC4581-01 marker to 69·41 % for the CiC5089-06 marker, which is in agreement with the expected PR values under tetrasomic segregation (Kamiri et al., 2011).
In the case of triploid and tetraploids obtained by somatic hybridization (Dambier et al., 2011; Grosser and Gmitter, 2011), the assignment of allelic configuration will be useful for revealing genome regions acquired from each parent, as well as potential chromosome fragment elimination or duplication.
Conclusions
This work demonstrates that the KASPar SNP genotyping technique, combined with the cluster analysis method we proposed, enables the efficient assignment of heterozygous allelic configuration within polyploid populations. This method is accurate, simple and cost-effective. It has been successfully applied to two citrus triploid populations arising from 2n gametes and interploid crosses. Moreover, correlation studies, cluster analysis and ANOVA support the usefulness of this method for performing relative quantitative studies, such as relative allele-specific expression analysis or, eventually, bulk segregant analysis.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank J. Romero, F. Ollitrault and A. Moreno for laboratory technical assistance, and J.A Pina and V. Lloris for managing plant material. We also thank Dr Y. Froelicher for providing haploid Chandler material. This work was supported by a grant [AGL2011-26490] from the Ministry of ‘Economía y Competividad’ – Fondo Europeo de Desarrollo Regional (FEDER) and a grant [Prometeo 2008/121] from the Generalitat Valenciana, Spain.
LITERATURE CITED
- Ahmadian A, Gharizadeh B, Gustafsson AC, et al. Single-nucleotide polymorphism analysis by pyrosequencing™. Analytical Biochemistry. 2000;280:103–110. doi: 10.1006/abio.2000.4493. [DOI] [PubMed] [Google Scholar]
- Akhunov E, Nicolet C, Dvorak J. Single nucleotide polymorphism genotyping in polyploid wheat with the Illumina GoldenGate assay. Theoretical and Applied Genetics. 2009;119:507–517. doi: 10.1007/s00122-009-1059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleza P, Juarez J, Hernandez M, Pina JA, Ollitrault P, Navarro L. Recovery and characterization of a Citrus clementina Hort. ex Tan. ‘Clemenules’ haploid plant selected to establish the reference whole Citrus genome sequence. BMC Plant Biology. 2009a;9(110) doi: 10.1186/1471-2229-9-110. http://dx.doi.org/10.1186/1471-2229-9-110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleza P, Juarez J, Ollitrault P, Navarro L. Production of tetraploid plants of non apomictic citrus genotypes. Plant Cell Reports. 2009b;28:1837–1846. doi: 10.1007/s00299-009-0783-2. [DOI] [PubMed] [Google Scholar]
- Aleza P, Juarez J, Cuenca J, Ollitrault P, Navarro L. Recovery of citrus triploid hybrids by embryo rescue and flow cytometry from 2x × 2x sexual hybridisation and its application to extensive breeding programs. Plant Cell Reports. 2010;29:1023–1034. doi: 10.1007/s00299-010-0888-7. [DOI] [PubMed] [Google Scholar]
- Aleza P, Froelicher Y, Schwarz S, et al. Tetraploidization events by chromosome doubling of nucellar cells are frequent in apomictic citrus and are dependent on genotype and environment. Annals of Botany. 2011;108:37–50. doi: 10.1093/aob/mcr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleza P, Juarez J, Cuenca J, Ollitrault P, Navarro L. Extensive citrus triploid hybrid production by 2x × 4x sexual hybridizations and parent-effect on the length of the juvenile phase. Plant Cell Reports. 2012a;31:1723–1735. doi: 10.1007/s00299-012-1286-0. [DOI] [PubMed] [Google Scholar]
- Aleza P, Juarez J, Hernández M, Ollitrault P, Navarro L. Implementation of extensive citrus triploid breeding programs based on 4x × 2x sexual hybridizations. Tree Genetics and Genomes. 2012b;8:1293. [Google Scholar]
