Abstract
Cardiac triacylglycerol (TG) catabolism critically depends on the TG hydrolytic activity of adipose triglyceride lipase (ATGL). Perilipin 5 (Plin5) is expressed in cardiac muscle (CM) and has been shown to interact with ATGL and its coactivator comparative gene identification-58 (CGI-58). Furthermore, ectopic Plin5 expression increases cellular TG content and Plin5-deficient mice exhibit reduced cardiac TG levels. In this study we show that mice with cardiac muscle-specific overexpression of perilipin 5 (CM-Plin5) massively accumulate TG in CM, which is accompanied by moderately reduced fatty acid (FA) oxidizing gene expression levels. Cardiac lipid droplet (LD) preparations from CM of CM-Plin5 mice showed reduced ATGL- and hormone-sensitive lipase-mediated TG mobilization implying that Plin5 overexpression restricts cardiac lipolysis via the formation of a lipolytic barrier. To test this hypothesis, we analyzed TG hydrolytic activities in preparations of Plin5-, ATGL-, and CGI-58-transfected cells. In vitro ATGL-mediated TG hydrolysis of an artificial micellar TG substrate was not inhibited by the presence of Plin5, whereas Plin5-coated LDs were resistant toward ATGL-mediated TG catabolism. These findings strongly suggest that Plin5 functions as a lipolytic barrier to protect the cardiac TG pool from uncontrolled TG mobilization and the excessive release of free FAs.
Keywords: adipose triglyceride lipase, cardiac lipid, energy metabolism
The ability to deposit triacylglycerol (TG) within specific cellular organelles is an evolutionary conserved process present in virtually every mammalian cell and in most microorganisms (1–3). TG storage within lipid droplets (LDs) not only represents an energy reservoir, but is also an important source for the generation of membrane and signaling lipids (4). However, excessive accumulation of lipids is a hallmark of many metabolic disorders including obesity, hepatic steatosis, and cardiac steatosis (5–7). Apart from that, fatty acid (FA) esterification and deposition within neutral lipids protect cells from the harmful excess of nonesterified FAs also referred to as lipotoxicity (8, 9). The LD surface is characterized by the presence of various hydrophobic proteins including members of the so-called PAT family (1, 10) (designation derived from perilipin, adipophilin, and tail-interacting protein of 47 kDa) and neutral lipid hydrolases, which are involved in TG breakdown and the release of FAs and glycerol.
TG mobilization from white and brown adipose tissue is a relatively well characterized process involving perilipin 1 (Plin1), adipose triglyceride lipase (ATGL), and hormone-sensitive lipase (HSL) among many other proteins and cofactors (11, 12). In contrast, much less is known about LD TG catabolism in nonadipose tissues, including muscle and liver, in which Plin1 is replaced by other PAT family members (7, 13). More recently, perilipin 5 (Plin5) was found to be highly present on LDs of oxidative tissues including cardiac, skeletal muscle, and liver (14, 15). Ectopic expression of Plin5 increased cellular TG levels and reduced FA oxidation (FAO) suggesting a role for Plin5 in energy catabolism (16–19). Interestingly, Plin5 colocalized with ATGL and its coactivator comparative gene indentification-58 (CGI-58) (17, 18), two critical players in the first and rate-limiting step of TG catabolism (20, 21). Furthermore, the interaction of Plin5 with ATGL decreased ATGL-mediated lipolysis implying that Plin5 participates in the regulation of ATGL activity (22). Mutations of both ATGL and CGI-58 are causative for the development of neutral lipid storage disease with a different clinical picture: ATGL mutations are exclusively linked to muscle TG accumulation and lethal cardiomyopathy in humans and mice (23, 24), whereas mutated CGI-58 alleles are primarily involved in the development of hepatic steatosis and a defect in skin barrier formation (25–27). The distinct expression pattern of Plin1 and Plin5 (in adipose and oxidative tissue, respectively) suggests that these proteins interact with LD-associated proteins and the lipolytic machinery in an organ-specific manner. A recent study showed that Plin5 recruits mitochondria to the proximity of LDs and that the inhibitory effect of Plin5 on basal LD-mediated FA release was partially reversed under stimulated conditions (17). Considering that Plin5 expression is induced by peroxisome proliferator-activated receptor (PPAR)α (16, 28), a nuclear hormone receptor that regulates the expression of numerous oxidative phosphorylation genes, it may suggest that Plin5 couples LD FA release to mitochondrial FAO. However, the global deletion of Plin5 provoked a relatively mild phenotype in mice (29), although LDs were virtually absent in cardiac muscle (CM), similar to the lack of LDs in transgenic mice with cardiac-specific ATGL overexpression (30). Thus, it appears feasible that Plin5 primarily shields LDs from uncontrolled TG mobilization but is not critical for ATGL-mediated lipolysis and FA channeling to mitochondria.
The aim of our study was to examine the consequences of cardiac muscle-specific overexpression of perilipin 5 (CM-Plin5) on heart lipid and energy metabolism. We found that Plin5 transgenic mice exhibit severe TG accumulation in CM and that Plin5-coated LDs are resistant to ATGL-mediated TG hydrolysis, suggesting that Plin5 acts as a lipolytic barrier to prevent uncontrolled TG mobilization.
RESEARCH DESIGN AND METHODS
Animals
Transgenic mice expressing mouse Plin5 cDNA under the control of the cardiomyocyte-specific α-myosin heavy chain (MHC) promoter (Myh6, GenBank accession number U71441) were generated by cloning full-length mouse Plin5 cDNA (amplified from CM cDNA using the 5′-ACA GGT CGA CAT GGA CCA GAG AGG TGA AGA CAC-3′ forward and 5′-ACA GGT CGA CTC AAT GAT GAT GAT GAT GAT GGA AGT CCA GCT CTG GCA TCA-3′ reverse primers) in the α-MHC promoter construct (31), kindly provided by J. Robbins as previously described (32). Characterized transgenic mice originated from a B6D2F2 background and were backcrossed four to five times on a C57BL6 background. Littermates were used for phenotyping and mice with CM-Plin5 were hemizygous with respect to the integrated transgene. Animals were housed in a specific pathogen-free facility and maintained on a regular light-dark cycle (14 h light, 10 h dark) with ad libitum access to a standard laboratory chow diet (4.5% fat; ssniff Spezialdiäten, Germany) and water. For tissue collection, mice were euthanized by cervical dislocation and excised tissues were immediately snap-frozen. Maintenance, handling, and tissue collection from mice was approved by the Austrian Federal Ministry for Science and Research and by the ethics committee of the University of Graz.
cDNA cloning and expression of recombinant proteins
Mouse Atgl, HSL (Lipe), CGI-58, and G0/G1 switch gene 2 (G0S2) were cloned in the pcDNA4/HisMaxC expression vector. Full length mouse Plin5 cDNA was amplified from CM cDNA (forward primer: 5′-AAG GTA CCA GAC CAG AGA GGT GAA GAC ACC ACC-3′ and reverse primer: 5′-GA CTC GAG TCA GAA GTC CAG CTC TGG CAT C-3′) and cloned into the pcDNA4/HisMaxC (Invitrogen Life Technologies) expression vector. COS-7 cells were grown in low glucose Dulbecco's Modified Eagle Medium (GIBCO) supplemented with 10% fetal calf serum, 100 μg/ml streptomycin, and 100 IU/ml penicillin at 37°C with 5% CO2 and 95% humidity. Cells were transfected with plasmids encoding respective cDNAs expressing plasmids using Metafectene reagent (Biontex) according to the manufacturer's instructions.
