Abstract
Preliminary studies of liver regeneration induced by partial hepatectomy (PHE) identified a substantial depletion of hepatic retinoid stores, by greater than 70%, in regenerating livers of wild-type C57Bl/6J mice. To understand this, we compared responses of wild-type and lecithin:retinol acyltransferase (Lrat)-deficient mice, which totally lack hepatic retinoid stores, to PHE. The Lrat-deficient livers showed delayed regeneration in the first 24 h after PHE. At 12 h after PHE, we observed significantly less mRNA expression for growth factors and cytokines implicated in regulating the priming phase of liver regeneration, specifically for Hgf and Tgfα, but not Tgfβ. Compared with wild-type mice, the changes in mRNA levels for p21 and cyclins E1, B1, and A2 mRNAs and for hepatocellular BrdU incorporation and mitoses were delayed (i.e., shifted to later times) in regenerating Lrat−/− livers. Concentrations of all-trans-retinoic acid were significantly lower in the livers of Lrat−/− mice following PHE, and this was accompanied by diminished expression of known retinoid-responsive genes. At later times after PHE, the rate of liver weight restoration for Lrat−/− mice was parallel to that of wild-type mice, although additional biochemical differences were observed. Thus, hepatic retinoid stores are required for maintaining expression of signaling molecules that regulate cell proliferation and differentiation immediately after hepatic injury, accounting for the delayed restoration of liver mass in Lrat−/− mice.
Keywords: vitamin A, retinyl ester, lipid droplet, cell proliferation, cell cycle, nuclear hormone receptor
Retinoids (vitamin A and its metabolites) are potent regulators of cell proliferation, differentiation, and apoptosis (1, 2). Retinoic acid, the major transcriptionally active retinoid species, is reported to regulate over 500 genes (3). The all-trans- and 9-cis-isomers of retinoic acid regulate transcription upon binding to one of their six cognate nuclear receptors, the retinoic acid receptors (RARα, RARβ, and RARγ) and the retinoid X receptors (RXRα, RXRβ, and RXRγ) (4, 5). All six of these ligand-dependent transcription factors are expressed within the liver (6). The liver accounts for approximately 70% of all retinoid that is present within the body of a healthy adult, with 70–80% of this hepatic retinoid being stored within the lipid droplets of hepatic stellate cells (HSC) (7, 8). Owing to the relationships between retinoid actions in cell proliferation, differentiation, and apoptosis (1), there is a need to understand whether hepatic retinoid stores may directly affect liver responses to acute and chronic injury. For instance, alcohol-induced hepatic injury is accompanied by a marked depletion of retinoid stores, as the HSCs rapidly lose their retinoid-containing lipid droplets (9). However, roles for these retinoid stores in hepatic disease development and its resolution are not established.
The adult liver possesses a remarkable capacity for regeneration, which allows it to restore quickly its health following acute injury (10–12). A distinguishing feature of liver regeneration is that all the molecular events required for this process are not restricted to a particular cell type, but rather are dependent on tight cooperation of extra- and intracellular factors derived from the different hepatic cell types, which include HSCs, hepatocytes, Kupffer cells, and sinusoidal endothelial cells (10–17). This results in the proliferation and restitution of normal liver structure and the return of proper liver function. Dietary studies and studies involving knockout animal models have shown that many factors can reduce the liver's capacity for healing following injury (18, 19). Several dietary studies exploring retinoid involvement in liver cell proliferation in response to hepatic injury have been reported (20–22). One of these focused on the effects of retinoid deficiency on liver epithelial cell proliferation in rats which had undergone common bile duct ligation (21). The authors of this study reported that retinoid deficiency was associated with enhanced bile duct epithelial cell proliferation (21). Hu et al. reported studies in which rats were fed a totally retinoid-deficient diet for up to 72 days prior to partial hepatectomy (PHE) (20), and Kimura et al. reported studies of the effects of retinoid supplementation (with retinol, retinoic acid, or a synthetic retinoid) for up to 14 days on the recovery of mice which had undergone PHE (22). Although both of these studies reported effects of retinoids on hepatic healing, neither of the studies reported hepatic or serum retinoid concentrations of the experimental animals (20, 22). Consequently, it is not possible to understand the true hepatic retinoid status of these animals or its effect on recovery from PHE.
PHE is an accepted experimental model used to study the multistep orchestrated processes important for liver regeneration. PHE results in synchronous induction of liver cell DNA replication and mitosis, in which approximately 95% of hepatic cells, which are normally quiescent, rapidly reenter the cell cycle. We have used a PHE model to explore the importance of hepatic retinoid stores in liver regeneration. We assessed the restoration of liver mass in age-, gender-, and diet-matched wild-type and congenic lecithin:retinol acyltransferase-deficient (Lrat−/−) mice. The Lrat−/− mice are unable to synthesize retinyl esters and consequently lack hepatic retinoid stores (23). However, Lrat−/− mice, when maintained on a standard retinoid-sufficient chow diet, are physiologically normal, even though their hepatic retinoid concentrations are only approximately 1% of those of matched chow fed wild-type mice (8, 23). Using this genetic model, we assessed the significance of hepatic retinoid stores to hepatic responses to injury. Our data indicate that these stores are required to support normal liver regeneration.
EXPERIMENTAL PROCEDURES
Animal husbandry and dietary regimens
All mice employed in our studies were treated and maintained according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and all experimental procedures were reviewed and approved by the Columbia University Institutional Animal Care and Use Committee. A detailed description of Lrat−/− mice is found elsewhere (23). The mice employed in our present studies were derived from these original mixed genetic background mice through 10 backcrosses, rendering the mice congenic in the C57BL/6J genetic background. During the breeding and lactation periods, all mice were maintained on breeder chow that contained 15 IU retinol/g diet (PicoLab Mouse Diet 20, PMI International). After weaning, the mice were placed on a standard chow diet that also contained 15 IU retinol/g diet.
Partial hepatectomy
Lrat−/− and wild-type mice (males weighing 20–25 g, 10–12 weeks of age) were subjected to 2/3 PHE to induce liver regeneration. Partial hepatectomy was performed during morning hours under anesthesia as described by Mitchell and Willenbring (24). The animals were maintained postoperatively on the standard chow diet and water, provided ad libitum. Briefly, mice were anesthetized using a cocktail consisting of 1 ml Ketaset, 0.5 ml xylazine, and 8.5 ml of PBS given at a dose of 10 μl/g body weight. The abdomen was opened to allow access to the liver, and the left lateral and medial lobes were ligated individually before removal. The surgery survival rate was greater than 95%, with no difference observed between the genotypes. Ten mice per group were euthanized at 0, 12, 24, 36, 48, 72 h and 7 days after PHE. Sham-operated mice were included as controls. At the time of euthanasia, the mice were weighed, blood was taken from the inferior vena cava, and the remnant liver was removed. The dissected livers were rapidly weighed, frozen in liquid N2 and stored at –80°C. Tissues were stored continuously without thawing at –80°C until analysis. Sections from the dissected livers were also fixed in 10% neutral buffered formalin for further histological analysis.
We harvested for groups of Lrat−/− and wild-type mice entire livers at 0 h, in an identical fashion as the later removal of residual liver from hepatectomized mice. This was done for use in assessing liver:body weight ratios which are reported in this article. The values reported for 0 h were obtained using livers collected to assess liver:body weight ratios.
Liver function test
Alanine aminotransferase (ALT) enzymatic activity was determined in mouse serum using a kit from Genzyme Diagnostics (Genzyme Diagnostics P.E.I. Inc., Canada), according to the manufacturer's instructions.
Histology, BrdU labeling, and immunohistochemical staining
For paraffin sections, livers were first fixed in neutral buffered formalin and then processed into paraffin blocks according to standard protocols (25). The embedded tissues were cut into 6 μm slices, mounted on charged adhesive slides, and dried overnight at 50°C. Slides were then deparaffinized in xylene and rehydrated in graded alcohol and distilled water. Representative histological sections of each specimen were stained with hematoxylin/eosin according to standard staining methods. Oil red O-stained liver sections were assessed for hepatocyte lipid accumulation. For cryosections, livers were fixed overnight in neutral buffered formalin and cryoprotected in 30% sucrose. Tissues were sectioned with a cryostat into 12 μm slices. After fixation, the sections were stained with oil red O (Sigma) according to standard methods (26).
DNA replication was assessed by nuclear incorporation of bromodeoxyuridine (BrdU) employing a BrdU antibody (Abcam, Cambridge, MA) and immunostained liver sections. For this purpose, 100 mg BrdU (Sigma-Aldrich Corp., St. Louis, MO) per kg body weight was injected intraperitoneally 2 h before harvesting the remnant regenerating livers. In each regenerating liver, we counted the number of BrdU-positive nuclei per field (corresponding to 1.5 mm2) and used this to calculate a BrdU labeling index.
HPLC separation and analyses for retinyl esters and retinol
Retinyl esters and retinol were extracted from serum and liver tissue under a dim yellow safety light. Briefly, livers were homogenized in 10 vol of PBS (10 mM sodium phosphate, pH 7.2, 150 mM sodium chloride) using a Polytron homogenizer (Brinkmann Instruments, Westbury, NY) set at half-maximal speed for 10 s. An aliquot of serum or tissue homogenate (a 200 µl aliquot) was then treated with an equal volume of absolute ethanol containing a known amount of retinyl acetate as an internal standard. The retinoid present in the homogenates were extracted into hexane. The hexane extract was dried under a stream of N2 and redissolved in benzene. For determination of liver and serum levels of retinol and retinyl esters, a reverse-phase HPLC method was employed using a 4.6 × 250 mm Ultrasphere C18 column (Beckman, Fullerton, CA) and a running solvent (acetonitrile/methanol/methylene chloride, 70:15:15 (v/v)) flowing at 1.8 ml/min. Retinol and retinyl esters were detected at 325 nm. Quantitation was based on comparisons of the area under the peaks and spectra for unknown samples to those of known amounts of standards. The recovery of internal standard was employed to correct for loss during extraction. HPLC grade solvents for HPLC analyses and extractions were purchased from Thermo Fisher Scientific (Pittsburgh, PA).