- Aleza P, Cuenca J, Juárez J, Navarro L, Ollitrault P. 12th International Citrus Congress. Valencia, Spain: Book of Abstracts; 2012c. Mechanism of 2n gametes formation and centromere mapping in citrus; pp. 37–38. 19–23 November 2012. [Google Scholar]
- Aleza P, Cuenca J, Juárez J, Ollitrault P, Navarro L. 12th International Citrus Congress. Valencia, Spain: Book of Abstracts; 2012d. Differences in the genetic structure of citrus triploid hybrids recovered from 2x × 2x and 4x × 2x sexual hybridisations; p. 49. 19–23 November 2012. [Google Scholar]
- Barcaccia G, Tavoletti S, Mariani A, Veronesi F. Occurrence, inheritance and use of reproductive mutants in alfalfa improvement. Euphytica. 2003;133:37–56. [Google Scholar]
- Bauer F, Elbers CC, Adan RAH, et al. Obesity genes identified in genome-wide association studies are associated with adiposity measures and potentially with nutrient-specific food preference. American Journal of Clinical Nutrition. 2009;90:951–959. doi: 10.3945/ajcn.2009.27781. [DOI] [PubMed] [Google Scholar]
- Berard A, Le Paslier MC, Dardevet M, et al. High-throughput single nucleotide polymorphism genotyping in wheat (Triticum spp.) Plant Biotechnology Journal. 2009;7:364–374. doi: 10.1111/j.1467-7652.2009.00404.x. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Albert TJ, Ladd-Acosta CM, et al. SNP-specific array-based allele-specific expression analysis. Genome Research. 2008;18:771–779. doi: 10.1101/gr.073254.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Christianson CM, Pai DA, Dunham MJ. Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics. 2006;173:1813–1816. doi: 10.1534/genetics.106.057927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L, Kohler C. Unreduced gamete formation in plants: mechanisms and prospects. Journal of Experimental Botany. 2011;62:1659–1668. doi: 10.1093/jxb/erq371. [DOI] [PubMed] [Google Scholar]
- Buteler MI, Jarret RL, LaBonte DR. Sequence characterization of microsatellites in diploid and polyploid Ipomoea. Theoretical and Applied Genetics. 1999;99:123–132. [Google Scholar]
- Byers RL, Harker DB, Yourstone SM, Maughan PJ, Udall JA. Development and mapping of SNP assays in allotetraploid cotton. Theoretical and Applied Genetics. 2012;124:1201–1214. doi: 10.1007/s00122-011-1780-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Lyon MT, O'Malley D, et al. Origin and frequency of 2n gametes in Citrus sinensis × Poncirus trifoliata and their reciprocal crosses. Plant Science. 2008;174:1–8. [Google Scholar]
- Chen W, Mingus J, Mammadov J, et al. Plant and Animal Genomes XVII Conference. San Diego, USA: 2010. KASPar: a simple and cost-effective system for SNP genotyping. January 9–13, 2010. [Google Scholar]
- Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes AJ, Chavarro MC, Blair MW. SNP marker diversity in common bean (Phaseolus vulgaris L.) Theoretical and Applied Genetics. 2011;123:827–845. doi: 10.1007/s00122-011-1630-8. [DOI] [PubMed] [Google Scholar]
- Cuenca J, Froelicher Y, Aleza P, Juarez J, Navarro L, Ollitrault P. Multilocus half-tetrad analysis and centromere mapping in citrus: evidence of SDR mechanism for 2n megagametophyte production and partial chiasma interference in mandarin cv ‘Fortune. Heredity. 2011;107:462–470. doi: 10.1038/hdy.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca J, Aleza P, Iborra E, Vicent A, Ollitrault P, Navarro L. 12th International Citrus Congress. Valencia, Spain: Book of Abstracts; 2012. Location of a chromosome region linked to Alternaria Brown Spot resistance from the evaluation of triploid mandarin populations; p. 42. 19–23 November 2012. [Google Scholar]
- Cuppen E. Genotyping by allele-specific amplification (KASPar) CSH Protocols. 2007;2007:172–173. doi: 10.1101/pdb.prot4841. [DOI] [PubMed] [Google Scholar]