Quantitative analysis of mRNA expression levels
Gene expression analysis was performed by quantitative reverse transcriptase polymerase chain reaction (RT-qPCR). Total RNA was extracted with the TRIzol reagent (Invitrogen) and treated with DNaseI (Invitrogen). For first strand cDNA synthesis, 1 μg of total RNA was reverse transcribed at 37°C for 1 h using random hexamer primer (Applied Biosystems) and Superscript II reverse transcriptase (Invitrogen). Primers used for RT-qPCR were designed to span exon-intron boundaries with an amplicon size of less than 150 bp and BLASTed for specificity. RT-qPCR reactions (20 μl) contained 8 ng of cDNA, 10 pM of each primer, and 10 μl of SYBR Green master mix (Fermentas) and were carried out using the ABI-StepOnePlus™ detection system (Applied Biosystems). Relative mRNA levels were quantified using the comparative ΔΔCT method with β-actin as reference gene. The following primer sequences were used for RT-PCR: β-actin forward, 5′-AGC CAT GTA CGT AGC CAT CCA-3′, reverse, 5′-TCT CCG GAG TCC ATC ACA ATG-3′ murine Plin5 forward, 5′-AGG GGA CTA GAC AAA TTG G-3′, reverse, 5′-GCT TCT CCG ACT TGC C-3′ Cpt1b (carnitine palmitoyltransferase 1β) forward, 5′-GGC ACC TCT TCT GCC TTT AC-3′, reverse, 5′-TTT GGG TCA AAC ATG CAG AT-3′ Ppara (peroxisome proliferator-activated receptor α) forward, 5′-GTA CCA CTA CGG AGT TCA CGC AT-3′, reverse, 5′-CGC CGA AAG AAG CCC TTA C-3′ Acox1 (acyl-CoA oxidase 1) forward, 5′-AGA TTG GTA GAA ATT GCT GCA AAA-3′, reverse, 5′-ACG CCA CTT CCT TGC TCT TC-3′ Acadm (acyl-CoA dehydrogenase, medium chain) forward, 5′-GAT GCA TCA CCC TCG TGT AAC-3′, reverse, 5′-AAG CCC TTT TCC CCT GAA-3′ Acadl (acyl-CoA dehydrogenase, long chain) forward, 5′-TTT CCG GGA GAG TGT AAG GA-3′, reverse, 5′-ACT TCT CCA GCT TTC TCC CA-3′ Acadvl (acyl-CoA dehydrogenase, very long chain) forward, 5′-ACC TTG CCA GGG CCT GAT-3′, reverse, 5′-TGG CCT GGT CAC CGG TAA-3′ Pdhk4 (pyruvate dehydrogenase kinase, isoenzyme 4) forward, 5′-ATC TAA CAT CAG AAT TAA ACC-3′, reverse, 5′-GGA ACG TAC ACA ATG TGG ATT G-3′ Ppargc1a (peroxisome proliferator-activated receptor γ coactivator 1α) forward, 5′-CCC TGC CAT TGT TAA GAC-3′, reverse, 5′-GC TGC TGT TCC TGT TTT C-3′ Ppargc1b (peroxisome proliferator-activated receptor γ coactivator 1β) forward, 5′-GAG GGC TCC GGC ACT TCC-3′, reverse, 5′-CGT ACT TGC TTT TCC CAG ATG-3′ Lpl forward, 5′-TCC AGC CAG GAT GCA ACA-3′, reverse, 5′-CCA CGT CTC CGA GTC CTC TCT-3′ Cd36 (cluster of differentiation 36) forward, 5′-GAA CCT ATT GAA GGC TTA CAT CC-3′, reverse, 5′-CCC AGT CAC TTG TGT TTT GAA C-3′ Pparg1 (peroxisome proliferator-activated receptor γ isoform 1) forward, 5′-AAC AAG ACT ACC CTT TAC TGA AAT TAC CA-3′, reverse, 5′-CAC AGA GCT GAT TCC GAA GTT G-3′.
Immunoblot analysis
Equal protein amounts (indicated in the corresponding figure legends) of tissue lysates, organelle preparations, and cell lysates were separated by SDS-PAGE and proteins were transferred onto a polyvinylidene fluoride membrane (Carl Roth, Karlsruhe, Germany). Blotted proteins were visualized using the following primary Abs: anti-ATGL (#2138; Cell Signaling Technology, Boston, MA), anti-Plin5 (#PAB12542; Abnova, Taipei City, Taiwan), anti-CGI-58 (#H00051099-M01; Abnova); anti-GAPDH (#2118; Cell Signaling Technology), anti-carnitine palmitoyltransferase (CPT)-1 (#98834; Santa Cruz Biotechnology, Santa Cruz, CA), anti-cytochrome c oxidase IV (COXIV) (#4844; Cell Signaling Technology), and anti-His (#27-4710-01; GE Healthcare, Waukesha, WI). Specifically bound immunoglobulins were detected in a second reaction using a horseradish peroxidase-conjugated anti-rabbit IgG Ab (Vector Laboratories, Burlingame, CA) or anti-mouse IgG Ab (GE Healthcare). Immunoblots were developed using the ECL Plus Western Blotting Detection System (GE Healthcare). Densitometric analyses were performed using ImageJ software (National Institutes of Health, Bethesda, MD).
Plasma parameters
Blood samples were collected by retro-orbital puncture from isoflurane-anesthetized mice. Plasma parameters were analyzed with commercially available kits from Wako, Sigma, and Thermo Fisher Scientific and plasma glucose levels were determined using the FreeStyle Freedom Lite® Blood Glucose Monitoring System (Abbott).
Tissue homogenization and lipid analysis
Snap-frozen hearts were homogenized in ice-cold lysis buffer A (0.25 M sucrose pH 7.0, 1 mM EDTA, 1 mM DTT, 20 μg/ml leupetine, 2 μg/ml antipain, 1 μg/ml pepstatin) using an Ultra Turrax Homogenizer (IKA). The homogenates were centrifuged at 20,000 g for 30 min at 4°C and the infranatants were collected. Protein concentrations were determined using the Bio-Rad Protein Assay reagent (Bio-Rad Laboratories GmbH) and lipid extractions were performed according to the method of Folch (33). Aliquots of the organic phase were evaporated and the lipid extracts were resuspended in ice-cold 1% Triton X-100 by brief sonication. TG concentrations were then measured using a colorimetric kit (Infinity TG reagent; Thermo Fisher Scientific).
TG hydrolase assay
TG hydrolase assays were performed as previously described (20). For the measurement of TG hydrolase activity cell lysates (1,000 g supernatant) or tissue extracts (10,000 g supernatant) were used. Stimulation of ATGL-mediated TG hydrolysis was performed by addition of purified murine GST-tagged CGI-58 (dissolved in 0.01% NP40) or COS-7 cell lysates containing His-tagged murine CGI-58 (20). Samples in a total volume of 100 μl buffer A were incubated with 100 μl substrate in a water bath at 37°C for 1 h. The micellar TG substrate contained 330 μM triolein, 3H-triolein as tracer, 45 μM phosphatidylcholine:phosphatidylinositol (PC:PI, 3:1), and was prepared by sonication (Virsonic 475; Virtis, Gardiner, NJ).
Labeling and preparation of LDs
COS-7 cells were transfected with beta-galactosidase (LacZ)- or Plin5-expression vectors as described before. To promote LD formation, one day posttransfection cells were incubated overnight in medium supplemented with 0.4 mM oleic acid complexed to essentially FA-free BSA in a molar ratio of 3:1 together with 4 mCi 3H-9,10-oleate/mmol as radioactive tracer. For isolation of LDs, cells were trypsinized, centrifuged, and washed three times with phosphate-buffered saline. Thereafter, cells were suspended in buffer A and disrupted by sonication (Virsonic 475; Virtis). Cell lysates were transferred to SW41 tubes, overlaid with buffer B (50 mM potassium phosphate pH 7.4, 100 mM KCl, 1 mM EDTA, 20 μg/ml leupetine, 2 μg/ml antipain, 1 μg/ml pepstatin), and centrifuged in an SW41 rotor (Beckman, Fullerton, CA) (2 h, 40,000 rpm, 4°C). LDs (visible as a white layer on top of the tube) were collected, transferred to a new tube, and concentrated by centrifugation (20,000 g, 15 min, 4°C) and removal of the underlying fluid. Subsequently, LDs were resuspended in buffer B by brief sonication.
Determination of TG hydrolase activity using purified LDs as substrate
LDs, prepared from COS-7 cells, were diluted to 0.05 μmol TG/100 μl (220 cpm/nmol) and 0.5% FA-free BSA in 100 mM potassium phosphate buffer (pH 7.0) was added. After incubation for 1 h, FA release from LDs was determined by extraction and determination of radioactivity essentially as described for the TG hydrolytic assay. LDs from cardiac tissue were isolated as described above and incubated with COS-7 cell lysates containing ATGL, HSL, CGI-58, or LacZ as control. LDs were diluted to a TG concentration of 0.4 μmol/100 μl and incubated in the presence of 0.5% FA-free BSA in 100 mM potassium phosphate buffer (pH 7.0) for 1 h at 37°C. The reaction was terminated by addition of 0.1% Triton X-100 followed by centrifugation at 20,000 g for 30 min. The lower phase was collected and FFAs were determined with a commercial kit (Wako Chemicals).
Analysis of TG levels of COS-7 cells expressing recombinant proteins
COS-7 cells were transfected with LacZ, Atgl, or Plin5 expression plasmids or cotransfected with both Atgl and Plin5 expression plasmids. To induce LD formation, COS-7 cells were loaded with 0.4 mM oleic acid and 4 mCi 3H-9,10-oleic acid/mmol as tracer overnight (20 h). Total lipids were extracted and separated by thin-layer chromatography using hexane/diethyl ether/acetic acid (70:29:1) as solvent. TG-corresponding bands were excised and radioactivity was measured by liquid scintillation counting. For the analysis of the time-dependent incorporation of 3H-labeled oleic acid into TG, LacZ and Plin5 expressing COS-7 cells were loaded with 0.4 mM oleic acid and 4 mCi 3H-9,10-oleic acid/mmol for time periods of 2, 4, 8, and 16 h. Lipids were extracted and analyzed as described above at the indicated time periods.