LC/MS/MS separation and analyses for retinoic acid and endocannabinoids
All solvents employed for sample extractions and liquid chromatography were LC/MS or LC grade and were purchased from Thermo Fisher Scientific (Pittsburgh, PA). All measurements were carried out on a Waters Xevo TQ MS ACQUITY UPLC system (Waters, Milford, MA). The system was controlled by MassLynx software version 4. 1 (Waters).
Retinoic acid was extracted using a two-step, acid-base extraction (27), with minor modifications. Briefly, 0.5 ml of 0.025 M KOH in ethanol was added to 250 μl of tissue homogenate, which contained 50 mg of wet tissue. Five nanograms of pentadeuterated all-trans-retinoic acid (atRA-d5) dissolved in absolute ethanol was added to each extract as an internal standard. The aqueous phase was extracted with 5 ml of hexane (Thermo Fisher). The organic phase containing nonpolar retinoids (retinol and retinyl esters) was removed. Thirty microliters of 4 M HCl was then added to the aqueous phase, and polar retinoids, including retinoic acid, were removed by extraction into 5 ml hexane. The hexane was removed under N2. Extracts were resuspended in 70 μl of acetonitrile (Thermo Fisher) and transferred to amber LC/MS vials (Waters). Only glass containers, pipettes, and calibrated syringes were used to handle and process retinoic acid. Samples were maintained at 4°C in the autosampler, and 5 μl was loaded onto a Waters ACQUITY UPLC HSS C18 column (2.1 mm inner diameter × 100 mm with 1.8 μm particles), preceded by a 2.1 × 5 mm guard column with the same packing material (Waters). The column was maintained at 40°C using a column heater. The flow rate was 300 μl/min in binary gradient mode with the following mobile phase gradient: initiated with 32% phase A [H2O (Thermo Fisher), containing 0.1% formic acid] and 68% mobile phase B [acetonitrile, containing 0.1% formic acid]; the gradient was maintained for 6.3 min. The acetonitrile content of the solvent was increased linearly to 85% over 6.4 min and maintained until 9.5 min, increased to 100% to wash the column for 2 min, and then decreased acetonitrile to 68%. All-trans-retinoic acid (atRA) eluted between 8.2 and 8.4 min. Positive ESI-MS/MS mass spectrometry was performed using the following parameters: capillary voltage 3.8 kV, source temperature 150°C, desolvation temperature 500°C, desolvation gas flow 800 l/h, collision gas flow 0.15 ml/min. Optimized cone voltage was 16 V, collision energy for multiple reaction monitoring mode (MRM) was 18 eV, and the following transitions were used: atRA for quantification, m/z 301.16→123.00; atRA for verification, m/z 301.16→205.03; and atRA-d5, m/z 306.15→127.03.
The extraction and LC/MS/MS protocols employed for the analysis of hepatic endocannabinoids employing a MRM methodology have been described in detail elsewhere (28).
Determination of tissue triglycerides
Approximately 100 mg of liver was placed into 50 ml tubes containing 5 ml of 1.0 M NaCl. The samples were homogenized using a Polytron Homogenizer at 19,000 rpm for 20 s. Subsequently, 10 ml of chloroform:methanol (2:1 v/v) was added to the liver homogenates. The samples were vortexed for 1 min to ensure that they were homogenous. After centrifuging the samples for 10 min at 800 g, the chloroform-containing lower phase was removed and placed into a glass tube. An additional 5 ml of chloroform:methanol was added to the remainder of the upper phase, and the samples were vortexed and centrifuged as above to ensure complete triglyceride recovery. The pooled chloroform phases were evaporated under N2. After the chloroform had completely evaporated, 1 ml of 2% Triton X-100 in chloroform was added into the samples. The samples were mixed well, and again the chloroform was evaporated under N2. Triglycerides were solubilized for colorimetric assay through addition of 1 ml of deionized water into the glass tubes. Using a Matrix Plus Chemistry Reference Kit, according to the manufacturer's instructions, a colorimetric triglyceride assay was performed in a 96-well plate for each liver extract. Color development was measured on a Multiskan Plus microtiter plate reader at 520 nm.
RNA preparation and quantitative real-time PCR
Total RNA was isolated from liver tissue, DNase I digested using a RNeasy minikit (Qiagen) according to the manufacturer's protocol, and quantitated at 260 nm using a Nanodrop spectrophotometer. For cDNA synthesis, 4 μg of total RNA (in a final volume of 20 μl) was first denatured at 65°C for 5 min. Subsequently, cDNA synthesis was carried out for 10 min at 25°C and 50 min at 50°C employing reverse transcriptase (SuperScript III, Invitrogen). The reaction was stopped at 85°C for 5 min, using a thermal cycler (Perkin Elmer, Waltham, MA). The primers employed for quantitative real-time PCR (qRT-PCR) analyses of target genes are provided in supplementary Table I.
As a reference housekeeping gene used to normalize mRNA expression, we employed 18S RNA. This gene gave excellent reproducibility, never varying in its Ct value by more than 0.5 units. qRT-PCR was performed in a total volume of 25 µl, including 40 ng of cDNA template, forward and reverse primers (100 nM each), and LightCycler 480 SYBR Green I Master (Roche) using a LightCycler 480 instrument (Roche). After initial enzyme activation (95°C for 10 min), 40 cycles (94°C for 10 s, 55°C for 30 s, 72°C for 30 s) were performed for the annealing/extension steps, and fluorescence was measured. A dissociation curve program was performed after each cycle. Expression of target genes was calculated based on the efficiency of each reaction and the crossing point deviation of each sample versus a control and expressed in comparison with the reference gene.
Statistical analysis
All data are presented as means ± SD. Student t-test was used to analyze data between the control and knockout strains. Differences with P < 0.05 were considered significant.
RESULTS
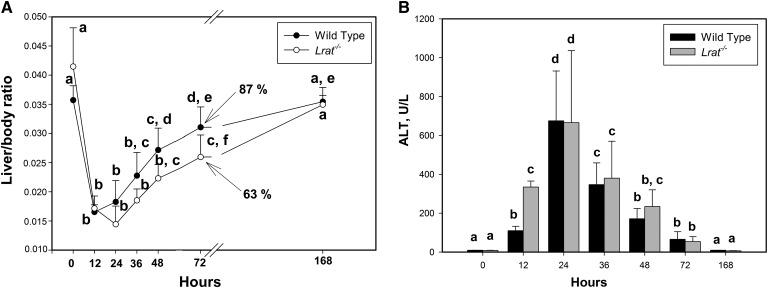
To gain understanding of the role of hepatic retinoid stores in liver regeneration, we assessed the restoration of liver mass, expressed as liver/body weight ratio, in wild-type and Lrat−/− mice at time intervals up to 7 days following PHE (Fig. 1A). As can be seen from Fig. 1A, there is a clear delay in mass restoration in the livers of Lrat−/− compared with wild-type mice. For wild-type mice, weight restoration commenced at 12 h after PHE, but not until 24 h did weight restoration commence for Lrat−/− mice. Thus, for all of the times studied, Lrat−/− livers showed a lower liver:body weight ratio until 7 days after PHE, when this ratio was the same for both the mutant and wild-type mice (Fig. 1A). No differences in postoperative morbidity, mortality, or behavior between the two strains were observed (the survival rate was more than 95% for both groups). Both wild-type and Lrat−/− mice showed the same level of serum ALT activity following PHE (Fig. 1B), except at the initial stages of liver regeneration 12 h after PHE, when the serum ALT activity was significantly greater for Lrat−/− than for wild-type mice. The ALT values of sham-operated group did not exceed 15 U/l for any of the times studied, and no significant differences between Lrat−/− and wild-type mice were observed at any time (supplementary Fig. I).
Fig. 1.
Liver mass restoration and injury in wild-type and Lrat−/− mice following PHE. (A) Ratio of liver weight to body weight at the time of euthanasia; numbers at 72 h indicate the percentage of liver mass restoration compared with time 0 values. Values marked with different letters (a, b, c, d, e) are significantly different, P < 0.05. (B) Serum ALT activity for wild-type and Lrat−/− mice following PHE. Values marked with different letters (a, b, c, d) are significantly different, P < 0.05. All values are given as the mean ± 1 SD, n = 10 for each time and genotype.