- Dambier D, Benyahia H, Pensabene-Bellavia G, et al. Somatic hybridization for citrus rootstock breeding: an effective tool to solve some important issues of the Mediterranean citrus industry. Plant Cell Reports. 2011;30:883–900. doi: 10.1007/s00299-010-1000-z. [DOI] [PubMed] [Google Scholar]
- De Jong WS, De Jong DM, Bodis M. A fluorogenic 5′ nuclease (TaqMan) assay to assess dosage of a marker tightly linked to red skin color in autotetraploid potato. Theoretical and Applied Genetics. 2003;107:1384–1390. doi: 10.1007/s00122-003-1420-z. [DOI] [PubMed] [Google Scholar]
- Douches DS, Maas DL. Comparison of FDR and SDR derived tetraploid progeny from 2x × 4x crosses using haploids of Solanum tuberosum L that produce mixed modes of 2n eggs. Theoretical and Applied Genetics. 1998;97:1307–1313. [Google Scholar]
- Douches DS, Quiros CF. Genetic strategies to determine the mode of 2n egg formation in diploid potatoes. Euphytica. 1988;38:247–260. [Google Scholar]
- Edwards D, Batley J. Plant genome sequencing: applications for crop improvement. Plant Biotechnology Journal. 2010;8:2–9. doi: 10.1111/j.1467-7652.2009.00459.x. [DOI] [PubMed] [Google Scholar]
- Esselink GD, Nybom H, Vosman B. Assignment of allelic configuration in polyploids using the MAC-PR (microsatellite DNA allele counting-peak ratios) method. Theoretical and Applied Genetics. 2004;109:402–408. doi: 10.1007/s00122-004-1645-5. [DOI] [PubMed] [Google Scholar]
- Gant V. Plant speciation. 2nd edn. New York: Columbia University Press; 1981. [Google Scholar]
- Garcia-Lor A, Ancillo G, Navarro L, Ollitrault P. Citrus (Rutaceae) SNP markers based on competitive allelespecific PCR; transferability across the Aurantioideae subfamily. Applications in Plant Sciences. 2013 doi: 10.3732/apps.1200406. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmitter FG, Jr, Chen C, Machado MA, et al. Citrus genomics. Tree Genetics and Genomes. 2012;8:611–626. [Google Scholar]
- Grant WB. A multicountry ecological study of risk-modifying factors for prostate cancer: apolipoprotein E ɛ4 as a risk factor and cereals as a risk reduction factor. Anticancer Research. 2010;30:189–200. [PubMed] [Google Scholar]
- Grosser JW, Gmitter FG., Jr. Protoplast fusion for production of tetraploids and triploids: applications for scion and rootstock breeding in citrus. Plant Cell, Tissue and Organ Culture. 2011;104:343–357. [Google Scholar]
- Gruber JD, Colligan PB, Wolford JK. Estimation of single nucleotide polymorphism allele frequency in DNA pools by using Pyrosequencing™. Human Genetics. 2002;110:395–401. doi: 10.1007/s00439-002-0722-6. [DOI] [PubMed] [Google Scholar]
- Guo M, Davis D, Birchler JA. Dosage effects on gene expression in a maize ploidy series. Genetics. 1996;142:1349–1355. doi: 10.1093/genetics/142.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Zinselmeier C, Habben J, Bowen BA, Smith OS. Allelic variation of gene expression in maize hybrids. The Plant Cell. 2004;16:1707–1716. doi: 10.1105/tpc.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzing LBK, Cook EH, Ledbetter DH. Allele-specific expression analysis by RNA-FISH demonstrates preferential maternal expression of UBE3A and imprint maintenance within 15q11–q13 duplications. Human Molecular Genetics. 2002;11:1707–1718. doi: 10.1093/hmg/11.15.1707. [DOI] [PubMed] [Google Scholar]
- Hutten RCB, Scholberg E, Huigen DJ, Hermsen JGT, Jacobsen E. Analysis of dihaploid induction and production ability and seed parent × pollinator interaction in potato. Euphytica. 1993;72:61–64. [Google Scholar]