Preparation of mitochondria and determination of CPT-1 activity
For isolation of mitochondria, cardiac tissues were minced and homogenized with a dounce homogenizer in ice-cold buffer C (0.25 M sucrose, 5 mM HEPES pH 7.7, 0.25 mM EDTA, 20 μg/ml leupetine, 2 μg/ml antipain, 1 μg/ml pepstatin). After centrifugation (1,300 g, 4°C, 15 min), the infranatant was collected and mitochondria were pelleted (11,000 g, 4°C, 20 min) and resuspended in buffer C. CPT-1 activities were measured according to an established protocol (34, 35).
Tissue LPL activity
Tissue LPL activity was measured essentially as described in Ref. 36.
Transmission electron microscopy
Mice were euthanized at the age of 10 weeks by an overdose of anesthetic (xylazine-ketamine; Sigma) and immediately perfused with 4% (wt/vol) paraformaldehyde in 0.1 M phosphate buffer pH 7.4, for 5 min. CM was dissected using a Zeiss OPI1 surgical microscope (Carl Zeiss). Small tissue fragments were fixed in 2.5% (wt/vol) glutaraldehyde and 2% (wt/vol) paraformaldehyde in 0.1 M phosphate buffer pH 7.4, for 2 h, postfixed in 2% (wt/vol) osmium tetroxide for 2 h at room temperature, dehydrated in graded series of acetone and embedded in a TAAB epoxy resin. Thin sections (70 nm thick) were contrasted with uranyl acetate and lead citrate. Images were taken using an FEI Tecnai G2 20 transmission electron microscope (FEI Eindhoven) with a Gatan UltraScan 1000 charge-coupled device camera. Acceleration voltage was 120 kV.
Statistical analysis
Data are presented as mean ± SD. Statistical significance was determined by the Student's unpaired two-tailed t-test. Group differences were considered significant for *P < 0.05, **P < 0.01, and ***P < 0.001.
RESULTS
Cardiac-specific Plin5 overexpression provokes severe cardiac steatosis
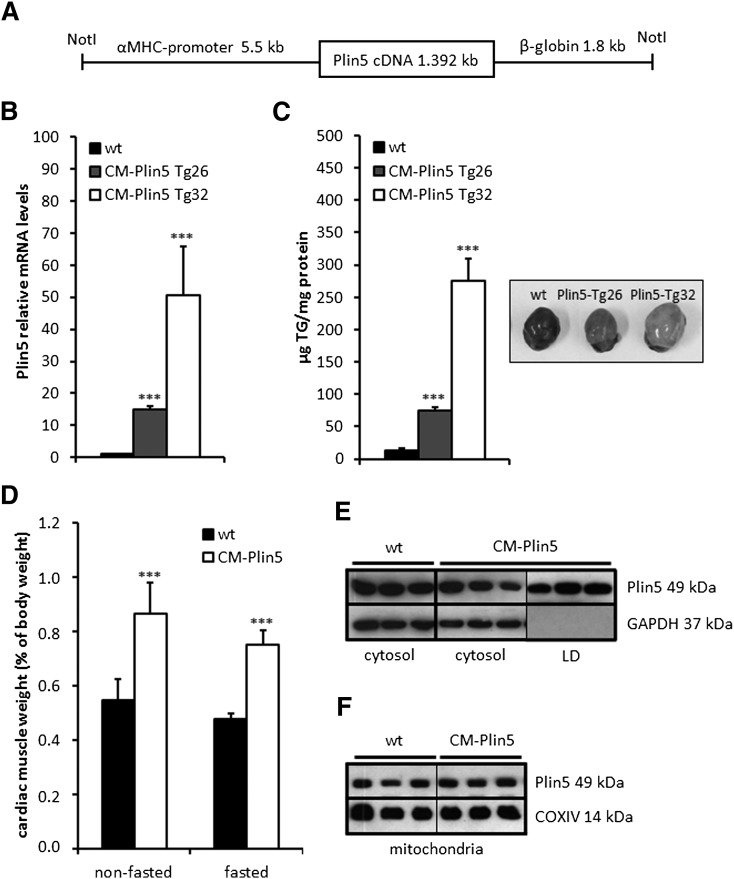
Cardiac-specific overexpression of Plin5 was achieved by cloning the murine Plin5 cDNA downstream of the α-MHC promoter and microinjection of the transgene DNA construct (Fig. 1A) into the pronucleus of mouse embryos. Measurement of Plin5 mRNA expression levels in CM of two founders showed a 14.8-fold and 50.6-fold increase in cardiac Plin5 mRNA levels of CM-Plin5 transgenic line (Tg)26 and Tg32, respectively, compared with wild type (wt) (Fig. 1B). CM-specific Plin5 overexpression caused massive cardiac steatosis (Fig. 1C, right) and the magnitude of TG accumulation (Fig. 1C, left) correlated with the degree of Plin5 mRNA expression levels in the transgenic lines. We then focused our characterization on Tg32 which is hereafter designated as CM-Plin5. The massive TG accumulation was also reflected by the relative increase of heart weight in relation to body weight of nonfasted and fasted CM-Plin5 mice (1.8- and 1.6-fold, respectively) compared with wt (Fig. 1D). Body weights were unchanged in CM-Plin5 mice compared with wt mice (22.1 ± 1.2 g vs. 21.8 ± 1.1 g in nonfasted mice and 20.8 ± 1.4 g vs. 20.2 ± 1.3 g in fasted mice, respectively). Next, we analyzed Plin5 protein levels in cytosolic, LD, and mitochondrial fractions of CM. Plin5 protein levels were similar in cytosolic preparations (Fig. 1E) of CM from CM-Plin5 and wt mice, respectively. After ultracentrifugation, an LD layer was visible only in CM homogenates of Plin5 transgenic mice which was withdrawn for Western blot analysis. Plin5 protein signals were abundant in these LD fractions of Plin5 transgenic mice (Fig. 1E, outermost panel, right). In contrast, Plin5 protein expression levels were similar in mitochondrial preparations of CM from both genotypes (Fig. 1F). As expected, Plin5 protein levels were unchanged in skeletal muscle (musculus quadriceps) of CM-Plin5 mice (supplementary Fig. I) implying that expression of the Plin5 cDNA under the control of the α-MHC promoter was CM-specific.
Fig. 1.
CM-Plin5 provokes massive TG accumulation in CM of transgenic mice. A: A scheme depicting the Plin5 transgene used for microinjection and the generation of transgenic mice. The mouse Plin5 cDNA was cloned downstream of the murine CM-specific α-MHC promoter. For mRNA stabilization, 1.8 kb of the nontranslated human β-globin gene were inserted at the 3′ end of the Plin5 cDNA. The transgene fragment was flanked by NotI restriction enzyme sides. B: Cardiac Plin5 mRNA expression levels were highly increased in 12-week-old mice of two different transgenic lines (indicated as CM-Plin5 Tg26 and CM-Plin5 Tg32, respectively) compared with wt Plin5 mRNA expression levels (n = 5; ***P < 0.001). C: TG levels were drastically increased in CM of 12-week-old transgenic mice and the extent of TG accumulation correlated with Plin5 mRNA expression levels of the two transgenic lines (n ≥ 4; ***P < 0.001 compared with wt). D: Hearts from 12-week-old CM-Plin5 mice (from transgenic line 32 which was established for further characterization) were significantly heavier compared with wt hearts of nonfasted and fasted mice (n ≥ 4; ***P < 0.001). E: Cytosolic Plin5 protein levels were not significantly changed in CM of CM-Plin5 mice compared with wt. LD fraction could be merely prepared from CM of CM-Plin5 mice and contained large amounts of Plin5 protein. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as loading control and cytosolic marker protein. F: Plin5 protein levels were comparable in mitochondria isolated from 12-week-old transgenic and wt mice, respectively. COXIV served as loading control for mitochondrial proteins. For Western blot analyses, equal protein amounts (10 μg) were separated by SDS-PAGE prior to blotting.
Plasma parameters of CM-Plin5 mice are virtually unchanged
Next we examined the impact of cardiac Plin5 overexpression on plasma parameters (Table 1). Plasma lipid and blood glucose levels of CM-Plin5 mice were similar compared with wt while plasma TG levels were moderately reduced (−17%) in nonfasted animals. These data implicate that cardiac Plin5 overexpression does not significantly affect systemic energy homeostasis.
TABLE 1.