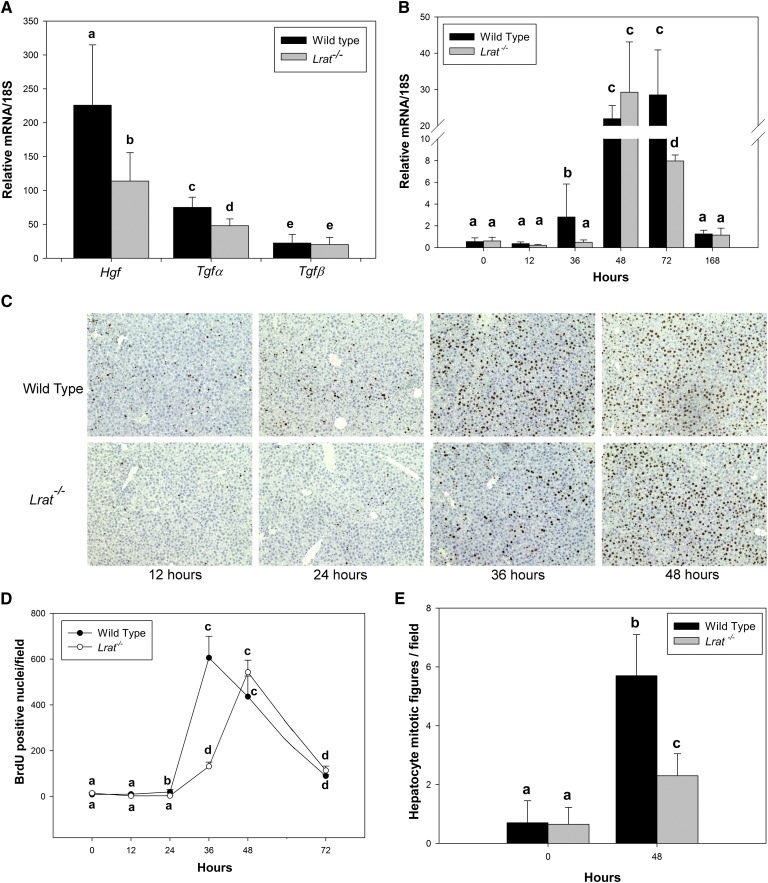
To understand the observed differences in liver mass restoration, we investigated the expression of key signaling molecules involved in cell cycle progression during liver regeneration. We assessed possible differences in hepatic expression of cytokines (TNFα, IL-6) and growth factors (HGF, TGFα, TGFβ) needed for priming and promoting the initial transition of the remaining quiescent cells into a proliferative state (10–12, 29, 30). At the initial period after PHE (12 h), we observed significantly lower mRNA expression in Lrat−/− mice for two growth factors implicated in regulating the priming phase of liver regeneration, specifically for Hgf and Tgfα, but no effect on Tgfβ was detected (Fig. 2A and supplementary Fig. II). No significant differences in cytokine (Il-6 and Tnfα) mRNA expression were observed (supplementary Fig. III). The quantitatively largest difference between wild-type and Lrat−/− mice was observed for Hgf mRNA expression, which is known to be associated with liver nonparenchymal cells, predominantly HSCs (11).
Fig. 2.
Hepatic cell cycle progression in livers of wild-type and Lrat−/− mice following PHE. (A) Hgf, Tgfα, Tgfβ mRNA expression in the livers of wild-type and Lrat−/− mice 12 h following PHE. Values marked with different letters (a, b, c, d, e) are statistically different, P < 0.05. (B) FoxM1 mRNA expression levels in livers of wild-type and Lrat−/− mice at different times following PHE. Values marked with different letters (a, b, c, d) are statistically different, P < 0.05. (C and D) Immunohistochemical analysis of hepatocellular BrdU incorporation (C) and numerical summary (D) of hepatocellular proliferation given as number of BrdU-positive nuclei per field corresponding to 1.5 mm2. Values marked with different letters (a, b, c, d) are statistically different, P < 0.05. E: Mean hepatocyte mitotic figures (metaphase plates) per field corresponding to 1.5 mm2. Values marked with different letters (a, b, c) are statistically different, P < 0.05. All values are given as the mean ± 1 SD, n = 10 for each time and genotype.
BrdU incorporation into regenerating Lrat−/− livers was also delayed compared with wild-type livers. Analysis of the regenerating livers showed significantly less hepatocellular BrdU incorporation for Lrat−/− mice 24 and 36 h after PHE (Fig. 2C, D). Moreover, the peak number of hepatocellular mitoses was significantly diminished, by over 2-fold, in Lrat−/− livers at their peak, 48 h after PHE (Fig. 2E). Collectively, these findings suggest that Lrat−/− livers experience a delay in DNA replication after PHE compared with wild-type mice. This observation is consistent with the delay in liver weight regain observed for Lrat−/− mice following PHE.
We also determined expression levels for a number of genes that control cell cycle progression, including cyclins D1, E1, A2, and B1 (31). Our qRT-PCR results given in Table 1 show that cyclin E1 mRNA expression was decreased in Lrat−/− livers 24 h after PHE, but significantly elevated at 72 h. This represents a shift in the temporal pattern of expression compared with wild-type mice. No statistically significant differences were observed in expression levels of cyclin D1 between Lrat−/− and wild-type mice. Cyclin A2 and B1 expression were also significantly diminished in Lrat−/− compared with wild-type mice, specifically 36 h after PHE (Table 1). Forkhead box transcription factor (FoxM1) mRNA expression (Fig. 2B), which is normally induced and promotes progression through G2 into the mitotic phase (32, 33), was also significantly decreased in Lrat−/− mice 36 h after PHE. This time corresponds to the time of S/G2 transition. Collectively, these data suggest that hepatic expression of factors that regulate the proliferative responses of cells in Lrat−/− liver is modified in a number of cell cycle periods. This undoubtedly contributes to the differences observed in liver mass restoration for Lrat−/− versus wild-type mice. Thus, compared with wild-type mice, the temporal patterns of cyclin E1, B1, and A2 mRNA expression (Table 1) and hepatocellular BrdU incorporation in Lrat−/− livers appear to be shifted to later times (Fig. 2B, C).
TABLE 1.
Cyclin mRNA expression in wild-type and livers at different times following PHE
| Cyclin D1 |
Cyclin E1 |
Cyclin A2 |
Cyclin B1 |
|||||
| Hour | Wild-type | Lrat−/− | Wild-type | Lrat−/− | Wild-type | Lrat−/− | Wild-type | Lrat−/− |
| 0 | 19.9 ± 6.1a | 16.7 ± 3.4a | 0.6 ± 0.3a | 1.2 ± 0.3a | 0.7 ± 0.3a | 0.6 ± 0.3a | 0.5 ± 0.3a | 0.3 ± 0.2a |
| 12 | 25.0 ± 19.9a | 25.1 ± 15.5a | 4.1 ± 1.9b, c | 2.8 ± 0.5b, c | 0,5 ± 0.2a | 0.3 ± 0.1a | 0.5 ± 0.2a | 0.1 ± 0.1a |
| 24 | 86.3 ± 44.1b | 52.6 ± 41.8a, b | 5.3 ± 1.8b, d | 1.8 ± 1.2a, c | 1.4 ± 0.5b, d | 0.8 ± 0.2c | 2.2 ± 1.3b | 1.3 ± 0.6a |
| 36 | 74.7 ± 20.4b | 55.1 ± 44.5a, b | 5.2 ± 3.1b, c, d | 3.4 ± 1.8b, c | 3.1 ± 1.3d | 0.8 ± 1.1c, b | 3.7 ± 1.2b | 1.0 ± 0.7b |
| 48 | 175.5 ± 32.8c | 189.2 ± 146.7b, c | 11.0 ± 5.5d | 13.2 ± 7.2d | 17.6 ± 9.5e | 31.7 ± 23.2e | 29.6 ± 21.1c | 39.8 ± 28.0c |
| 72 | 65.8 ± 29.0b | 72.8 ± 27.5b, c | 3. 0 ± 1.1b | 7.2 ± 2.9d | 15.5 ± 6.3e | 15. 5 ± 3.5e | 26.5 ± 14.8c | 26.1 ± 4.3c |
| 168 | 45.7 ± 27.1a, b | 70.9 ± 19.6b, c | 0.9 ± 0.5a | 1.6 ± 0.4a, b, c | 1.2 ± 0.3b, c, d | 1.1 ± 0.5b, c, d | 1.0 ± 0.6a, b | 1.3 ± 0.7a, b |
Relative mRNA/18S values expressed as the mean ± 1 SD, n = 10 for each time and genotype. Values for each cyclin marked with different letters (a, b, c, d) are significantly different, P < 0.05.