- Ishikawa S, Komura D, Tsuji S, et al. Allelic dosage analysis with genotyping microarrays. Biochemical and Biophysical Research Communications. 2005;333:1309–1314. doi: 10.1016/j.bbrc.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Julier B, Flajoulot S, Barre P, et al. Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biology. 2003;3(9) doi: 10.1186/1471-2229-3-9. http://dx.doi.org/10.1186/1471-2229-3-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiri M, Stift M, Srairi I, et al. Evidence for non-disomic inheritance in a Citrus interspecific tetraploid somatic hybrid between C. reticulata and C. limon using SSR markers and cytogenetic analysis. Plant Cell Reports. 2011;30:1415–1425. doi: 10.1007/s00299-011-1050-x. [DOI] [PubMed] [Google Scholar]
- Kirov G, Nikolov I, Georgieva L, Moskvina V, Owen MJ, O'Donovan MC. Pooled DNA genotyping on Affymetrix SNP genotyping arrays. BMC Genomics. 2006;7(27) doi: 10.1186/1471-2164-7-27. http://dx.doi.org/10.1186/1471-2164-7-27 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok PY. Methods for genotyping single nucleotide polymorphisms. Annual Review of Genomics and Human Genetics. 2001;2:235–258. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- Landergott U, Naciri Y, Schneller JJ, Holderegger R. Allelic configuration and polysomic inheritance of highly variable microsatellites in tetraploid gynodioecious Thymus praecox agg. Theoretical and Applied Genetics. 2006;113:453–465. doi: 10.1007/s00122-006-0310-6. [DOI] [PubMed] [Google Scholar]
- Lavebratt C, Sengul S, Jansson M, Schalling M. Pyrosequencing™-based SNP allele frequency estimation in DNA pools. Human Mutation. 2004;23:92–97. doi: 10.1002/humu.10292. [DOI] [PubMed] [Google Scholar]
- Lo HS, Wang ZN, Hu Y, et al. Allelic variation in gene expression is common in the human genome. Genome Research. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Houlihan LM, Harris SE, et al. Association of existing and new candidate genes for anxiety, depression and personality traits in older people. Behavior Genetics. 2010;40:518–532. doi: 10.1007/s10519-009-9326-4. [DOI] [PubMed] [Google Scholar]
- Luro F, Rist D, Ollitrault P. Proceedings of the International Symposium on Molecular Markers for Characterizing Genotypes and Identifying Cultivars in Horticulture. Montpellier: France; 2001. Evaluation of genetic relationships in citrus genus by means of sequence tagged microsatellites. [Google Scholar]
- MacQueen J. Proceedings of 5th Berkeley Symposium on Mathematical Statistics and Probability. Berkeley: University of California Press; 1967. Some methods for classification and analysis of multivariate observations; pp. 287–297. [Google Scholar]
- Main BJ, Bickel RD, McIntyre LM, Graze RM, Calabrese PP, Nuzhdin SV. Allele-specific expression assays using Solexa. BMC Genomics. 2009;10(422) doi: 10.1186/1471-2164-10-422. http://dx.doi.org/10.1186/1471-2164-10-422 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden JE, Schwager SJ, May B. Single-locus inheritance in the tetraploid treefrog Hyla versicolor with an analysis of expected progeny ratios in tetraploid organisms. Genetics. 1987;116:299–311. doi: 10.1093/genetics/116.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins FA, Carneiro PCS, Guimaraes CT, Magalhaes JV, Carneiro JES, Cruz CD. Distinction between plant samples according to allele dosage by semiquantitative polymerase chain reaction. Genetics and Molecular Research. 2009;8:319–327. doi: 10.4238/vol8-1gmr585. [DOI] [PubMed] [Google Scholar]
- McQuown E, Gall GAE, May B. Characterization and inheritance of six microsatellite loci in lake sturgeon. Transactions of the American Fisheries Society. 2002;131:299–307. [Google Scholar]
