Abstract
Lipid droplets (LD) are dynamic storage organelles that are involved in lipid homeostasis. Hepatitis C virus (HCV) is closely associated with LDs. HCV Core and nonstructural (NS) proteins colocalize with LDs and presumably are involved in virion formation at that site. We demonstrated that HCV NS4B, an integral membrane protein in endoplasmic reticulum (ER), strongly targeted LDs. Confocal imaging studies showed that NS4B localized at the margins of LDs. Biochemical fractionation of HCV-replicating cells suggested that NS4B existed in membranes associated with LDs rather than on the LD surface membrane itself. The N- and C-terminal cytosolic domains of NS4B showed targeting of LDs, with the former being much stronger. In both domains, activity was present in the region containing an amphipathic α-helix, in which 10 hydrophobic residues were identified as putative determinants for targeting LDs. JFH1 mutants with alanine substitutions for the hydrophobic residues were defective for virus replication. W43A mutant with a single alanine substitution showed loss of association of NS4B with LDs and severely reduced release of infectious virions compared with wild-type JFH1. NS4B plays a crucial role in virus replication at the site of virion formation, namely, the microenvironment associated with LDs.
Keywords: alanine substitution, confocal imaging, JFH1, RNA transfection
Cellular lipid droplets (LD) are dynamic organelles connecting storage, metabolism, and dynamics of lipids in eukaryotic cells (1–3). LDs consist of core neutral lipids, mainly triglycerides and cholesteryl esters, and the surrounding phospholipid monolayer. The lipids stored in LDs are used for various cellular events, such as metabolism as an energy source, membrane synthesis, steroid and eicosanoid synthesis, lipoprotein formation, and protein modification. LDs are thought to be generated from endoplasmic reticulum (ER) and move in a microtubule-dependent, bidirectional manner, interacting with other organelles, such as ER, mitochondria, and peroxisomes.
Recently, a close association between LDs and hepatitis C virus (HCV) was revealed (4). HCV infects about 150 million to 200 million people worldwide, and chronic infection is associated with liver steatosis, cirrhosis, and hepatocellular carcinoma. HCV is a positive- and single-stranded RNA virus with about 9,600 nucleotides of genome (5), which codes 10 HCV proteins: Core, envelope 1 (E1), envelope 2 (E2), p7, nonstructural protein (NS)2, NS3, NS4A, NS4B, NS5A, and NS5B. All of these proteins are essential for replication of HCV. Both the 5′ and 3′ untranslated regions (UTR) of the genome are also essential for viral replication: the 5′ UTR has an internal ribosome entry site (6), and the 3′ UTR contains a highly conserved structure, the 3′ X (7, 8). The infectious viral particles were found to contain the VLDL components, such as triglycerides, cholesterol, and apoB and E, and are therefore called “lipoviroparticles” (9–12). HCV infects cells by utilizing the LDL receptor and scavenger receptor B1 in addition to other receptor molecules, such as CD81, claudin 1, and occludin (13). Through the life cycle of HCV, namely, from viral entry to virion release, cellular lipids and lipid metabolism, especially in lipoproteins, play important roles (10–12). HCV modulates cellular lipid metabolism to facilitate its replication: for example, HCV upregulates lipogenesis, resulting in accumulation of triglycerides in the increased mass of LDs. Clinically, liver steatosis is found in about half of chronic HCV patients. Liver steatosis occurs also in transgenic mice harboring the HCV Core protein (14). Inhibition of microsomal triglyceride transfer protein activity and VLDL secretion by HCV protein(s) is also an important mechanism of the lipid accumulation in hepatocytes (11). Several studies have shown that many HCV-derived components, such as Core (15) and NS proteins [NS2 (16), NS3, NS4A, NS4B, NS5A (17), and NS5B] and HCV RNAs, also localize at sites close to LDs (4). Core and NS5A are thought to mediate association of these HCV components with LDs (4); however, the molecular mechanisms by which NS proteins associate with LDs remain unclear. Ultrastructural studies showed accumulation of virion-like structures around LDs (18). Although several cellular and viral proteins localize on the LD membrane, consensus signals for LD targeting are not defined (1). Recent reports have shown that LD-targeting activity resides in amphipathic helices (19–23) or in hydrophobic regions (24–26).
Here, we present evidence that HCV NS4B strongly targets LDs. NS4B is a 261-amino-acid, multifunctional protein consisting of N- and C- terminal cytosolic domains and a central membrane domain harboring at least four transmembrane helices (27). NS4B is an integral membrane protein in ER (28) that can induce alteration of the membrane structure to form a membranous web (29), in which HCV RNA replication is thought to occur (30–32). We provide evidence to suggest how ER-resident protein NS4B can interact with LDs. Imaging studies, including a series of mutation analyses, revealed the regions responsible for LD targeting of NS4B. The hydrophobic amino acid residues in amphipathic α-helices are critical for LD targeting. The JFH1 (33) mutant harboring alanine substitutions for the hydrophobic residues of NS4B was defective for virus replication. In a single alanine substitution mutant, W43A, we observed that loss of LD targeting of NS4B caused a severe defect in virus replication. Our results strongly suggest that NS4B functions at sites close to LDs, which is critical for replication of HCV.
MATERIALS AND METHODS
Cell culture system
Oc cells (34) and OR6 HCV-RNA-replicating cells (35) were cultured in collagen (type I)-coated dishes (Iwaki, Chiba, Japan) with DMEM (Sigma) supplemented with 10% FBS, penicillin, and streptomycin.
Antibodies
The antibodies used in this study were those against Core (CP11; MBL, Nagoya, Japan), NS3 (R212), NS4B (52-1, detection for HCV-O NS4B), NS4AB (RR12, detection for JFH1 NS4B), NS5A (8926; BioAcademia, Osaka, Japan), NS5B (NS5B-6, BioAcademia), adipose differentiation-related protein (ADRP; Abcam), FLAG (Sigma), myc-tag (MBL), mCherry (Clontech), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Millipore), and calnexin (BD Transduction Laboratories). The anti-NS3 (R212), anti-NS4B (52-1), and anti-NS4AB (RR12) antibodies were gifts from Dr. M. Kohara (Tokyo Metropolitan Institute of Medical Science, Japan). Rabbit polyclonal antibodies specific to JFH1 NS3, NS5A, and NS5B were raised against bacterially expressed glutathione S-transferase (GST)-NS3 (amino acids 165–631), GST-NS5A (amino acids 25–466), and GST-NS5B (amino acids 1–570), respectively. Alexa-Fluor-555-conjugated anti-rabbit IgG (Invitrogen), Alexa-Fluor-568-conjugated anti-mouse IgG (Invitrogen), and rhodamine-conjugated anti-mouse IgG (Cappel) antibodies were also used.
Expression plasmids
HCV sequences for expression experiments (NS4B, NS5A and Core) were derived from type 1b HCV strain O [HCV-O, DDBJ/EMBL/GenBank accession number AB191333 (34)]. The HCV fragment was amplified by PCR using the restriction-site-tagged primers with an appropriate template plasmid that contained the desired HCV sequence. To create fusion constructs with fluorescent protein, the BglII-EcoRI-digested PCR fragment was ligated with BglII-EcoRI-digested pmCherry-C1 or pEGFP-C1 vector (Clontech). In another case, the NheI-HindIII-digested PCR fragment was ligated with NheI-HindIII-digested pcDNA3.1(+) vector (Invitrogen) to create expression constructs. To create a mutant fragment, primers that had desired mutated sequences were used for PCR, and usually two appropriate PCR fragments, namely, “-N” and “-C,” were connected by overlapping PCR. Sequences of the primers and their usage, and which fragment the primer sets amplified, are described in supplementary Tables I and II, respectively. pDsRed2-ER, which expressed an ER marker protein, was obtained from Clontech. For construction of enhanced green fluorescent protein (EGFP)-ADRP and Cherry-ADRP, ADRP cDNA was synthesized from Superscript III (Invitrogen) with a primer 5′-CCACAGCATGCACTAGTGAT-3′ from total RNAs prepared from Oc cells with Trizol reagent (Life Technologies) according to the manufacturer's protocol. The ADRP fragment was amplified using the restriction-site-tagged primers 5′-TTTAGATCTATGGCATCCGTTGCAGTTGA-3′ and 5′-TTTGAATTCTTAATGAGTTTTATGCTCAGATCGC-3′, digested with BglII and EcoRI, and ligated with the BglII-EcoRI-digested pmCherry-C1 or pEGFP-C1 vector.
Transient expression experiment
Oc cells (105 cells/well) were seeded on a glass-based, 24-well culture plate (EZView, Iwaki), and after being cultured for 16 h, the cells were transfected with expression plasmids with the aid of Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Usually, 1.6 μg plasmid was used for each well with 1:2.5–3 ratio to the lipofection reagent. When two kinds of plasmids were cotransfected into cells, 1.0–1.6 μg of the plasmid was used.
