Abstract
Micrometric membrane lipid segregation is controversial. We addressed this issue in attached erythrocytes and found that fluorescent boron dipyrromethene (BODIPY) analogs of glycosphingolipids (GSLs) [glucosylceramide (BODIPY-GlcCer) and monosialotetrahexosylganglioside (GM1BODIPY)], sphingomyelin (BODIPY-SM), and phosphatidylcholine (BODIPY-PC inserted into the plasma membrane spontaneously gathered into distinct submicrometric domains. GM1BODIPY domains colocalized with endogenous GM1 labeled by cholera toxin. All BODIPY-lipid domains disappeared upon erythrocyte stretching, indicating control by membrane tension. Minor cholesterol depletion suppressed BODIPY-SM and BODIPY-PC but preserved BODIPY-GlcCer domains. Each type of domain exchanged constituents but assumed fixed positions, suggesting self-clustering and anchorage to spectrin. Domains showed differential association with 4.1R versus ankyrin complexes upon antibody patching. BODIPY-lipid domains also responded differentially to uncoupling at 4.1R complexes [protein kinase C (PKC) activation] and ankyrin complexes (in spherocytosis, a membrane fragility disease). These data point to micrometric compartmentation of polar BODIPY-lipids modulated by membrane tension, cholesterol, and differential association to the two nonredundant membrane:spectrin anchorage complexes. Micrometric compartmentation might play a role in erythrocyte membrane deformability and fragility.
Keywords: lipid domains, plasma membrane, compartmentation, confocal imaging, cholesterol, membrane tension, spherocytosis, membrane:spectrin anchorage complexes
Membrane lipid bilayers, long viewed as homogenous solvents for membrane proteins (1), actually show lateral heterogeneity at two different scales, transient nanometric assemblies in “lipid rafts” (2–6) versus submicrometric/mesoscale domains evidenced not only on artificial vesicles (3, 7–11) but also on living cells (12–23). The major structural lipids at the mammalian plasma membrane (PM) are: i) glycerophospholipids, hereafter represented by phosphatidylcholine (PC); ii) sphingolipids (SLs) including SM and glycosphingolipids (GSLs), an heterogeneous family from the simple glucosylceramide (GlcCer) to complex monosialotetrahexosylganglioside (GM1) (for a review, see Ref. 24); and iii) cholesterol, a crucial fluidity regulator without a protruding polar head. The wedge-like shapes of SLs and cholesterol allow them to come in very close apposition via van der Waals forces and this proximity is proposed to account for their spontaneous clustering into lipid rafts (25). This nanometric short-lived lateral asymmetry led to the first revision of the original Singer-Nicolson model (2, 26).
In addition, compartmentation of polar lipids into stable submicrometric PM domains in living cells was originally implied from restricted lateral mobility of fluorescent PC evidenced by fluorescence recovery after photobleaching (FRAP) of increasingly large fields (13). It was further supported by high-resolution tracking of single fluorescent phosphatidylethanolamine, which revealed lasting confinement into, with intermittent hopping between, mesoscale compartments (12, 14). Although the concept remains debated (7), micrometric membrane lipid domains have been visualized with a variety of fluorescent lipids on erythrocytes (19, 20), blood platelets (16), and nucleated cells (15, 17–19). Micrometric fluorescent lipid domains were found to depend on cholesterol (19, 27) and to show distinct phase behavior in response to changes of temperature (19, 20).
We recently compared by vital imaging the lateral organization of fluorescent boron dipyrromethene (BODIPY) lipid analogs of GlcCer, SM, and PC at the outer PM leaflet of Chinese hamster ovary (CHO) cells and human red blood cells (RBCs) (19, 20). These studies visualized well-defined brilliant spots corresponding to submicrometric domains (∼0.5–1 μm; i.e., well-above the resolution limit of confocal microscopy), with 5–8-fold enrichment over the rest of the labeled cell surface. In CHO cells, the three analogs segregated into distinct domains, exhibiting decreasing packing (GSLsBODIPY > BODIPY-SM > BODIPY-PC without detectable flip-flop from the outer to the inner leaflet, and covering together up to ∼70% of the cell surface. As shown by inhibition of SM biosynthesis and sphingomyelinase surface cleavage, BODIPY-SM domains depended on endogenous SM, supporting the hypothesis that fluorescent patches could reflect genuine lasting micrometric compartmentation, complementary to the transient nanometric rafts.
The remarkable deformability of RBCs allows squeezing into narrow blood capillaries and the narrower pores of spleen sinusoids, where they undergo >10,000 “quality controls” in their 120-day lifetime. Two properties are essential for RBC stability: a uniquely high cholesterol content (∼40 mol% vs. ∼30 mol% in fibroblasts vs. ∼15 mol% in blood platelets) and a strong membrane:cytoskeleton anchorage. Cholesterol itself is a key regulator of two membrane biophysical properties: i) membrane fluidity via lipid ordering, and ii) membrane deformability via modulation of PM protein interactions at the cortical cytoskeleton interface (28). Interactions between the PM bilayer and the underlying spectrin network dominate elastic properties of RBCs, thanks to two nonredundant 4.1R- and ankyrin-based complexes (29, 30) (see supplementary Fig. I). These provide RBCs with a 20-fold higher adhesive energy than fibroblasts (31). Linkage via 4.1R complexes is regulated by protein kinase C (PKC)-dependent phosphorylation. Ankyrin-based complexes are frequently defective in spherocytosis, a genetic disease impairing resistance to shear stress and hypotonicity. When spherocytosis leads to severe hemolytic anemia, blood counts can be restored by spleen removal (splenectomy).
The present investigation aimed at testing the relevance of micrometric domains labeled by fluorescent lipids at the outer PM leaflet of RBCs for endogenous lipids and analyzing the mechanisms governing their biogenesis. RBCs are the simplest and very robust living cell model, where membrane composition (32) and cytoskeletal control (29, 33, 34) are best characterized and are crucial for cell plasticity and high resistance to shear stress. Analysis of RBC PM is compounded by neither vesicular trafficking nor metabolic lipid turnover. Pharmacological tools and genetic deficiencies in patients allow dissection of the reciprocal interactions between membrane lipids and spectrin. Using RBCs attached onto poly-l-lysine, we first observed that fluorescent GSLs and endogenous GM1, which can be decorated by the fluorescent pentameric cholera toxin B subunit, segregate into visible domains. We next observed the differential effects on the three classes of fluorescent lipid micrometric domains (BODIPY-GlcCer, BODIPY-SM, and BODIPY-PC) of various controlled manipulations: i) stretching, ii) cholesterol depletion, iii) membrane:cytoskeleton uncoupling at 4.1R complexes upon acute PKC activation, and iv) uncoupling at ankyrin complexes in spherocytosis. This was confirmed by preferential association of micrometric domains with antibody-patched components of the two anchorage complexes. These observations further support the differential clustering of polar lipids at the RBC PM up to micrometric domains that can be manipulated by membrane tension and oppositely controlled by cholesterol and membrane:cytoskeleton anchorage. A dual level of lateral asymmetry, transient nanometric rafts and stable micrometric domains, most likely play a crucial role in RBC deformability and vulnerability to membrane fragility diseases such as spherocytosis.
MATERIALS AND METHODS
RBC isolation and immobilization
This study was approved by the Medical Ethics Institutional Committee; each donor gave written informed consent. RBCs were isolated from three healthy volunteers, two with their spleen (named C#2 and C#3) and one splenectomized for traumatic rupture (C#1); and from two splenectomized adult spherocytotic patients (P#1 and P#2). Blood was collected by venopuncture into dry EDTA (K+ salt)-coated tubes, diluted 1:10 in medium (DMEM containing 25 mM glucose and 25 mM HEPES) and washed twice by centrifugation at 133 g for 2 min and resuspension. For spreading onto poly-l-lysine-coated coverslips (PLK-coverslips), RBCs were plated at ∼20 · 106 cells/ml onto 2 cm2 coverslips precoated with 0.1 mg/ml 70–150 kDa PLK (Sigma) at 20°C for exactly 4 min after which the suspension was removed and replaced with fresh medium in which RBCs were allowed to spread for another 4 min (unless otherwise stated). This caused a variable level of stretching exploited in Fig. 1A.
Fig. 1.
