Abstract
Although the synthesis pathways of intracellular triacylglycerol (TAG) species have been well elucidated, assessment of the contribution of an individual pathway to TAG pools in different mammalian organs, particularly under pathophysiological conditions, is difficult, although not impossible. Herein, we developed and validated a novel bioinformatic approach to assess the differential contributions of the known pathways to TAG pools through simulation of TAG ion profiles determined by shotgun lipidomics. This powerful approach was applied to determine such contributions in mouse heart, liver, and skeletal muscle and to examine the changes of these pathways in mouse liver induced after treatment with a high-fat diet. It was clearly demonstrated that assessment of the altered TAG biosynthesis pathways under pathophysiological conditions can be readily achieved through simulation of lipidomics data. Collectively, this new development should greatly facilitate our understanding of the biochemical mechanisms underpinning TAG accumulation at the states of obesity and lipotoxicity.
Keywords: data interpretation, high-fat diet, mass spectrometry, shotgun lipidomics
Triglycerides are an important class of lipids that largely exist as triacylglycerol (TAG) species, although a small amount of ether-linked triglyceride species are also present in biological systems (1). The main biological role of TAG species is to serve as energy storage depots. As metabolic syndrome becomes increasingly prevalent, the excess levels of TAG mass that manifest in the organs have been frequently blamed as a causal factor of pathophysiological complications associated with those organs (2). The term of “lipotoxicity” has been generally used to describe this epidemiological phenomenon (3, 4). Numerous technologies and animal models have been created and applied to study the biochemical mechanism(s) underpinning lipotoxicity (4–9).
The demand for systematic identification and quantification of global cellular lipids for the aforementioned purpose has greatly facilitated the rapid spread of lipidomics research in recent years (10–12). Although extensive utilization of bioinformatics and computational algorithms for accurate identification and quantification of individual lipid molecular species and classes (13–17) is still a key node in lipidomics, the greatest challenge of bioinformatics for lipidomics research currently is the interpretation of alterations in biological functions resulting in adaptive or pathological changes in lipid metabolism and/or homeostasis (18).
One of the most powerful platforms in lipidomics practice is multidimensional mass spectrometry-based shotgun lipidomics (MDMS-SL) (15, 19). At its current stage of development, MDMS-SL enables us to analyze over 30 lipid classes and hundreds to thousands of individual lipid species, collectively representing more than 95% of the total content of a cellular lipidome directly from the lipid extracts of limited biological source materials (15, 19). However, it has remained a challenge to utilize the vast datasets associated with the MDMS-SL platform, e.g., to develop a bioinformatic approach for the interpretation of lipidomic networks of TAG biosynthesis and thus reveal their association with lipotoxicity under pathophysiological conditions. The development of such an approach would significantly advance the practical use of the platform for investigating the association of altered TAG mass with any (patho)physiological changes.
Recently, we have successfully developed dynamic simulation approaches for understanding the remodeling processes of cardiolipin, phosphatidylcholine (PC), and phosphatidylethanolamine species (20, 21) based on the known biological pathways and MDMS-SL-derived data. The parameters derived from the simulation can be used to assess the enzymatic activities that meditate the extent of remodeling of those lipid classes. Herein, we developed a novel simulation approach following a similar reasoning to determine the contributions of individual TAG biosynthesis pathways to TAG pools, thereby recapitulating the enzymatic activities involved in TAG biosynthesis. Moreover, the compositions of individual TAG species, including all isomeric and isobaric ones, were automatically determined from the simulation. The accuracy of this simulation approach was validated by comparisons of a variety of fatty acyl profiles derived from simulation to those acquired from neutral loss scans (NLS) of the corresponding FAs from TAG species as previously described (22) as well as by direct product ion analysis of individual TAG ions. In this study, we applied this novel approach to determine the differential contributions of different biosynthesis pathways to the TAG pools present in mouse heart, liver, and skeletal muscle, and to dissect alterations in the biosynthesis of mouse liver TAG species induced by feeding with a high-fat (HF) diet. We believe that development of this bioinformatic approach for simulation of TAG ion profiles will not only allow us to determine alterations in TAG biosynthesis in health and disease but also greatly facilitate a better understanding of the molecular mechanism(s) underpinning lipotoxicity.
MATERIALS AND METHODS
Materials
All phospholipid species and heptadecanoeyl-CoA used as internal standards were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Triheptadecanoeylglycerol (T17:1 TAG) was purchased from Nu Chek Inc. (Elysian, MN). All of the solvents were obtained from Burdick and Jackson (Honeywell International Inc., Muskegon, MI). All other chemical reagents were at least analytical grade or the best grade available and obtained from Fisher Scientific (Pittsburgh, PA), Sigma-Aldrich Chemical Co. (St. Louis, MO), or as indicated.
Animal studies and tissue collection
The protocols for animal experiments were conducted in accordance with the National Institutes of Health guidelines for humane treatment of animals and were approved by the Animal Studies Committee of Sanford-Burnham Medical Research Institute. One-month-old mice were fed with either a HF diet composed of 43% of calories from fat (TD 01381, Harlan Teklad, Madison, WI) containing TAG composed of long chain FAs (mostly 16:0 and 18:1) or the standard mouse chow (Rodent Chow 5053, Purina Mills Inc.) for four weeks. At the indicated age, animals were euthanized by asphyxiation with CO2. Tissue samples, including heart, liver, and skeletal muscle, were immediately harvested, perfused with ice-cold, diluted (10×) PBS, blotted with Kimwipes, and freeze-clamped at the temperature of liquid nitrogen. The tissue wafers were pulverized into a fine powder with a stainless-steel mortar and pestle and stored at −80°C for lipid analysis.
Analysis of lipids by mass spectrometry
Lipids were extracted from mouse tissue powders by a modified Bligh and Dyer method as previously described (23). Internal standards for quantification of individual molecular species of lipid classes were added prior to lipid extraction (23). MDMS-SL analyses were performed with a QqQ mass spectrometer (Thermo Fisher Scientific TSQ Vantage, San Jose, CA) equipped with an automated nanospray device (Triversa Nanomate, Advion Biosciences, Ithaca, NY) and operated with Xcalibur software as previously described (24, 25). Identification and quantification of lipid molecular species were performed using an automated software program as previously described (15). Lipid extraction for acyl-CoA analysis, and identification and quantification of acyl-CoA species by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry were performed as we previously described (20). The results are included in supplementary Table I for comparison.