Immunofluorescence
After 24 h transfection, the cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.01% digitonin for 30 min, and stained with Bodipy 493/503 as described previously (36). When antigens were visualized by indirect immunofluorescence, the fixed permeabilized cells were blocked with 1% BSA in TBS (20 mM Tris-HCl, 0.15 M NaCl, pH 7.4) and reacted with an appropriate dilution of primary antibody in 1% BSA-TBS for 1 h at room temperature. After three washes with TBS, the cells were incubated with an Alexa-Fluor-dye-conjugated (Invitrogen) or rhodamine-conjugated (Cappel) secondary antibody with 1,000-fold dilution in 1% BSA-TBS for 30 min. Cell nuclei were stained with Hoechst 33342 for 20 min (nuclei were stained in most of the experiments but not always shown in the figures). The fluorescent images were collected by confocal laser scanning microscope FV1000 (Olympus, Tokyo, Japan) in a sequential scanning mode.
Immunoblotting
Proteins were resolved on 8 or 12% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare). The membranes were rinsed in water and then soaked in TBS-0.1% Tween 20 (TBST) for 30 min to enhance detection sensitivity (37). The membranes were again rinsed in water and blocked with PVDF Blocking Reagent (Toyobo, Osaka, Japan) for 1 h at room temperature. The membranes were reacted with an appropriate dilution of primary antibody in Can Get Signal Immunoreaction Enhancer Solution 1 (Toyobo) for 1 h at room temperature. After washing with TBST (five times for 10 min each), the membranes were reacted with an appropriate dilution of horseradish-peroxidase-conjugated secondary antibody in Can Get Signal Immunoreaction Enhancer Solution 2 (Toyobo) for 30 min at room temperature. After washing with TBST (five times for 10 min each), the blots were visualized by an ECL Plus immunoblot detection system (GE Healthcare).
Construction of JFH1 mutant
We generated JFH1 (33) (DDBJ/EMBL/GenBank accession number AB047639) mutant constructs that contained alanine substitutions for hydrophobic residues in the NS4B region as follows: 4Bmtmt, W43A, V46A, W50A, F57A, I61A, L64A, I242A, L246A, L249A, and I253A; 4Bmt0, W43A, V46A, W50A, F57A, I61A, and L64A; 4Bmt1, W43A, V46A, and W50A; 4Bmt2, F57A, I61A, and L64A; and 4Bmt3, I242A, L246A, L249A, and I253A. Mutants harboring single alanine substitutions, W43A, V46A and W50A, were also created. To create a mutant fragment, primers with the desired mutated sequences were used for PCR, and two or three appropriate PCR fragments (“-N” and “-C” or “-N1,” “-N2,” and “-C”) were connected by overlapping PCR. The resultant mutant fragment was digested with NsiI and RsrII and ligated with NsiI-RsrII-digested pJFH1 (33). Sequences of the primers and their usage and which fragment the primer sets amplified are described in supplementary Tables III and IV, respectively.
In vitro transcription and electroporation of viral RNA into Oc cells
pJFH1 and its mutant plasmids were linearized with XbaI followed by treatment with mung bean nuclease (Toyobo) according to the manufacturer's protocol. RNA was transcribed in vitro from the linearized constructs using MEGAscript T7 kit (Ambion) and purified with MEGAclear kit (Ambion) according to the instruction manual. RNA integrity was checked by agarose gel electrophoresis. Prior to electroporation, Oc cells were trypsinized and resuspended in complete DMEM. The cells were washed twice with PBS and resuspended in buffer R (Invitrogen) at a concentration of 107 cells/ml. JFH1 and mutant RNA was introduced into the Oc cells using the NEON transfection system (Invitrogen) according to the instruction manual as follows. For titration of virus production, 0.5 μg RNA was introduced into 105 cells in a 10 μl tip supplied by the manufacturer with two pulses of 30 ms at 1,150 V. Usually, four shots of the electroporated cells (4 × 105 cells) are pooled in 4 ml prewarmed DMEM supplemented with 10% FBS and seeded into 12-well plates (1 ml for each well). For preparation of LDs, 5 μg RNA was introduced into 106 cells in a 100 μl tip supplied by the manufacturer, with two pulses of 30 ms at 1,150 V. Three shots of the electroporated cells (3 × 106 cells) were pooled in 30 ml prewarmed DMEM supplemented with 10% FBS and seeded into dishes with a diameter of 10 cm (10 ml each). At 48 h posttransfection, LDs were prepared as described below.
Cell fractionation by step-wise sucrose density gradient
OR6 cells were fractionated in sucrose density gradient as described previously (38) with minor modifications. Twelve hours before preparation, 100 μM oleate was added to the culture medium. Cells (2 × 106 to 3 × 106 cells) were homogenized in 0.5 ml buffer A [10 mM tricine, 3 mM EDTA, and Complete Protease Inhibitor Cocktail (Roche), pH 7.4] containing 0.25 M sucrose with 30 strokes of a glass Dounce homogenizer using a tight-fitting pestle. The lysate was centrifuged for 10 min at 1,000 g at 4°C. The sucrose concentration of the postnuclear supernatant (PNS) was adjusted to 26% by adding buffer A containing 60% sucrose. In a 5 ml ultracentrifuge tube (5PA; Hitachi Koki, Tokyo, Japan), 0.34 ml buffer A containing 51% sucrose and 0.75 ml buffer A containing 43% and 35% sucrose were layered. Subsequently, 0.63 ml 26% sucrose-PNS was layered on top of this. Following this, 0.75 ml buffer A containing 18% and 10% sucrose was layered sequentially onto the PNS fraction. Finally, 0.97 ml of buffer A containing 2% sucrose was loaded on top. The step-wise gradient was centrifuged at 32,000 rpm, at 4°C for 90 min using an S52ST rotor (Hitachi Koki). Following centrifugation, the samples (0.5 ml each) were collected from the bottom.
Preparation of LDs
LDs were prepared as described previously (4) with minor modifications. Twelve hours before preparation, 100 μM oleate was added to the culture medium. Samples and buffers were handled on ice or at 4°C through the procedures below. Cells (∼3 × 106 OR6 or Oc cells transfected with JFH1 or mutant RNA) were pelleted by centrifugation at 3000 rpm for 5 min at 4°C. The pellet was resuspended in 1.2 ml hypotonic buffer (50 mM HEPES, 1 mM EDTA, 2 mM MgCl2, and Complete Protease Inhibitor Cocktail, pH 7.4) and incubated for 10 min. The suspension was homogenized with 30 strokes of a glass Dounce homogenizer using a tight-fitting pestle, followed by addition of 120 μl 10× sucrose buffer [0.2 M HEPES, 1.2 M potassium acetate, 40 mM Mg(oAc)2, and 50 mM DTT, pH 7.4]. The nuclei were removed by centrifugation at 2,000 rpm for 10 min at 4°C. The supernatant was collected and centrifuged at 16,000 g for 10 min at 4°C. The supernatant (S16, 1.25 ml) was mixed with 1.25 ml 1.04 M sucrose in isotonic buffer (50 mM HEPES, 100 mM KCl, 2 mM MgCl2, and Complete Protease Inhibitor Cocktail). The solution was set at the bottom of 5 ml 5PA ultracentrifuge tubes, and 2.5 ml isotonic buffer was loaded onto the sucrose mixture. The gradient was centrifuged at 100,000 g in an S52ST rotor for 45 min at 4°C. After centrifugation, the LD fraction on the top of the gradient solution was collected as the first LD fraction (LD1). After 0.1 ml of the fraction was removed for analysis, the volume of the LD1 fraction was then adjusted to 1.25 ml with isotonic buffer and the fraction was mixed with 1.25 ml isotonic buffer containing 1.04 M sucrose. The fraction was set at the bottom of the ultracentrifuge tube and centrifuged again at 100,000 g as described above. The LD fraction on the top of the gradient solution was collected as the second LD fraction (LD2).
Titration of HCV infection
Culture supernatant of RNA-transfected cells was filtrated (0.45 μm pore filter) and assayed for infectivity titer by the end-point dilution method. Before 16 h inoculation, Oc cells were seeded into 96-well plates at a density of 8 × 103/well. Samples were serially diluted 10-fold in complete growth medium, and 20 μl (when necessary, 100 μl) was inoculated into Oc cells in each well in duplicate. After three days of incubation, the cells were immunostained by anti-Core antibody (CP11). Positive foci were counted, and the infectivity titer was calculated from the average number of foci counted in the last and second-to-last wells of the dilution series that still contained positive foci. The virus titer was expressed as focus-forming units per milliliter of supernatant.