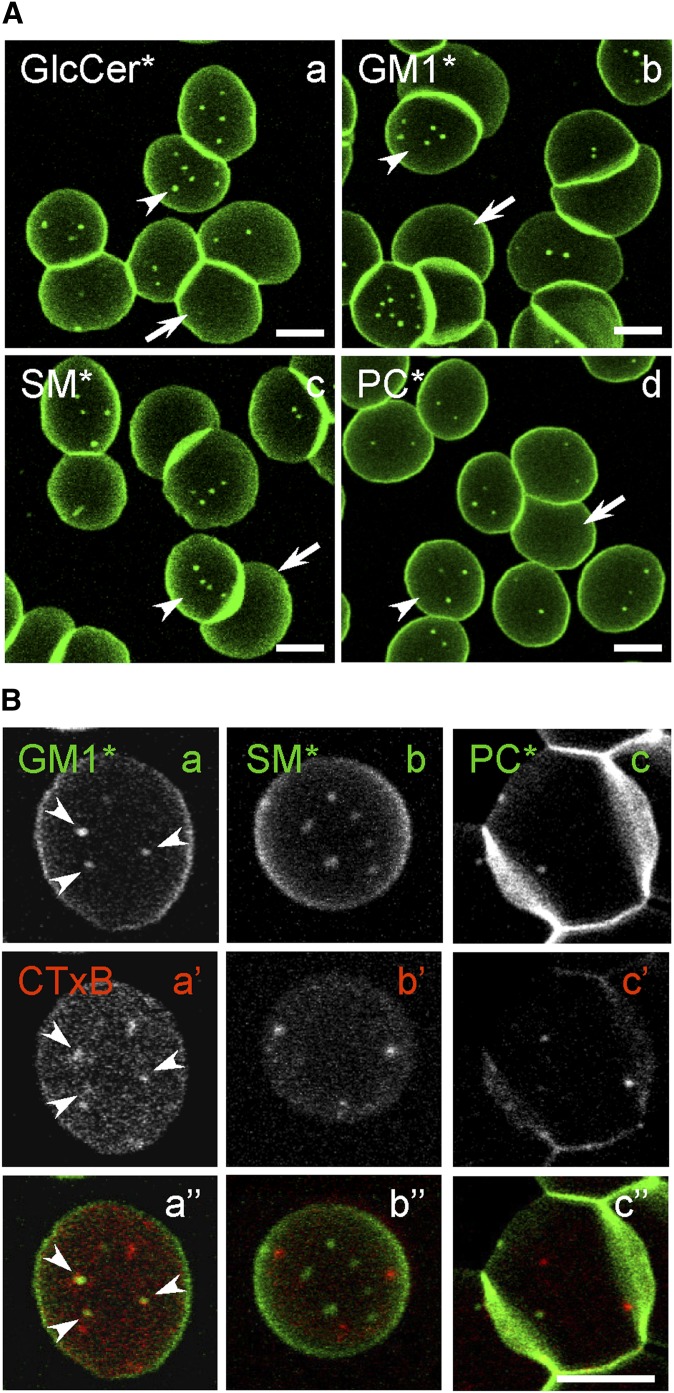
In adherent RBCs, fluorescent exogenous membrane lipids and cholera toxin binding to endogenous GM1 labeled submicrometric domains. A: Single vital imaging of three classes of polar BODIPY-lipid analogs (*). Freshly isolated RBCs (from a normal donor) were allowed to attach onto high-molecular mass (70–150 kDa) PLK-coverslips for 4 min and spread for another 4 min, then briefly labeled with GSLs* [BODIPY-GlcCer (a), GM1* (b)], BODIPY-SM (c), or BODIPY-PC (d), rinsed and immediately imaged. Notice that each lipid analog forms several micrometric domains on partially spread (<8 μm; arrowheads) but not on fully spread RBCs (>8 μm; arrows). For general views, see supplementary Fig. IV (upper row). B: Double vital imaging of BODIPY-lipid domains versus endogenous GM1 (CTxB labeling). RBCs were sequentially labeled in suspension with the indicated BODIPY-lipids (upper row; green at merge) then with Alexa568-CTxB (middle row; red at merge) in the continued presence of BODIPY-lipids. Cells were then attached onto PLK-coverslips before additional spreading, washed and imaged as in Fig. 1A (sequential recording in the green and red channels, with settings adjusted to best match signal intensities), then merged (lower row). Notice that CTxB patches largely colocalize with GM1* (arrowheads at left) but not with BODIPY-SM and BODIPY-PC domains. All scale bars, 5 μm. *, BODIPY.
RBC vital imaging, FRAP, and scanning electron microscopy
In most experiments, RBCs were labeled with BODIPY-lipids after spreading on PLK-coverslips. Briefly, cells were rinsed in DMEM and labeled at 20°C for 15 min with: i) 0.5–1 μM BODIPY- lipids in DMEM containing equimolar defatted BSA (DF-BSA; Sigma) (19); ii) 5 μM Vybrant DiIC18 (Invitrogen) in DMEM with 1 mg/ml DF-BSA (DMEM/BSA); or iii) sequentially BODIPY-SM then DiIC18. For vital imaging of endogenous GM1, RBCs were sequentially labeled in suspension at 37°C for 15 min with BODIPY-lipids then with 4 μg/ml Alexa568-cholera toxin B (CTxB) subunit (Invitrogen) in the continued presence of BODIPY-lipids in DMEM/BSA. RBCs were then spread on PLK-coverslips and washed twice with DMEM/BSA. For confocal imaging, coverslips were placed bottom-up into Lab-Tek chambers and examined with a Zeiss LSM510 confocal microscope using a plan-Apochromat 63× NA 1.4 oil immersion objective in a thermostatically controlled cabinet set at 37 ± 1°C (XL/LSM incubator, Zeiss; Tempcontrol 37-2, PeCon) (19, 20). FRAP and scanning electron microscopy were performed as described (19, 20).
Pharmacological treatments
For cholesterol depletion, RBCs were preincubated in suspension with 0–0.25 mM methyl-β-cyclodextrin (mβCD) (Sigma) at 37°C for 30 min, pelleted, and resuspended in DMEM. RBCs were then either plated onto PLK-coverslips for imaging or lipids were extracted (35) and cholesterol was measured using the Amplex Red Cholesterol kit (Invitrogen) with omission of cholesterol esterase (19). To phosphorylate 4.1R complexes by PKC, RBCs were treated on PLK-coverslips in DMEM containing 20 nM calyculin A (CalA) for 4 min, washed 3 times, reincubated with CalA and 5 μM PMA (both from Sigma) for 5 min, then labeled with BODIPY-lipids and imaged in the continued presence of PMA and CalA for <15 min.
RBC immunolabeling
After spreading on PLK-coverslips, RBCs were labeled with BODIPY-lipids and washed as above. After blocking for 30 min with 1% BSA and 0.1% l-lysine (w/v, BSA/lysine), RBCs were incubated for 1 h with rabbit monoclonal antibody to glycophorin C together with mouse monoclonal antibody to CD47 (both from Abcam), washed 6 times in BSA/lysine, incubated for 1 h with the appropriate Alexa-secondary antibodies (5 μg/ml of each), washed 6 times with PBS, then transferred for 5 min into ice-cold dimethylsuberimidate fixative [extemporaneously prepared at 15 mM in 0.135 M borate buffer containing MgCl2, pH 8.0 (36)] or 4% formaldehyde, rinsed, and mounted in Mowiol in the dark for 24 h. Preparations were examined with an LSM510 confocal microscope as above.
Western blotting
RBC ghosts were prepared at 4°C as published (37), then immediately solubilized in 0.3 M Tris/HCl, pH 6.8, 10% SDS, 50% glycerol, 0.1 M DTT, bromophenol blue, and protease inhibitors and analyzed by western blotting using rabbit antibodies to phosphoS726-α adducin (Abcam), human erythroid ankyrin (Abcam), or aquaporin-1 (Chemicon International) or pan-spectrin mouse monoclonal antibodies (Abcam), followed by HRP-conjugated secondary antibodies and chemoluminescence (ECL kit, PerkinElmer) with Kodak X-Omat blue films and high resolution scanning (Epson Perfection 4990 Photo).
Thin layer chromatography
BODIPY-lipids were inserted in the RBC membrane at 0.75–1 μM. After washing, all lipids (endogenous and inserted BODIPY) were extracted, separated by thin layer chromatography (TLC) in chloroform:methanol:15 mM CaCl2 (65:35:8; v/v/v) (38) and revealed by charring densitometry after staining with 10% cupric sulfate in 8% orthophosphoric acid (39). Band intensity of inserted fluorescent BODIPY-lipids was quantified and expressed by reference to the sum of major lipids (cholesterol, PC, phosphatidylethanolamine, ceramide, and SM) from the same sample, after correction for band intensity of corresponding endogenous lipid.
Statistical analyses
Values are mean ± SEM. Statistical significance of comparisons was tested by Student's t-test.
RESULTS
Spontaneous clustering of GSLsBODIPY, BODIPY-SM, and BODIPY-PC into submicrometric domains on RBCs partially spread onto PLK-coverslips
Labeling of RBCs partially spread on PLK-coverslips with trace levels of BODIPY analogs of GlcCer, SM, or PC (respectively, 0.7 ± 0.5%, 1.1 ± 0.7%, and 1.0 ± 0.3% of RBC major membrane lipids; mean ± SEM pooled from 3 independent experiments) revealed submicrometric domains readily visible by confocal microscopy (Fig. 1A). Increasing BODIPY-SM concentration from 0.5 μM to 3 μM changed neither the number nor the size of micrometric domains (data not shown). In most spread cells (projected diameter >8 μm; arrows in Fig. 1A), lipid analogs were still inserted into the PM but no longer clustered into visible domains. When compared to CHO cells where membrane tension is much lower, fluorescent lipid domains of RBCs were overall less abundant, showed a more uniform size, and had a more round shape, but showed comparable enrichment (>5-fold) over the rest of the labeled cell surface. For each class of BODIPY-lipids, relative domain abundance was undistinguishable between conventional donors and a healthy donor splenectomized for traumatic spleen rupture, but domain number consistently varied according to the lipid analog (4.8 ± 0.1 for GlcCerBODIPY; 3.8 ± 0.2 for BODIPY-SM 3.2 ± 0.3 for BODIPY-PC mean ± SEM of 984–1,323 RBCs pooled from 6 to 10 independent experiments).
Labeling of endogenous GM1 by cholera toxin also reveals submicrometric domains that colocalize with GM1BODIPY
In view of the lasting controversy on the very existence of micrometric domains for endogenous lipids in living cells, as opposed to labeling by fluorescent lipid analogs with a grafted BODIPY acyl tail, we first asked whether endogenous GM1 decorated by the fluorescent CTxB subunit (Alexa 568-CTxB) also segregates into visible domains. Of note, distribution of this probe was reported to be patchy in erythrocytes at both room temperature and 37°C (40). As shown in Fig. 1B (second row), CTxB-labeled submicrometric patches that largely colocalized with GM1BODIPY (arrowheads at left) segregated from BODIPY-SM and BODIPY-PC domains (Fig. 1Bb”, c”). The latter observation also indicated that patches revealed by CTxB were not induced by exogenous GM1BODIPY insertion. These results supported the interpretation that micrometric BODIPY lipid domains could reflect endogenous lipid compartmentation, as previously suggested for BODIPY-SM domains in CHO cells based on endogenous SM depletion (19, 20).