- Meaburn E, Butcher LM, Schalkwyk LC, Plomin R. Genotyping pooled DNA using 100K SNP microarrays: a step towards genomewide association scans. Nucleic Acids Research. 2006;34:e28. doi: 10.1093/nar/gnj027. http://dx.doi.org/10.1093/nar/gnj027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A, Lewis SJ, Smith GD, et al. PTGS2-899G>C and prostate cancer risk: a population-based nested case–control study (ProtecT) and a systematic review with meta-analysis. Prostate Cancer and Prostatic Diseases. 2009;12:296–300. doi: 10.1038/pcan.2009.18. [DOI] [PubMed] [Google Scholar]
- Neve B, Froguel P, Corset L, Vaillant E, Vatin V, Boutin P. Rapid SNP allele frequency determination in genomic DNA pools by Pyrosequencing™. Biotechniques. 2002;32:1138–1142. doi: 10.2144/02325dd03. [DOI] [PubMed] [Google Scholar]
- Nijman IJ, Kuipers S, Verheul M, Guryev V, Cuppen E. A genome-wide SNP panel for mapping and association studies in the rat. BMC Genomics. 2008;9(95) doi: 10.1186/1471-2164-9-95. http://dx.doi.org/10.1186/1471-2164-9-95 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollitrault P, Dambier D, Luro F, Froelicher Y. Ploidy manipulation for breeding seedless triploid citrus. Plant Breeding Review. 2008;30:323–352. [Google Scholar]
- Ollitrault P, Terol J, Chen C, et al. A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. BMC Genomics. 2012a;13(593) doi: 10.1186/1471-2164-13-593. http://dx.doi.org/10.1186/1471-2164-13-593 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollitrault P, Terol J, Garcia-Lor A, et al. SNP mining in C. clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genomics. 2012b;13(13) doi: 10.1186/1471-2164-13-13. http://dx.doi.org/10.1186/1471-2164-13-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Quarrie SA, Lazic-Jancic V, Kovacevic D, Steed A, Pekic S. Bulk segregant analysis with molecular markers and its use for improving drought resistance in maize. Journal of Experimental Botany. 1999;50:1299–1306. [Google Scholar]
- Rickert AM, Premstaller A, Gebhardt C, Oefner PJ. Genotyping of SNPs in a polyploid genome by pyrosequencing™. Biotechniques. 2002;32 doi: 10.2144/02323rr01. 592–593; 596–598; 600. [DOI] [PubMed] [Google Scholar]
- Ritland K. Estimation of gene frequency and heterozygosity from pooled samples. Molecular Ecology Notes. 2002;2:370–372. [Google Scholar]
- Rockman MV, Kruglyak L. Genetics of global gene expression. Nature Reviews Genetics. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- Ronald J, Akey JM, Whittle J, Smith EN, Yvert G, Kruglyak L. Simultaneous genotyping, gene-expression measurement, and detection of allele-specific expression with oligonucleotide arrays. Genome Research. 2005;15:284–291. doi: 10.1101/gr.2850605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso ML, Burleson S, Maupin L, Rainey K. Development of breeder-friendly markers for selection of MIPS1 mutations in soybean. Molecular Breeding. 2011;28:127–132. [Google Scholar]
- Rozowsky J, Abyzov A, Wang J, et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Molecular Systems Biology. 2011;7:522. doi: 10.1038/msb.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh B, Allario T, Dambier D, Ollitrault P, Morillon R. Tetraploid citrus rootstocks are more tolerant to salt stress than diploid. Comptes Rendus Biologies. 2008;331:703–710. doi: 10.1016/j.crvi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Sapolsky RJ, Hsie L, Berno A, Ghandour G, Mittmann M, Fan JB. High-throughput polymorphism screening and genotyping with high-density oligonucleotide arrays. Genetic Analysis-Biomolecular Engineering. 1999;14:187–192. doi: 10.1016/s1050-3862(98)00026-6. [DOI] [PubMed] [Google Scholar]
- Shaw SH, Carrasquillo MM, Kashuk C. Allele frequency distributions in pooled DNA samples: applications to mapping complex disease genes. Genome Research. 1998;8:111–123. doi: 10.1101/gr.8.2.111. [DOI] [PubMed] [Google Scholar]