RESULTS
The intrinsic localization of NS4B to LDs
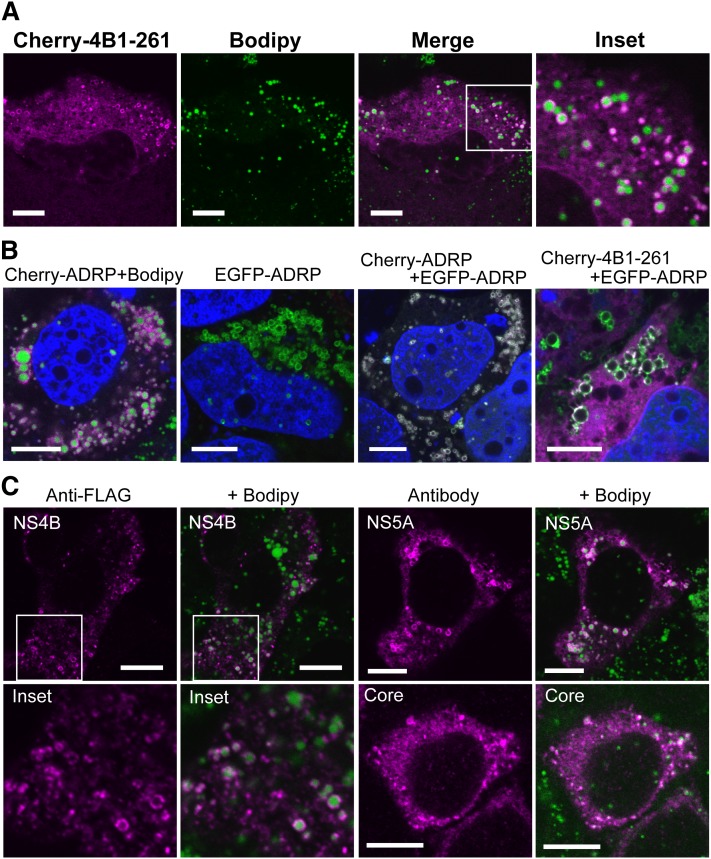
To clarify whether NS4B associated with LDs by itself and the mechanisms of LD targeting by NS4B, we performed fluorescent imaging using confocal microscopy (Fig. 1 and supplementary Fig. I). NS4B was fused with mCherry or EGFP, and subcellular localization of the fusion proteins was observed simultaneously with that of LDs in Oc cells. LDs were visualized by Bodipy 493/503. We confirmed that the native forms of mCherry or EGFP, found in both cytosol and the nucleus, showed no colocalization with LDs (supplementary Fig. I-A). When the full-length NS4B was fused with the color proteins (Cherry-4B1–261 and EGFP-4B1–262), a major portion of NS4B showed a membranous, reticular pattern of localization in both cases, suggesting the localization to ER. Upon coexpression with an ER marker construct pDsRed2-ER (DsRed-ER), which contained the ER-targeting sequence of calreticulin and the ER retention signal KDEL, localization of NS4B was consistent with that of the ER marker (supplementary Fig. I-A). This suggested that the full-length NS4B resided in ER membrane as previously reported (28, 39). However, we found a small level of ring-shaped localization of NS4B surrounding the green circular fluorescence of Bodipy, which may correspond to the margin of LDs (Fig. 1A and supplementary Fig. I-B). This ring-shape localization pattern merged with that obtained with anti-ADRP antibody (data not shown), suggesting that NS4B localized to the surface of LDs (LD membrane itself or ER membranes associated with LDs). To confirm the colocalization of NS4B with ADRP, we made fusion constructs of ADRP with color fluorescent proteins and coexpressed them with NS4B (Fig. 1B). The fusion constructs of ADRP (Cherry-ADRP and EGFP-ADRP) showed clear ring-shape localization, and the images of Cherry-ADRP and EGFP-ADRP were completely merged, indicating their localization to the LD membranes (Fig. 1B). When Cherry-4B1–261 was coexpressed with EGFP-ADRP, a portion of Cherry-4B1–261 showed ring-shape localization that was consistent with that of EGFP-ADRP, suggesting colocalization of the fusion proteins (Fig. 1B and supplementary Fig. I-C). Thus, a portion of expressed NS4B was thought to localize on the surface of LDs. To represent the degree of LD targeting for each construct, we categorized the NS4B-expressing cells (for at least 50 transfectants) into the following four categories: exclusive or dominant localization of the protein to LDs in the observed section (category I); apparent (but not dominant) localization to LDs (category II); slight localization to LDs (with 1–5 rings surrounding LDs seen in the section, category III); and no localization to LDs (category IV). For Cherry-4B1–261 transfectant, about 4 and 15% of the cells were classified into categories I and II, respectively (the results of the count are described in Table 1). Thus, NS4B colocalized with LDs in about 20% of cells.
Fig. 1.
Full-length NS4B localizes to LDs. (A) Localization of Cherry-4B1–261. Oc cells were transfected with Cherry-4B1–261 for 24 h followed by staining with Bodipy493/503. (A, inset) magnified image. Scale bars, 10 μm. (B) Colocalization of Cherry-4B1–262 with ADRP. From left, Cherry-ADRP, EGFP-ADRP, Cherry-ADRP with EGFP-ADRP (cotransfection), and Cherry-4B1–261 with EGFP-ADRP (cotransfection) was transfected into Oc cells. Scale bars, 10 μm. (C) Localization of NS4B, NS5A, and Core protein. Oc cells were transfected with pcDNA-4B1–261-FLAG (left), pcDNA-5A-myc (right, top), or pcDNA-CORE (right, bottom) for 24 h. After fixation, the antigens were visualized with anti-FLAG (NS4B), anti-Myc (NS5A), or anti-Core antibodies and Alexa-Fluor-dye-labeled secondary antibodies. Scale bars, 10 μm. Inset, magnified image.
TABLE 1.
LD-targeting activity of various NS4B constructs
| Category (% Cells) |
||||
| Construct | I | II | III | IV |
| Cherry-4B1–261a | 3.7 ± 1.5 | 15 ± 7 | 11 ± 4 | 71 ± 9 |
| Cherry-4B1–73a | 71 ± 12 | 29 ± 12 | 0 | 0 |
| Cherry-4B74–191 | 0 | 0 | 0 | 100 |
| Cherry-4B192–261a | 14 ± 7 | 34 ± 5 | 22 ± 9 | 30 ± 8 |
| Cherry-4B1–49 | 0 | 0 | 0 | 100 |
| Cherry-4B26–73 | 15 | 45 | 21 | 19 |
| Cherry-4B1–25 | 0 | 0 | 0 | 100 |
| Cherry-4B26–49 | 0 | 0 | 0 | 100 |
| Cherry-4B50–73 | 0 | 2 | 21 | 77 |
| Cherry-4B192–239 | 0 | 0 | 2 | 98 |
| Cherry-4B218–261 | 0 | 0 | 10 | 90 |
| Cherry-4B192–217 | 0 | 0 | 0 | 100 |
| Cherry-4B218–239 | 0 | 0 | 0 | 100 |
| Cherry-4B240–261 | 0 | 0 | 8 | 92 |
| pcDNA-4B1–261-FLAG | 0 | 3 | 5 | 93 |
The degree of LD targeting for each cell is expressed as category I, II, III, or IV. Each construct was transfected into Oc cells, and the percentages of cells with each category are shown.
Results are mean ± SD from three independent experiments.
We next expressed full-length NS4B fused with an epitope tag FLAG (amino acid sequence, DYKDDDDK) (pcDNA4B1–261-FLAG) and detected it using an anti-FLAG antibody (this is because we could not obtain antibodies that detected NS4B in HCV-O via immunofluorescence). In most cases, the stain showed differently sized dots, but no clear reticular pattern was obtained. However, we also found that, in some cells, the small dots of NS4B surrounded LDs, suggesting LD localization of FLAG-tagged NS4B (Fig. 1C). Thus, NS4B localizes to LDs by itself. A similar pattern of LD localization was observed for HCV NS5A [tagged with myc (EQKLISEEDL) at the C terminus] and Core, which are known to associate with the LD surface (Fig. 1C). We also found that NS5AΔ1–30, which lacks the N-terminal amphipathic α-helix (amino acids 1–30), did not localize to LDs (data not shown). We conclude that NS4B has an intrinsic activity to target the surface of LDs.