Spontaneous formation of BODIPY lipid submicrometric domains depends on membrane tension
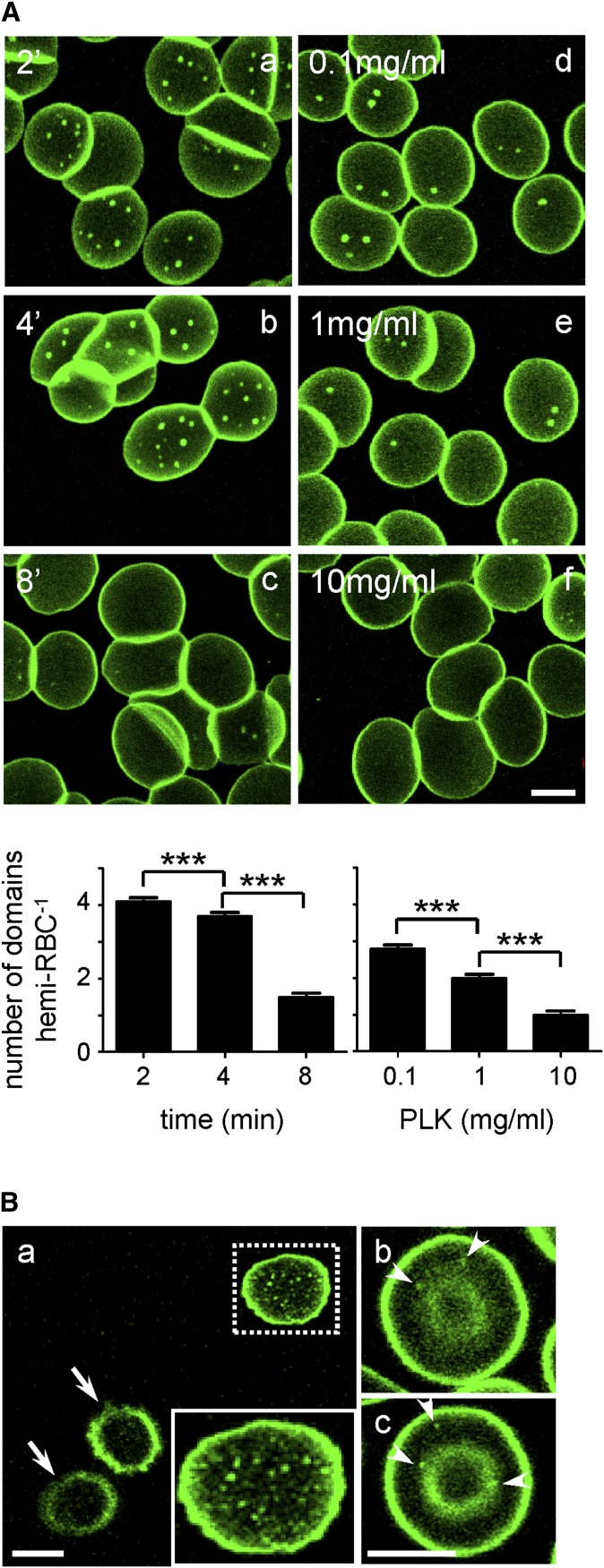
The consistent decrease of BODIPY lipid domain abundance upon RBC spreading suggested a role for membrane tension in BODIPY lipid domain formation/biogenesis. To clarify this role, we tested the relation between membrane stretching due to cell spreading and BODIPY-lipids clustering into submicrometric membrane domains, by titration of the adhesion time onto PLK-coverslips and PLK concentration. Both parameters increased spreading and abrogated domains (illustrated for BODIPY-GlcCer in Fig. 2A). In contrast, the number of domains was greatly increased at very low PLK concentration (0.01 mg/ml; Fig. 2Ba), but large variation in size precluded rigorous quantification. Likewise, micrometric BODIPY-lipid domains could be evidenced on loosely attached stomatocytes/discocytes (Fig. 2Bb, c), ruling out artifact due to PLK-coverslip spreading. We concluded that BODIPY-lipid micrometric domains depended on membrane tension.
Fig. 2.
Spontaneous formation of lipid submicrometric domains is regulated by membrane tension. A: Extended spreading onto PLK-coverslips by increased adhesion time (a–c) or increased PLK concentration (d–f) abrogate micrometric BODIPY (*) lipid domains. Upper panel, representative confocal images. Freshly isolated normal RBCs were attached for 4 min either onto coverslips precoated with 0.1 mg/ml 70–150 kDa PLK with additional spreading time as indicated (a–c), or onto coverslips coated with the indicated concentrations of 1–5 kDa PLK before additional spreading for 4 min (d–f), then labeled with BODIPY-GlcCer (as an example). Notice the highest number of BODIPY-GlcCer domains for shortest times and lowest concentrations of PLK (a, b, d, e) and their disappearance upon extended stretching (c, f). Scale bar, 5 μm. Lower panel, morphometry. Number of BODIPY-GlcCer domains are mean ± SEM of 93–1,144 RBCs from 3 to 6 independent experiments at left and of 37–346 RBCs from 2 independent experiments at right. B: Fluorescent lipids label submicrometric domains on loosely-attached RBCs. RBCs were attached either onto 0.01 mg/ml 1–5 kDa PLK-coverslips for 4 min before additional spreading for 4 min (a), or onto plastic IBIDI chambers coated or not (b) with 1–5 kDa PLK (c) and labeled with BODIPY-GlcCer. Although most RBCs appear as echinocytes [arrows in (a)], the rare flat RBCs show numerous domains with variable size at very low PLK concentration [inset in (a)]. Domains can also be found on barely-attached discocytes/stomatocytes [arrowheads in (b, c)]. Scale bars, 5 μm. *** p < 0.001.
The high cholesterol level of RBCs promotes BODIPY-SM and BODIPY-PC domains
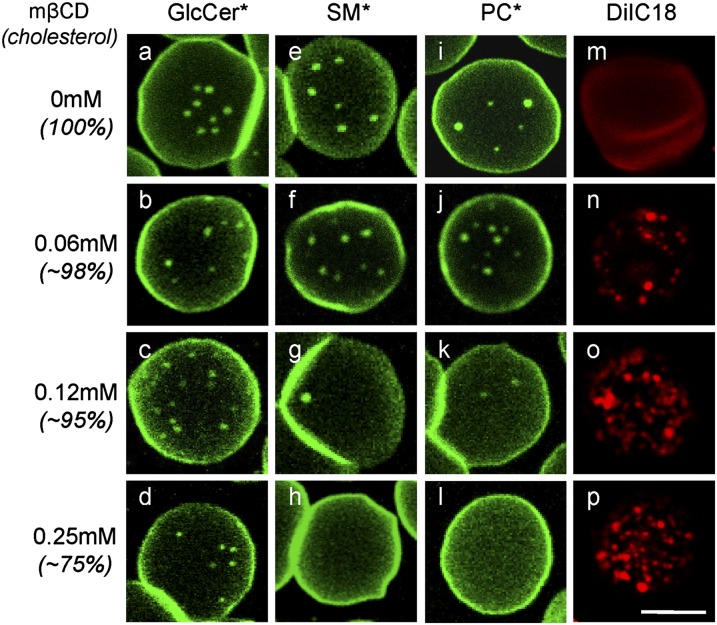
We next turned our attention to cholesterol because of its particular abundance at the RBC PM where it could regulate membrane tension by modulating both lateral membrane fluidity and transversal anchorage to the cytoskeleton (28, 41). Cholesterol abundance can be precisely decreased by controlled extraction with empty mβCD. An ∼25% cholesterol depletion was obtained by incubating RBCs in suspension with 0.25 mM mβCD (supplementary Fig. IIIAa), without hemolysis (supplementary Fig. IIIAb) or a detectable effect on subsequent BODIPY-lipid insertion (supplementary Fig. IIIAc). This moderate cholesterol depletion totally abrogated BODIPY-SM (Fig. 3g, h) and BODIPY-PC domains (Fig. 3k, l) but BODIPY-GlcCer domains were largely preserved (Fig. 3a–d; for general views, see supplementary Fig. IIIB). Thus, cholesterol depletion specifically impaired the less-packed BODIPY-PC and BODIPY-SM domains and spared the most-packed BODIPY-GlcCer domains, packing of domains being previously suggested by the propensity of BODIPY-lipids to form excimers (20). To further test whether cholesterol depletion could instead favor other domains, we looked at the artificial dialkylindocarbocyanine DiIC18, a well-established membrane probe that homogenously labels untreated RBCs (19, 42), but which is recognized to preferentially partition into the liquid-disordered phase in giant unilamellar vesicles, segregated from the liquid-ordered phase decorated by fluorescent cholera toxin that binds ganglioside GM1 (8). Using a minimal mβCD concentration (0.06 mM) that barely affected total cholesterol (supplementary Fig. IIIAa), DiIC18 already started concentrating into bright submicrometric patches (Fig. 3n–p). At low mβCD concentrations that combined preservation of BODIPY-SM domains with induction of DiIC18 patches, the two types of assemblies were fully segregated (supplementary Fig. IIIC). We concluded that the high cholesterol level of RBCs favored BODIPY-SM and BODIPY-PC micrometric domain formation, also implying that local cholesterol variations could finely modulate their stability.
Fig. 3.
Moderate cholesterol depletion suppresses BODIPY-SM and BODIPY-PC, but not BODIPY-GlcCer, domains and induces DiIC18 patches. Normal RBCs were treated in suspension with the indicated mβCD concentrations (for level of depletion and lack of toxicity, see supplementary Fig. IIIA), then attached onto PLK-coverslips for 4 min, spread for an additional 4 min, labeled with BODIPY-GlcCer (a–d), BODIPY-SM (e–h), BODIPY-PC (i–l), or DiIC18 (m–p), rinsed, and imaged as in Fig. 1A. Notice the disappearance of BODIPY-SM and BODIPY-PC domains under moderate (∼25%) cholesterol depletion (g, h, k, l) contrasting with better preservation of BODIPY-GlcCer domains (a–d), and conversely, induction of DiIC18 patches (m–p). Scale bar, 5 μm. For general views, see supplementary Fig. IIIB. *, BODIPY.
BODIPY-lipid domains are immobile clusters of highly mobile constituents
To analyze the stability of BODIPY-lipid domains and their constituents, we next combined time-lapse imaging with FRAP (19, 20). Prolonged imaging revealed that domains were remarkably immobile in the adherent RBCs (examples in Fig. 4A). Moreover, bleached domains were regenerated exactly at their original position. Domain stability contrasted with the very fast (t1/2 ∼10 sec) and high recovery of their constituents (Fig. 4B), indicating that domains were stable large-scale assemblies of highly dynamic lipids or small clusters nondetectable by conventional confocal microscopy. We thus looked at a relation between domain stability and membrane anchorage to the underlying spectrin network, by addressing the respective contribution of the two nonredundant 4.1R- and ankyrin-based complexes.