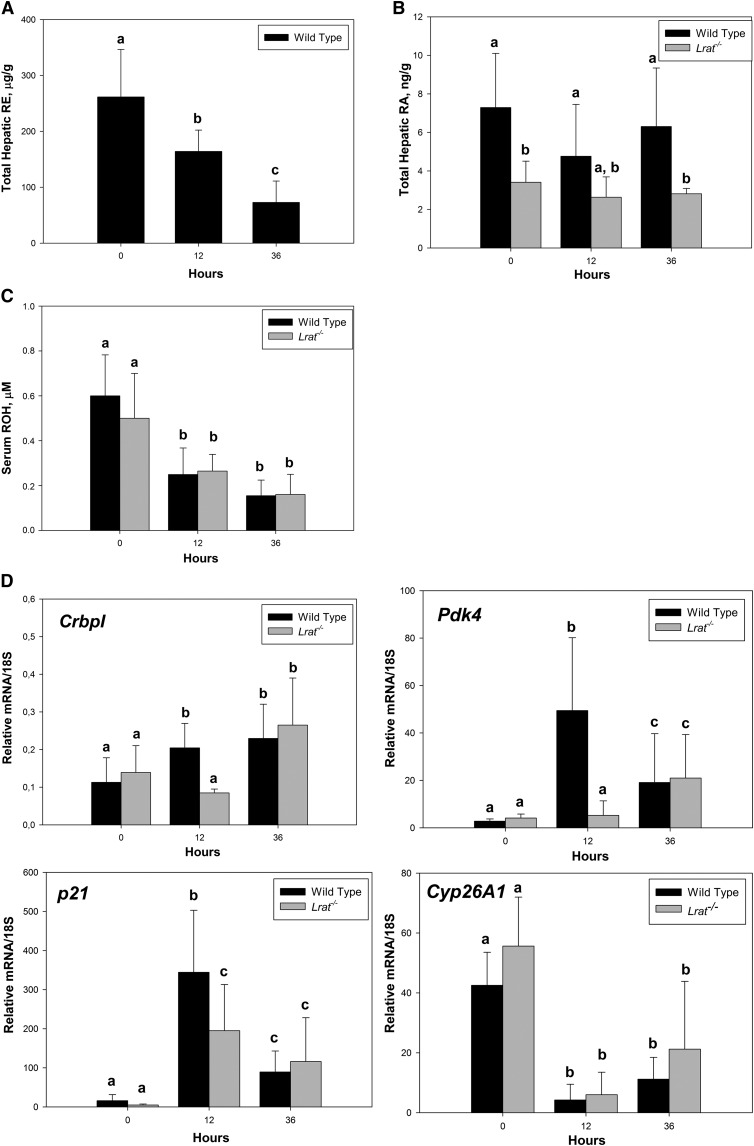
Liver regeneration was accompanied by a quantitatively large decline of retinyl ester concentrations in regenerating wild-type livers (Fig. 3A). The mean value of the total amount of retinoid storage form decreased by 38% (from 261.5 µg/g to 164.1 µg/g) 12 h after PHE and declined further to only 28% (72.8 µg/g) of the initial value at 36 h. Hepatic retinyl ester and serum retinol concentrations were not observed to decline in sham-operated animals across all experimental periods. (As noted above, Lrat−/− mice are unable to synthesize retinyl esters.) We did not observe a change in hepatic retinol levels (supplementary Fig. IV), whereas serum retinol levels progressively declined for both Lrat−/− and wild-type mice (Fig. 3C). This suggests an early demand for retinoids during the priming phase of liver regeneration (from 0 to 36 h after PHE), which stimulates retinoid release from the retinyl ester form, possibly to allow for the formation of retinoic acid. We did not detect an increase of hepatic retinoic acid concentrations by LC/MS/MS measurements for livers of either Lrat−/− or wild-type mice for the first 36 h after PHE (Fig. 3B). However, retinoic acid concentrations at all times were approximately 50% lower in the livers of Lrat−/− compared with wild-type mice. We also asked whether there might be differences in expression patterns for hepatic genes whose expression is known to be responsive to retinoic acid (Fig. 3D). Levels of mRNA expression for Cyp26A1, which encodes a protein that catalyzes catabolism of retinoic acid, were markedly diminished for both wild-type and Lrat−/− livers 12 and 36 h after PHE, but no significant differences were observed between the wild-type and Lrat−/− mice. Expression of the CrbpI gene, which encodes a protein proposed to be important in retinoic acid formation, was significantly lower for livers of Lrat−/− versus wild-type mice 12 h after PHE, but not at other times. Levels of mRNA for Pdk4, a potent regulator of hepatic carbohydrate metabolism, and for p21, a regulator of cell proliferation, were significantly elevated in livers of both Lrat−/− and wild-type mice both 12 and 36 h after PHE, but the fold elevation in expression for these two genes in Lrat−/− mice was significantly less than that observed for wild-type mice. Collectively, these findings support the notion that there is considerable demand for retinoids during the early priming phase of liver regeneration, which is likely needed to support retinoid-dependent transcriptional regulation.
Fig. 3.
Hepatic total retinyl ester and retinoic acid concentrations, serum retinol levels, and retinoic acid-responsive gene expression in the livers of wild-type and Lrat−/− mice at early times following PHE. (A) Total retinyl ester (RE) concentrations were determined for liver lobes from wild-type mice at time 0 and at early stages of liver regeneration (12 and 36 h following PHE). Values marked with different letters (a, b, c) are significantly different, P < 0.05. (B) Hepatic retinoic acid (RA) concentrations were determined by LC/MS/MS for liver lobes from wild-type and Lrat−/− mice at time 0 and at initial stages of liver regeneration (12 and 36 h following PHE). Values marked with different letters (a, b) are significantly different, P < 0.05. (C) Serum retinol levels for wild-type and Lrat−/− mice at 0, 12, and 36 h following PHE. Values marked with different letters (a, b) are statistically different, P < 0.05. (D) Gene expression levels for known retinoic acid-responsive genes (CrbpI, Pdk4, p21, and Cyp26A1) determined by qRT-PCR for RNA obtained from liver lobes harvested at time 0 from wild-type and Lrat−/− mice and in the initial stages of liver regeneration (12 and 36 h following PHE). Values marked with different letters (a, b, c) are significantly different, P < 0.05. All values are given as the mean ± 1 SD, n = 10 for each time and genotype.
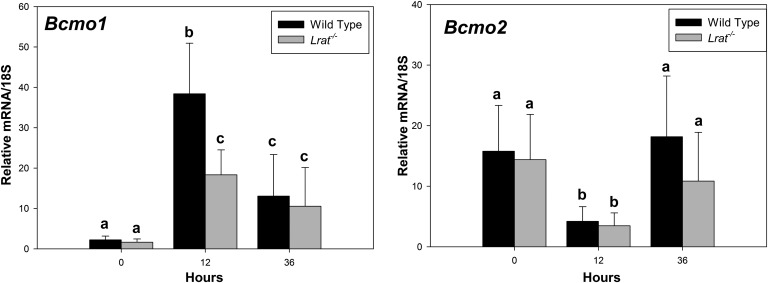
Transcriptionally active retinoid metabolites can be formed not only following the hydrolysis of retinyl esters but also through the oxidative cleavage of their carotenoid precursors. HSCs have been shown to be able to accumulate β-carotene (34), which can be converted to retinoic acid following its symmetric oxidative cleavage catalyzed by β-carotene 15,15′-monooxygenase (BCMO1). During the regeneration process, we observed a nearly 20-fold upregulation in Bcmo1 mRNA expression for both Lrat−/− and wild-type mice, 12 h after PHE (Fig. 4). We also observed a downregulation in mRNA expression of β-carotene 9′,10′-monooxygenase (BCMO2), an oxygenase involved in carotenoid oxidative degradation, but not in retinoic acid synthesis (Fig. 4). These observations are consistent with a need for carotenoid central cleavage products in PHE.
Fig. 4.
Expression of carotenoid oxygenase (Bcmo1 and Bcmo2) mRNAs in livers of wild-type and Lrat−/− mice following PHE. Gene expression values were determined by qRT-PCR for RNA obtained from liver lobes harvested at time 0 from wild-type and Lrat−/− mice and at 12 and 36 h following PHE. Values marked with different letters (a, b, c) are significantly different, P < 0.05. All values are given as the mean ± 1 SD, n = 10 for each time and genotype.
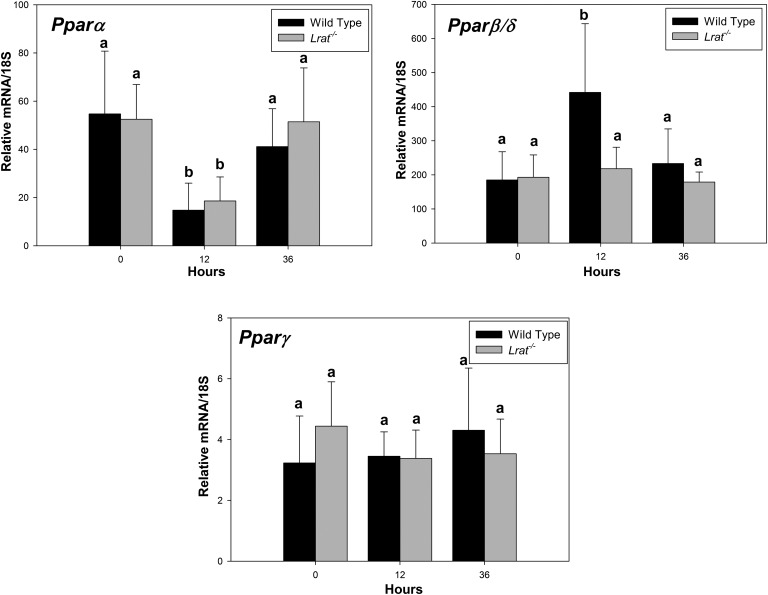
A characteristic feature of liver regeneration is the accumulation of lipid droplets in hepatocytes of regenerating livers. This process is known to be a characteristic of hepatocyte proliferation during regeneration (35, 36). We examined whether Lrat−/− mice and the absence of retinoid stores affect this process following PHE through histological analyses (supplementary Fig. V) and measurement of triglyceride content (Fig. 5). Lipid droplets accumulated in the livers of both genotypes starting from 12 h after PHE. No neutral lipid accumulation was observed in sham-operated controls for either genotype (supplementary Fig. VI). Triglyceride measurements showed that hepatic triglyceride content was significantly lower in Lrat−/− compared with wild-type mice 36 and 72 h after PHE (Fig. 5). To better understand these differences, we investigated the expression pattern of peroxisome proliferator-activated receptor (PPAR) isoforms during liver regeneration. These lipid-sensing transcription factors regulate downstream genes that regulate lipid metabolism and function as heterodimeric partners with RXRs. Twelve hours after PHE, we observed a significant downregulation in Pparα expression for both Lrat−/− and wild-type mice. We also observed at 12 h after PHE a significant upregulation in Pparβ/δ mRNA expression for wild-type but not for Lrat−/− mice (Fig. 6.). No differences in Pparγ mRNA expression were observed at any time for either genotype.
Fig. 5.