Subcellular distribution of NS4B in OR6 cells
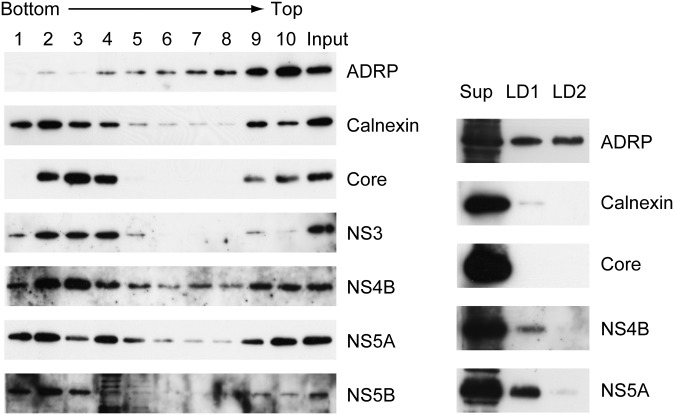
We investigated whether intact NS4B targets LDs in the presence of other HCV proteins. We could not obtain antibodies applicable to immunofluorescent study for HCV-O; therefore, we adopted a biochemical approach. Subcellular fractionation of OR6 HCV-RNA-replicating cells was performed and distribution of HCV antigens was examined by immunoblotting. In OR6 cells, genome-length RNA of HCV-O (fused with Renilla luciferase gene for monitoring of HCV genome replication) was stably replicated; hence, intact functional HCV antigens were expressed in the cells. Distributions of cellular and HCV antigens in fractions obtained from OR6 cells by discontinuous sucrose density gradient (38) are shown in Fig. 2 (left). ADRP was detected most abundantly in the top fraction (fraction 10) followed by fraction 9, suggesting that LDs were enriched in fraction 10 and partly in fraction 9. Calnexin, a marker protein of ER, was found mostly concentrated in fraction 2, showing that a major portion of ER was fractionated into fraction 2. A small portion of calnexin was also enriched in fraction 9 (and 10), suggesting that this fraction contained ER membranes associated with LDs. HCV antigens (Core, NS3, NS4B, NS5A, and NS5B) were detected most abundantly in fraction 2, suggesting that these HCV antigens were associated with ER membranes. However, a small portion of HCV antigens, especially Core, NS4B and NS5A, were clearly detected in fractions 10 and 9. This suggested that a substantial amount of HCV antigens, including NS4B, were present in LD-associated ER membranes or in the LD membrane itself in OR6 cells.
Fig. 2.
Localization of HCV antigens to LDs in OR6 cells. (Left) Distribution of cellular and HCV antigens in fractions obtained by discontinuous sucrose density gradient ultracentrifugation (51, 43, 35, 26, 18, 10, and 2% sucrose from bottom to top). OR6 cells were fractionated. The 1/100 vol of each fraction (1–10) and 1/1,000 vol of original PNS (Input) were run on 12% SDS-PAGE followed by immunoblot analysis. (Right) Localization of HCV antigens to LD preparations. LDs were prepared from OR6 cells. LD1, LD preparation obtained from first ultracentrifugation; LD2, final LD preparation obtained from second ultracentrifugation; Sup (S16), supernatant after 16,000 g centrifugation. Fractions were analyzed by immunoblotting. The amount of each fraction applied for immunoblotting was adjusted to give approximately equal signal intensity for ADRP. The apparent molecular weights of the indicated proteins were: ADRP, 52 kDa; calnexin, 90 kDa; Core, 21 kDa; NS3, 70 kDa; NS4B, 27 kDa; NS5A, 56/58 kDa; and NS5B, 66 kDa.
We also prepared LDs from OR6 cells using sucrose density gradient fractionation (by two rounds of discontinuous sucrose gradient ultracentrifugation) as previously described (4) (Fig. 2, right). LD1 was a crude LD preparation after the first ultracentrifugation, and LD2 was the final LD preparation from the second ultracentrifugation. To evaluate the enrichment of antigens to LDs, the amount of each fraction used for immunoblotting was adjusted to give approximately equal signal intensities for ADRP. LD1 seemed to contain a small amount of ER proteins, presumably LD-associated, because a small amount of calnexin was detected in LD1. A small amount of NS4B was detected in LD1 (and a small amount in LD2 with a long chemiluminescence lifetime; data not shown). NS5A was clearly detected in LD1, and a small amount in LD2 as expected. However, Core protein was not detected in either LD preparation, even with longer exposure.
NS4B of JFH1 strain targets LDs in RNA-transfected cells
HCV strain JFH1 (subtype 2a) is the first effective replication/infection system of HCV (33). When the genome-length JFH1 RNA is transfected into HuH-7 and its derivative cells, virus replication occurs in cells and infectious virion is released into the culture medium. We transfected JFH1 RNA into Oc cells and prepared and analyzed the LDs by immunoblotting (supplementary Fig. II). NS3, NS4B, NS5A, and NS5B proteins were all positive in both LD1 and LD2. NS4B was apparently enriched in LD1 as compared with the ratio of calnexin content between Sup (S16) and LD1, but it was much less abundant in LD2 than in LD1. This distribution profile suggested that NS4B was localized to LD-associated ER membranes rather than to the LD membranes themselves. Core protein was not detected in LD preparations as in those of OR6 cells; instead, it was precipitated to the bottom of the ultracentrifugation tube.
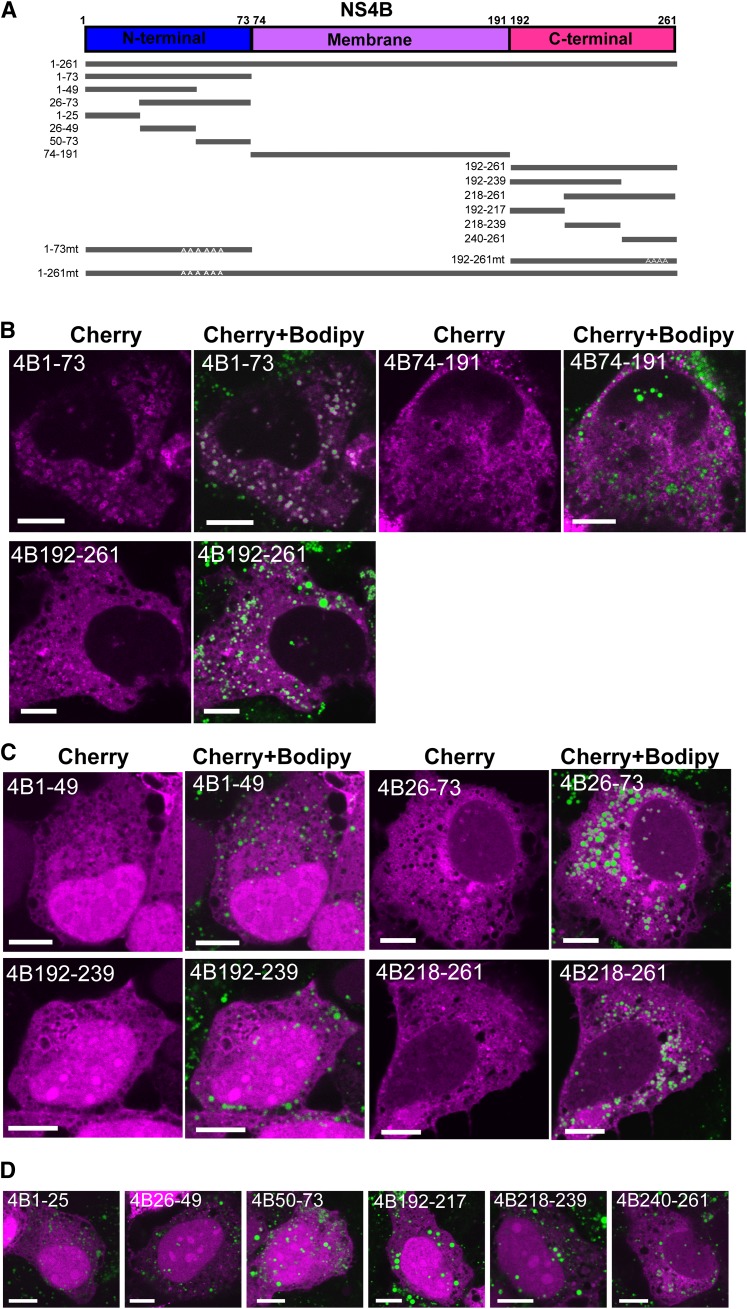
NS4B has two independent LD-targeting regions in the N- and C-terminal cytosolic domains
To determine which regions or motifs in NS4B were responsible for the targeting to LDs, various NS4B constructs of HCV-O were prepared as fusion proteins with mCherry (Fig. 3A). The results were similar for mCherry and EGFP constructs; therefore, we chose mCherry as a tag protein together with Bodipy 493/503 to stain LDs. We determined the activity of the N- and C-terminal cytosolic domains and the central membrane domain (Fig. 3B and Table 1). Crystal structure analysis is not yet possible for NS4B; therefore, the boundary of the N-terminal and central membrane domains was not determined. The boundary of the central and C-terminal domains has been determined from the secondary structure prediction as being between 191 and 192 (27). We tentatively defined the domains as amino acids 1–73, 74–191, and 192–261, respectively, and expressed them in Oc cells. We found that the N-terminal domain (Cherry-4B1–73) showed a strong LD localization. For Cherry-4B1–73, about 70% of the cells were counted as category I and 30% as II; therefore, in nearly all of the cells, the N-terminal domain of NS4B showed LD localization. When the membrane domain (Cherry-4B74–191) was expressed in cells, many ring-shaped localizations were seen, but they had no relation with LDs (Fig. 3B). This ring-shape structure implies that NS4B activity can alter the membrane structure of ER, forming a “membranous web” (29).