Fig. 4.
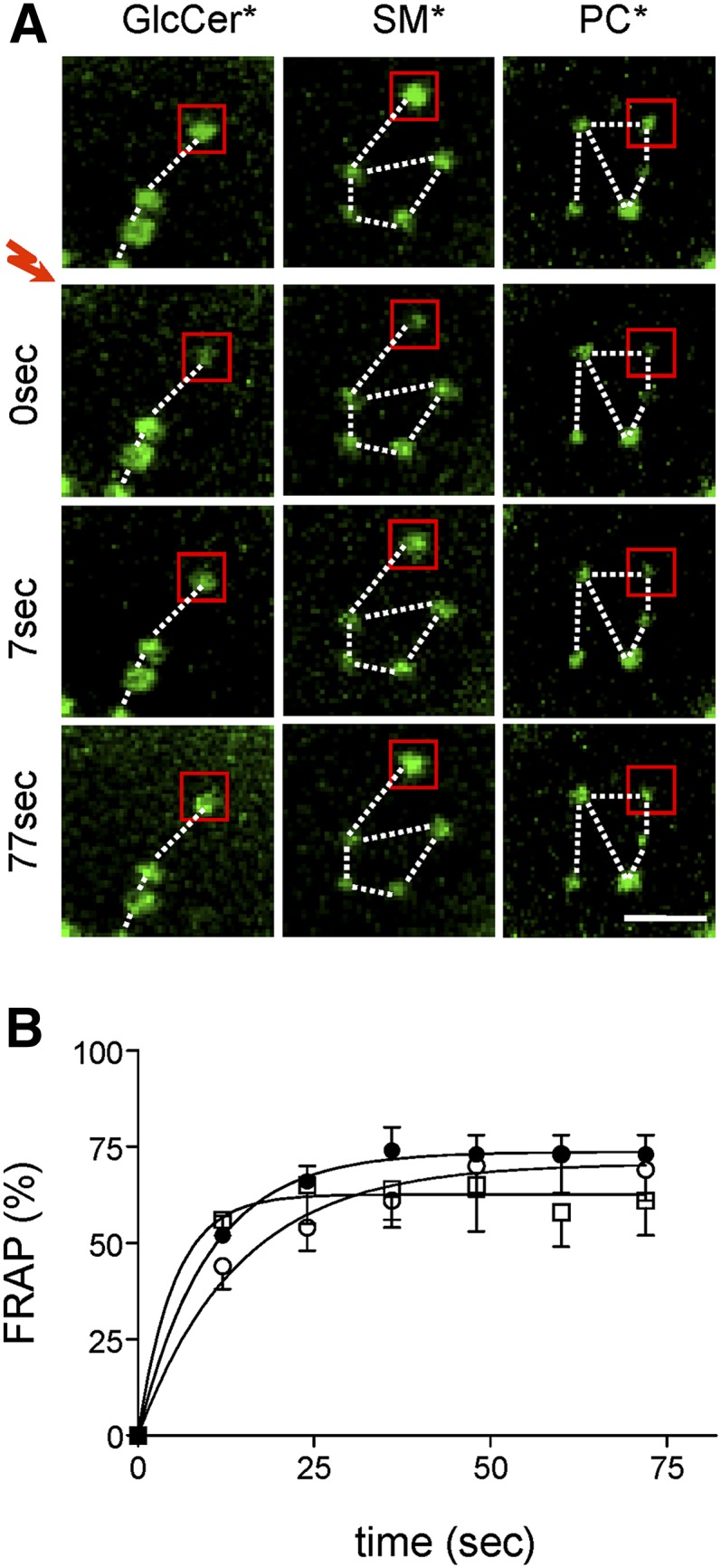
Micrometric BODIPY-lipid domains assume fixed positions but rapidly exchange their constituents. Normal RBCs were attached onto PLK-coverslips, spread, and labeled with BODIPY-GlcCer [left column in (A); open squares in (B)], BODIPY-SM [central column in (A); closed circles in (B)], or BODIPY-PC [right in (A); open circles in (B)], then processed for FRAP at 37°C in 5 μm2 fields centered on a micrometric domain [exemplified by red squares in (A)]. A: Representative vital imaging. Red flash indicates bleaching of areas shown by red squares. Scale bar, 2 μm. Notice domain immobility (fixed positions as in constellations, dotted lines). B: Quantification. Fluorescence recovery is expressed as percentage of signal before photobleaching, after correction for residual fluorescence immediately after bleaching. Curves are derived by monoexponential fitting. Notice the fast (t1/2 ∼10 s) and extensive fluorescence recovery of SL* (BODIPY-GlcCer, BODIPY-SM) domains in bleached areas, indicating high mobility of constituents. *, BODIPY.
Membrane:spectrin anchorage at 4.1R complexes restricts BODIPY-GlcCer and BODIPY-SM domains
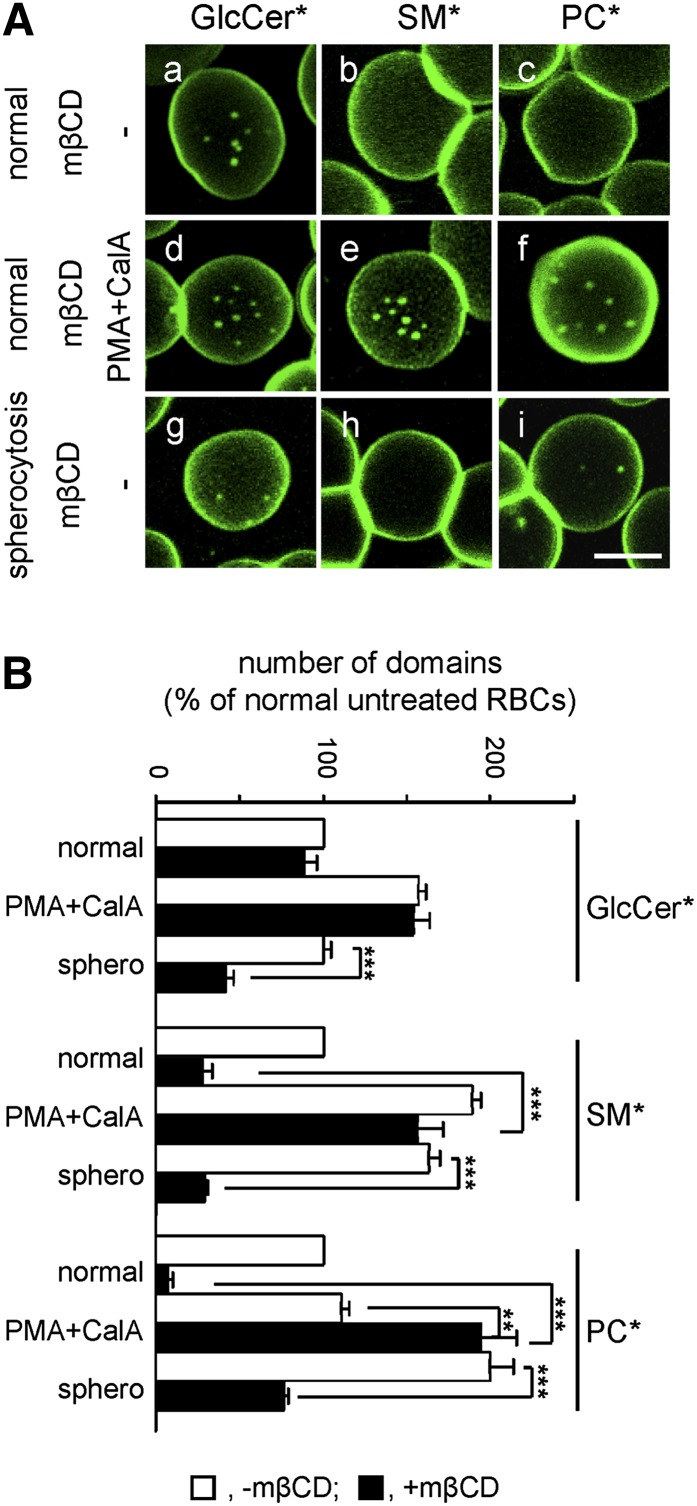
Membrane:spectrin anchorage via 4.1R complexes can be acutely and selectively uncoupled by acute phosphorylation upon PKC activation via phorbol 12-myristate 13-acetate (PMA) combined with CalA as phosphatase inhibitor (43). As shown in supplementary Fig. IIB, this treatment rapidly induced a robust and stable phosphorylation of α-adducin, a key component of the 4.1R complex (see supplementary Fig. I) (44). Scanning electron microscopy further disclosed circumferential retraction (arrowhead in supplementary Fig. IIC, right) and central bulging (arrow), compatible with decreased spreading and membrane tension upon partial release from the spectrin network, with preservation of a featureless smooth RBC surface. PKC activation increased the abundance of BODIPY-GlcCer domains by ∼50% (Fig. 5Ad) and BODIPY-SM domains by ∼90% (Fig. 5Ae) without a detectable effect on BODIPY-PC domains (Fig. 5Af). General views are provided in supplementary Fig. IVb, e, h and quantification is shown at Fig. 5B. Thus, because uncoupling of membrane:spectrin anchorage at 4.1R complexes selectively increased the number of BODIPY-GlcCer and BODIPY-SM domains, we concluded that, in untreated cells, 4.1R normally restricts the spontaneous propensity for BODIPY-GlcCer and BODIPY-SM clustering.
Fig. 5.