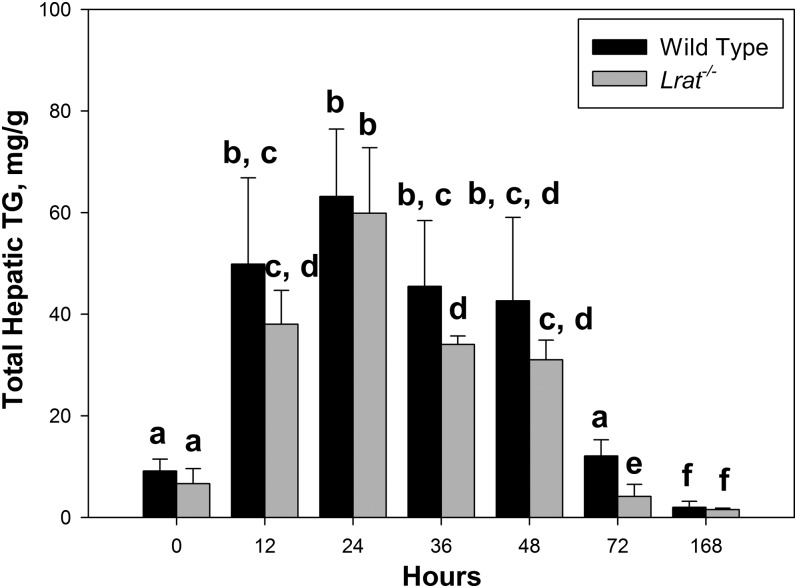
Hepatic triglyceride accumulation in livers of wild-type and Lrat−/− mice following PHE. Hepatic triglyceride concentrations for wild-type and Lrat−/− mice at 0, 12, 24, 36, 48, 72, and 168 h after PHE. Values marked with different letters (a, b, c, d, e, f) are significantly different, P < 0.05. All values are given as the mean ± 1 SD, n = 10 for each time and genotype.
Fig. 6.
Pparα, Pparγ, and Pparβ/δ mRNA levels in livers of wild-type and Lrat−/−mice following PHE. Ppar mRNA expression was determined by qRT-PCR for RNA obtained from livers of wild-type and Lrat−/− mice at 0, 12, and 36 h after PHE. Values marked with different letters (a, b) are significantly different, P < 0.05. All values are given as the mean ± 1 SD, n = 10 for each time and genotype.
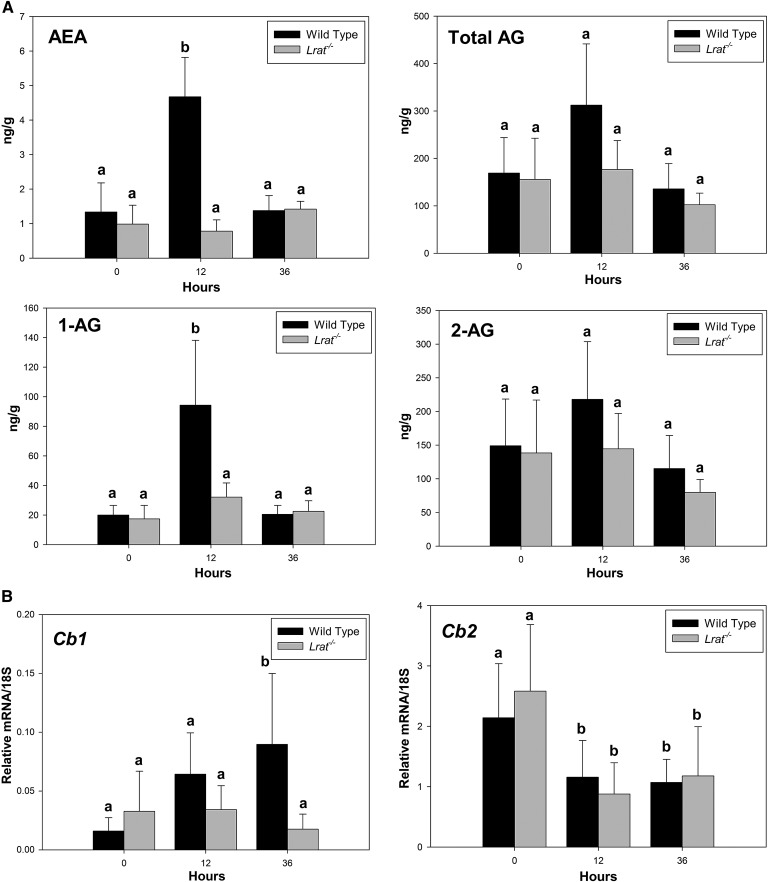
We also assessed possible changes in the endocannabinoid system, since endocannabinoids are known to act as potent promitogenic factors during liver regeneration (37, 38). Moreover, retinoids are reported to regulate endocannabinoid receptor expression, and it has been proposed that this is required for assuring proper healing from hepatic injury (39). Consequently, we assessed hepatic concentrations of the canonical endocannabinoids, arachidonoyl ethanolamide (AEA) and 1- and 2-arachidonoyl glycerol (1-AG and 2-AG) by LC/MS/MS, as well as mRNA expression for the two endocannabinoid receptors, Cb1 and Cb2, during liver regeneration (Fig. 7). Twelve hours after hepatic surgery, the remnant liver lobes of hepatectomized wild-type mice showed an increase in AEA and 1-AG, but not in total AG (Fig. 7A). These changes were accompanied by a statistically significant increase in Cb1 mRNA for wild-type mice at 36 h PHE and a decrease in Cb2 mRNA expression (Fig. 7B). Interestingly, no statistically significant changes in endocannabinoid concentrations were observed for Lrat−/− livers (Fig. 7A). These data underscore the interactions between the retinoid and endocannabinoid systems and their coordinated involvement in the hepatic regenerative response.
Fig. 7.
Hepatic endocannabinoid concentrations and cannabinoid receptor mRNA expression in livers of wild-type and Lrat−/−mice following PHE. (A) Hepatic endocannabinoid concentrations [AEA, 1-AG, 2-AG, and total AG (1-AG + 2-AG)] were determined for livers from wild-type and Lrat−/−mice at 0, 12, and 36 h after PHE. Values represented as bar graphs marked with different letters (a, b) are significantly different, P < 0.05. (B) Gene expression levels determined by qRT-PCR for the Cb1 and Cb2 endocannabinoid receptors. Values marked with different letters (a, b) are significantly different, P < 0.05. All values are given as the mean ± 1 SD, n = 10 for each time and genotype.
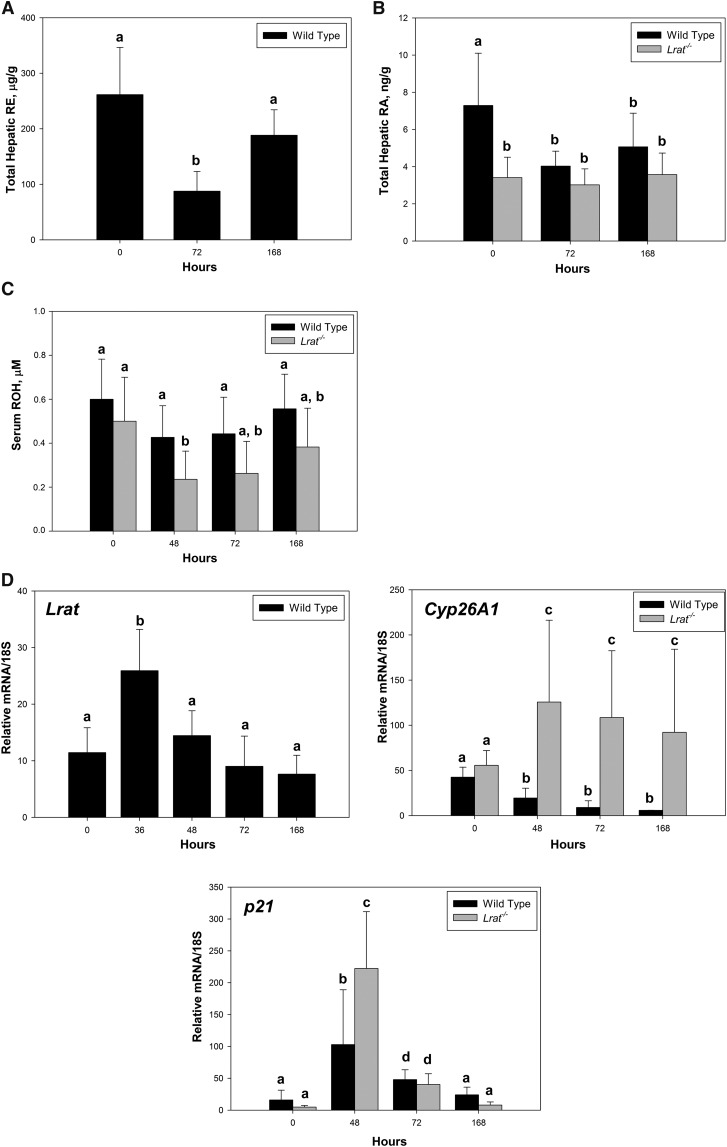
The central focus of our research was on the early stages of liver regeneration, within the first 36 h after PHE. However, to better understand retinoid involvement in this process, we also assessed retinoid levels and retinoid-related gene expression at later times (72 and 168 h) after PHE. These were different from those observed in initial phases after PHE. Hepatic retinyl ester and serum retinol levels rebounded at these later times, while hepatic retinoic acid levels remained constant (Fig. 8A–C). Consistent with the rebound in hepatic retinyl ester levels, for wild-type mice, Lrat mRNA expression levels were significantly elevated 36 h after PHE, but these levels progressively declined through 168 h (Fig. 8D). Interestingly, Cyp26A1 mRNA expression remained high throughout the later times for Lrat−/− mice, but for wild-type mice, these were lower than those observed at 0 h. For Lrat−/− and wild-type mice, expression levels of p21 were different at baseline 48 and 72 h, but not at 168 h after PHE. Significantly greater elevation in Cyp26A1 expression was observed for Lrat−/− than for wild-type livers at 48 h.