Fig. 3.
Two independent target sites for LDs are present in NS4B. (A) Schematic representation of the intact and mutant NS4B. NS4B proteins were N-terminally tagged with mCherry protein. Numbers indicate the amino acid position in NS4B, and mt denotes the alanine substitution construct. (B) Localization of the N-terminal, central, and C-terminal domains of NS4B. Oc cells were transfected with Cherry-4B1–73, Cherry-4B74–191, or Cherry-4B192–261 for 24 h, and then fixed and stained with Bodipy 493/503. (C, D) Localization of the subdivided N- and C-terminal domains of NS4B. (B–D) Scale bars, 10 μm.
The C-terminal domain (Cherry-4B192–261) also showed LD localization, but to a lesser extent than that of the N-terminal region: ∼10% of the cells were category I, and 30% were category II. To identify the detailed LD-binding region, each terminal region was further divided into three, and their localization or that of the region spanning the two parts (two thirds of the terminal domain) was determined (Fig. 3C, D and Table 1). For the N-terminal region, Cherry-4B26–73 showed significant LD targeting. Within the amino acid residues 26–73, Cherry-4B26–49 showed no activity, whereas Cherry-4B50–73 was slightly active. Although the degree of localization was much lower than that of Cherry-4B1–73, the N-terminal site for LD localization was identified in the C-terminal half (or one third) of the N-terminal domain (Table 1). Regarding the C-terminal region, Cherry-4B218–261 and Cherry-4B240–261 were slightly active, suggesting that the C-terminal site is in the C-terminal half (or one third) of the C-terminal domain, although the region had little activity by itself (Table 1). Thus, NS4B had two independent regions for targeting LDs.
Hydrophobic amino acid residues in amphipathic α-helices in NS4B are critical for LD targeting
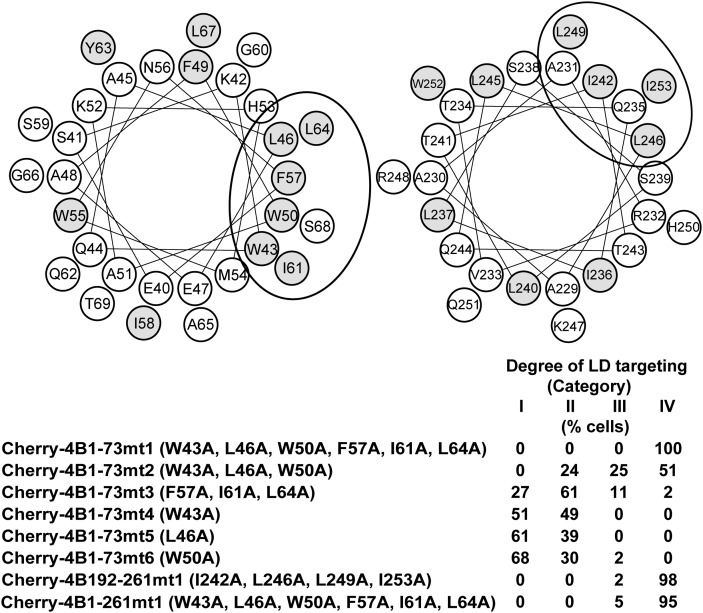
For both the N- and C-terminal sites, an amphipathic α-helix has been identified: amino acids 40–69 for the N-terminal region, and 229–253 for the C-terminal region. Both helices are thought to mediate membrane association of NS4B (40, 41). Amphipathic helices are also known to mediate LD association (19–23); therefore, we hypothesized that the amphipathic helices, especially the hydrophobic amino acids within them, were important for LD targeting of NS4B. From helical wheel projection analysis (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi, provided by D. Armstrong et al.), we noticed that 10 hydrophobic residues (43W, 46L, 50W, 57F, 61I, and 64L for the N-terminal amphipathic helix; 242I, 246L, 249L, and 253I for the C-terminal amphipathic helix) were present on the hydrophobic faces of the helices (Fig. 4). To establish whether these residues were involved in LD targeting, we made various alanine substitutions for the residues. The influence of mutations was evaluated by reduction of LD localization as compared with the mother constructs (Fig. 4).
Fig. 4.
Hydrophobic amino acid residues in amphipathic α-helices are critical for targeting LDs. (Top) Helical wheel projection analysis (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi) for amphipathic α-helices in the N-terminal (left) and C-terminal (right) domains. Hydrophobic residues (W, F, Y, L, I) are shown as gray circles. The ellipse shows a putative hydrophobic face. (Bottom) Inhibition of LD-targeting activity by alanine substitutions for the hydrophobic residues. Localization to LDs was determined for each mutant, and the degree of LD targeting is presented by the percentages of cells classified into categories I–IV. Category I, expressed protein localized to LDs exclusively or dominantly; category II, expressed protein localized to LDs apparently but not dominantly (at least six ring-shape localizations surrounding LDs are seen in the section); category III, expressed protein slightly localized to LDs (1–5 ring-shape localizations surrounding LDs are seen in the section); category IV, expressed protein did not localize to LDs.
When all of the six residues in the N-terminal amphipathic helix were substituted by alanine (Cherry-4B1–73mt1), the LD-targeting activity of Cherry-4B1–73 completely disappeared. As a result of a six-residue interval between 50W and 57F, the hydrophobic residues could be segregated into two clusters of three hydrophobic residues each. When the three residues of the cluster were substituted simultaneously, the influence on the LD-targeting activity was stronger by mutating the upstream cluster (Cherry-4B1–73mt2, containing W43A, L46A, and W50A) than the downstream cluster (Cherry-4B1–73mt3, containing F57A, I61A, and L64A) (Fig. 4). The single residue substitution in the upstream cluster, W43A, L46A, or W50A, did not significantly affect the LD-targeting activity of Cherry-4B1–73 (Cherry-4B1–73mt4, mt5, and mt6). The results indicated that the hydrophobic residues in the amphipathic helix were required for the LD-targeting activity in the N-terminal domain of NS4B, in which the residues in the upstream cluster (43W, 46L, and 50W) had a relatively stronger effect in a synergistic manner.
Regarding the C-terminal domain of NS4B, we made alanine substitutions for all four residues simultaneously (Cherry-4B192–261mt1, containing I242A, L246A, L249A, and I253A), because the activity of the mother Cherry-4B192–261 was relatively low (Table 1). This mutant showed no activity for LD targeting. Although the degree of contribution from each residue was not clear, these hydrophobic amino acids were considered to be critical for the LD-targeting activity of the C-terminal domain. Similarly, we made six alanine substitutions simultaneously (W43A, L46A, W50A, F57A, I61A, and L64A) in a full-length NS4B construct (Cherry-4B1–261mt1), because of the relatively low targeting activity of the mother construct Cherry-4B1–261. This alanine mutant showed no apparent activity for LD targeting, indicating that the hydrophobic residues were critical for LD targeting of full-length NS4B.
When the expression of cherry fusion proteins was checked by immunoblotting, each construct showed its predicted molecular weight, although differences in expression level were seen among the different constructs (supplementary Fig. III). Cherry-4B1–73 and its mutants showed dual bands by unknown mechanisms; however, this seemed to be unrelated to the degree of LD-targeting for each protein. It should be noted that the 10 amino acid residues described here (43W, 46L, 50W, 57F, 61I, 64L, 242I, 246L, 249L, and 253I) were highly conserved among the wide variety of subtypes of HCV, supporting their critical role in the virus life cycle (supplementary Fig. IV).