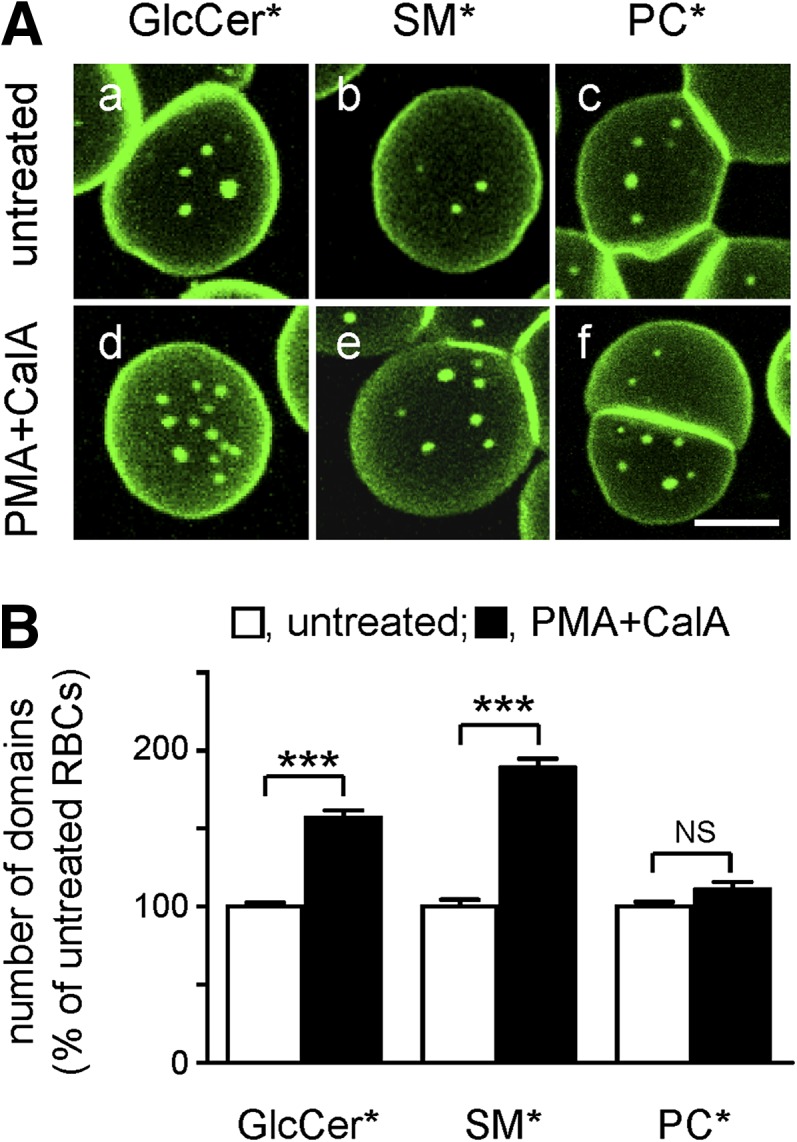
Anchorage via 4.1R-based complexes restricts BODIPY-GlcCer and BODIPY-SM, but not BODIPY-PC, domains. A: Representative vital imaging. RBCs from a normal donor were attached onto PLK-coverslips and spread as in Fig. 1, then either kept untreated (a–c) or treated with PMA as PKC activator and the phosphatase inhibitor CalA to uncouple membrane:cytoskeleton at 4.1R complexes (d–f), labeled with BODIPY-GlcCer, BODIPY-SM, or BODIPY-PC and imaged (in the continued presence of PMA+CalA if appropriate). Scale bar, 5 μm. For general views, see supplementary Fig. IVb, e, h. B: Morphometry. Abundance of BODIPY-GlcCer, BODIPY-SM, and BODIPY-PC domains on untreated (open bars) and PMA+CalA-treated RBCs (filled bars) is expressed as the percentage of untreated cells and are mean ± SEM of: i) 296 (untreated) and 361 (PMA+CalA) RBCs from 3 independent experiments for BODIPY-GlcCer domains; ii) 256 and 471 from 5 experiments for BODIPY-SM domains; and iii) 238 and 117 from 2 experiments for BODIPY-PC domains. *, BODIPY; NS, not significant. *** p < 0.001.
Membrane:spectrin anchorage at ankyrin complexes restricts BODIPY-SM and BODIPY-PC domains
In parallel, to evaluate the role of ankyrin-based complexes, we examined the organization of BODIPY-lipids in RBCs from two splenectomized spherocytotic patients (P#1 and P#2) versus two control adults of comparable age, one splenectomized for traumatic spleen rupture (C#1) and one not (C#2). P#2 suffered from a previously described single point mutation (c.3157C>T) at exon 28 of the ankyrin-1 gene (45). This mutation generates a stop codon within the minimal ZU5-ANK binding domain for β-spectrin (46) (supplementary Fig. IIF). Although no mutation was found in any of the 42 exons of the ankyrin-1 gene of P#1, RBCs from both patients showed a comparable ∼2-fold lower ankyrin level without obvious spectrin or band 3 deficiency (supplementary Fig. IID, E), and exhibited a similar osmotic fragility (supplementary Fig. IIG).
RBCs from the two spherocytotic patients showed normal abundance of BODIPY-GlcCer domains (illustrated for P#1 in Fig. 6Ad) but increased (∼60–100%) abundance of BODIPY-SM and BODIPY-PC domains (illustrated for P#1 in Fig. 6Ae, f). General views are provided in supplementary Fig. IVc, f, i and quantification is shown in Fig. 6B. These results indicated that ankyrin-based complexes preferentially restricted BODIPY-SM and BODIPY-PC clustering into submicrometric domains.
Fig. 6.
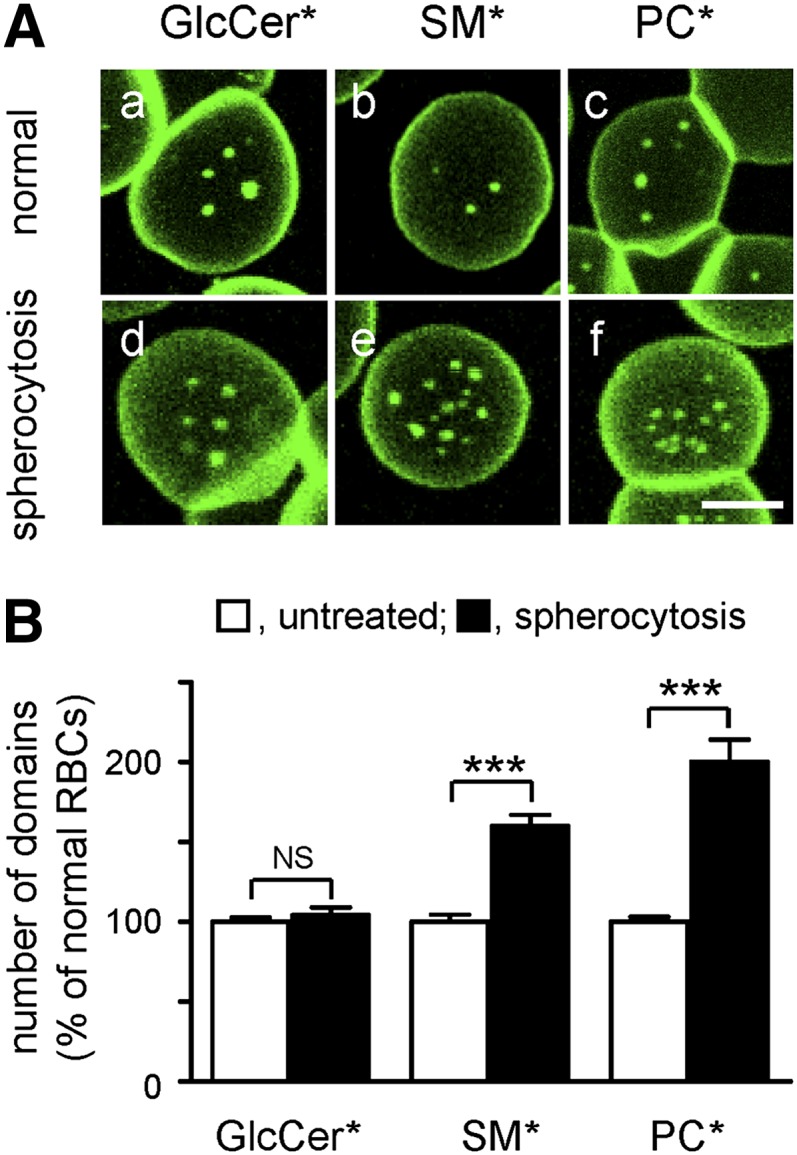
Anchorage via ankyrin-based complexes restricts BODIPY-SM and BODIPY-PC, but not BODIPY-GlcCer, domains. A: Representative vital imaging. RBCs from a normal donor (normal; a–c) versus a spherocytotic patient (spherocytosis; d–f), both splenectomised, were attached onto PLK-coverslips, spread, and labeled with BODIPY-GlcCer, BODIPY-SM, or BODIPY-PC for vital imaging. Scale bar, 5 μm. For general views, see supplementary Fig. IV, lower row. B: Morphometry. Abundance of BODIPY-GlcCer, BODIPY-SM, and BODIPY-PC domains on normal (open bars) and spherocytotic RBCs (filled bars) is expressed by reference to normal RBCs and are mean ± SEM of: i) 200 and 558 RBCs for BODIPY-GlcCer domains; ii) 114 and 454 RBCs for BODIPY-SM domains; and iii) 114 and 398 RBCs for BODIPY-PC domains, each from 2 independent experiments. *, BODIPY; NS, not significant. *** p < 0.001.
High cholesterol level promotes BODIPY-SM and BODIPY-PC domains via 4.1R complexes and BODIPY-GlcCer domains via ankyrin complexes
Conversely, to test whether BODIPY-GlcCer domains would instead be modulated by the cholesterol:ankyrin interplay, a similar experiment was performed on P#1 spherocytotic RBCs. Whereas BODIPY-GlcCer domains of normal RBCs largely resisted mβCD (see above, Fig. 3a–d), they were sensitized by spherocytosis to cholesterol depletion (Fig. 7Ag; quantification in Fig. 7B). In contrast, mβCD equally decreased BODIPY-SM and BODIPY-PC domains both in normal and spherocytotic RBCs (Fig. 7Ab, c, h, i; quantification at Fig. 7B). Thus, cholesterol:ankyrin interplay could promote BODIPY-GlcCer domains, suggesting preferential association of BODIPY-GlcCer domains with ankyrin complexes.