Plasma parameters of CM-Plin5 mice were similar compared with that of wt mice except for a moderate decrease of TG in fasted transgenic mice.
| Nonfasted |
Fasted |
|||
| wt | CM-Plin5 | wt | CM-Plin5 | |
| Glucose (mg/dl) | 181.2 ± 21.5 | 168.4 ± 25.3 | 103.2 ± 4.1 | 98.2 ± 9.0 |
| FFA (mmol/l) | 0.60 ± 0.06 | 0.69 ± 0.12 | 1.32 ± 0.36 | 1.15 ± 0.32 |
| Glycerol (mmol/l) | 0.41 ± 0,08 | 0.40 ± 0.14 | 0.43 ± 0.07 | 0.46 ± 0.10 |
| TG (mg/dl) | 102.2 ± 13.6 | 81.2 ± 3.4* | 65.6 ± 11.0 | 72.8 ± 11.2 |
| TC (mg/dl) | 102.6 ± 18.6 | 91.4 ± 7.4 | 92.1 ± 16.5 | 84.0 ± 17.6 |
Various parameters were assayed with a glucometer and commercial kits in plasma samples obtained from nonfasted and fasted 12-week-old female mice (n ≥ 4, *P < 0.05). Comparable values were obtained from male mice (data not shown). TC, total cholesterol.
Impaired ATGL- and HSL-mediated FA release of LD preparations from CM of CM-Plin5 mice
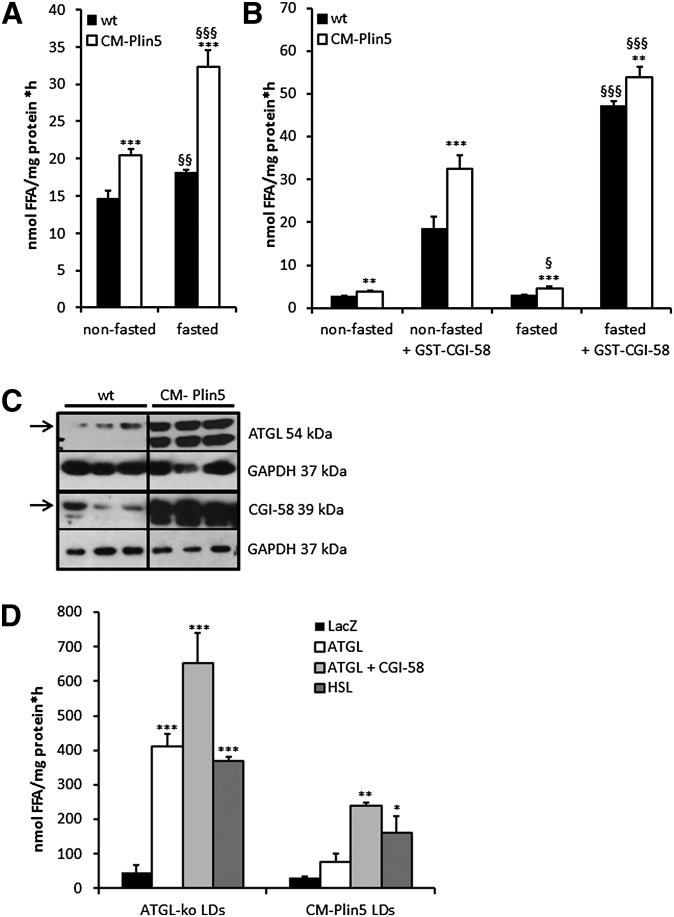
To examine whether the marked TG accumulation in CM of Plin5 transgenic mice involves changes in cardiac lipolysis, we performed TG hydrolytic assays using an established micellar triolein substrate emulsified with phospholipids. Interestingly, TG hydrolytic activities were markedly increased in CM tissue extracts derived from nonfasted and fasted CM-Plin5 mice (1.4-fold in the nonfasted and 1.8-fold in the fasted state, respectively) compared with wt tissue extracts (Fig. 2A). Addition of the ATGL coactivator CGI-58 significantly increased TG hydrolytic activities in cardiac extracts of both genotypes independent of the nutritional state (Fig. 2B). Notably, the highest differences in TG hydrolytic activities were observed in cardiac tissue extracts of nonfasted CM-Plin5 mice compared with wt mice upon addition of recombinant CGI-58 (1.8-fold). Next, we examined whether the observed changes in cardiac TG hydrolytic activities of CM-Plin5 mice involve differences in ATGL and/or CGI-58 protein expression levels. Western blot analyses revealed a marked increase in ATGL and CGI-58 protein levels in cardiac homogenates of transgenic mice (Fig. 2C). This finding indicates that the increased TG hydrolytic activities measured in CM extracts of transgenic mice (Fig. 2A) are due to augmented protein expression of ATGL and its coactivator CGI-58. Because CM-Plin5 mice exhibited massive TG accumulation in CM, we assumed that the increased in vitro TG hydrolytic activities may not reflect the in vivo situation, where Plin5 is abundantly present on the LD surface. To address this issue, we prepared LDs from CM of transgenic mice and used these LD preparations as a substrate for TG hydrolysis and measured the FA release in the presence of ATGL, ATGL together with CGI-58, HSL, and LacZ as control. Because LDs are in extremely low abundance in CM of wt mice, we used LDs from CM of ATGL-knockout mice instead. The comparison of the FA release of LD preparations from CM-Plin5 and ATGL-deficient mice is particularly intriguing with regard to the severe TG accumulation in CM of both genotypes. TG hydrolysis of LD preparations of both genotypes was induced by the addition of cell lysates containing ATGL, ATGL together with CGI-58, HSL, and LacZ which served as reference (Fig. 2D). Yet, FA release was in general markedly lower in LD preparations from CM of Plin5 mice upon addition of lysates containing ATGL (−81.4%), ATGL combined with CGI-58 (−63.5%), and HSL (−56.7%) compared with that of cardiac LD preparations from ATGL-deficient mice. These differences in LD TG mobilization upon addition of exogenous lipases are a strong indication that Plin5 could function as a lipolytic barrier and block the access of lipases to the TG substrate of Plin5-enriched LDs.
Fig. 2.
Measurement of tissue TG hydrolytic activities and FAs released from cardiac LDs. A: Cardiac TG hydrolase activities were increased in nonfasted and fasted 12-week-old Plin5 transgenic mice compared with the activity of wt tissues (n = 5; ***P < 0.001 versus wt; §§P < 0.01; §§§P < 0.001 versus nonfasted). B: Addition of recombinant GST-tagged CGI-58 significantly increased TG hydrolytic activities in both genotypes (n = 5; **P < 0.01; ***P < 0.001 versus wt; §P < 0.05; §§§P < 0.001 fasted versus nonfasted). C: Protein expression levels of ATGL and its coactivator CGI-58 were markedly elevated in CM of CM-Plin5 mice compared with wt (12-week-old mice). Analyzed samples contained 30 μg protein. Specific signals for ATGL and CGI-58 are indicated as arrows. GAPDH served as protein loading control. D: FA release of LD preparations isolated from CM of Plin5 transgenic and ATGL-deficient mice when incubated with COS-7 cell lysates containing LacZ, ATGL, ATGL + CGI-58, and HSL. The ATGL- and HSL-mediated FA release was significantly reduced in LD preparations of Plin5 transgenic mice compared with that of LDs from ATGL-deficient mice (n = 3). Data are shown as mean + SD. *P < 0.05; **P < 0.01; and ***P < 0.001 versus LDs incubated with LacZ containing lysates.