JFH1 mutants harboring alanine substitutions abolishing LD-targeting activity of NS4B are defective for replication
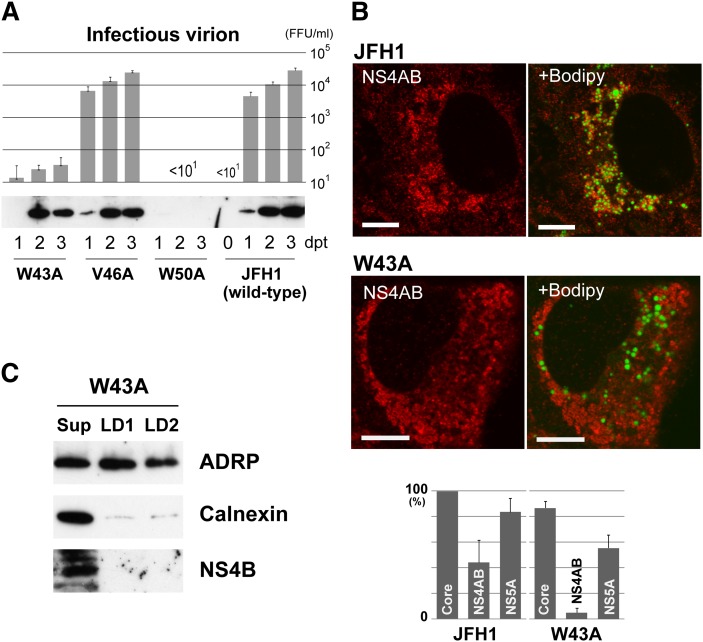
To clarify the importance of LD targeting of NS4B for replication of HCV, we examined JFH1 mutants that contained alanine substitutions of hydrophobic residues critical for LD targeting in the NS4B region. The 10 hydrophobic residues, 43W, 46V (L in strain O), 50W, 57F, 61I, 64L, 242I, 246L, 249L, and 253I, were divided into three clusters as described above, and mutants were created for each cluster of three or four simultaneous alanine substitutions: 4Bmt1, W43A, V46A, and W50A; 4Bmt2, F57A, I61A, and L64A; and 4Bmt3, I242A, L246A, L249A, and I253A. When the wild-type and mutant RNAs were transfected into Oc cells, wild-type JFH1 released an increasing number of infectious virions (Fig. 5A), whereas the cluster mutants (4Bmt1, 4Bmt2, and 4Bmt3) produced no detectable infectious virions. For all these mutants, no HCV antigens were detected by immunoblotting in transfected cells (Core, NS4B, and NS5A were tested; data not shown). The cluster mutants were defective for virus replication. We then concentrated on the first N-terminal cluster and created single alanine substitution mutants for the three hydrophobic residues, W43A, V46A, and W50A, because the cluster was considered to be most effective for LD targeting of NS4B as described above. When RNAs were transfected into Oc cells, the mutants released different numbers of infectious virions (Fig. 5A). As compared with wild-type, W43A released only 1/300 to 1/1,000 of infectious virions, and V46A released comparable amounts of virions. W50A released no detectable infectious virions. Interestingly, W43A mutant, as well as V46A, expressed comparable amounts of HCV antigens as did wild-type, as confirmed by immunoblotting (Fig. 5A). In contrast, expression of HCV antigens was negligible for W50A (only faint bands were seen with longer exposure; data not shown). Although the W50A mutant was of interest to clarify the mechanisms of the defect, further analysis was not performed.
Fig. 5.
JFH1 mutants harboring single alanine substitutions for hydrophobic residues of NS4B are defective for replication. (A) Effect on virus replication of alanine substitutions in NS4B region of JFH1. (Top) Titration of infectious virions released from transfectants of JFH1 or mutant RNAs. Culture supernatant of each transfectant was assayed for infectivity at the indicated days post transfection (dpt). FFU, focus forming unit. Mean values from three or four independent experiments are shown; error bars denote SD. (Bottom) Expression of NS4B in the transfectants of JFH1 or mutant RNAs. (B) Localization of HCV antigens and the changes caused by W43A mutation. (Top) Change in localization of NS4AB (NS4B and/or NS4A). Transfectants with JFH1 or W43A RNA were stained with anti-NS4AB antibody, followed by staining with Bodipy 493/503. Scale bars, 10 μm. (Bottom) Comparison of LD association for HCV antigens between W43A- and JFH1-RNA-transfected cells. The percentages of positive cells for LD association of each antigen are shown. When association was seen for at least six LDs in a section, the transfectant was counted as positive. Mean values from three independent experiments are shown; error bars denote SD. (C) Localization of NS4B to LD preparations obtained from W43A-RNA-transfected cells. LD fractions were prepared and analyzed as in Fig. 2, right.
We then analyzed by confocal microscopy LD targeting of NS4B for W43A mutant, comparing it with that of the wild-type (Fig. 5B and supplementary Fig. V). We could not obtain antibodies for immunofluorescent detection of JFH1 NS4B; therefore, we used anti-NS4AB polyclonal antibody, which reacted with both NS4A and NS4B proteins. Distributions of NS4B (and/or 4A) and other antigens were observed by confocal microscopy for transfectants of wild-type and W43A (Fig. 5B and supplementary Fig. V). For JFH1, NS4B (and/or 4A) showed a reticular staining pattern, but a considerable amount of stain was concentrated around LDs and some showed broken (incomplete) ring-shape staining surrounding the LDs. In contrast, for W43A, NS4B (and/or 4A) showed a reticular pattern or small bubble-shape staining, but the stain had no relation to the LDs. Core protein showed clear ring-shape staining at the margins of the LDs in both wild-type (supplementary Fig. V) and W43A. However, for the latter, there was a slight increase in the percentage of transfectants without ring-shape staining of Core (but with reticular pattern alone). NS5A showed the staining patterns seen for both the Core and NS4AB. For both wild-type (supplementary Fig. V) and W43A, we observed clear or broken ring-shape staining surrounding the LDs, and the stain concentrated around LDs. We also tried imaging with anti-NS3 and anti-NS5B antibodies, which were used for immunoblotting of JFH1 antigens; however, the results of staining were not informative. The percentages of transfectants positive for LD association of antigens were compared for wild-type and W43A (Fig. 5B, bottom). In W43A, LD association of NS4B (and/or 4A) was markedly decreased as compared with wild-type, whereas that of Core and NS5A was mildly decreased. For V46A mutant, percentages of positive transfectants were 97, 28, and 79%, for Core, NS4AB, and NS5A, respectively.
Finally, we purified LD fractions from W43A transfectant and analyzed for HCV antigens by immunoblotting (Fig. 5C and supplementary Fig. VI). In contrast to LD fractions from wild-type (supplementary Fig. II), calnexin content in LD1 and LD2 was comparable, suggesting that the W43A mutation in NS4B affects membrane micro-architecture around LDs. This suggestion was supported by drastic changes in distribution of NS4B (and/or 4A) around LDs, where it seemed as if the distribution pattern of the ER membranes themselves was altered by the mutation of NS4B (Fig. 5B). In both LD1 and LD2, NS4B was not detected by immunoblotting, suggesting that NS4B harboring the W43A mutation could not interact with LDs (Fig. 5C). The other HCV antigens, NS3, NS5A, and NS5B, were detected in both LD1 and LD2 (supplementary Fig. VI). Also in W43A, the Core protein was not detected in LD fractions but in the ultracentrifugation pellet.
DISCUSSION
In the present study, we demonstrated that HCV NS4B has the ability to target LDs. In the imaging analysis using Cherry constructs, some expressed NS4B showed ring-shape structures at the margins of LDs, suggesting that NS4B was present near to LD membranes. This was confirmed by the colocalization results of NS4B with ADRP in ring-shape structures. Biochemical approaches also showed LD targeting of NS4B. It was important to establish whether LD targeting of intact NS4B protein occurred in the presence of other HCV proteins, because the HCV proteins, including NS4B, physically and functionally interact with each other during virus replication. To clarify this, we used OR6 cells in which genome-length HCV-O RNA was stably replicating, hence, functional HCV proteins were present. We found that NS4B existed in the crude LD fraction (LD1 in Fig. 2, right), suggesting that NS4B was localized to the membranes associated with LDs. Analysis of LD fractions from JFH1-RNA-transfected cells also suggested that NS4B localized to such LD-associated ER membranes rather than on the LD surface membranes themselves (supplementary Fig. II).