Fig. 7.
High cholesterol promotes BODIPY-SM and BODIPY-PC domains via 4.1R complexes and BODIPY-GlcCer domains via ankyrin complexes. A: Representative vital imaging. RBCs from a normal donor (normal; a–f) versus a spherocytotic patient (P#1; spherocytosis; g–i), both splenectomised, were preincubated in suspension with 0.25 mM mβCD as in Fig. 3, attached onto PLK-coverslips, spread, labeled with BODIPY-GlcCer (a, d, g), BODIPY-SM (b, e, h), or BODIPY-PC (c, f, i), then either kept in fresh medium (a–c, g–i) or further incubated with PMA+CalA (d–f) as in Fig. 5. Scale bar, 5 μm. B: Morphometry. Abundance of BODIPY-GlcCer, BODIPY-SM, or BODIPY-PC domains in the absence (open bars) or presence of mβCD (filled bars) is expressed by reference to untreated normal RBCs and are mean ± SEM of: i) 37–361 RBCs from 1 to 5 independent experiments for BODIPY-GlcCer domains; ii) 50–471 RBCs from 2 to 6 experiments for BODIPY-SM domains; and iii) 35–300 RBCs from 2 to 6 experiments for BODIPY-PC domains. *, BODIPY. ** p < 0.01; *** p < 0.001.
Glycophorin C (4.1R complex) overlaps with BODIPY-SM domains and circumscribes BODIPY-PC domains
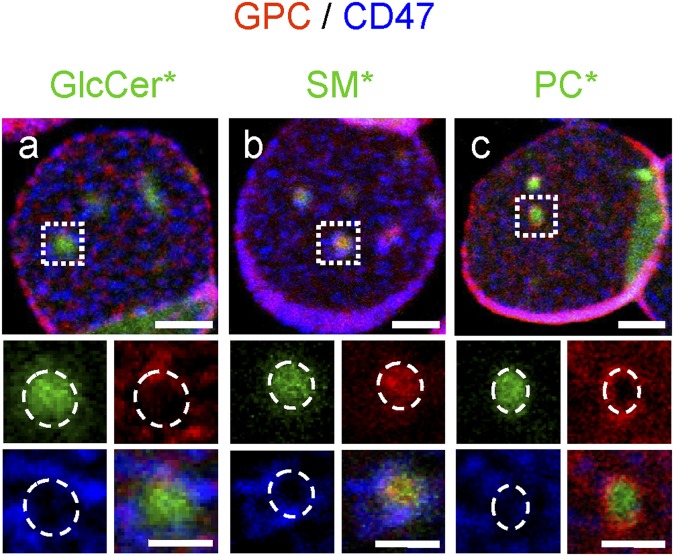
To look more directly for preferential protein:lipid interactions at the outer PM leaflet, we tested the association of BODIPY-lipid domains with antibody-induced patches of the two anchorage complexes by targeting glycophorin C (GPC) for the 4.1R-based and CD47 for ankyrin-based complexes (see supplementary Fig. I). Patches were previously observed for CD47 (47, 48). GPC and CD47 patches were clearly segregated from one another (compare red and blue labeling in Fig. 8) and differentially associated with BODIPY-lipid domains. BODIPY-SM domains largely colocalized with GPC patches (4.1R complex; red in Fig. 8b), whereas BODIPY-PC domains were instead closely circumscribed by GPC patches (red in Fig. 8c). In contrast, CD47 patches were never as tightly linked to BODIPY-lipid domains, but frequently seemed in closer proximity to BODIPY-GlcCer and BODIPY-SM (ankyrin complex; blue in Fig. 8a, b) than to BODIPY-PC domains (Fig. 8c). We concluded that differential association of BODIPY-lipid domains with patched proteins of the 4.1R and ankyrin complexes supported the concept of preferential protein:lipid interaction, as predicted from differential inactivation of the two anchorage complexes upon cholesterol depletion.
Fig. 8.
Differential association of micrometric BODIPY-lipid domains with patched glycophorin C and CD47. RBCs from a normal donor were attached onto PLK-coverslips, labeled with BODIPY-lipid (green), then GPC (4.1R complex; red) and CD47 (ankyrin complex; blue) were patched by antibodies. Notice that GPC patches selectively overlapped BODIPY-SM domains (b), while circumscribing BODIPY-PC domains (c). In contrast, CD47 patches were never as tightly linked to lipid-domains, but frequently seemed in closer proximity to BODIPY-GlcCer and BODIPY-SM (a, b) than to BODIPY-PC domains (c). Scale bars, 2 μm; at insets, 1 μm. Dashed circles at insets indicate lipid* domain boundaries. *, BODIPY.
DISCUSSION
Overview
The classical model of the organization of lipids in biological membranes as a homogenous bilayer has been abandoned due to clear evidence of lateral heterogeneity and clustering mechanisms (2, 7, 49). However, knowledge remains scarce on: i) the actual level and stability of lateral membrane lipid organization in living cells (i.e., transient nanometric rafts vs. more stable submicrometric/mesoscale domains); ii) integration of various assembly/stabilization/restriction mechanisms; and iii) potential roles in disease, due to limitations of available tools and methods, and to the diversity of cells studied so far. Here we addressed the biological relevance, mechanisms, and relation to a membrane fragility disease of micrometric domains resulting from spontaneous partitioning of fluorescent (BODIPY) analogs of three major classes of polar lipids, GSLs (GlcCer and GM1), SM, and PC, when inserted at trace levels into the outer membrane of partially spread RBCs as a simple, robust, and powerful experimental system. Our observations yield new insights on the role of tension, cholesterol enrichment, and preference for proteins of two nonredundant anchorage complexes in micrometric lipid organization.
To what extent do BODIPY-lipid domains reflect endogenous lateral compartmentation?
The validity of exogenous fluorescent polar lipid analogs based on partial replacement of an acyl chain by a BODIPY fluorophore of comparable length as bona fide reporters of endogenous membrane lipids has long been questioned (50). However, polar heads are also important for lateral lipid segregation so that their substitution for alternative fluorophore labeling is not without potential pitfalls. The impact of sphingolipid headgroups on lateral lipid organization has been demonstrated in artificial membranes (51). Moreover, we recently reported that BODIPY-GlcCer/GM1BODIPY and BODIPY-SM, built on the very same BODIPY-ceramide backbone and differing only by their respective polar head, segregated into distinct domains exhibiting different properties (20). We further reported that BODIPY-SM micrometric domains spontaneously formed in CHO cells, irrespective of direct BODIPY-SM surface insertion versus secondary exocytosis after Golgi synthesis from BODIPY-ceramide and selectively vanished upon depletion of endogenous SM upon biosynthetic inhibition or surface cleavage, indirectly suggesting that BODIPY-SM domains somehow reflected some endogenous SM compartmentation in a living cell (19). Now, we show that direct labeling of endogenous GM1 at the RBC PM by fluorescent CTxB yielded submicrometric domains similar to those seen after insertion of exogenous GM1BODIPY, and that both are largely colocalized. In addition, differential modulation of the three BODIPY-lipid domains in RBCs by temperature (19, 20), stretching, cholesterol, and membrane:cytoskeleton anchorage not only points to a key role of lipid phases and membrane tension in domains biogenesis/maintenance, but also rules out that domains are simply fluorescent lipid aggregates. We and others further noticed that the size and stability of BODIPY-lipid domains evidenced in RBCs strongly resembled those recorded at the surface of yeast expressing appropriate green fluorescent protein-tagged membrane proteins (21, 52, 53). Thus, while available data do not allow us to conclude that micrometric BODIPY-lipid domains faithfully reflect the level of the organization of corresponding endogenous lipids, convergent evidence indicates that both must be linked. The level of fidelity of BODIPY-lipids deserves to be further evaluated, e.g., by comparison with the labeling of endogenous lipids using monovalent toxin-derived probes combined with super-resolution microscopy (6), even though this approach is not suited for dynamic vital imaging. We will now discuss mechanistic information gained from the study of micrometric BODIPY-lipid domains.
BODIPY-lipid domains are regulated by membrane tension
We first found that membrane tension strongly impacted on the number and size of visible BODIPY domains formed by the three main classes of polar BODIPY-lipids. Domains were numerous and polydisperse when RBCs were barely attached, declined to a stable low number with monodisperse size at partial spreading, and vanished upon maximal stretching. Conversely, when membrane tension was relaxed by uncoupling either of the two spectrin anchorage complexes, the number of BODIPY domains increased, albeit to a different extent, according to the BODIPY-lipid class as will be discussed below. Biophysical studies should address the mechanical parameters governing the relation between membrane tension and BODIPY domains cohesion and size in RBCs.
BODIPY-lipid domains are differentially promoted by cholesterol
Upon moderate cholesterol depletion, BODIPY-SM and BODIPY-PC domains disappeared, but BODIPY-GlcCer domains were largely preserved. This indicates that cholesterol can finely tune the lateral heterogeneity of membrane lipids in a living cell, not only to stabilize nanometric rafts but also up to a micrometric scale. It remains to be clarified to what extent the very stable micrometric GSLBODIPY and BODIPY-SM domains of RBCs result from the coalescence of nanometric rafts, due to the particularly high cholesterol level and strong membrane:cytoskeleton anchorage.
We suggest that the higher vulnerability of BODIPY-SM and BODIPY-PC domains to cholesterol depletion may reflect a looser intrinsic packing, as already indicated by their weaker propensity than BODIPY-GlcCer domains to generate excimers in CHO cells (20). Conversely, the relative resistance of BODIPY-GlcCer domains to mβCD likely reflects their higher intrinsic packing. Refractoriness of BODIPY-GlcCer domains to mβCD cannot be explained by selective inaccessibility (e.g., by cholesterol shielding under extended polar heads of complex GSLs), because we found that BODIPY-GlcCer domains were sensitive to mβCD in spherocytotic RBCs where lipid composition was unaltered (54). Taking together our comparative data on excimers (20) and sensitivity to mβCD (this paper), we suggest that the lower the packing of BODIPY lipid domains, the higher their dependence on cholesterol.