Tandem MS analyses of TAG lithium adducts in the product ion mode were performed by using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an automated nanospray apparatus (Advion Bioscience, Ithaca, NY). The NanoMate apparatus was controlled by Chipsoft 8.3.1 software. An ionization voltage of 1.20 kV and a gas pressure of 2.00 psi were employed for the MS analyses. All mass spectra were recorded under Xcalibur 2.2.48 software with an AGC value of 1 × 106 in the full MS mode for up to 120 ms, in which the mass resolution of the analyzer was set at 140,000 (m/Δm, fwhm at m/z 200), and the product ion spectra were performed with higher-energy collisional dissociation (HCD) at 30.0 eV and gas pressure of 1 mT.
Model for simulation of TAG ion profiles
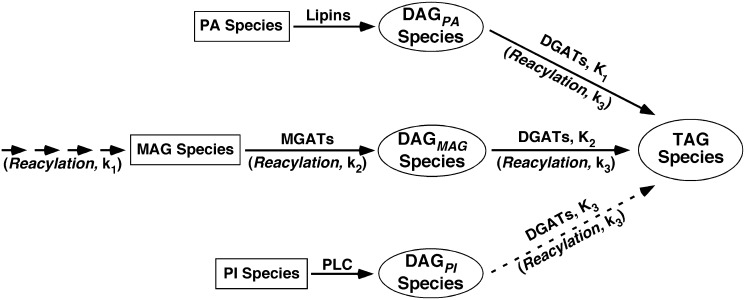
To quantitatively describe the TAG biosynthesis process, we developed an algorithm which simulated the ion profiles of TAG species determined by MDMS-SL. The simulation was based on the known TAG biosynthesis pathways of diacylglycerol (DAG) reacylation (26) (Fig. 1). These pathways include the DAG pools resulting from i) the dephosphorylation of phosphatidic acid (PA) (DAGPA); ii) the reacylation of monoacylglycerol (MAG) species (DAGMAG), which could result from multiple sources including dephosphorylation of lysophosphatidic acid, TAG/DAG hydrolysis with lipase activities, and reacylation from glycerol (26, 27); and iii) to a minimal extent, the hydrolysis of phosphatidylinositol (PI) species through phospholipase C (PLC) activities (DAGPI). It should be pointed out that phosphatidylinositol-4,5-diphosphate [PI(4,5)P2] species are the known substrates of PLC with a high turnover rate. Therefore, PLC hydrolysis of PI(4,5)P2 may result in a larger amount of DAG mass than that yielded from PI hydrolysis. However, the DAG species from PI(4,5)P2 largely serve as signaling molecules instead of TAG synthesis. Moreover, since the DAG composition resulting from PI(4,5)P2 species is generally similar to that from PI species, the contribution of PI(4,5)P2 hydrolysis pathway to TAG synthesis can be covered by the representation of PI-DAG pathway. In our simulation, we assumed that the cellular acyl-CoA profile was similar to the fatty acyl composition of the individual TAG pool. This assumption was validated by measurement of acyl-CoA profiles of mouse heart, liver, and skeletal muscle (supplementary Table I). Therefore, each of the TAG ion profiles was simulated with one of three weighting parameters, K1, K2, and K3, which were iterated utilizing PA composition, fatty acyl composition of TAG species, and PI composition (all of which were derived from MDMS-SL analysis), respectively, (see below). These K parameters represented the relative contributions of the different DAG pools resulting from the PA, MAG, and PI pathways to TAG synthesis.
Fig. 1.
Illustration of triacylglycerol biosynthesis model for simulation of triacylglycerol ion profiles. TAG species are de novo synthesized with reacylation of DAG species of different pools produced mainly through dephosphorylation of PA (DAGPA) and reacylation of MAG (DAGMAG) as well as, to a lesser degree (as indicated with a broken line arrow), through hydrolysis of PI with PLC activities (DAGPI). The contributions of these pathways to the TAG pools were determined through simulation of individual TAG ion profile with parameters of K1, K2, and K3, respectively, which are the probabilities of individual DAG pools being reacylated to TAG. In addition, the parameters of k1, k2, and k3 were used in the sn-1, 2, and 3 reacylation steps of TAG species in the forms of exp(-k1.×j), exp(k2.×j) and exp(-k3.×j), respectively, where k1 and k3 represents a simulated decay constant, k2 represents a simulated enhancing constant, and xj is the number of double bonds present in the corresponding fatty acyl chain. MGAT and DGAT denote MAG and DAG acyltransferases, respectively. The multiple arrows at the k1 step indicate that MAG species could yield from a variety of sources (see text).
In each of the reacylation steps as indicated (Fig. 1), the corresponding acyltransferase acylated a hydroxy group of glycerol at positions 1, 2, and 3 to yield MAG, DAG, and TAG, respectively, with fatty acyl-CoA based on the fatty acyl composition derived from TAG pools. We introduced a parameter of ks (s = 1, 2, and 3) at each step in the form of exp(-k1.xj), exp(k2.xj), or exp(-k3.xj), respectively, where both k1 and k3 represents simulated decay constants, k2 represents a simulated enhancing constant, and xj is the number of double bonds present in the jth fatty acyl chain. The parameters and their formats represent the selectivity of fatty acyl chains by the corresponding acyltransferase to reflect that saturated fatty acyls are enriched at the sn-1 and 3 positions, whereas unsaturated fatty acyls are largely acylated to the sn-2 position (28). Therefore, the levels of individual TAG species resulting from the PA-DAG pathway were calculated as:
| (1) |
and the TAG pool resulting from the PA-DAG pathway was calculated as:
| (2) |
where (PA)i and (FA)j are the compositions of the ith PA species and of the jth fatty acyl chain, respectively. Similarly, the TAG pools resulting from MAG-DAG and PI-DAG pathways were also calculated as:
| (3) |
and
| (4) |
respectively. Therefore, the normalized levels of individual TAG pools resulting from the PA-DAG, MAG-DAG, and PI-DAG pathways were calculated as:
| (5) |
| (6) |
and
| (7) |
respectively, Thus, the normalized total TAG level of an organ was the sum of individual TAG pools as:
| (8) |
with the restraint:
| (9) |
Accordingly, the levels of individual TAG ions including all isobaric and isomeric species were the sum of the levels of all individual TAG species with identical nominal mass. The criterion for our simulation was to achieve a TAG ion profile that maximized the linear correlation coefficient compared with that determined experimentally.