Plin5 blocks lipase access to the LD TG moiety but does not specifically inhibit ATGL enzymatic activity
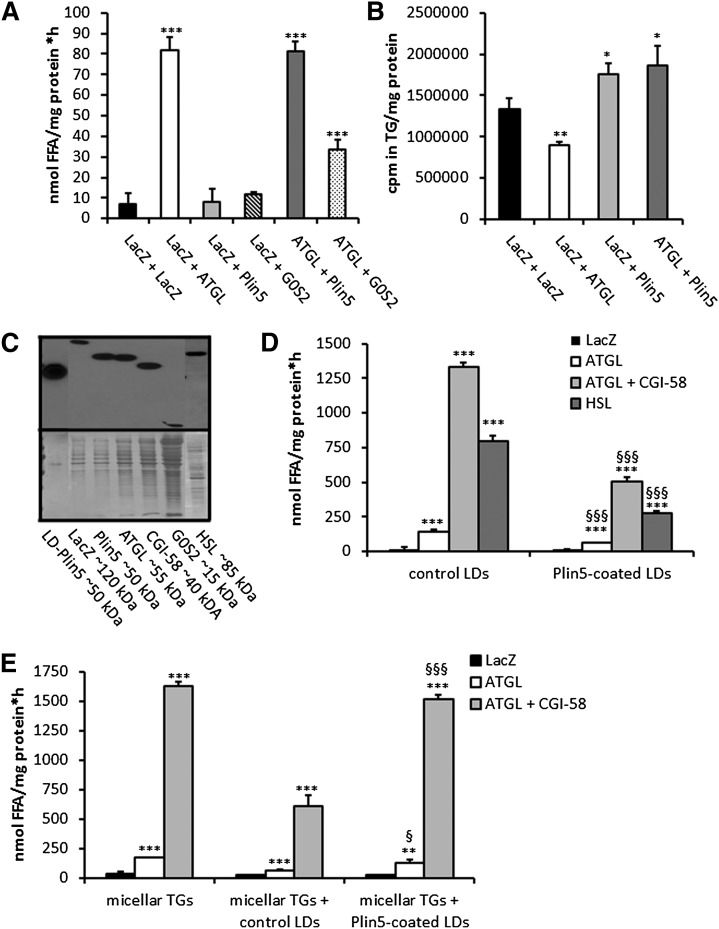
To confirm our hypothesis that Plin5 functions as a lipolytic barrier to preserve the LD TG pool from unrestricted TG catabolism, we used a cell culture approach and examined TG hydrolysis and homeostasis under various conditions. First, we examined the in vitro impact of Plin5 on basal and ATGL-mediated TG hydrolysis using again the artificial 3H-labeled triolein substrate. TG hydrolytic activities were measured in a combination of cell lysates containing LacZ, ATGL, and Plin5. We also included COS-7 cell lysates containing G0S2, a known inhibitory protein of ATGL (37, 38). TG hydrolytic activities substantially increased in cell preparations containing ATGL (up to 11.6-fold compared with the LacZ control) independent of whether ATGL-containing cell lysates were mixed with lysates containing LacZ or Plin5 (Fig. 3A). In contrast, ATGL TG hydrolytic activity was markedly inhibited (−58.5%) by the addition of G0S2 cell lysates. These findings demonstrate that ATGL enzymatic activity per se is not affected by the presence of Plin5 in in vitro assays. To investigate the in vivo impact of Plin5 on ATGL-mediated TG hydrolysis, we cotransfected COS-7 cells with LacZ and ATGL- or Plin5-expressing plasmids, loaded cells with oleic acid and 3H-labeled oleic acid as tracer, and analyzed the incorporation of radioactivity into TG (Fig. 3B). COS-7 cells transfected with ATGL showed reduced incorporation of radioactivity into the TG pool (−33.0%) compared with LacZ, due to increased TG mobilization. In contrast, Plin5-transfected cells showed moderately increased incorporation of radioactivity into cellular TG (1.3-fold) and this effect was unchanged even if these cells were cotransfected with ATGL (1.4-fold). The increased radioactivity present in the TG fraction of Plin5-expressing cells could originate from increased lipogenesis or impaired TG catabolism. To address this question, we loaded LacZ- and Plin5-transfected COS-7 cells again with oleic acid and 3H-labeled oleic acid and measured radioactivity levels in TG at several time points over a period of 16 h. As shown in supplementary Fig. II, the incorporation of radioactivity constantly increased to a similar extent in LacZ- and Plin5-transfected cells indicating that Plin5 is not involved in the lipogenic pathway.
Fig. 3.
Ectopic Plin5 expression does not affect ATGL enzymatic activity but establishes a lipolytic barrier. A: TG hydrolase activity assays of mixtures of COS-7 cell lysates containing LacZ, ATGL, Plin5, and G0S2, respectively. TG hydrolytic activities were assayed with a micellar 3H-labeled triolein substrate. Data are shown as mean + SD of n = 3. ***P < 0.001 versus LacZ control. B: COS-7 cells transfected with lacZ, ATGL, and/or Plin5 were loaded with 400 μM oleic acid and 3H-labeled oleic acid as tracer and intracellular 3H-incorporation into the TG pool was determined. Data are shown as mean + SD of n = 3. *P < 0.05; **P < 0.01 versus LacZ controls. C: Western blot analyses of Plin5 protein levels in LDs prepared from Plin5-transfected COS-7 cells (LD-Plin5) and protein levels of LacZ, ATGL, CGI-58, G0S2, and HSL in cell lysates of COS-7 cells transfected with the respective cDNA expression plasmids. Analyzed cell lysates contained 10 μg protein. D: TG hydrolysis using isolated LDs as substrate. LDs were isolated from COS-7 cells transfected either with LacZ or Plin5 and loaded with oleic acid and 3H-labeled oleic acid as tracer. LD preparations were incubated with ATGL, ATGL + CGI-58, or HSL-containing COS-7 cell lysates and the release of 3H-FAs were measured. Data are mean + SD of n = 3. ***P < 0.001 versus LacZ-containing lysates and §§§P < 0.001 indicates significances among control versus Plin5-coated LDs. E: COS-7 cell lysates containing either LacZ, ATGL, or ATGL + CGI-58 were incubated with 3H-labeled triolein substrate. In some cases, LDs isolated from LacZ or Plin5-transfected cells were added to determine whether these LDs can compete with the TG substrate. Addition of LDs of LacZ, but not of Plin5 transfected cells largely inhibited hydrolysis of the 3H-triolein substrate suggesting that Plin5-coated LDs cannot compete with the micellar TG substrate. Data are shown as mean + SD of n = 3. **P < 0.01 and ***P < 0.001 versus LacZ samples. §P < 0.01 and §§§P < 0.001 indicate significances of control versus Plin5-enriched LDs.
Taken together, these data suggest that Plin5 protects TG from ATGL-mediated hydrolysis in vivo presumably by limiting the access of lipases to TG stores. To validate this hypothesis, we tested whether Plin5-coated LDs are resistant toward in vitro ATGL-mediated TG hydrolysis. We also included HSL in the experiment to address whether Plin5 may represent a general lipolytic barrier or if it is specifically shielding/inhibiting ATGL. Therefore, we incubated LacZ- and Plin5-transfected COS-7 cells with oleic acid and 3H-labeled oleic acid to induce TG synthesis and LD formation. Western blot experiments confirmed that the recombinant proteins were present in the investigated cell lysates and that LDs isolated from Plin5-transfected COS-7 cells contained large amounts of Plin5 (Fig. 3C). Then, in vitro assays were performed using isolated control LDs or LDs coated with Plin5 as substrate after the addition of lysates containing various recombinant proteins. FAs were efficiently released from control LDs (prepared from LacZ-transfected COS-7 cells incubated with 3H-labeled oleic acid) when incubated with an ATGL-containing lysate (7.3-fold), lysates containing ATGL and CGI-58 (70.1-fold), and a lysate containing HSL (40.9-fold) as compared with FA release upon addition of LacZ-containing lysates (Fig. 3D). Notably, FA release from Plin5-coated LDs was markedly reduced compared with that of control LDs when incubated with lysates containing ATGL (−16.6%), ATGL and CGI-58 (−62.4%), or HSL (−65.9%) strongly suggesting that Plin5 represents a lipolytic barrier that hinders lipase access to the TG substrate.
To investigate whether Plin5 interferes with ATGL-mediated TG hydrolysis per se, we performed TG hydrolytic activity assays using again the artificial micellar 3H-labeled TG substrate. We assumed that addition of control LDs would competitively lower TG hydrolysis of the artificial TG substrate, while if Plin5 shields the access of lipases to TG, addition of Plin5-coated LDs would have less or even no effect on TG lipolysis. In fact, addition of control (non-radioactively labeled) LD preparations to the artificial 3H-triolein substrate lowered TG hydrolysis of lysates containing ATGL or ATGL and CGI-58 by about 50% (Fig. 3E). In contrast, TG hydrolytic activities of lysates containing ATGL or ATGL and CGI-58 were mildly affected upon addition of Plin5-coated LDs, indicating that Plin5 in fact substantially limits the access to the TG substrate.