BODIPY-lipid domains are differentially restricted by the two nonredundant anchorage complexes to spectrin
Fixed positions lasting for at least a half hour and reappearance after bleaching at the exact same place indicated that BODIPY-lipid domains at the outer membrane leaflet were strongly linked to the underlying spectrin network, suggesting dependence on transmembrane components of the two anchorage complexes. Acute uncoupling at 4.1R complexes selectively increased the abundance of GlcCerBODIPY and BODIPY-SM but not BODIPY-PC domains, indicating that the network of 4.1R-based anchors in untreated RBCs preferentially restricts BODIPY-GlcCer and BODIPY-SM coalescence. Alternatively, ankyrin deficiency in two unrelated spherocytotic patients selectively increased the abundance of BODIPY-SM and BODIPY-PC but did not enhance BODIPY-GlcCer domains, indicating that ankyrin preferentially restricts BODIPY-SM and BODIPY-PC coalescence.
BODIPY-lipid domains show preferential protein association and are finely tuned by cholesterol and anchorage interplay
The large colocalization of patched glycophorin C within BODIPY-SM domains and its close association with the borders of BODIPY-PC domains suggested that assembly of BODIPY-SM and BODIPY-PC domains, which are both restricted by ankyrin complexes (see above), could instead be promoted by 4.1R complexes in two different ways, “within” the domains or “around” them. Combination of cholesterol depletion with differential anchorage uncoupling provided additional information. The reappearance (and even doubling, as compared with untreated cells) of BODIPY-SM and BODIPY-PC domains in cholesterol-depleted RBCs upon acute uncoupling of 4.1R complexes, but not in ankyrin-deficient cells, suggested that cholesterol selectively modulates membrane lipid:4.1R interactions, thereby favoring BODIPY-SM and BODIPY-PC domains, in agreement with previous reports demonstrating that cholesterol not only regulates lateral membrane fluidity, but also regulates transversal membrane:cytoskeleton anchorage (28, 41). Cholesterol may regulate membrane:cytoskeleton coupling via its strong direct interaction with band 3, as shown in mixed phospholipid:cholesterol monolayers (55, 56). Cholesterol has also been shown to indirectly affect spectrin:phospholipid interactions in a similar system (57). In addition, because cholesterol depletion was shown to disrupt the lateral organization of phosphatidylinositol-4,5-biphosphate (PIP2) at the PM (58, 59), and because 4.1R contains a PIP2-binding motif (60), it was tempting to speculate that cholesterol could impact on 4.1R:lipid interaction. By complementarity, the selective decrease of BODIPY-GlcCer domains by mβCD in spherocytosis, but not in normal RBCs, would suggest that BODIPY-GlcCer domains are promoted by cholesterol at ankyrin-based complexes. Taken together, our observations revealed that the three types of BODIPY-lipid domains showed differential preference for the two types of nonredundant anchorage complexes, as shown by copatching experiments and functional destabilization studies based on cholesterol depletion and selective anchorage uncoupling. It should be possible to clarify the issue of BODIPY lipid:protein preference based on biochemical studies after differential domain budding into microvesicles and fractionation (see below).
Current models for micrometric BODIPY-lipid domain biogenesis, functions, and role in disease
Biological membrane fluidity depends on several levels of heterogeneity: i) complex composition covering a broad Tm range; ii) differential clustering at the nanometric versus the micrometric scale; and iii) transversal anchorage to a dynamic cortical cytoskeleton that can be modulated. For RBCs, we here showed that a combinatorial interplay of moderate variations in (local?) cholesterol concentrations with differential preference to, or restriction by, the two anchorage complexes and their regulated linkage to the spectrin network can finely tune three types of micrometric assemblies of polar membrane BODIPY-lipids. One can thus interpret BODIPY-lipid domain assembly in erythrocytes as essentially governed by two complementary mechanisms that were recently proposed to generate dynamic lateral heterogeneity (7, 49): i) differential intrinsic lipid cohesion, and ii) regulated linkage to the spectrin network thanks to preferential interaction with two nonredundant anchorage complexes and providing either internal stabilization (e.g., BODIPY-SM or peripheral retention (e.g., BODIPY-PC Finally, domain expansion would be limited by fences and membrane tension.
The significance of micrometric BODIPY-lipid domains for RBC deformability and membrane fragility diseases may be viewed by two opposite, yet not mutually exclusive models. In the first one, packed domains would favor membrane resilience by providing a reservoir when lipid spacing is needed for membrane deformation, e.g., for crossing spleen sinusoid pores, somehow analogous to the recruitment of caveolae upon endothelial cell stretching (61, 62). In the second model, domain formation would reflect a propensity to local rupture, favoring release of microvesicles (63) known as microspherocytes in spherocytosis. Based on this well-known fact, we recently found that controlled perturbations can trigger differential budding from the three types of BODIPY-lipid domains in RBCs, which may pave the way to definition of their respective biochemical composition and biophysical properties.
Supplementary Material
Acknowledgments
The authors thank A. Cominelli, A. Debue, and J. Van Hees for expert technical assistance.
Footnotes
Abbreviations:
- BODIPY
- boron dipyrromethene
- CalA
- calyculin A
- CHO
- Chinese hamster ovary
- CTxB
- cholera toxin B
- FRAP
- fluorescence recovery after photobleaching
- GlcCer
- glucosylceramide
- GM1
- monosialotetrahexosylganglioside
- GPC
- glycophorin C
- GSL
- glycosphingolipid
- mβCD
- methyl-β-cyclodextrin
- PC
- phosphatidylcholine
- PIP2
- phosphatidylinositol-4,5-biphosphate
- PKC
- protein kinase C
- PLK
- poly-l-lysine
- PLK-coverslips
- poly-l-lysine-coated coverslips
- PM
- plasma membrane
- RBC
- red blood cell
- SL
- sphingolipid
This work was supported by grants from the Université catholique de Louvain, Fonds de la Recherche Scientifique/Fonds National de la Recherche Scientifique (F.R.S./FNRS), Région Wallonne, Région Bruxelloise, Loterie Nationale, IUAP, and the Salus Sanguinis Foundation (all Belgium). The corresponding author declares, on the behalf of all authors, that there were no financial, personal, or professional interests that could be construed to have influenced this paper.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of eight figures.
REFERENCES
- 1.Singer S. J., Nicolson G. L. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175: 720–731. [DOI] [PubMed] [Google Scholar]
- 2.Lingwood D., Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science. 327: 46–50. [DOI] [PubMed] [Google Scholar]
- 3.Lingwood D., Ries J., Schwille P., Simons K. 2008. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc. Natl. Acad. Sci. USA. 105: 10005–10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pike L. J. 2006. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47: 1597–1598. [DOI] [PubMed] [Google Scholar]
- 5.Fujita A., Cheng J., Hirakawa M., Furukawa K., Kusunoki S., Fujimoto T. 2007. Gangliosides GM1 and GM3 in the living cell membrane form clusters susceptible to cholesterol depletion and chilling. Mol. Biol. Cell. 18: 2112–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizuno H., Abe M., Dedecker P., Makino A., Rocha S., Ohno-Iwashita Y., Hofkens J., Kobayashi T., Miyawaki A. 2011. Fluorescent probes for superresolution imaging of lipid domains on the plasma membrane. Chem Sci. 2: 1548–1553. [Google Scholar]
- 7.Bagatolli L. A., Ipsen J. H., Simonsen A. C., Mouritsen O. G. 2010. An outlook on organization of lipids in membranes: searching for a realistic connection with the organization of biological membranes. Prog. Lipid Res. 49: 378–389. [DOI] [PubMed] [Google Scholar]
- 8.Kahya N., Scherfeld D., Bacia K., Poolman B., Schwille P. 2003. Probing lipid mobility of raft-exhibiting model membranes by fluorescence correlation spectroscopy. J. Biol. Chem. 278: 28109–28115. [DOI] [PubMed] [Google Scholar]
- 9.Baumgart T., Hess S. T., Webb W. W. 2003. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 425: 821–824. [DOI] [PubMed] [Google Scholar]
- 10.Goñi F. M., Alonso A. 2009. Effects of ceramide and other simple sphingolipids on membrane lateral structure. Biochim. Biophys. Acta. 1788: 169–177. [DOI] [PubMed] [Google Scholar]
- 11.Busto J. V., Sot J., Requejo-Isidro J., Goni F. M., Alonso A. 2010. Cholesterol displaces palmitoylceramide from its tight packing with palmitoylsphingomyelin in the absence of a liquid-disordered phase. Biophys. J. 99: 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kusumi A., Suzuki K. G., Kasai R. S., Ritchie K., Fujiwara T. K. 2011. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem. Sci. 36: 604–615. [DOI] [PubMed] [Google Scholar]
- 13.Yechiel E., Edidin M. 1987. Micrometer-scale domains in fibroblast plasma membranes. J. Cell Biol. 105: 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiwara T., Ritchie K., Murakoshi H., Jacobson K., Kusumi A. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157: 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh R. D., Liu Y., Wheatley C. L., Holicky E. L., Makino A., Marks D. L., Kobayashi T., Subramaniam G., Bittman R., Pagano R. E. 2006. Caveolar endocytosis and microdomain association of a glycosphingolipid analog is dependent on its sphingosine stereochemistry. J. Biol. Chem. 281: 30660–30668. [DOI] [PubMed] [Google Scholar]
- 16.Gousset K., Wolkers W. F., Tsvetkova N. M., Oliver A. E., Field C. L., Walker N. J., Crowe J. H., Tablin F. 2002. Evidence for a physiological role for membrane rafts in human platelets. J. Cell. Physiol. 190: 117–128. [DOI] [PubMed] [Google Scholar]