Finally, the level of a fatty acyl of a TAG ion was calculated first by summing the levels of all TAG species with identical nominal mass carrying the fatty acyl and then dividing by 3. A factor of 2 or 3 was multiplied if a TAG species contained two or three of an identical fatty acyl.
Validation of the simulated TAG ion profiles
The accuracy of the simulated TAG ion profiles (thereby supporting the rationale of the hypothesized model used for simulation) was not only determined with a good linear correlation coefficient as aforementioned but also could strictly be validated by two approaches. It was demonstrated that determination of individual TAG species, including isobaric and isomeric ones, by MDMS-SL was achieved by using the cross peaks of a TAG ion with those of NLS of all potentially existing fatty acyl chains present in TAG species (22). These cross peaks in two-dimensional mass spectrometry represent the fragment ions of the precursor TAG ion (see Results). Therefore, good matches between the fatty acyl compositional profiles extracted from the simulation described above and those acquired from MDMS-SL analysis provided a critical validation of our simulation approach, the hypothesized model, as well as the determined TAG species. Accordingly, validation was performed through examination of the linear correlation coefficients of the simulated individual fatty acyl profiles with the corresponding ones determined by NLS.
Alternatively, the fatty acyl profile of an individual TAG ion could be directly examined by tandem mass spectrometric analysis of the TAG ion of interest in the product ion mode. However, it should be noticed that this latter approach has a few limitations, including: 1) limited dynamic range; therefore, the existence of very low abundance fatty acyls is unable to be detected and the fatty acyl profiles of low abundance TAG ions are hard to be validated; 2) it is not quantitative without correction for differential fragmentation; and 3) it is complicated with the presence of other fragments, such as those from neutral losses of lithium fatty carboxylates.
Miscellaneous
Protein concentration was determined utilizing a bicinchoninic acid protein assay kit (Pierce, Rockford, IL) using BSA as a standard. All data were presented as the means ± SD of at least three separate animals. Statistical significance was determined by a two-tailed Student t-test in comparisons with control, in which *P < 0.05 and **P < 0.01.
RESULTS
Representative simulation of TAG ion profiles present in mouse heart, liver, and skeletal muscle
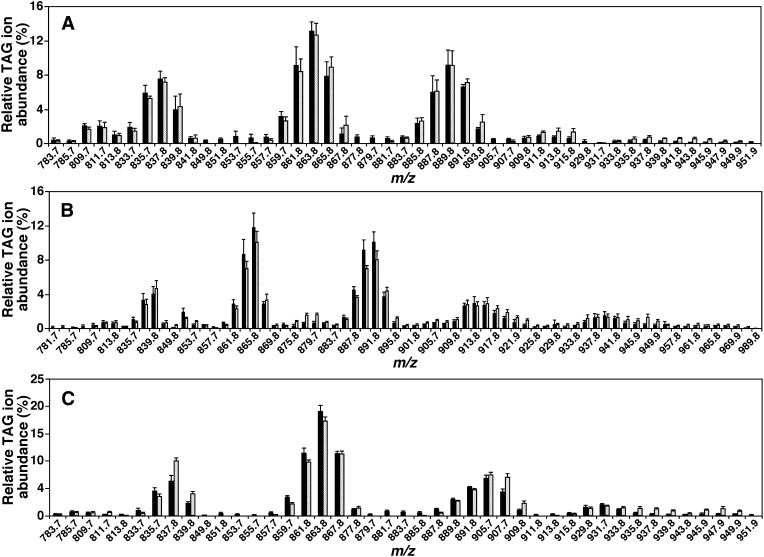
To exemplify the performance of simulation algorithm with the hypothesized model (Fig. 1), we analyzed the mass content of individual lipid species of PA, PI, and TAG classes from mouse heart, liver, and skeletal muscle by MDMS-SL (supplementary Table I). The composition of fatty acyls present in TAG pools of these organs was also derived from the levels of TAG species (supplementary Table I). By finely iterating the parameters of K1, K2, and K3 with a step size of 0.001, as well as the parameters of k1, k2, and k3 with a step size of 0.01, a correlation coefficient (γ) of better than (e.g., skeletal muscle and heart) or close to (e.g., liver) 0.95 between the simulated and MS-determined TAG ion profiles was obtained (Table 1). Simulated TAG ion profiles were well matched to those experimentally determined utilizing the hypothetical model and lipidomics data with the indicated parameters (Fig. 2).
TABLE 1.
Simulated parameters representing the contributions of TAG biosynthesis pathways and the selectivity of reacylation manifest in mouse organs
| Sample | K1 | K2 | K3 | k1 | k2 | k3 | γ |
| Skeletal muscle | 0 | 1.000 | 0 | 0.11 ± 0.09 | 0.05 ± 0.05 | 0.01 ± 0.01 | 0.977 ± 0.007 |
| Heart | 0.145 ± 0.033 | 0.825 ± 0.043 | 0.030 ± 0.014 | 0.01 ± 0.01 | 0 | 0 | 0.960 ± 0.015 |
| Liver | 0.493 ± 0.031 | 0.507 ± 0.031 | 0 | 0.47 ± 0.15 | 0.23 ± 0.05 | 0 | 0.943 ± 0.007 |
The determined TAG levels of mouse skeletal muscle, heart, and liver by MDMS-SL (supplementary Table I) were simulated with the TAG biosynthesis pathways as illustrated in Fig. 1. The parameters of K1, K2, and K3 represent the contributions of individual pathways (i.e., PA-, MAG-, and PI-DAG pathways, respectively) to the TAG pools. The parameters of k1, k2, and k3 represent the degrees of acyltransferase selectivity to the different fatty acyl chains at the sn-1, 2, and 3 positions of TAG species, respectively (see text for details).
Fig. 2.