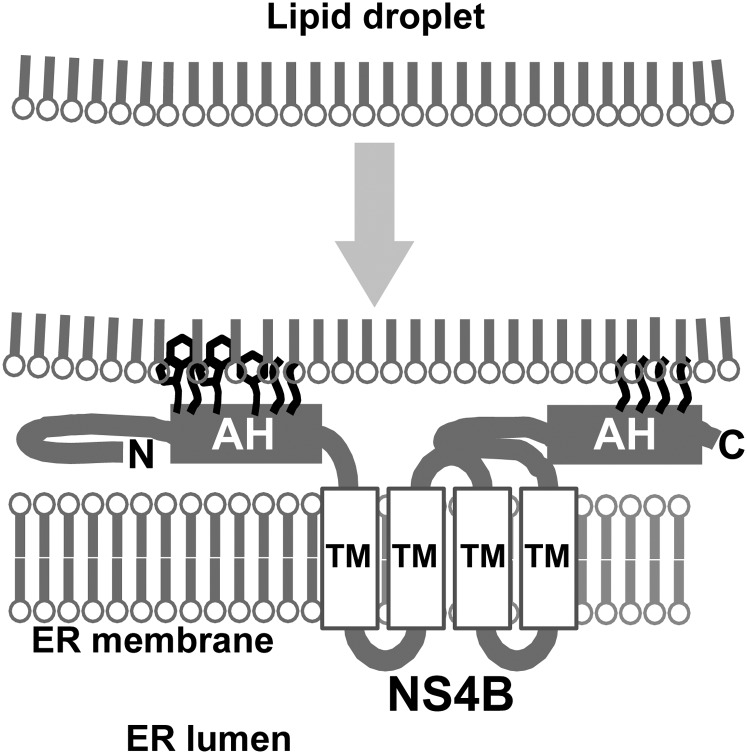
In cellular LD-targeting proteins, the targeting mechanism is thought to be complex and multifactorial (1). However, two major required features of amino acid sequence have been proposed: one is the amphipathic α-helix (19–23), and the other is a rather undefined hydrophobic region (24–26). Some LD-targeting proteins, such as caveolin-1 and associated with lipid droplet protein 1 (ALDI) (24, 42), showed dual localization to LDs and ER in relation with cellular lipid dynamics. One simple explanation may be that the LD-targeting sequences also have ER-targeting activity. LD biogenesis is believed to be initiated from the accumulation of lipids between the two membrane leaflets of the ER (1–3); therefore, another explanation is that proteins initially target the ER membranes and are then assorted on the emerging LD membranes by lateral diffusion with the aid of additional, yet undefined mechanisms. The latter targeting model has also been proposed for the HCV Core, in which the truncated form of the Core protein targets LDs more efficiently (22). However, this LD-assorting mechanism is unlikely for NS4B, because NS4B has at least four transmembrane helices by which the protein is integrated into the ER bilayer membrane (43). It should be noted that LDs are surrounded by lipid monolayers. Taking together the proposed sorting mechanisms of other LD-targeting proteins with the result that LD-targeting sites were present in each cytosolic domain, a likely model is that NS4B is present in the ER and that both cytosolic terminal domains interact simultaneously with the LD surface membrane through the hydrophobic residues in amphipathic helices; NS4B tethers or catches the emerging LDs on or very near to the ER (Fig. 6). The results from biochemical analysis of LD preparations, in which NS4B seemed to be present in LD-associated membranes rather than on the LD surface membranes themselves, support this model. To clarify whether NS4B and ADRP were present in the identical membrane structure, we also performed fluorescence recovery after photobleaching (FRAP) analysis at the site of colocalization of Cherry-4B1–261 and EGFP-ADRP. Unfortunately, definitive data have not been obtained, because of the time resolution constraints of our equipment. We think that further investigation is needed to prove our hypothesis.
Fig. 6.
Schematic representation of the hypothetical role of NS4B. NS4B tethers LDs on ER through the hydrophobic residues in the amphipathic helices (AH). N and C indicate the N- and C-terminal of NS4B, respectively. TM, transmembrane helix.
Our imaging study also revealed that hydrophobic residues in amphipathic helices [43W, 46L (V in JFH1), 50W, 57F, 61I, and 64L in the N-terminal domain; 242V, 246L, 249L, and 253I in the C-terminal domain] were important for LD targeting of NS4B. Mutational study of these residues in JFH1 replication/infection system revealed their role in LD targeting by NS4B and the significance of LD targeting in replication of infectious virions. The cluster mutants (three or four residues simultaneously mutated to alanine) were defective in virus replication. The amphipathic helices are multifunctional; for example, they are involved in oligomerization of NS4B (31, 44), formation of membranous web (31, 44) and replication complex (31, 45), and membrane association (40, 41). Therefore, multiple inhibitory mechanisms may be acting in the cluster mutants to cause severe defects.
Examination of single alanine substitution mutants focused on the residues in the first cluster (W43, V46, and W50) and clarified their role in virus replication. The results from W43A and W50A mutants indicated that both tryptophan residues are critical for the production of infectious virions. In contrast to W43A, the W50A mutant severely impaired the expression of HCV antigens. This might have resulted from a severe defect in RNA replication, or the residue 50W might be involved in the synthesis or processing of HCV polyproteins or in protein stability, in addition to known functions in NS4B. The defective mutant W43A was key to demonstrating the relationship between the hydrophobic residues and LD targeting of NS4B and the significance of LD targeting in virus replication. As shown in Fig. 5, we could not detect LD association of W43A NS4B, indicating that disruption of LD association of NS4B resulted in reduced infectious virion release. This conclusion was supported by the results for V46A mutant, in which LD association of NS4B was apparently not affected and infectious virion release was comparable to that of wild-type JFH1. The residue 46V might not be important for LD targeting in strain JFH1.
In contrast to the JFH1 system, the impact of W43A mutation was unclear from the overexpression results using a truncated form of NS4B (Fig. 4). According to our model (Fig. 6), NS4B mediates association of LDs with ER membranes. When LDs move away from the site of association, a large force may act to disrupt the interaction between NS4B and LD membranes, and the residue 43W may be important to maintain the interaction as it is highly hydrophobic. The interactions of each of the hydrophobic residues involved in LD targeting depend on the adopted conformations of NS4B and those of molecules interacting with NS4B (HCV proteins, cellular proteins, or lipids); the changes in the mutual positioning of these molecules can induce steric hindrance. The 43W residue might be critical in such microenvironments. In contrast, the truncated form of NS4B (amino acids 1–73) may target LDs directly in a less regulated manner. This interaction may not be related to ER membranes and, hence, may not be affected by LD movements. The 43W residue influence is less than that of the intact NS4B.
We used an anti-NS4AB antibody; therefore, it remains to be resolved which antigens were observed in our imaging study (Fig. 5B and supplementary Fig. V). NS4A is a small protein consisting of 54 amino acids and is closely associated with NS3 to form NS3–4A complex. NS3–4A complex is thought to associate with ER membranes through the N-terminal hydrophobic α-helix of NS4A as a membrane anchor (46). Thus, it is unlikely that the W43A mutation of NS4B directly elicits solely the loss of association between NS3–4A and LDs as seen in Fig. 5B. At present, it is also not known whether the NS3–4A complex has an activity by itself to target LDs, although NS3 harbors an amphipathic helix α0 that mediates membrane association (46). The most straightforward interpretation is that both NS4B and NS4A (and NS3) are present on ER membranes close to LDs, via the function of NS4B, and that W43A mutation causes loss of association with (or proximity to) LDs of both these proteins (but mostly NS4B).
The reason why LD targeting by NS4B is critical for virus replication is of interest. The microenvironment of LDs and ER membranes is thought to initiate HCV virion assembly, in which the nascent genome meets and associates with the Core protein to form nucleocapsids (10). Upon or after capsid formation, some particles may acquire an envelope (47), probably by utilizing ER membranes containing envelope proteins. Simultaneously, the virions might associate (or be coated) with components resembling VLDL, such as triglycerides, probably supplied from LDs, and apolipoproteins to become lipoviroparticles. Although the process of lipoviroparticle formation remains unclear, NS4B may function to establish such a microenvironment. NS4B can undergo a major conformational change, in which the N-terminal domain translocates to the luminal side of the ER (43), suggesting that the LD-targeting domain is masked by this event. The translocation was reduced in the presence of NS5A (48), suggesting a functional switch for NS4B. NS4B is indeed multifunctional because of its amphipathic helices, and in addition, it is thought to interact with other NS proteins (49), to hydrolyze GTP (50), and to modulate lipid metabolism (51). It is, therefore, of particular interest how, when, and where these activities have emerged and been regulated. A simple explanation may be that each activity simply emerges depending on the position or location of the NS4B molecule and what exists around it. The amphipathic helices of NS4B may function as ER anchors, as long as LDs are not present near the ER.
Supplementary Material
Acknowledgments
The authors thank Drs. K. Shimizu, Y. Shimotai, M. Hijikata, and T. Hayashida for valuable comments. Technical assistance by K. Toyosawa is gratefully acknowledged.
Footnotes
Abbreviations:
- ADRP
- adipose differentiation-related protein
- EGFP
- enhanced green fluorescent protein
- ER
- endoplasmic reticulum
- GST
- glutathione S-transferase
- HCV
- hepatitis C virus
- LD
- lipid droplet
- NS
- nonstructural (protein)
- PNS
- post-nuclear supernatant
- PVDF
- polyvinylidene difluoride
- UTR
- untranslated region
This work was supported by the Strategic Research Base Development Program for Private Universities subsidized by the Ministry of Education, Culture, Sports, Science and Technology, Japan (since 2010); by Nihon University Multidisciplinary Research Grant (2010–2011); and by grants-in-aid for research on hepatitis from the Ministry of Health, Labour, and Welfare of Japan.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six figures and one table.