Fig. 8.
Hepatic total retinyl ester and retinoic acid concentrations, serum retinol levels and retinoic acid-responsive gene expression in the livers of wild-type and Lrat−/− mice at later times following PHE. (A) Total retinyl ester (RE) concentrations were determined for livers from wild-type mice at late stages of liver regeneration (72 and 168 h) following PHE. Values with different letters (a, b) are significantly different, P < 0.05. (B) Hepatic retinoic acid (RA) concentrations were determined for livers from wild-type and Lrat−/− mice at late stages of liver regeneration (72 and 168 h) following PHE. Values marked with different letters (a, b) are significantly different, P < 0.05. (C) Serum retinol levels for wild-type and Lrat−/− mice at 48, 72, and 168 h following PHE. Values marked with different letters (a, b) are significantly different, P < 0.05. (D) Gene expression levels for known retinoic acid-responsive genes (Lrat, Cyp26A1, and p21) determined by qRT-PCR. Values marked with different letters (a, b, c, d) are significantly different, P < 0.05. All values are given as the mean ± 1 SD, n = 10 for each time and genotype.
DISCUSSION
Our studies, employing a genetic mouse model that lacks hepatic retinoid stores, establish that retinoid stores are required for assuring optimal healing of the liver upon PHE. Hepatic total retinoid levels for the Lrat−/− mice fed a chow diet are about 1% of those of matched wild-type mice, and these animals are healthy and do not display any clinical signs of retinoid insufficiency when maintained on a standard chow diet (23, 40, 41). No retinyl ester is detectable in the livers of Lrat−/− mice, although these livers still possess retinol, albeit at concentrations which are about 5% of those of wild-type mice (see supplementary Fig. IV). Thus, although without retinoid stores, the livers of Lrat−/− mice are retinoid sufficient (23, 40, 41). The retinoic acid that is present in the livers of Lrat−/− mice is synthesized primarily from either very recently ingested dietary retinoid or retinol recycled to the liver from peripheral tissues. Because of the total lack of hepatic retinyl ester stores and the low hepatic retinol concentrations in Lrat−/− mice, the capacity of these livers to synthesize retinoic acid to maintain normal concentrations is markedly diminished upon PHE (see Fig. 3). This, we propose, fundamentally underlies the observed delay in the proliferative regenerative response of Lrat−/− mice within the first 12–24 h following hepatectomy (see supplementary Fig. VII). These mice are simply unable to synthesize sufficient retinoic acid to meet the strong demand placed on the liver immediately after PHE. Thus, hepatic retinoid stores are needed for ensuring optimal retinoic acid synthesis/levels needed within the liver and for ensuring appropriate hepatic retinoic acid-responsive gene expression.
Transcriptionally active retinoid metabolites are formed not only from retinol and retinyl esters but also through the oxidative cleavage of their carotenoid precursors that are present in the diet, including small amounts present in rodent chow diets. HSCs have been shown to be able to accumulate β-carotene (34), which can be converted to retinoic acid following its symmetric oxidative cleavage catalyzed by β-carotene 15,15′-monooxygenase (BCMO1). During the regeneration process, we observed a nearly 20-fold upregulation in Bcmo1 mRNA expression for both Lrat−/− and wild-type mice, 12 h after PHE (Fig. 4). We also observed a downregulation in mRNA expression of β-carotene 9′,10′-monooxygenase (BCMO2), an oxygenase involved in carotenoid oxidative degradation, but not in retinoic acid synthesis (Fig. 4). These observations support the notion that there is a need during hepatic regeneration for BCMO1 activity and synthesis of carotenoid central cleavage products for use in retinoic acid synthesis.
The requirement for proper retinoic acid signaling to allow for normal liver regeneration is underscored by studies of hepatocyte-specific RXRα-null mice (42). When RXRα is ablated, there is reduced hepatocyte lifespan, which is accompanied by premature hepatocyte death and the appearance of necrotic areas (43). RXRα ablation also results in delayed hepatocyte proliferation following PHE (42). Several studies in rodent models showed a stimulatory effect of retinoic acid administration on liver mass restoration and BrdU incorporation in regenerating livers following PHE (22, 44, 45). The literature also indicates that, during hepatic regeneration, there is an increase in mRNA and protein expression of CRBPI (46) and CRABPI (47), which are known to be encoded by retinoid-responsive genes, as well as increased expression of the retinoid receptor RXRα (43). Our data (Fig. 3D) also show an increase in CrbpI gene expression in wild-type mice 12 and 36 h after PHE, in addition to the increases in expression for the retinoic acid-responsive Pdk4 and p21 genes (48, 49). However, levels of CrbpI, Pdk4, and p21 expression are significantly lower (P < 0.05) in Lrat−/− compared with wild-type liver, 12 h after PHE. This undoubtedly reflects the lower retinoic acid concentrations present in regenerating Lrat−/− livers and contributes to the delay in liver regeneration observed in these mutant mice. This supports our proposal that retinoic acid is not sufficiently synthesized to bring about a full induction of these genes in Lrat−/− mice. Cyp26A1 encodes an enzyme that degrades excessive retinoic acid, and its expression is induced by excessive retinoic acid concentrations. Elevated Cyp26A1 expression is routinely taken as an indication of excessive retinoic acid concentrations or toxicity (50, 51). At initial times, we observed no differences between Lrat−/− and wild-type expression of Cyp26A1 and conclude that retinoic acid concentrations are not being sensed by the livers as being excessive at these times. Collectively, these differences in gene expression arise because wild-type mice are able to draw on hepatic retinoid stores for retinoic acid synthesis, whereas the Lrat−/− mice lack retinoid stores and are unable to immediately respond like wild-type mice by increasing retinoic acid synthesis to support the need for increased retinoic acid-responsive transcription.
Delayed liver regeneration in Lrat−/− mice is associated with impaired expression of cell cycle regulators. Retinoic acid has been shown in many different in vitro and in vivo model systems to be required for maintaining normal cell proliferation. Among cell cycle regulating genes, only p21, a known cell cycle progression regulator, is directly transactivated by a liganded RAR via a retinoic acid response element in its promoter (49). P21 acts importantly in the formation of higher order complexes of cyclins and cyclin-dependent kinases, functioning either as a positive or negative regulator of cell proliferation (52). Several published studies have reported an upregulation of p21 mRNA and protein expression during early times after hepatectomy (53). We observed a similar effect on p21 expression for wild-type mice but observed significantly diminished induction of p21 expression 12 h after PHE for livers of Lrat−/− mice. Published studies of p21−/− mice (54) suggest that p21 plays a growth inhibitory role during liver regeneration. Although P21 is known as a negative regulator of cell cycle progression, our observation of upregulation of p21 expression during the liver cell G0–G1 transition led us to conclude that it has an important role during this period. This is consistent with literature reporting that binding of P21 protein to cyclin D1-CDK4 stabilizes this protein-protein complex and facilitates its nuclear import, without necessarily inhibiting cyclin D-associated kinase activity. Moreover, p21 is induced when quiescent fibroblasts and T lymphocytes are stimulated to proliferate by mitogenic signals. This apparent paradox regarding the regulatory actions of P21 (proliferative versus antiproliferative) can be understood from observations that P21-cyclin-CDK complexes harvested from proliferating cells retain kinase activity that is extinguished by addition of more P21. The conclusion that was drawn from these observations is that conversion of active complexes to inactive ones is achieved by changing the ratio of P21 to cyclin-CDK, such that active complexes contain a single P21 molecule, whereas inactive ones include multiple P21 subunits (52). Thus, the effects of P21 on cell proliferation are concentration dependent. Our data are consistent with the later proposal regarding P21 actions in stimulating proliferation and not with conclusions drawn from study of p21−/− mice or of mice which greatly overexpress P21 in hepatocytes.
With regard to cyclin expression, our findings are similar to those of Ledda-Columbano et al. who reported that hepatocyte proliferation in retinoic acid-fed mice is associated with increased hepatic levels of cyclins D1, E, and A (44) and studies performed by Yang et al. showing the alteration in the expression and regulation of cyclin D1/cyclin-dependent kinase (Cdk)4, cyclin E1/Cdk2, cyclin A2/Cdk2, and cyclin B1/Cdk1 in regenerating RXRα-null livers. In these later studies, hepatocyte RXRα-deficiency also affected basal, as well as regeneration-induced cyclin E1 expression levels. For Lrat−/− mice, with diminished hepatic retinoic acid availability, we observed significantly diminished mRNA levels of cyclins E1, A2, and B1 within the first 36 h after PHE, compared with wild-type mice. We did not however observe a significant difference in cyclin D1 expression in regenerating Lrat−/− liver. It should be noted though that many of the cell growth-related proteins, including the cyclins, are regulated during the cell cycle through changes in their phosphorylation status without dramatic changes in the level of the protein. Thus, the differences we observed in expression levels for cyclins E1, A2, and B1 may contribute to the delayed liver mass restoration observed for Lrat−/− mice; however, more in-depth analyses will be required to establish this possibility.
Levels and temporal patterns of growth factor expression are altered in Lrat−/− compared with wild-type mice. This includes significantly (P < 0.05) diminished expression of Hgf and Tgfα 12 h after PHE, and diminished FoxM1 expression 36 h after PHE. Notably, a strong correlation between the presence of retinoid-containing lipid droplets in HSCs (ones that are absent in Lrat−/− mice) and the ability of HSCs to express Hgf has been observed for both animal and human studies (55–57). It has been shown that preactivated or semiactivated HSCs lose their ability to express Hgf mRNA, thus affecting normal hepatocyte proliferation and liver regeneration (55–57). Our data also clearly show an upregulation in Pdk4 expression during initial periods (12 h) of liver regeneration in wild-type mice. The metabolic action of Pdk4, a gene which is known to be retinoic acid-responsive (58), may be required for effectively shifting the hepatic preference for energy substrates toward lipids, under the condition of hypoglycemia caused by PHE (supplementary Fig. VIII). We propose that the observed differences in expression for these important regulatory molecules also contribute toward the delayed restoration of liver mass when hepatic retinoid stores are absent.