Comparisons of the ion profiles of triacylglycerol species yielded from simulation with those acquired from MS analysis. Lipid extracts of mouse heart, liver, and skeletal muscle were prepared with a modified Bligh-Dyer procedure (23). TAG ion profiles displayed as lithiated TAG species (solid bars) of mouse heart (A), liver (B), and skeletal muscle (C) were obtained from NLS of all potentially present fatty acyl chains in TAG species in the presence of a small amount of lithium hydroxide in the positive-ion mode as previously described (22). The corresponding TAG ion profiles simulated (hatched bars) with the simulated parameters listed in Table 1 were displayed for comparison.
These results indicate that the MAG-DAG pathway (i.e., K2) is the sole or major synthesis pathway contributing to the TAG pool present in mouse skeletal muscle or heart, respectively, while both MAG-DAG and PA-DAG (i.e., K1) pathways equally contribute to the TAG pool present in mouse liver (Table 1). Moreover, the contribution of the PI hydrolysis pathway (i.e., K3) to those TAG pools might be negligible. The simulation results further indicate that the degree of selective reacylation with DGAT activity (i.e., k3) during TAG synthesis in all three examined organs was negligible, whereas considerable selectivity of other reacylation steps (i.e., k1 and k2) were exhibited in mouse liver (Table 1).
Validation of the simulation approach for determination of TAG ion profiles
As each individual TAG ion represents a combination of numerous isomeric and isobaric TAG species, a good match between the simulated and experimentally determined TAG ion profiles is a necessary condition for simulation, but it does not sufficiently guarantee that the levels of all those isomeric and isobaric species of an individual TAG ion also match accurately. Therefore, these simulated TAG species must be validated by an independent method.
Although it is not impossible to quantitatively analyze the composition of isobaric/isomeric TAG species after resolution by multiple column steps, the current field lacks an effective approach to definitively demonstrate the existence of these species. The identities of TAG species have only been indirectly determined through tandem MS analysis of the fatty acyl components of an individual TAG ion in combination with the total numbers of carbon atoms and double bonds, which are the known restrictions of individual TAG species after determination of the TAG ion m/z (22, 29, 30). Accordingly, both product ion analysis of individual TAG ions and MDMS-SL analysis of an entire TAG profile could be used to validate the determined TAG ion profile from simulation.
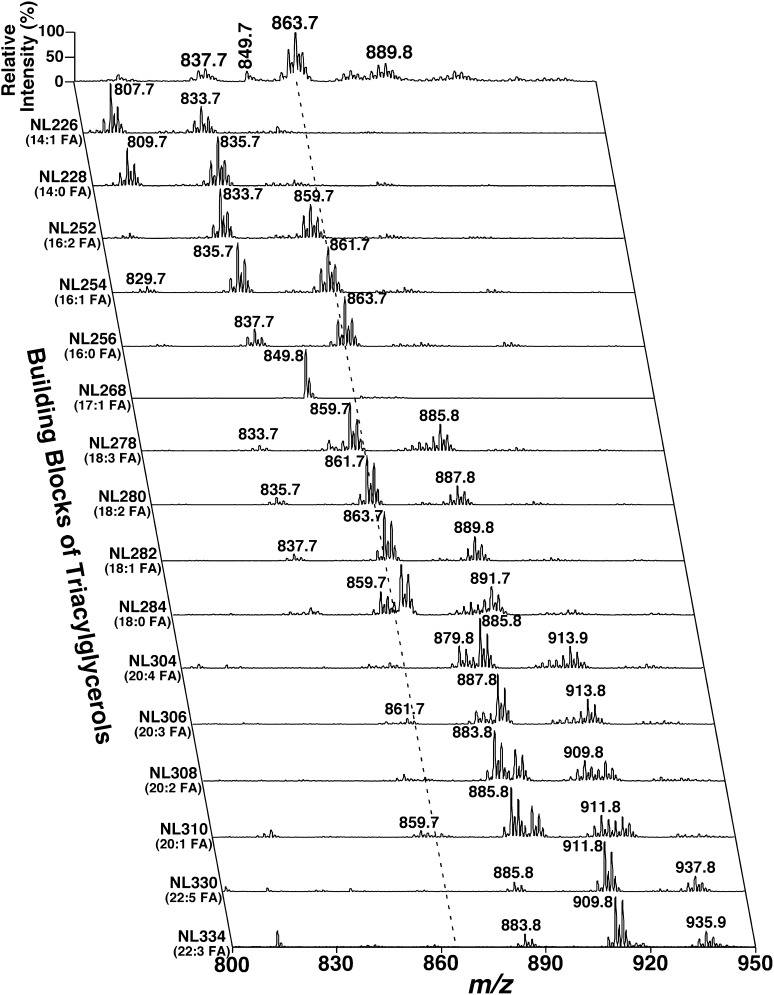
It has been previously demonstrated that individual isomeric and isobaric TAG species can be identified and quantified from the cross peaks of a TAG ion with the ions present in NLS of all potentially existing FA chains of TAG species (22). Fig. 3 shows an example of the analysis of mouse liver TAG species, wherein a broken line indicates that the TAG ion at m/z 863.7 crosses with the peaks present in the spectra of NL252 (16:2 FA), NL254 (16:1 FA), NL256 (16:0 FA), NL278 (18:3 FA), NL280 (18:2 FA), NL282 (18:1 FA), NL284 (18:0 FA), NL306 (20:3 FA), NL308 (20:2 FA), etc. These cross-peak intensities represent the abundance of the fragment ions of m/z 863.7 TAG ion after separate losses of the indicated fatty acids. The intensities of all these cross peaks are tabulated (supplementary Table II). A good simulation may be validated only if all of the cross peaks derived from the simulation match well with those acquired from MDMS-SL analysis (i.e., all NL spectra). This validation also affirms the accuracy of our simulation approach and hypothesized TAG biosynthesis model.
Fig. 3.
Representative two-dimensional mass spectrometric analyses of triacylglycerol species in a mouse liver lipid extract. NLS of all naturally existing fatty acyl chains (i.e., the building blocks of TAG species) of a mouse liver lipid extract were used to determine the identities of each lithiated TAG molecular ion, deconvolute isomeric/isobaric species, and quantify individual TAG species by comparisons with a selected internal standard (i.e., T17:1 TAG, shown in NL268). Collision activation was performed with collision energy of 35 eV and gas pressure of 1 mT on a triple quadrupole mass spectrometer (TSQ Vantage, Thermo Fisher Scientific, San Jose, CA). All displayed mass spectral traces are normalized to the base peak in each trace of NLS.