REFERENCES
- 1.Digel M., Ehehalt R., Füllekrug J. 2010. Lipid droplets lighting up: insights from live microscopy. FEBS Lett. 584: 2168–2175. [DOI] [PubMed] [Google Scholar]
- 2.Beller M., Thiel K., Thul P. J., Jäckle H. 2010. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett. 584: 2176–2182. [DOI] [PubMed] [Google Scholar]
- 3.Ohsaki Y., Cheng J. L., Suzuki M., Shinohara Y., Fujita A., Fujimoto T. 2009. Biogenesis of cytoplasmic lipid droplets: From the lipid ester globule in the membrane to the visible structure. Biochim. Biophys. Acta. 1791: 399–407. [DOI] [PubMed] [Google Scholar]
- 4.Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9: 1089–1097. [DOI] [PubMed] [Google Scholar]
- 5.Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA. 87: 9524–9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66: 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka T., Kato N., Cho M. J., Shimotohno K. 1995. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 215: 744–749. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T., Kato N., Cho M. J., Sugiyama K., Shimotohno K. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70: 3307–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLauchlan J. 2009. Lipid droplets and hepatitis C virus infection. Biochim. Biophys. Acta. 1791: 552–559. [DOI] [PubMed] [Google Scholar]
- 10.Bartenschlager R., Penin F., Lohmann V., André P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 19: 95–103. [DOI] [PubMed] [Google Scholar]
- 11.Negro F. 2010. Abnormalities of lipid metabolism in hepatitis C virus infection. Gut. 59: 1279–1287. [DOI] [PubMed] [Google Scholar]
- 12.Herker E., Ott M. 2011. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol. Metab. 22: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ploss A., Evans M. J. 2012. Hepatitis C virus host cell entry. Curr. Opin. Virol. 2: 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moriya K., Yotsuyanagi H., Shintani Y., Fujie H., Ishibashi K., Matsuura Y., Miyamura T., Koike K. 1997. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J. Gen. Virol. 78: 1527–1531. [DOI] [PubMed] [Google Scholar]
- 15.Moradpour D., Englert C., Wakita T., Wands J. R. 1996. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology. 222: 51–63. [DOI] [PubMed] [Google Scholar]
- 16.Jirasko V., Montserret R., Lee J. Y., Gouttenoire J., Moradpour D., Penin F., Bartenschlager R. 2010. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 6: e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi S. T., Polyak S. J., Tu H., Taylor D. R., Gretch D. R., Lai M. M. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology. 292: 198–210. [DOI] [PubMed] [Google Scholar]
- 18.Roingeard P., Hourioux C., Blanchard E., Prensier G. 2008. Hepatitis C virus budding at lipid droplet-associated ER membrane visualized by 3D electron microscopy. Histochem. Cell Biol. 130: 561–566. [DOI] [PubMed] [Google Scholar]
- 19.Christianson J. L., Boutet E., Puri V., Chawla A., Czech M. P. 2010. Identification of the lipid droplet targeting domain of the Cidea protein. J. Lipid Res. 51: 3455–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinson E. R., Cresswell P. 2009. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc. Natl. Acad. Sci. USA. 106: 20452–20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poppelreuther M., Rudolph B., Du C., Grossmann R., Becker M., Thiele C., Ehehalt R., Fuellekrug J. 2012. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J. Lipid Res. 53: 888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulant S., Montserret R., Hope R. G., Ratinier M., Targett-Adams P., Lavergne J. P., Penin F., McLauchlan J. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281: 22236–22247. [DOI] [PubMed] [Google Scholar]
- 23.Unterstab G., Gosert R., Leuenberger D., Lorentz P., Rinaldo C. H., Hirsch H. H. 2010. The polyomavirus BK agnoprotein co-localizes with lipid droplets. Virology. 399: 322–331. [DOI] [PubMed] [Google Scholar]
- 24.Ingelmo-Torres M., Gonzalez-Moreno E., Kassan A., Hanzal-Bayer M., Tebar F., Herms A., Grewal T., Hancock J. F., Enrich C., Bosch M., et al. 2009. Hydrophobic and basic domains target proteins to lipid droplets. Traffic. 10: 1785–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian V., Garcia A., Sekowski A., Brasaemle D. L. 2004. Hydrophobic sequences target and anchor perilipin A to lipid droplets. J. Lipid Res. 45: 1983–1991. [DOI] [PubMed] [Google Scholar]
- 26.Zehmer J. K., Bartz R., Liu P., Anderson R. G. 2008. Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J. Cell Sci. 121: 1852–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gouttenoire J., Penin F., Moradpour D. 2010. Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory. Rev. Med. Virol. 20: 117–129. [DOI] [PubMed] [Google Scholar]
- 28.Hügle T., Fehrmann F., Bieck E., Kohara M., Krausslich H. G., Rice C. M., Blum H. E., Moradpour D. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology. 284: 70–81. [DOI] [PubMed] [Google Scholar]
- 29.Egger D., Wölk B., Gosert R., Bianchi L., Blum H. E., Moradpour D., Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76: 5974–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferraris P., Beaumont E., Uzbekov R., Brand D., Gaillard J., Blanchard E., Roingeard P. Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection. Cell. Mol. Life Sci.Epub ahead of print. November 25, 2012; 10.1007/s00018-012-1213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul D., Romero-Brey I., Gouttenoire J., Stoitsova S., Krijnse-Locker J., Moradpour D., Bartenschlager R. 2011. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J. Virol. 85: 6963–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero-Brey I., Merz A., Chiramel A., Lee J. Y., Chlanda P., Haselman U., Santarella-Mellwig R., Habermann A., Hoppe S., Kallis S., et al. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 8: e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Krausslich H. G., Mizokami M., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11: 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda M., Abe K., Dansako H., Nakamura T., Naka K., Kato N. 2005. Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem. Biophys. Res. Commun. 329: 1350–1359. [DOI] [PubMed] [Google Scholar]
- 35.Naka K., Ikeda M., Abe K., Dansako H., Kato N. 2005. Mizoribine inhibits hepatitis C virus RNA replication: effect of combination with interferon-alpha. Biochem. Biophys. Res. Commun. 330: 871–879. [DOI] [PubMed] [Google Scholar]
- 36.Ohsaki Y., Maeda T., Fujimoto T. 2005. Fixation and permeabilization protocol is critical for the immunolabeling of lipid droplet proteins. Histochem. Cell Biol. 124: 445–452. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka T., Ito T., Furuta M., Eguchi C., Toda H., Wakabayashi-Takai E., Kaneko K. 2002. In situ phage screening. A method for identification of subnanogram tissue components in situ. J. Biol. Chem. 277: 30382–30387. [DOI] [PubMed] [Google Scholar]
- 38.Fujimoto Y., Onoduka J., Homma K. J., Yamaguchi S., Mori M., Higashi Y., Makita M., Kinoshita T., Noda J., Itabe H., et al. 2006. Long-chain fatty acids induce lipid droplet formation in a cultured human hepatocyte in a manner dependent of Acyl-CoA synthetase. Biol. Pharm. Bull. 29: 2174–2180. [DOI] [PubMed] [Google Scholar]
- 39.Boleti H., Smirlis D., Dalagiorgou G., Meurs E. F., Christoforidis S., Mavromara P. 2010. ER targeting and retention of the HCV NS4B protein relies on the concerted action of multiple structural features including its transmembrane domains. Mol. Membr. Biol. 27: 50–74. [DOI] [PubMed] [Google Scholar]
- 40.Gouttenoire J., Montserret R., Kennel A., Penin F., Moradpour D. 2009. An amphipathic alpha-helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association. J. Virol. 83: 11378–11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gouttenoire J., Castet V., Montserret R., Arora N., Raussens V., Ruysschaert J. M., Diesis E., Blum H. E., Penin F., Moradpour D. 2009. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J. Virol. 83: 6257–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turró S., Ingelmo-Torres M., Estanyol J. M., Tebar F., Fernandez M. A., Albor C. V., Gaus K., Grewal T., Enrich C., Pol A. 2006. Identification and characterization of associated with lipid droplet protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 7: 1254–1269. [DOI] [PubMed] [Google Scholar]
- 43.Lundin M., Monné M., Widell A., Von Heijne G., Persson M. A. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77: 5428–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gouttenoire J., Roingeard P., Penin F., Moradpour D. 2010. Amphipathic alpha-helix AH2 is a major determinant for the oligomerization of hepatitis C virus nonstructural protein 4B. J. Virol. 84: 12529–12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aligo J., Jia S. A. Z., Manna D., Konan K. V. 2009. Formation and function of hepatitis C virus replication complexes require residues in the carboxy-terminal domain of NS4B protein. Virology. 393: 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brass V., Berke J. M., Montserret R., Blum H. E., Penin F., Moradpour D. 2008. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc. Natl. Acad. Sci. USA. 105: 14545–14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gastaminza P., Dryden K. A., Boyd B., Wood M. R., Law M., Yeager M., Chisari F. V. 2010. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J. Virol. 84: 10999–11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundin M., Lindström H., Grönwall C., Persson M. A. 2006. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J. Gen. Virol. 87: 3263–3272. [DOI] [PubMed] [Google Scholar]
- 49.Dimitrova M., Imbert I., Kieny M. P., Schuster C. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77: 5401–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Einav S., Elazar M., Danieli T., Glenn J. S. 2004. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J. Virol. 78: 11288–11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park C. Y., Jun H. J., Wakita T., Cheong J. H., Hwang S. B. 2009. Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway. J. Biol. Chem. 284: 9237–9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.