Endocannabinoid homeostasis was altered during hepatic regeneration for livers lacking retinoid stores. We were specifically interested in exploring retinoid/endocannabinoid interactions during hepatic regeneration because interactions between these two lipid signaling families, involving retinoic acid regulation of expression of the endocannabinoid receptor Cb1 in the liver, have been reported in the literature (39). Consequently, we assessed whether endocannabinoid concentrations and/or endocannabinoid receptor levels might be altered in Lrat−/− mice recovering from PHE. Hepatic levels of the endocannabinoid AEA are significantly lower 12 h after PHE in Lrat−/− compared with wild-type liver (Fig. 7). Expression levels of Cb1 mRNA are significantly lower in Lrat−/− mice 36 h after PHE. This is potentially important for understanding our data since activation of the CB1 receptor in the liver by newly synthesized AEA has been shown to promote liver regeneration by controlling the expression of cell-cycle regulators that drive M phase progression (37). Moreover, it has been shown that FoxM1 mRNA and protein levels are regulated by AEA via the CB1 receptor (37). As far as we are aware, there are no published data regarding the presence of retinoic acid-responsive elements in genes encoding endocannabinoid-metabolizing enzymes. Interestingly, hepatic triglyceride concentrations were not different for wild-type and Lrat−/− mice, aside from 36 h after PHE when triglyceride levels in Lrat−/− livers were approximately 75% of those of wild-type mice. Hepatic Pparα and Pparγ mRNA expression levels, transcription factors proposed to be important in liver regeneration, were not different for Lrat−/− and wild-type mice following PHE. Somewhat surprisingly, significantly lower (P < 0.05) Pparβ/δ mRNA expression was observed for Lrat−/− compared with wild-type mice 12 h after PHE. This nuclear receptor has been proposed to be able to use all-trans-retinoic acid as a physiologic ligand (59).
Although hepatic regeneration is initially delayed in Lrat−/− mice compared with wild-type mice 12–24 h after PHE, liver weights of these mice increase at rates that appear to be parallel to those of the wild-type mice. Although our primary focus was on the early time period after PHE (within 36 h after PHE), we assessed a number of retinoid-related parameters at later times. These data indicate that Lrat−/− mice are eventually able to synthesize sufficient retinoic acid to allow for a proliferative, albeit delayed, response to commence with a rate similar to that of wild-type mice. Most of the retinoid used for this purpose must be derived from recent dietary intake. Also, there may be increased recycling of retinol from peripheral tissues back to the liver. It is well established in the literature that retinol can be returned from the periphery to the liver (60). As seen in Fig. 8, hepatic retinoic acid levels are not different for wild-type and Lrat−/− mice 72 and 168 h after PHE. However, unlike the earlier times, Cyp26A1 expression levels are now significantly elevated in Lrat−/− compared with wild-type mice. Fig. 8 also shows for wild-type mice that, at 168 h after PHE, hepatic retinyl ester stores have greatly increased over earlier times. This is likely because the liver is now predisposed to accumulating retinyl ester via LRAT action in esterifying retinol acquired from the diet or from peripheral tissues, rather than converting it to retinoic acid. However, the increased fluxes of retinol arriving at the liver from the diet and periphery cannot be stored in Lrat−/− mice, owing to the absence of LRAT, and must be catabolized and wasted. Retinoid catabolism involves its conversion to retinoic acid and subsequent oxidative catabolism of retinoic acid by CYP26A1. Consequently, Cyp26A1 expression is elevated at later times in Lrat−/− mice to allow the mice to accommodate the increased flux of retinol to the liver. Moreover, at these later times, there is a progressive decline in expression levels of a number of retinoic acid-responsive genes (Fig. 8D). Thus, at early stages of liver regeneration, there appears to be a very strong demand or need for retinoic acid to support retinoic acid-dependent signaling, but this demand recedes later as the liver nears complete mass restoration, allowing again for accumulation of hepatic retinoid stores.
In summary, the central finding of our studies is that the absence of hepatic retinoid stores delays the onset of liver weight restoration following PHE. Our data also provide a rationale for understanding why there is a delay in the proliferative response when hepatic retinoid stores are unavailable. This is outlined in Fig. 9. Because Lrat−/− mice have insufficient hepatic retinoid stores, they are unable to synthesize sufficient retinoic acid to meet needs in regulating retinoid-responsive gene expression. This affects genes involved in mediating proliferative responses and results in a lag in the proliferative response, including diminished expression of a number of genes that are importantly involved in cell cycle progression and its regulation (Hgf, Tgfα, p21, cyclin E1, cyclin A2, cyclin B1, and FoxM1). We also observed alterations in hepatic concentrations of the endocannabinoid AEA and Cb1 receptor levels, which also have roles in ensuring normal liver regeneration. Collectively, our findings point to a need for retinoic acid derived from endogenous synthesis to allow optimal hepatic wound healing.
Fig. 9.
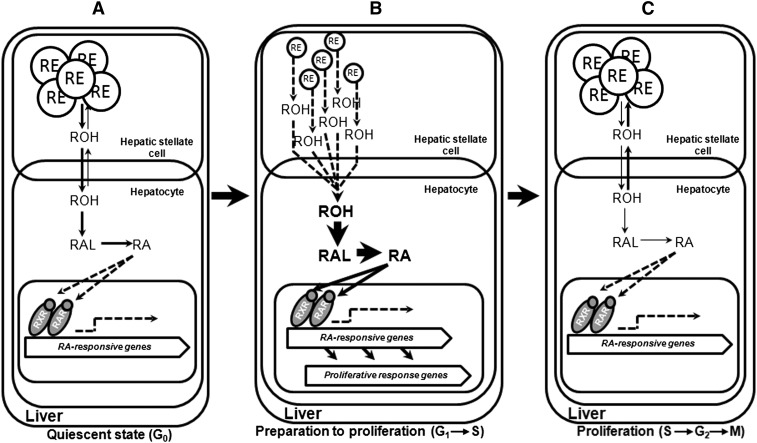
A schematic representation of retinoid metabolism and actions during liver regeneration. (A) In a healthy adult liver, 70–80% of hepatic retinoid is stored within the lipid droplets of hepatic stellate cells (HSC) in the form of retinyl ester (RE). Following hydrolysis, the released retinol (ROH) is transported to the hepatocytes, where it can undergo enzymatic conversion into retinoic acid (RA), which functions as a ligand for its specific nuclear hormone receptors (RARs and RXRs), affecting the expression of retinoic acid-responsive genes. (B) After acute liver injury, caused by partial hepatectomy in our experiments, liver cells shift from the quiescent state (G0) toward a proliferative one in order to restore liver mass. Liver regeneration is accompanied by a massive utilization and decline of retinyl ester levels, suggesting a large demand for retinoids during the early priming phase of hepatic healing. Retinoic acid is known to be required for signaling proliferative response genes to allow for normal liver regeneration. (C) At later stages of liver mass restoration, following the transition of cells into S phase, there is less demand for retinoic acid and its formation. A rebound in hepatic retinyl ester concentrations occurs, as well as a progressive decline in expression levels of retinoic acid-responsive genes.
Supplementary Material
Footnotes
Abbreviations:
- AEA
- arachidonoyl ethanolamide
- 1-AG
- 1-arachidonoylglycerol
- 2-AG
- 2-arachidonoylglycerol
- ALT
- alanine aminotransferase
- BrdU
- bromodeoxyuridine
- FoxM1
- forkhead box protein M1
- HGF
- hepatocyte growth factor
- HSC
- hepatic stellate cell
- IL
- interleukin
- LRAT
- lecithin:retinol acyltransferase
- PHE
- partial hepatectomy
- PPAR
- peroxisome proliferator-activated receptor
- qRT-PCR
- quantitative real-time PCR
- RAR
- retinoic acid receptor
- RXR
- retinoid X receptor
- TGF
- transforming growth factor
- TNF
- tumor necrosis factor
This work was supported by National Institutes of Health Grants RC2 AA019413, R01 DK068437, R01 DK079221, and UL1 RR024156.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of nine figures and one table.