- 17.Hao M., Mukherjee S., Maxfield F. R. 2001. Cholesterol depletion induces large scale domain segregation in living cell membranes. Proc. Natl. Acad. Sci. USA. 98: 13072–13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaus K., Gratton E., Kable E. P., Jones A. S., Gelissen I., Kritharides L., Jessup W. 2003. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc. Natl. Acad. Sci. USA. 100: 15554–15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyteca D., D'Auria L., Der Smissen P. V., Medts T., Carpentier S., Monbaliu J. C., de Diesbach P., Courtoy P. J. 2010. Three unrelated sphingomyelin analogs spontaneously cluster into plasma membrane micrometric domains. Biochim. Biophys. Acta. 1798: 909–927. [DOI] [PubMed] [Google Scholar]
- 20.D'Auria L., Van der Smissen P., Bruyneel F., Courtoy P. J., Tyteca D. 2011. Segregation of fluorescent membrane lipids into distinct micrometric domains: evidence for phase compartmentation of natural lipids? PLoS ONE. 6: e17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grossmann G., Malinsky J., Stahlschmidt W., Loibl M., Weig-Meckl I., Frommer W. B., Opekarova M., Tanner W. 2008. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 183: 1075–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klose C., Ejsing C. S., Garcia-Saez A. J., Kaiser H. J., Sampaio J. L., Surma M. A., Shevchenko A., Schwille P., Simons K. 2010. Yeast lipids can phase-separate into micrometer-scale membrane domains. J. Biol. Chem. 285: 30224–30232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusumi A., Fujiwara T. K., Chadda R., Xie M., Tsunoyama T. A., Kalay Z., Kasai R. S., Suzuki K. G. 2012. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson's fluid-mosaic model. Annu. Rev. Cell Dev. Biol. 28: 215–250. [DOI] [PubMed] [Google Scholar]
- 24.van Meer G., Voelker D. R., Feigenson G. W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramstedt B., Slotte J. P. 2002. Membrane properties of sphingomyelins. FEBS Lett. 531: 33–37. [DOI] [PubMed] [Google Scholar]
- 26.Edidin M. 2003. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 32: 257–283. [DOI] [PubMed] [Google Scholar]
- 27.Bali R., Savino L., Ramirez D. A., Tsvetkova N. M., Bagatolli L., Tablin F., Crowe J. H., Leidy C. 2009. Macroscopic domain formation during cooling in the platelet plasma membrane: an issue of low cholesterol content. Biochim. Biophys. Acta. 1788: 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun M., Northup N., Marga F., Huber T., Byfield F. J., Levitan I., Forgacs G. 2007. The effect of cellular cholesterol on membrane-cytoskeleton adhesion. J. Cell Sci. 120: 2223–2231. [DOI] [PubMed] [Google Scholar]
- 29.Salomao M., Zhang X., Yang Y., Lee S., Hartwig J. H., Chasis J. A., Mohandas N., An X. 2008. Protein 4.1R-dependent multiprotein complex: new insights into the structural organization of the red blood cell membrane. Proc. Natl. Acad. Sci. USA. 105: 8026–8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baines A. J. 2010. The spectrin-ankyrin-4.1-adducin membrane skeleton: adapting eukaryotic cells to the demands of animal life. Protoplasma. 244: 99–131. [DOI] [PubMed] [Google Scholar]
- 31.Sheetz M. P., Sable J. E., Dobereiner H. G. 2006. Continuous membrane-cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct. 35: 417–434. [DOI] [PubMed] [Google Scholar]
- 32.Zachowski A. 1993. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem. J. 294: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svoboda K., Schmidt C. F., Branton D., Block S. M. 1992. Conformation and elasticity of the isolated red blood cell membrane skeleton. Biophys. J. 63: 784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett V., Healy J. 2008. Organizing the fluid membrane bilayer: diseases linked to spectrin and ankyrin. Trends Mol. Med. 14: 28–36. [DOI] [PubMed] [Google Scholar]
- 35.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 36.Marinetti G. V., Baumgarten R., Sheeley D., Gordesky S. 1973. Cross-linking of phospholipids to proteins in the erythrocyte membrane. Biochem. Biophys. Res. Commun. 53: 302–308. [DOI] [PubMed] [Google Scholar]
- 37.Prausnitz M. R., Lau B. S., Milano C. D., Conner S., Langer R., Weaver J. C. 1993. A quantitative study of electroporation showing a plateau in net molecular transport. Biophys. J. 65: 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Z. J., Singh R. D., Sharma D. K., Holicky E. L., Hanada K., Marks D. L., Pagano R. E. 2006. Distinct mechanisms of clathrin-independent endocytosis have unique sphingolipid requirements. Mol. Biol. Cell. 17: 3197–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsh J. B., Weinstein D. B. 1966. Simple charring method for determination of lipids. J. Lipid Res. 7: 574–576. [PubMed] [Google Scholar]
- 40.Mikhalyov I., Samsonov A. 2011. Lipid raft detecting in membranes of live erythrocytes. Biochim. Biophys. Acta. 1808: 1930–1939. [DOI] [PubMed] [Google Scholar]
- 41.Byfield F. J., Aranda-Espinoza H., Romanenko V. G., Rothblat G. H., Levitan I. 2004. Cholesterol depletion increases membrane stiffness of aortic endothelial cells. Biophys. J. 87: 3336–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferru E., Giger K., Pantaleo A., Campanella E., Grey J., Ritchie K., Vono R., Turrini F., Low P. S. 2011. Regulation of membrane-cytoskeletal interactions by tyrosine phosphorylation of erythrocyte band 3. Blood. 117: 5998–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Betz T., Lenz M., Joanny J. F., Sykes C. 2009. ATP-dependent mechanics of red blood cells. Proc. Natl. Acad. Sci. USA. 106: 15320–15325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manno S., Takakuwa Y., Mohandas N. 2005. Modulation of erythrocyte membrane mechanical function by protein 4.1 phosphorylation. J. Biol. Chem. 280: 7581–7587. [DOI] [PubMed] [Google Scholar]
- 45.Gallagher P. G. 2005. Hematologically important mutations: ankyrin variants in hereditary spherocytosis. Blood Cells Mol. Dis. 35: 345–347. [DOI] [PubMed] [Google Scholar]
- 46.Ipsaro J. J., Huang L., Mondragon A. 2009. Structures of the spectrin-ankyrin interaction binding domains. Blood. 113: 5385–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dahl K. N., Westhoff C. M., Discher D. E. 2003. Fractional attachment of CD47 (IAP) to the erythrocyte cytoskeleton and visual colocalization with Rh protein complexes. Blood. 101: 1194–1199. [DOI] [PubMed] [Google Scholar]
- 48.King M. J., Jepson M. A., Guest A., Mushens R. 2011. Detection of hereditary pyropoikilocytosis by the eosin-5-maleimide (EMA)-binding test is attributable to a marked reduction in EMA-reactive transmembrane proteins. Int. J. Lab. Hematol. 33: 205–211. [DOI] [PubMed] [Google Scholar]
- 49.Ziółkowska N. E., Christiano R., Walther T. C. 2012. Organized living: formation mechanisms and functions of plasma membrane domains in yeast. Trends Cell Biol. 22: 151–158. [DOI] [PubMed] [Google Scholar]
- 50.Kuerschner L., Ejsing C. S., Ekroos K., Shevchenko A., Anderson K. I., Thiele C. 2005. Polyene-lipids: a new tool to image lipids. Nat. Methods. 2: 39–45. [DOI] [PubMed] [Google Scholar]
- 51.Fidorra M., Heimburg T., Bagatolli L. A. 2009. Direct visualization of the lateral structure of porcine brain cerebrosides/POPC mixtures in presence and absence of cholesterol. Biophys. J. 97: 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malínská K., Malinsky J., Opekarova M., Tanner W. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell. 14: 4427–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malinsky J., Opekarova M., Tanner W. 2010. The lateral compartmentation of the yeast plasma membrane. Yeast. 27: 473–478. [DOI] [PubMed] [Google Scholar]
- 54.Reed C. F., Swisher S. N. 1966. Erythrocyte lipid loss in hereditary spherocytosis. J. Clin. Invest. 45: 777–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klappauf E., Schubert D. 1977. Band 3-protein from human erythrocyte membranes strongly interacts with cholesterol. FEBS Lett. 80: 423–425. [DOI] [PubMed] [Google Scholar]
- 56.Schubert D., Boss K. 1982. Band 3 protein-cholesterol interactions in erythrocyte membranes. Possible role in anion transport and dependency on membrane phospholipid. FEBS Lett. 150: 4–8. [DOI] [PubMed] [Google Scholar]
- 57.Diakowski W., Ozimek L., Bielska E., Bem S., Langner M., Sikorski A. F. 2006. Cholesterol affects spectrin-phospholipid interactions in a manner different from changes resulting from alterations in membrane fluidity due to fatty acyl chain composition. Biochim. Biophys. Acta. 1758: 4–12. [DOI] [PubMed] [Google Scholar]
- 58.Kwik J., Boyle S., Fooksman D., Margolis L., Sheetz M. P., Edidin M. 2003. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA. 100: 13964–13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pike L. J., Miller J. M. 1998. Cholesterol depletion delocalizes phosphatidylinositol bisphosphate and inhibits hormone-stimulated phosphatidylinositol turnover. J. Biol. Chem. 273: 22298–22304. [DOI] [PubMed] [Google Scholar]
- 60.An X., Zhang X., Debnath G., Baines A. J., Mohandas N. 2006. Phosphatidylinositol-4,5-biphosphate (PIP2) differentially regulates the interaction of human erythrocyte protein 4.1 (4.1R) with membrane proteins. Biochemistry. 45: 5725–5732. [DOI] [PubMed] [Google Scholar]
- 61.Sinha B., Koster D., Ruez R., Gonnord P., Bastiani M., Abankwa D., Stan R. V., Butler-Browne G., Vedie B., Johannes L., et al. 2011. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 144: 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gauthier N. C., Masters T. A., Sheetz M. P. 2012. Mechanical feedback between membrane tension and dynamics. Trends Cell Biol. 22: 527–535. [DOI] [PubMed] [Google Scholar]
- 63.Muralidharan-Chari V., Clancy J. W., Sedgwick A., D'Souza-Schorey C. 2010. Microvesicles: mediators of extracellular communication during cancer progression. J. Cell Sci. 123: 1603–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.