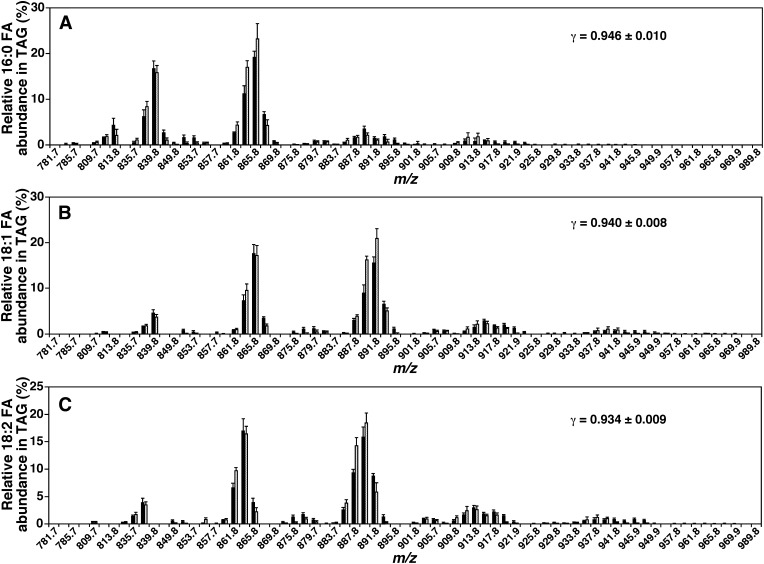
A good match [i.e., a linear correction coefficient (γ) of approximately 0.95] was essentially found between the individual FA profiles obtained from simulation and MDMS-SL analysis for all of the NLS of fatty acyl chains from all examined TAG datasets. Fig. 4 shows some representative comparisons of the abundant fatty acyl chains present in the mouse heart TAG pool. The comparisons not only validate the simulated TAG profiles but also suggest that the hypothetical model and all aforementioned assumptions for simulation (Fig. 1) are reasonable to the extent of the simulated accuracy (see Discussion).
Fig. 4.
Comparisons of the representative fatty acyl profiles of mouse myocardial TAG species yielded from simulation with those acquired from MS analysis. Lipid extracts of mouse heart were prepared with a modified Bligh-Dyer procedure (23). Fatty acyl (FA) profiles of 16:0 (palmitate, A), 18:1 (oleate, B), and 18:2 (linoleate, C) determined by neutral loss scanning of the corresponding fatty acid masses as previously described (22) (solid bars) were compared with those derived from simulation (hatched bars). The linear correlation coefficients between the corresponding data sets were indicated.
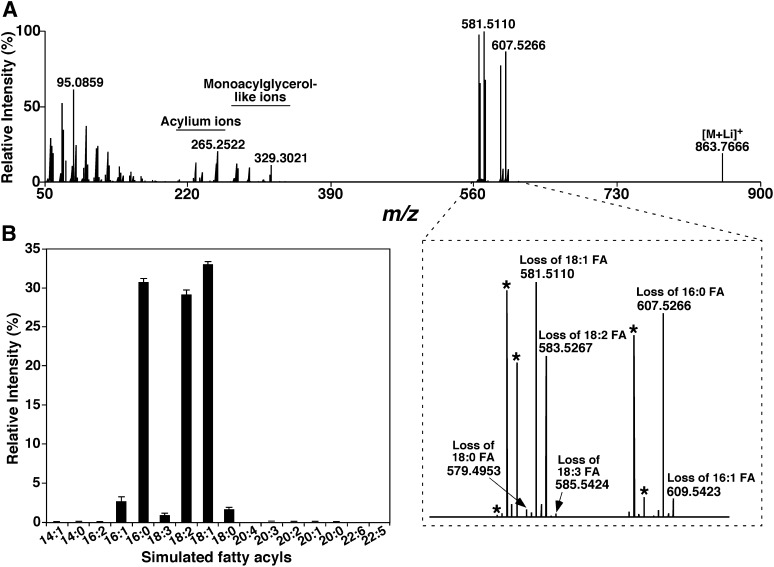
In an alternative approach, the presence of the simulated fatty acyls of a particular TAG ion could be validated by product ion analysis of this TAG ion. For example, the product ion mass spectrum of the TAG ion at m/z 863.7 present in mouse liver lipid extracts displays numerous fragment ions corresponding to neutral losses of FAs or lithium fatty carboxylates (Fig. 5A, inset), as well as fatty acylium ions (Fig. 5). The presence of the simulated FAs in the product ion mass spectrum further validates the simulation (Fig. 5B).
Fig. 5.
Representative product ion analysis of triacylglycerol lithium adducts in the positive-ion mode. Tandem MS analysis of a lithiated TAG ion at m/z 863.7666 was performed on a Thermo Scientific Q-Exactive mass spectrometer utilizing a precursor isolation window of 0.4 Th by the quadrupole mass filter. Collision activation was carried out with higher energy collisional dissociation in the HCD cell at 30.0 eV and gas pressure of 1 mT. The resultant fragment ions were analyzed in the Orbitrap. The peaks indicated with asterisks are the fragment ions corresponding to the neutral losses of lithium fatty carboxylates from the TAG molecular ion.
Determination of isomeric and isobaric TAG species from simulation
At the current stage of development of lipidomics, it is difficult, although not impossible (31), to identify the TAG regioisomers resulting from linkages of the fatty acyl chains to different glycerol positions. But MDMS-SL allows us to readily determine the different isomers containing variable fatty acyl chains that constitute individual TAG ions (22). For example, the lithiated TAG ion at m/z 863.7 was crossed with the NL spectra of 16:2, 16:1, 16:0, 18:3, 18:2, 18:1, 18:0, 20:3, and 20:2 fatty acyl chains, indicating the presence of these fatty acyls with abundance of the peak intensities (Fig. 3). Since a lithiated TAG ion at m/z 863.7 should predominantly contain species with a total of 55 carbon atoms (3 from glycerol and 52 from fatty acyl chains) with 3 double bonds (with a theoretical mass of 863.7674 Da), isomeric TAG species of 16:2-18:1-18:0, 16:1-16:0-20:2, 16:0-16:0-20:3, 16:1-18:1-18:1, 16:0-18:1-18:2, 16:0-18:0-18:3, 16:1-18:1-18:1, and other low abundance species were manually identified with these criteria as previously described (22). Unfortunately, automation of this identification process does not exist. Therefore, this process was very labor-intensive and inaccurate. The process was further complicated by the possibility that isomeric TAG species may be isobaric with the TAG species containing 56 carbon atoms with 10 double bonds (with a theoretical mass of 863.6735 Da) if present, as the nominal resolution mass spectrometer used for MDMS-SL analysis was not capable of resolving them.