- Sjoling A, Walentinsson A, Nordlander C, et al. Assessment of allele dosage at polymorphic microsatellite loci displaying allelic imbalance in tumors by means of quantitative competitive-polymerase chain reaction. Cancer Genetics and Cytogenetics. 2005;157:97–103. doi: 10.1016/j.cancergencyto.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Soltis D, Soltis P. Molecular data and the dynamic nature of polyploidy. Critical Reviews in Plant Sciences. 1993;12:243–273. [Google Scholar]
- Steer S, Abkevich V, Gutin A, et al. Genomic DNA pooling for whole-genome association scans in complex disease: empirical demonstration of efficacy in rheumatoid arthritis. Genes and Immunity. 2007;8:57–68. doi: 10.1038/sj.gene.6364359. [DOI] [PubMed] [Google Scholar]
- Tabone T, Mather DE, Hayden MJ. Temperature Switch PCR (TSP): robust assay design for reliable amplification and genotyping of SNPs. BMC Genomics. 2009;10(580) doi: 10.1186/1471-2164-10-580. http://dx.doi.org/10.1186/1471-2164-10-580 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor GM, Kubisiak TL, Klopfenstein NB, Hall RB, McNabb HS., Jr. Bulked segregant analysis identifies molecular markers linked to Melampsora medusae resistance in Populus deltoides. Phytopathology. 2000;90:1039–1042. doi: 10.1094/PHYTO.2000.90.9.1039. [DOI] [PubMed] [Google Scholar]
- Tai G, DeJong H. A comparison of performance of tetraploid progenies produced by diploid and their vegetatively doubled (tetraploid) counterpart parents. Theoretical and Applied Genetics. 1997;94:303–308. [Google Scholar]
- Terol J, Conesa A, Colmenero JM, et al. Analysis of 13000 unique Citrus clusters associated with fruit quality, production and salinity tolerance. BMC Genomics. 2007;8(31) doi: 10.1186/1471-2164-8-31. http://dx.doi.org/10.1186/1471-2164-8-31 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol J, Naranjo MA, Ollitrault P, Talon M. Development of genomic resources for Citrus clementina: characterization of three deep-coverage BAC libraries and analysis of 46,000 BAC end sequences. BMC Genomics. 2008;9(423) doi: 10.1186/1471-2164-9-423. http://dx.doi.org/10.1186/1471-2164-9-423 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE, Gort G, Vosman B. Genotype calling in tetraploid species from bi-allelic marker data using mixture models. BMC Bioinformatics. 2011;12(172) doi: 10.1186/1471-2105-12-172. http://dx.doi.org/10.1186/1471-2105-12-172 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Blackstone N, Cartwright P, et al. Surveys of gene families using polymerase chain-reaction – PCR selection and PCR drift. Systematic Biology. 1994;43:250–261. [Google Scholar]
- Wasson J, Skolnick G, Love-Gregory L, Permutt MA. Assessing allele frequencies of single nucleotide polymorphisms in DNA pools by Pyrosequencing™ technology. Biotechniques. 2002;32 doi: 10.2144/02325dd04. 1144–1146; 1148; 1150. [DOI] [PubMed] [Google Scholar]
- Wendel J, Doyle J. Polyploidy and evolution in plants. In: Henry R, editor. Plant diversity and evolution. Wallingford, UK: CABI Publishing; 2005. pp. 97–117. [Google Scholar]
- Wenger JW, Schwartz K, Sherlock G. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genetics. 2010;6:e1000942. doi: 10.1371/journal.pgen.1000942. http://dx.doi.org/10.1371/journal.pgen.1000942 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JE, Douches DS, Freyre R. Use of half-tetrad analysis to discriminate between 2 types of 2n egg formation in a potato haploid. Genome. 1992;35:741–745. [Google Scholar]
- Yang HC, Fann CSJ. Association mapping using pooled DNA. Methods in Molecular Biology. 2007;379:161–175. doi: 10.1007/978-1-59745-389-9_12. [DOI] [PubMed] [Google Scholar]
- Zhang X, Borevitz JO. Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics. 2009;182:943–954. doi: 10.1534/genetics.109.103499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.