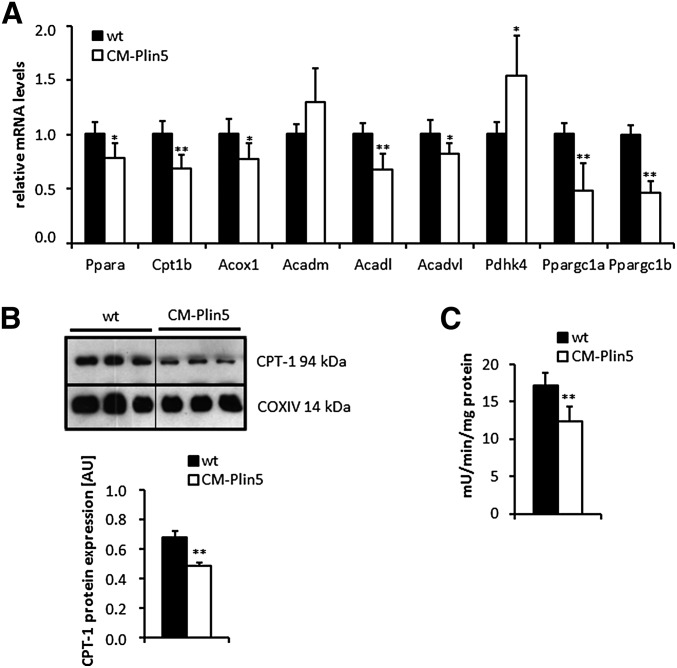
Moderately decreased FAO gene expression levels and reduced mitochondrial CPT-1 activity in CM of CM-Plin5 mice
Because mice lacking ATGL exhibit severe cardiac TG accumulation linked to defective PPARα-activated FAO gene expression, we hypothesized that cardiac Plin5 overexpression may similarly affect FAO gene expression levels. To address this hypothesis, we measured mRNA expression levels of PPARα, PPARα (and PPARβ/δ) target genes, and peroxisome proliferator-activated receptor γ coactivator (PGC)-1α and PGC-1β in CM of overnight fasted mice (Fig. 4A). Cardiac mRNA expression levels of Ppara (−22%) and several established PPARα and PPARβ/δ target genes including Cpt1b (−32%), Acox1 (−23%), Acadl (−32%), and Acadvl (−18%) were moderately reduced in CM of CM-Plin5 mice compared with levels found in wt mice. Furthermore, expression levels of Ppargc1a and Ppargc1b were significantly reduced (−52% and −54%, respectively) (Fig. 4A) in CM of CM-Plin5 mice compared with controls. Nonetheless, mRNA levels of Acadm and Pdhk4 (+55%) were similar, or even increased, in transgenic mice compared with controls (Fig. 4A). To further examine the impact of cardiac Plin5 overexpression on mitochondrial energy metabolism, we measured CPT-1 protein levels (Fig. 4B) and CPT-1 activity (Fig. 4C) in mitochondrial preparations derived from CM-Plin5 transgenic and wt mice, respectively. Mitochondrial CPT-1 protein signals and activity levels were decreased (−28.4% and −28.5%, respectively) in CM of transgenic mice suggesting decreased mitochondrial FA uptake.
Fig. 4.
Measurement of cardiac mRNA levels of established PPARα and PPARβ/δ target genes and mitochondrial CPT-1 activity. A: mRNA expression levels of PPARα, selected PPARα and PPARβ/δ target genes, PGC-1α and PGC-1β were determined by RT-qPCR in CM RNA prepared from 12-week-old fasted Plin5 transgenic and wt mice, respectively (n ≥ 5). *P < 0.05 and **P < 0.01 versus wt samples. B: Western blot and densitometric analysis of CPT-1 protein expression levels in mitochondria preparations of 15-week-old fasted mice. COXIV served as loading control (**P < 0.01). Ten micrograms of mitochondrial protein were separated by SDS-PAGE prior to blotting. C: CM-specific Plin5 overexpression significantly decreased CPT-1 activity in mitochondria preparations from CM of CM-Plin5 mice compared with that of controls (n ≥ 5; **P < 0.01 versus WT mice).
Marked divergences in cardiomyocyte morphology and LD size in cardiac tissue of CM-Plin5 mice compared with that of ATGL-deficient mice
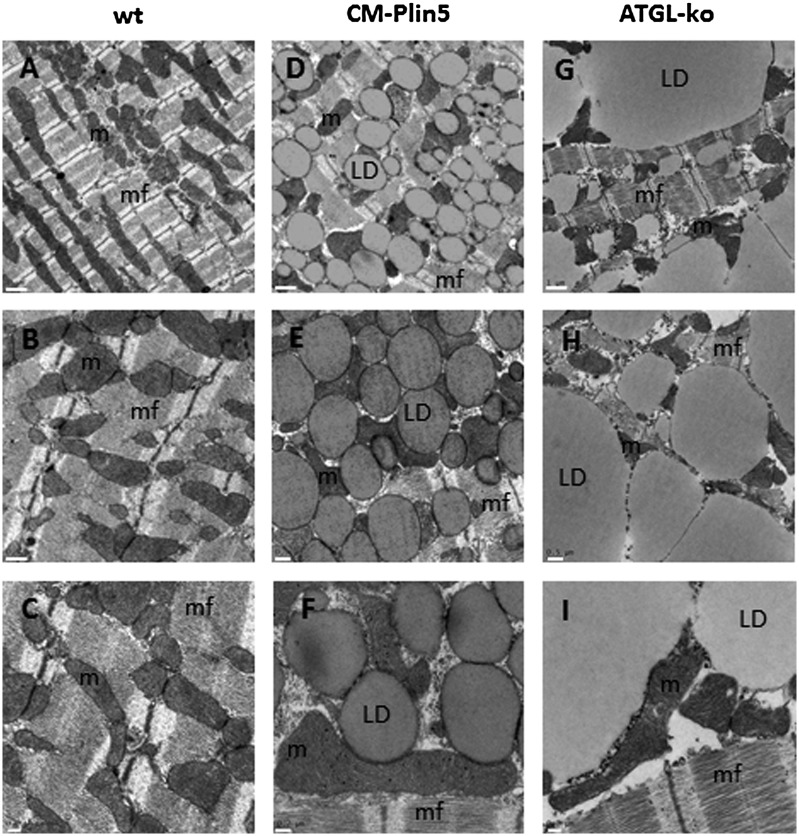
The severe TG accumulation in CM of CM-Plin5 and ATGL-deficient mice together with impaired FAO gene expression in both mouse models prompted us to examine cardiomyocyte morphology in WT, CM-Plin5, and ATGL-deficient mice, respectively. Therefore, mice were euthanized with anesthetic and immediately perfused with paraformaldehyde. Thin sections from fixed cardiac tissue were stained with uranylacetate and lead citrate and examined by transmission electron microscopy. At first view, there were obvious differences in cardiomyocyte morphology of all examined genotypes. While LDs were not present in CM sections of wt tissue (Fig. 5A–C), there was a pronounced increase of uniform and hypertrophied LDs which were homogenously dispersed in cardiomyocytes of CM-Plin5 mice (Fig. 5D–F). Notably, ATGL-deficient cardiomyocytes contained LDs of all sizes including giant LDs, and the cellular architecture appeared atrophic (Fig. 5G–I). A higher resolution revealed that cardiac mitochondria of CM-Plin5 mice were tightly attached to LDs (Fig. 5E, F) and the increased number of LDs seemed not to affect cellular integrity as assumed for ATGL-deficient cardiomyocytes. Furthermore, mitochondrial appearance and cristae structure were similar in CM-Plin5 tissue sections compared with WT mice. In contrast, ATGL-deficient cardiomyocytes exhibited changes in mitochondrial shape and cristae structure (see Fig. 5H, I). The marked differences in cardiomyocyte morphology of CM-Plin5 mice compared with ATGL-deficient mice suggest that the cardiac phenotype of CM-Plin5 mice is not as severe as in the hearts of mice globally lacking ATGL.
Fig. 5.
Transmission electron microscopy of CM sections revealed marked differences in cardiac morphology of Plin5 transgenic and ATGL-deficient mice. Cardiac tissue was prepared from mice after perfusion with paraformaldehyde. Fixed tissue sections were stained and examined by transmission electron microscopy. Cardiac sections from wt (A–C) showed a typical intermyofibrillar network and clusters of mitochondria. CM-Plin5 cardiac sections (D–F) showed hypertrophied LDs of similar size homogenously dispersed throughout the cytoplasm which seems not to interfere with the intermyofibrillar network. Most obviously, LDs were tightly associated with mitochondria. In contrast, cardiac sections obtained from ATGL-deficient mice [ATGL-knockout (ko) (G–I)] showed the accumulation of LDs of varying sizes including giant droplets. Overall, the cardiomyocyte architecture as well as the shape of mitochondria seemed to be markedly hampered by the giant droplets. Scale bars: 1 μm upper panel, 0.5 μm middle panel, 0.2 μm lower panel. m, mitochondria; mf, intermyofibrillar network.
Reduced mRNA expression levels of lipoprotein lipase, CD36, and PPARγ in CM of Plin5 transgenic mice
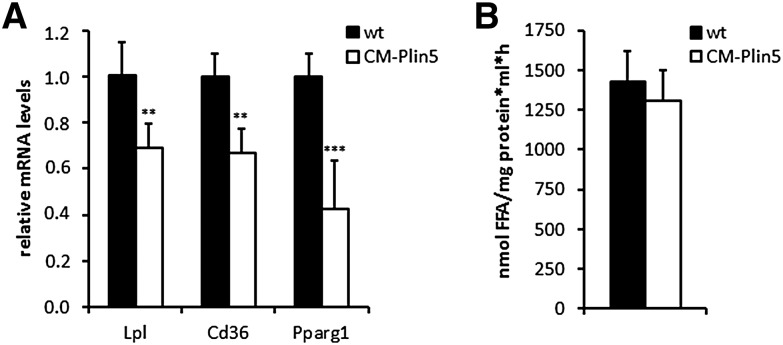
Finally, we examined whether changes in cardiac FA uptake and lipogenesis contribute to TG accumulation in CM of CM-Plin5 mice. Therefore, we measured mRNA expression levels of genes implicated in FA uptake and lipogenesis. LPL and the FA transporter CD36 are critical players in cardiac FA uptake (39) and the expression of both genes is regulated by PPARγ. As shown in Fig. 6, mRNA expression levels of lpl (−31%), cd36 (−33%) and pparg1 (−68%) are significantly reduced in CM of CM-Plin5 mice compared with levels found in wt mice. In contrast, LPL activities were similar in CM of fasted Plin5 transgenic mice compared with wt indicating that the marked increase in cardiac TG levels of CM-Plin5 mice does not involve changes in cardiac FA uptake and lipogenic pathways.