In our simulation, the probable prevalence of each individual isomeric and isobaric species was calculated from each TAG biosynthesis pathway, whereas the sum of these isomeric and isobaric species was used to compare the intensity of a corresponding TAG ion determined by MDMS-SL. Therefore, development of this simulation algorithm automatically projected the content of individual isomeric and isobaric species of each detected TAG ion. Accordingly, 919, 1795, and 823 TAG species were readily projected in this study from the simulation of TAG ion profiles of mouse skeletal muscle, heart, and liver, respectively. Clearly, the numbers of these simulated TAG species were much larger than those manually determined (22, 32). As an example, a list of the identified and quantified TAG species from the simulation of mouse myocardial TAG profiles is given (supplementary Table III).
It should be emphasized that the simulation approach only determined the composition of individual TAG species. However, the mass levels of individual simulated TAG species may be readily calculated from the total TAG mass levels determined by MDMS-SL if necessary.
Determination of the altered mouse liver TAG biosynthesis after feeding with a HF diet
It is well known that after feeding with a HF diet, the uptake and storage of fat increase in liver (33). We speculated that our simulation should be able to recapitulate alterations in contributions of the different TAG synthesis pathways to liver TAG pool after HF diet feeding.
To test this hypothesis, we treated mice with a HF diet for a short period (four weeks) and determined the content and composition of TAG species present in the liver by MDMS-SL. We found that liver TAG content only slightly increased (from 442.65 ± 48.97 nmol/mg protein when mice were fed with a standard chow to 473.57 ± 42.94 nmol/mg protein with a HF diet). However, the composition of fatty acyl chains was significantly changed in different directions (Table 2). The composition of fatty acyl chains corresponding to those endogenously synthesized (e.g., 14:1, 16:1, and 18:1) was significantly reduced whereas the composition of essential FAs representing uptake and storage (e.g., 18:2, 20:4, 22:3, 22:4, 22:5, and 22:6) significantly increased (Table 2). These results indicate the reduction of de novo FA synthesis.
TABLE 2.
Fatty acyl composition in liver TAG pools of mice fed with a standard chow or a high-fat diet
| Fatty Acyl | Standard Chow | High-Fat Diet |
| 14:1 | 0.28 ± 0.05 | 0.17 ± 0.03** |
| 14:0 | 0.94 ± 0.08 | 0.74 ± 0.07** |
| 16:2 | 0.38 ± 0.05 | 0.38 ± 0.04 |
| 16:1 | 7.42 ± 0.91 | 3.86 ± 0.40** |
| 16:0 | 23.48 ± 0.82 | 24.11 ± 0.30 |
| 18:3 | 2.77 ± 0.20 | 2.96 ± 0.21 |
| 18:2 | 19.61 ± 1.06 | 22.69 ± 1.10** |
| 18:1 | 29.98 ± 0.97 | 25.77 ± 1.05** |
| 18:0 | 5.38 ± 0.09 | 5.55 ± 0.34 |
| 20:4 | 1.66 ± 0.06 | 2.53 ± 0.22** |
| 20:3 | 0.69 ± 0.05 | 0.93 ± 0.16* |
| 20:2 | 0.69 ± 0.07 | 1.00 ± 0.09** |
| 20:1 | 1.30 ± 0.04 | 1.69 ± 0.17** |
| 20:0 | 0.74 ± 0.08 | 0.92 ± 0.17 |
| 21:0 | 0.21 ± 0.02 | 0.42 ± 0.18 |
| 22:6 | 1.61 ± 0.16 | 2.25 ± 0.13** |
| 22:5 | 0.59 ± 0.05 | 0.87 ± 0.03** |
| 22:4 | 0.34 ± 0.04 | 0.56 ± 0.07** |
| 22:3 | 0.94 ± 0.09 | 1.31 ± 0.08** |
| 22:2 | 0.39 ± 0.03 | 0.56 ± 0.03** |
| 22:1 | 0.29 ± 0.05 | 0.41 ± 0.03* |
| 22:0 | 0.30 ± 0.05 | 0.33 ± 0.05 |
The mass levels of individual fatty acyl chains were determined by neutral losses of individual fatty acid mass in MDMS-SL as previously described (15, 22). The m:n indicates the FA containing a total of m carbon atoms and n double bond. Data represent the means ± SD of at least three separate animals. *P < 0.05 and **P < 0.01 compared with those from a standard chow.
When we simulated the TAG ion profiles of liver extracts of mice fed with a HF diet, we found that the contribution of MAG-DAG pathway (i.e., K2) to the liver TAG pool increased approximately 10% (i.e., from 0.507 ± 0.031 with a standard chow to 0.554 ± 0.030 with a HF diet, P < 0.05), whereas the reacylation selectivity is virtually identical (i.e., k1, from 0.47 ± 0.15 to 0.49 ± 0.04; k2, from 0.23 ± 0.08 to 0.20 ± 0.01; no change with k3) between the conditions. These findings, in combination with the reduction of de novo FA synthesis, indicate that mechanisms to deposit excess energy into the TAG pool through uptake of FAs dominate in mouse liver after feeding with a HF diet. This result is consistent with those of previous observations (33).
DISCUSSION
In the current study, we developed and validated a novel approach for the simulation of the synthesis of TAG species and determined the contributions of individual biosynthesis pathways to the TAG pool using lipidomics data. The work was significant on two levels. First, to the best of our knowledge, this was the first study to substantiate the contributions of individual TAG biosynthesis pathways to TAG pools, as well as the first to assess substrate selectivity in TAG biosynthesis by using a bioinformatics approach. Second, with the simulation, the mass levels of individual TAG species, including those of isomeric and isobaric ones, could be automatically assessed, greatly facilitating the accuracy and speed of lipidomic analysis of TAG species. It is clear that this bioinformatic tool in combination with lipidomics analysis could allow us to reveal potential biochemical mechanisms underpinning TAG accumulation under different pathophysiological conditions, which is of particular significance at a time when obesity and lipotoxicity are becoming epidemic.
As a proof of concept, we assessed the contributions of different TAG biosynthesis pathways to the TAG pools of mouse heart, liver, and skeletal muscle utilizing this simulation approach. The results are consistent with what people have generally recognized, i.e., that TAG pools in skeletal muscle serve as a storage depot of free FAs through the MAG-DAG pathway, whereas de novo synthesis of TAG species through the PA-DAG pathway greatly contributes to mouse liver TAG pool. Increased contribution of the MAG-DAG pathway to the liver TAG pool after mice were fed with a HF diet strongly supports this notion (34).
The simulated data would be best validated by direct comparisons between the cellular activities of enzymes involving individual TAG biosynthesis pathways and those derived from simulation. However, assessment of these individual enzymatic activities can usually be performed only under optimal experimental conditions for a particular enzyme. The comparison of activities determined directly from those enzymatic assays remains challenging, and although isotope-labeling experiments may provide some insights into FA distribution and the turnover rates of de novo TAG synthesis, it is very difficult to clearly dissect the contributions of individual biosynthesis pathways to a TAG pool through such an approach. Accordingly, in the current study, we employed MS approaches by either comparing all simulation-derived FA profiles to those obtained by NLS of individual FA from TAG species to validate simulated data or by comparing simulated FA distributions with those obtained from product ion analysis of a particular TAG ion. Surprisingly, we found that the simulation-derived FA profiles matched all the corresponding FA profiles obtained from MS analysis very well, suggesting the hypothetical model and assumptions for simulation are reasonably accurate under experimental conditions.
The match of TAG ion profiles between simulation and MS determination was achieved with a linear correlation coefficient of approximately 0.95 (Table 1). If desired, the correlation coefficient could be further improved with two additions. First, hydrolysis of other classes of phospholipids (particularly PC) through PLC could be included in the simulation model (Fig. 1). Although we believe that the contribution of this pathway to a TAG pool should be relatively small, it could serve as a fine-tune step for simulation and would introduce the ether-linked species from the PC pool to the TAG pool, the presence of which was previously demonstrated (1). Second, addition of NLS of FAs containing odd-numbered carbon atoms in MDMS-SL analysis should lead to better simulation of low abundance TAG species containing such fatty acyls, as this would allow for the simulation of the TAG ions containing those FA(s).
It should be explicitly stated that the simulation model was simplified in at least two aspects. We assumed the existence of reacylation selectivity. In biological systems, this selectivity could result from the remodeling of TAG species after synthesis through the selective activity of one or more particular lipases. Moreover, the MAG species could be derived from multiple sources, including dephosphorylation of lysophosphatidic acid and TAG/DAG hydrolysis with lipase activities (26), in addition to possible direct acylation of glycerol (27).
A huge number of TAG species, the majority of which were of low or very low abundance, were obtained from simulation (e.g., supplementary Table III). The presence of such a huge number of TAG species might be physiologically correct, but is very difficult to validate experimentally due to technological limitations. Rather, the question is whether it is meaningful to have such a number of species in biological systems. Although we cannot exclude the possibility that the metabolite(s) of these low abundance TAG species play an important role in biological functions, we believe that these species are largely the resultant products of nonselective TAG biosynthesis.
In summary, we developed and validated a novel bioinformatic approach for simulation of TAG ion profiles, which allowed us to mathematically analyze biosynthesis pathways through the interpretation of lipidomics data. We assessed the contributions of different biosynthesis pathways to TAG pools in mouse heart, liver, and skeletal muscle and examined changes to these pathways in mouse liver induced following treatment with a HF diet. It was proved that bioinformatic simulation provides a powerful vehicle to determine altered TAG biosynthesis pathways under pathophysiological conditions. We believe that this type of assessment could provide deep insights into the biochemical mechanisms underpinning the TAG accumulation that is characteristic throughout the development of obesity and lipotoxicity.
Supplementary Material
Footnotes
Abbreviations:
- DAG
- diacylglycerol
- HF
- high fat
- MAG
- monoacylglycerol
- MDMS-SL
- multidimensional mass spectrometry-based shotgun lipidomics
- m:n
- acyl chain containing m carbon atoms and n double bonds
- NLS
- neutral loss scanning
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PI
- phosphatidylinositol
- PI(4,5)P2
- phosphatidylinositol-4,5-diphosphates
- PLC
- phospholipase C
- TAG
- triacylglycerol
This work was supported by National Institutes of Health Grant R01-AG-31675 and intramural institutional research funds.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables.