Fig. 6.
mRNA expression levels of LPL, CD36, and PPARγ were reduced in CM of Plin5 transgenic mice whereas LPL activities were comparable to that of WT mice. A: mRNA expression levels were measured by RT-qPCR in cardiac tissue RNA derived from 12-week-old Plin5 transgenic and wt mice, respectively (n ≥ 4). **P < 0.01 and ***P < 0.001 versus WT mice. B: For the measurement of cardiac LPL-activities, hearts from overnight-fasted mice were surgically removed and minced in a medium containing heparin. LPL-activity of the supernatant was measured in duplicates. Values are shown as mean + SD of tissue samples from 4 mice of each genotype.
DISCUSSION
Defective lipolysis caused by mutations in ATGL alleles strongly affected cardiac energy catabolism in humans and mice and led to severe cardiac steatosis and dysfunction (23, 32). More recently, Plin5, a member of the PAT family, was shown to interact with ATGL and its coactivator CGI-58 (18, 22, 40) suggesting a role for Plin5 in the regulation of cardiac lipolysis. This assumption was also corroborated by the phenotype of Plin5-deficient mice which exhibited an almost complete depletion of cardiac LDs (29).
Here we show that mice with cardiac-specific overexpression of Plin5 showed massive cardiac TG accumulation, similar to the cardiac phenotype of ATGL-deficient mice. This phenotype suggests that ATGL-mediated lipolysis is impaired by Plin5 overexpression and inspired us to carefully examine cardiac TG hydrolysis in this transgenic mouse model. Surprisingly, in in vitro assays, we measured increased TG hydrolytic activities in heart extracts of Plin5 transgenic mice using an artificial triolein substrate (41). This finding was even more challenging with respect to the report that mice with cardiac-specific ATGL overexpression exhibit a severe decline in cardiac TG levels (30) suggesting that results of our in vitro assay may not reflect in vivo cardiac lipolysis of CM-Plin5 mice. In accordance with this assumption, we found that isolated LDs from CM of Plin5 transgenic mice are a less effective substrate for exogenously added ATGL as compared with LDs isolated from CM of ATGL-deficient mice. This reduced ability of ATGL to hydrolyze TG of LDs derived from CM of Plin5 transgenic mice could be a consequence of Plin-5 directly inhibiting ATGL enzymatic activity, or from the more general formation of a lipolytic barrier that hinders lipase access to the TG substrate.
Employing recombinant proteins and cell experiments, we examined the direct effect of Plin5 and Plin5-coated LDs on ATGL activity in in vitro assays as well as in living cells. We found that Plin5 apparently acts as a general lipolytic barrier on the LD surface, because: i) ATGL- and HSL-mediated TG catabolism was impaired in Plin5-enriched LDs; ii) ATGL TG hydrolytic activity, as determined in in vitro assays, was not affected by addition of Plin5 containing lysates alone; iii) ATGL-mediated TG hydrolysis of an artificial triolein substrate was not affected by the addition of Plin5-coated LDs whereas control LDs competed with the hydrolysis of the artificial substrate; and iv) in living cells, overexpression of Plin5 led to increased TG accumulation even if ATGL was coexpressed. To summarize, these data suggest that Plin5 forms a lipolytic barrier at the LD surface thereby impairing the access of ATGL and HSL to the TG substrate. Whether this Plin5-mediated barrier function mechanistically involves the inhibition of ATGL enzymatic activity per se when Plin5 and ATGL colocalize and interact on the LD surface needs further experimental clarification.
Cardiac steatosis of ATGL-deficient mice was mainly caused by a severe defect in PPARα-activated gene expression and the TG accumulation could be reversed by treatment with a PPARα agonist (32). Interestingly, the mRNA expression levels of selected PPARα and PPARβ/δ target genes which have been implicated in mitochondrial FA uptake and oxidation, were moderately decreased in CM of CM-Plin5 mice when compared with the pronounced defect in PPARα-activated gene expression in CM of ATGL-deficient mice (32). Furthermore, cardiomyocyte morphology strongly differed in CM of Plin5 transgenic mice compared with the cellular architecture of ATGL-deficient cardiomyocytes. Cardiac Plin5 overexpression provoked the accumulation of hypertrophied LDs of similar sizes which were homogenously distributed throughout the cytosol in tight association with mitochondria. In contrast, ATGL-deficient cardiomyocytes appeared atrophic and showed a severe accumulation of heterogenous LDs, including giant droplets which strongly interfered with the myofibrillar architecture and mitochondrial shape. These divergences in cardiac morphology strongly imply that Plin5 overexpression does not severely impair ATGL-mediated TG catabolism and hence may moderately affect cardiac function. In accordance with this assumption, we actually have no evidence that Plin5 overexpression provokes an early lethal heart dysfunction reported for mice lacking ATGL (data not shown).
Ectopic Plin5 expression variably increased TG levels in several cell lines and affected mRNA expression levels of genes implicated in FAO (16, 19). Notably, the induction of Plin5 expression particularly impaired TG mobilization under nonstimulated conditions and this effect could be partially reversed via forskolin-induced protein kinase A stimulation which was paralleled by increased Plin5 phosphorylation (22). Accordingly, stimulation of cardiac lipolysis may lead to Plin5 phosphorylation/modification and a relaxation of the lipolytic barrier. Given that cardiac Plin5 overexpression mainly impairs lipolysis under nonfasted conditions, a more moderate cardiac phenotype would be predicted compared with the severe cardiac dysfunction of mice globally lacking ATGL. Along that line, we cannot exclude that changes in LD protein composition (42) and/or Plin5 interaction partners including CGI-58 and ATGL in response to increased energy demands possibly affect Plin5 modification and its binding to interaction partners thereby allowing lipase access to the TG substrate. Finally, Plin5 protein concentrations may generally differ among LDs and thus variably affect TG mobilization leading to increased TG accumulation of a distinct LD population thereby not generally impairing TG catabolism. In line with such an assumption, Plin5-coated LDs did not affect the TG hydrolytic activity of the micellar TG substrate.
Plin5 protein is also present on mitochondria where the protein promotes the tight association of mitochondria and LDs thereby spatially connecting LD TG mobilization and FA release to mitochondrial FA uptake and oxidation (15, 17, 43). Notably, virtually every LD present in CM sections of Plin5 transgenic mice was in close proximity to one or more mitochondria strongly supporting the assumption that Plin5 recruits and tightly attaches mitochondria to the LD surface. Whether this observed phenomenon has an adverse or even beneficial impact on TG mobilization and/or mitochondrial function in cardiomyocytes of Plin5 transgenic mice is currently unknown and an important question to be addressed in further studies.
In summary, the present study reveals an important role of Plin5 in cardiac TG homeostasis via the formation of a lipolytic barrier. Accordingly, mutations linked to increased Plin5 expression may also be involved in the development of cardiac steatosis and dysfunction in humans.
Supplementary Material
Acknowledgments
The authors thank B. Juritsch, B. Seisser, and A. Steiner for their excellent technical assistance.
Footnotes
Abbreviations:
- Acadl
- acyl-CoA dehydrogenase, long chain
- Acadm
- acyl-CoA dehydrogenase, medium chain
- Acadvl
- acyl-CoA dehydrogenase, very long chain
- Acox1
- acyl-CoA oxidase 1
- ATGL
- adipose triglyceride lipase
- Cd36
- cluster of differentiation 36
- CGI-58
- comparative gene indentification-58
- CM
- cardiac muscle
- CM-Plin5
- cardiac muscle-specific overexpression of perilipin 5
- COXIV
- cytochrome c oxidase IV
- CPT
- carnitine palmitoyltransferase
- Cpt1b
- carnitine palmitoyltransferase 1β
- FAO
- fatty acid oxidation
- G0S2
- G0/G1 switch gene 2
- HSL
- hormone-sensitive lipase
- LacZ
- beta-galactosidase
- LD
- lipid droplet
- MHC
- myosin heavy chain
- PAT
- perilipin, adipophilin, and TIP47
- Pdhk4
- pyruvate dehydrogenase kinase, isoenzyme 4
- PGC
- peroxisome proliferator-activated receptor γ coactivator
- Plin1
- perilipin 1
- Plin5
- perilipin 5
- PPAR
- peroxisome proliferator-activated receptor
- Ppara
- peroxisome proliferator-activated receptor α
- Pparg1
- peroxisome proliferator-activated receptor γ isoform 1
- Ppargc1a
- peroxisome proliferator-activated receptor γ coactivator 1α
- Ppargc1b
- peroxisome proliferator-activated receptor γ coactivator 1β
- RT-qPCR
- quantitative reverse transcriptase polymerase chain reaction
- TG
- triacylglycerol
- Tg
- transgenic line
- wt
- wild type
This work was supported by the Austrian Ministry for Science and Research and the grants FWF project DK-MCD W1226, P20602-B05, and SFB Lipotox F30-B05, which are funded by the Austrian Science Fund [Fonds zur Förderung der wissenschaftlichen Forschung (FWF)].