REFERENCES
- 1.Bartz R., Li W. H., Venables B., Zehmer J. K., Roth M. R., Welti R., Anderson R. G., Liu P., Chapman K. D. 2007. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 48: 837–847. [DOI] [PubMed] [Google Scholar]
- 2.Moller D. E., Kaufman K. D. 2005. Metabolic syndrome: a clinical and molecular perspective. Annu. Rev. Med. 56: 45–62. [DOI] [PubMed] [Google Scholar]
- 3.Unger R. H., Orci L. 2000. Lipotoxic diseases of nonadipose tissues in obesity. Int. J. Obes. Relat. Metab. Disord. 24: S28–S32. [DOI] [PubMed] [Google Scholar]
- 4.Garbarino J., Sturley S. L. 2009. Saturated with fat: new perspectives on lipotoxicity. Curr. Opin. Clin. Nutr. Metab. Care. 12: 110–116. [DOI] [PubMed] [Google Scholar]
- 5.Unger R. H., Scherer P. E. 2010. Gluttony, sloth and the metabolic syndrome: a roadmap to lipotoxicity. Trends Endocrinol. Metab. 21: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalscheuer R., Luftmann H., Steinbuchel A. 2004. Synthesis of novel lipids in Saccharomyces cerevisiae by heterologous expression of an unspecific bacterial acyltransferase. Appl. Environ. Microbiol. 70: 7119–7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamisaka Y., Tomita N., Kimura K., Kainou K., Uemura H. 2007. DGA1 (diacylglycerol acyltransferase gene) overexpression and leucine biosynthesis significantly increase lipid accumulation in the Deltasnf2 disruptant of Saccharomyces cerevisiae. Biochem. J. 408: 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birse R. T., Choi J., Reardon K., Rodriguez J., Graham S., Diop S., Ocorr K., Bodmer R., Oldham S. 2010. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 12: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ackerman D., Gems D. 2012. The mystery of C. elegans aging: an emerging role for fat. Distant parallels between C. elegans aging and metabolic syndrome? Bioessays. 34: 466–471. [DOI] [PubMed] [Google Scholar]
- 10.Han X., Gross R. W. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412. [DOI] [PubMed] [Google Scholar]
- 11.Dennis E. A. 2009. Lipidomics joins the omics evolution. Proc. Natl. Acad. Sci. USA. 106: 2089–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niemelä P. S., Castillo S., Sysi-Aho M., Oresic M. 2009. Bioinformatics and computational methods for lipidomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2855–2862. [DOI] [PubMed] [Google Scholar]
- 13.Seppänen-Laakso T., Oresic M. 2009. How to study lipidomes. J. Mol. Endocrinol. 42: 185–190. [DOI] [PubMed] [Google Scholar]
- 14.Fahy E., Cotter D., Byrnes R., Sud M., Maer A., Li J., Nadeau D., Zhau Y., Subramaniam S. 2007. Bioinformatics for lipidomics. Methods Enzymol. 432: 247–273. [DOI] [PubMed] [Google Scholar]
- 15.Yang K., Cheng H., Gross R. W., Han X. 2009. Automated lipid identification and quantification by multi-dimensional mass spectrometry-based shotgun lipidomics. Anal. Chem. 81: 4356–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheelock C. E., Goto S., Yetukuri L., D'Alexandri F. L., Klukas C., Schreiber F., Oresic M. 2009. Bioinformatics strategies for the analysis of lipids. Methods Mol. Biol. 580: 339–368. [DOI] [PubMed] [Google Scholar]
- 17.Song H., Ladenson J., Turk J. 2009. Algorithms for automatic processing of data from mass spectrometric analyses of lipids. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2847–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrester J. S., Milne S. B., Ivanova P. T., Brown H. A. 2004. Computational lipidomics: a multiplexed analysis of dynamic changes in membrane lipid composition during signal transduction. Mol. Pharmacol. 65: 813–821. [DOI] [PubMed] [Google Scholar]
- 19.Han X., Yang K., Gross R. W. 2012. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 31: 134–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiebish M. A., Bell R., Yang K., Phan T., Zhao Z., Ames W., Seyfried T. N., Gross R. W., Chuang J. H., Han X. 2010. Dynamic simulation of cardiolipin remodeling: greasing the wheels for an interpretative approach to lipidomics. J. Lipid Res. 51: 2153–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarringhalam K., Zhang L., Kiebish M. A., Yang K., Han X., Gross R. W., Chuang J. 2012. Statistical analysis of the processes controlling choline and ethanolamine glycerophospholipid molecular species composition. PLoS ONE. 7: e37293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han X., Gross R. W. 2001. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 295: 88–100. [DOI] [PubMed] [Google Scholar]
- 23.Christie W. W., Han X. 2010. Lipid Analysis: Isolation, Separation, Identification and Lipidomic Analysis. 4th edition. Woodhead Publishing, Cambridge, UK. [Google Scholar]
- 24.Kiebish M. A., Han X., Cheng H., Lunceford A., Clarke C. F., Moon H., Chuang J. H., Seyfried T. N. 2008. Lipidomic analysis and electron transport chain activities in C57BL/6J mouse brain mitochondria. J. Neurochem. 106: 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han X., Yang K., Gross R. W. 2008. Microfluidics-based electrospray ionization enhances intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: development of an automated high throughput platform for shotgun lipidomics. Rapid Commun. Mass Spectrom. 22: 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coleman R. A., Lee D. P. 2004. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43: 134–176. [DOI] [PubMed] [Google Scholar]
- 27.Rushdi A. I., Simoneit B. R. 2006. Abiotic condensation synthesis of glyceride lipids and wax esters under simulated hydrothermal conditions. Orig. Life Evol. Biosph. 36: 93–108. [DOI] [PubMed] [Google Scholar]
- 28.Christie W. W. 2011. Triacylgycerols. Part 1. Structure and composition. AOCS Libid Library. Accessed February 7, 2013 at http://lipidlibrary.aocs.org/lipids/tag1/index.htm.
- 29.Krank J., Murphy R. C., Barkley R. M., Duchoslav E., McAnoy A. 2007. Qualitative analysis and quantitative assessment of changes in neutral glycerol lipid molecular species within cells. Methods Enzymol. 432: 1–20. [DOI] [PubMed] [Google Scholar]
- 30.Schwudke D., Oegema J., Burton L., Entchev E., Hannich J. T., Ejsing C. S., Kurzchalia T., Shevchenko A. 2006. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal. Chem. 78: 585–595. [DOI] [PubMed] [Google Scholar]
- 31.Hsu F-F., Turk J. 1999. Structural characterization of triacylglycerols as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisionally activated dissociation on a triple stage quadrupole instrument. J. Am. Soc. Mass Spectrom. 10: 587–599. [DOI] [PubMed] [Google Scholar]
- 32.Cheng H., Guan S., Han X. 2006. Abundance of triacylglycerols in ganglia and their depletion in diabetic mice: implications for the role of altered triacylglycerols in diabetic neuropathy. J. Neurochem. 97: 1288–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gropler M. C., Harris T. E., Hall A. M., Wolins N. E., Gross R. W., Han X., Chen Z., Finck B. N. 2009. Lipin 2 is a liver-enriched phosphatidate phosphohydrolase enzyme that is dynamically regulated by fasting and obesity in mice. J. Biol. Chem. 284: 6763–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yen C. L., Cheong M. L., Grueter C., Zhou P., Moriwaki J., Wong J. S., Hubbard B., Marmor S., Farese R. V., Jr 2009. Deficiency of the intestinal enzyme acyl CoA:monoacylglycerol acyltransferase-2 protects mice from metabolic disorders induced by high-fat feeding. Nat. Med. 15: 442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.