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six figures and one table.
REFERENCES
- 1.Bickel P. E., Tansey J. T., Welte M. A. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta. 1791: 419–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kühnlein R. P. 2012. Lipid droplet-based storage fat metabolism in Drosophila. J. Lipid Res. 53: 1430–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mak H. Y. 2012. Lipid droplets as fat storage organelles in Caenorhabditis elegans. J. Lipid Res. 53: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zechner R., Zimmermann R., Eichmann T. O., Kohlwein S. D., Haemmerle G., Lass A., Madeo F. 2012. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15: 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schweiger M., Lass A., Zimmermann R., Eichmann T. O., Zechner R. 2009. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am. J. Physiol. Endocrinol. Metab. 297: E289–E296. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg A. S., Coleman R. A., Kraemer F. B., McManaman J. L., Obin M. S., Puri V., Yan Q. W., Miyoshi H., Mashek D. G. 2011. The role of lipid droplets in metabolic disease in rodents and humans. J. Clin. Invest. 121: 2102–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okumura T. 2011. Role of lipid droplet proteins in liver steatosis. J. Physiol. Biochem. 67: 629–636. [DOI] [PubMed] [Google Scholar]
- 8.Schaffer J. E. 2003. Lipotoxicity: when tissues overeat. Curr. Opin. Lipidol. 14: 281–287. [DOI] [PubMed] [Google Scholar]
- 9.Listenberger L. L., Han X., Lewis S. E., Cases S., Farese R. V., Jr, Ory D. S., Schaffer J. E. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA. 100: 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Londos C., Sztalryd C., Tansey J. T., Kimmel A. R. 2005. Role of PAT proteins in lipid metabolism. Biochimie. 87: 45–49. [DOI] [PubMed] [Google Scholar]
- 11.Brasaemle D. L. 2007. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48: 2547–2559. [DOI] [PubMed] [Google Scholar]
- 12.Lass A., Zimmermann R., Oberer M., Zechner R. 2011. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog. Lipid Res. 50: 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul A., Chan L., Bickel P. E. 2008. The PAT family of lipid droplet proteins in heart and vascular cells. Curr. Hypertens. Rep. 10: 461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalen K. T., Dahl T., Holter E., Arntsen B., Londos C., Sztalryd C., Nebb H. I. 2007. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim. Biophys. Acta. 1771: 210–227. [DOI] [PubMed] [Google Scholar]
- 15.Wang H., Sztalryd C. 2011. Oxidative tissue: perilipin 5 links storage with the furnace. Trends Endocrinol. Metab. 22: 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Croce M. A., Gropler M. C., Varma V., Yao-Borengasser A., Rasouli N., Kern P. A., et al. 2006. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. 55: 3418–3428. [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Sreenevasan U., Hu H., Saladino A., Polster B. M., Lund L. M., Gong D. W., Stanley W. C., Sztalryd C. 2011. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52: 2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z. 2009. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J. Biol. Chem. 284: 3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Song Y., Zhang L. J., Gu Y., Li F. F., Pan S. Y., Jiang L. N., Liu F., Ye J., Li Q. 2012. LSDP5 enhances triglyceride storage in hepatocytes by influencing lipolysis and fatty acid beta-oxidation of lipid droplets. PLoS ONE. 7: e36712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman syndrome. Cell Metab. 3: 309–319. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., et al. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 306: 1383–1386. [DOI] [PubMed] [Google Scholar]
- 22.Wang H., Bell M., Sreenevasan U., Hu H., Liu J., Dalen K., Londos C., Yamaguchi T., Rizzo M. A., Coleman R., et al. 2011. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J. Biol. Chem. 286: 15707–15715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., et al. 2006. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 312: 734–737. [DOI] [PubMed] [Google Scholar]
- 24.Fischer J., Lefevre C., Morava E., Mussini J. M., Laforet P., Negre-Salvayre A., Lathrop M., Salvayre R. 2007. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat. Genet. 39: 28–30. [DOI] [PubMed] [Google Scholar]
- 25.Lèfevre C., Jobard F., Caux F., Bouadjar B., Karaduman A., Heilig R., Lakhdar H., Wollenberg A., Verret J. L., Weissenbach J., et al. 2001. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am. J. Hum. Genet. 69: 1002–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igal R. A., Rhoads J. M., Coleman R. A. 1997. Neutral lipid storage disease with fatty liver and cholestasis. J. Pediatr. Gastroenterol. Nutr. 25: 541–547. [DOI] [PubMed] [Google Scholar]
- 27.Radner F. P., Streith I. E., Schoiswohl G., Schweiger M., Kumari M., Eichmann T. O., Rechberger G., Koefeler H. C., Eder S., Schauer S., et al. 2010. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58). J. Biol. Chem. 285: 7300–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi T., Matsushita S., Motojima K., Hirose F., Osumi T. 2006. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 281: 14232–14240. [DOI] [PubMed] [Google Scholar]
- 29.Kuramoto K., Okamura T., Yamaguchi T., Nakamura T. Y., Wakabayashi S., Morinaga H., Nomura M., Yanase T., Otsu K., Usuda N., et al. 2012. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J. Biol. Chem. 287: 23852–23863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kienesberger P. C., Pulinilkunnil T., Sung M. M., Nagendran J., Haemmerle G., Kershaw E. E., Young M. E., Light P. E., Oudit G. Y., Zechner R., et al. 2012. Myocardial ATGL Overexpression Decreases the Reliance on Fatty Acid Oxidation and Protects against Pressure Overload-Induced Cardiac Dysfunction. Mol. Cell. Biol. 32: 740–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramaniam A., Jones W. K., Gulick J., Wert S., Neumann J., Robbins J. 1991. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J. Biol. Chem. 266: 24613–24620. [PubMed] [Google Scholar]
- 32.Haemmerle G., Moustafa T., Woelkart G., Buttner S., Schmidt A., van de Weijer T., Hesselink M., Jaeger D., Kienesberger P. C., Zierler K., et al. 2011. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat. Med. 17: 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Folch J., Lees M., Sloane Stanley G. H. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 34.Bieber L. L., Abraham T., Helmrath T. 1972. A rapid spectrophotometric assay for carnitine palmitoyltransferase. Anal. Biochem. 50: 509–518. [DOI] [PubMed] [Google Scholar]
- 35.Bieber L. L., Markwell M. A. 1981. Peroxisomal and microsomal carnitine acetyltransferases. Methods Enzymol. 71: 351–358. [DOI] [PubMed] [Google Scholar]
- 36.Levak-Frank S., Radner H., Walsh A., Stollberger R., Knipping G., Hoefler G., Sattler W., Weinstock P. H., Breslow J. L., Zechner R. 1995. Muscle-specific overexpression of lipoprotein lipase causes a severe myopathy characterized by proliferation of mitochondria and peroxisomes in transgenic mice. J. Clin. Invest. 96: 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X., Lu X., Lombes M., Rha G. B., Chi Y. I., Guerin T. M., Smart E. J., Liu J. 2010. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 11: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweiger M., Paar M., Eder C., Brandis J., Moser E., Gorkiewicz G., Grond S., Radner F. P., Cerk I., Cornaciu I., et al. 2012. G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. J. Lipid Res. 53: 2307–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldberg I. J., Eckel R. H., Abumrad N. A. 2009. Regulation of fatty acid uptake into tissues: lipoprotein lipase- and CD36-mediated pathways. J. Lipid Res. 50: S86–S90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z., Zhou L. 2011. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J. Biol. Chem. 286: 5126–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doolittle M. H., Reue K., editors 1998. Lipase and Phospholipase Protocols. Humana Press, Totowa, NJ. 112–117. [Google Scholar]
- 42.Bartholomew S. R., Bell E. H., Summerfield T., Newman L. C., Miller E. L., Patterson B., Niday Z. P., Ackerman W. E., 4th, Tansey J. T. 2012. Distinct cellular pools of perilipin 5 point to roles in lipid trafficking. Biochim. Biophys. Acta. 1821: 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosma M., Minnaard R., Sparks L. M., Schaart G., Losen M., de Baets M. H., Duimel H., Kersten S., Bickel P. E., Schrauwen P., et al. 2012. The lipid droplet coat protein perilipin 5 also localizes to muscle mitochondria. Histochem. Cell Biol. 137: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.