REFERENCES
- 1.Gudas L. J. 2012. Emerging roles for retinoids in regeneration and differentiation in normal and disease states. Biochim. Biophys. Acta. 1821: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noy N. 2010. Between death and survival: retinoic acid in regulation of apoptosis. Annu. Rev. Nutr. 30: 201–217. [DOI] [PubMed] [Google Scholar]
- 3.Balmer J. E., Blomhoff R. 2002. Gene expression regulation by retinoic acid. J. Lipid Res. 43: 1773–1808. [DOI] [PubMed] [Google Scholar]
- 4.Chawla A., Repa J. J., Evans R. M., Mangelsdorf D. J. 2001. Nuclear receptors and lipid physiology: opening the X-files. Science. 294: 1866–1870. [DOI] [PubMed] [Google Scholar]
- 5.Duong V., Rochette-Egly C. 2011. The molecular physiology of nuclear retinoic acid receptors. From health to disease. Biochim. Biophys. Acta. 1812: 1023–1031. [DOI] [PubMed] [Google Scholar]
- 6.Wagner M., Zollner G., Trauner M. 2011. Nuclear receptors in liver disease. Hepatology. 53: 1023–1034. [DOI] [PubMed] [Google Scholar]
- 7.Goodman D. W., Huang H. S., Shiratori T. 1965. Tissue distribution and metabolism of newly absorbed vitamin A in the rat. J. Lipid Res. 6: 390–396. [PubMed] [Google Scholar]
- 8.Blaner W. S., O'Byrne S. M., Wongsiriroj N., Kluwe J., D'Ambrosio D. M., Jiang H., Schwabe R. F., Hillman E. M., Piantedosi R., Libien J. 2009. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim. Biophys. Acta. 1791: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leo M. A., Lieber C. S. 1999. Alcohol, vitamin A, and beta-carotene: adverse interactions, including hepatotoxicity and carcinogenicity. Am. J. Clin. Nutr. 69: 1071–1085. [DOI] [PubMed] [Google Scholar]
- 10.Fausto N., Campbell J. S., Riehle K. J. 2006. Liver regeneration. Hepatology. 43: S45–S53. [DOI] [PubMed] [Google Scholar]
- 11.Taub R. 2004. Liver regeneration: from myth to mechanism. Nat. Rev. Mol. Cell Biol. 5: 836–847. [DOI] [PubMed] [Google Scholar]
- 12.Michalopoulos G. K. 2010. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am. J. Pathol. 176: 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurinna S., Barton M. C. 2011. Cascades of transcription regulation during liver regeneration. Int. J. Biochem. Cell Biol. 43: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Utoh R., Tateno C., Kataoka M., Tachibana A., Masumoto N., Yamasaki C., Shimada T., Itamoto T., Asahara T., Yoshizato K. 2010. Hepatic hyperplasia associated with discordant xenogeneic parenchymal-nonparenchymal interactions in human hepatocyte-repopulated mice. Am. J. Pathol. 177: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mabuchi A., Mullaney I., Sheard P., Hessian P., Zimmermann A., Senoo H., Wheatley A. M. 2004. Role of hepatic stellate cells in the early phase of liver regeneration in rat: formation of tight adhesion to parenchymal cells. Comp. Hepatol. 3(Suppl. 1): S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mabuchi A., Mullaney I., Sheard P. W., Hessian P. A., Mallard B. L., Tawadrous M. N., Zimmermann A., Senoo H., Wheatley A. M. 2004. Role of hepatic stellate cell/hepatocyte interaction and activation of hepatic stellate cells in the early phase of liver regeneration in the rat. J. Hepatol. 40: 910–916. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W., Chen X. P., Zhang W. G., Zhang F., Xiang S., Dong H. H., Zhang L. 2009. Hepatic non-parenchymal cells and extracellular matrix participate in oval cell-mediated liver regeneration. World J. Gastroenterol. 15: 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ezaki H., Yoshida Y., Saji Y., Takemura T., Fukushima J., Matsumoto H., Kamada Y., Wada A., Igura T., Kihara S., et al. 2009. Delayed liver regeneration after partial hepatectomy in adiponectin knockout mice. Biochem. Biophys. Res. Commun. 378: 68–72. [DOI] [PubMed] [Google Scholar]
- 19.DeAngelis R. A., Markiewski M. M., Taub R., Lambris J. D. 2005. A high-fat diet impairs liver regeneration in C57BL/6 mice through overexpression of the NF-kappaB inhibitor, IkappaBalpha. Hepatology. 42: 1148–1157. [DOI] [PubMed] [Google Scholar]
- 20.Hu Z., Fujio K., Marsden E. R., Thorgeirsson S. S., Evarts R. P. 1994. Hepatic regeneration in vitamin A-deficient rats: changes in the expression of transforming growth factor alpha/epidermal growth factor receptor and retinoic acid receptors alpha and beta. Cell Growth Differ. 5: 503–508. [PubMed] [Google Scholar]
- 21.Weiss B., Barshack I., Onaca N., Goldberg I., Berkovich Z., Melzer E., Jonas A., Reifen R. 2010. Vitamin A deficiency associated with enhanced proliferation of bile duct epithelial cells in the rat. Isr. Med. Assoc. J. 12: 82–86. [PubMed] [Google Scholar]
- 22.Kimura M., Watanabe M., Ishibashi N., Yanagida S., Ogihara M. 2010. Acyclic retinoid NIK-333 accelerates liver regeneration and lowers serum transaminase activities in 70% partially hepatectomized rats, in vivo. Eur. J. Pharmacol. 643: 267–273. [DOI] [PubMed] [Google Scholar]
- 23.O'Byrne S. M., Wongsiriroj N., Libien J., Vogel S., Goldberg I. J., Baehr W., Palczewski K., Blaner W. S. 2005. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT). J. Biol. Chem. 280: 35647–35657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell C., Willenbring H. 2008. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 3: 1167–1170. [DOI] [PubMed] [Google Scholar]
- 25.Fischer A. H., Jacobson K. A., Rose J., Zeller R. 2008. Paraffin embedding tissue samples for sectioning. CSH Protoc. 2008: pdb.prot4989. [DOI] [PubMed] [Google Scholar]
- 26.Penney D. P., Powers J. M., Frank M., Willis C., Churukian C. 2002. Analysis and testing of biological stains–the Biological Stain Commission Procedures. Biotech. Histochem. 77: 237–275. [DOI] [PubMed] [Google Scholar]
- 27.Kane M. A., Folias A. E., Wang C., Napoli J. L. 2008. Quantitative profiling of endogenous retinoic acid in vivo and in vitro by tandem mass spectrometry. Anal. Chem. 80: 1702–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clugston R. D., Jiang H., Lee M. X., Piantedosi R., Yuen J. J., Ramakrishnan R., Lewis M. J., Gottesman M. E., Huang L. S., Goldberg I. J., et al. 2011. Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: a targeted lipidomic and gene expression study. J. Lipid Res. 52: 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borowiak M., Garratt A. N., Wustefeld T., Strehle M., Trautwein C., Birchmeier C. 2004. Met provides essential signals for liver regeneration. Proc. Natl. Acad. Sci. USA. 101: 10608–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su A. I., Guidotti L. G., Pezacki J. P., Chisari F. V., Schultz P. G. 2002. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc. Natl. Acad. Sci. USA. 99: 11181–11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chauhan A., Lorenzen S., Herzel H., Bernard S. 2011. Regulation of mammalian cell cycle progression in the regenerating liver. J. Theor. Biol. 283: 103–112. [DOI] [PubMed] [Google Scholar]
- 32.Wang X., Kiyokawa H., Dennewitz M. B., Costa R. H. 2002. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. USA. 99: 16881–16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalin T. V., Ustiyan V., Kalinichenko V. V. 2011. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle. 10: 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shmarakov I., Fleshman M. K., D'Ambrosio D. N., Piantedosi R., Riedl K. M., Schwartz S. J., Curley R. W., Jr, von Lintig J., Rubin L. P., Harrison E. H., et al. 2010. Hepatic stellate cells are an important cellular site for beta-carotene conversion to retinoid. Arch. Biochem. Biophys. 504: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newberry E. P., Kennedy S. M., Xie Y., Luo J., Stanley S. E., Semenkovich C. F., Crooke R. M., Graham M. J., Davidson N. O. 2008. Altered hepatic triglyceride content after partial hepatectomy without impaired liver regeneration in multiple murine genetic models. Hepatology. 48: 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shteyer E., Liao Y., Muglia L. J., Hruz P. W., Rudnick D. A. 2004. Disruption of hepatic adipogenesis is associated with impaired liver regeneration in mice. Hepatology. 40: 1322–1332. [DOI] [PubMed] [Google Scholar]
- 37.Mukhopadhyay B., Cinar R., Yin S., Liu J., Tam J., Godlewski G., Harvey-White J., Mordi I., Cravatt B. F., Lotersztajn S., et al. 2011. Hyperactivation of anandamide synthesis and regulation of cell-cycle progression via cannabinoid type 1 (CB1) receptors in the regenerating liver. Proc. Natl. Acad. Sci. USA. 108: 6323–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tam J., Liu J., Mukhopadhyay B., Cinar R., Godlewski G., Kunos G. 2011. Endocannabinoids in liver disease. Hepatology. 53: 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhopadhyay B., Liu J., Osei-Hyiaman D., Godlewski G., Mukhopadhyay P., Wang L., Jeong W. I., Gao B., Duester G., Mackie K., et al. 2010. Transcriptional regulation of cannabinoid receptor-1 expression in the liver by retinoic acid acting via retinoic acid receptor-gamma. J. Biol. Chem. 285: 19002–19011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batten M. L., Imanishi Y., Maeda T., Tu D. C., Moise A. R., Bronson D., Possin D., Van Gelder R. N., Baehr W., Palczewski K. 2004. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J. Biol. Chem. 279: 10422–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L., Gudas L. J. 2005. Disruption of the lecithin:retinol acyltransferase gene makes mice more susceptible to vitamin A deficiency. J. Biol. Chem. 280: 40226–40234. [DOI] [PubMed] [Google Scholar]
- 42.Yang X., Guo M., Wan Y. J. 2010. Deregulation of growth factor, circadian clock, and cell cycle signaling in regenerating hepatocyte RXRalpha-deficient mouse livers. Am. J. Pathol. 176: 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imai T., Jiang M., Kastner P., Chambon P., Metzger D. 2001. Selective ablation of retinoid X receptor alpha in hepatocytes impairs their lifespan and regenerative capacity. Proc. Natl. Acad. Sci. USA. 98: 4581–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ledda-Columbano G. M., Pibiri M., Molotzu F., Cossu C., Sanna L., Simbula G., Perra A., Columbano A. 2004. Induction of hepatocyte proliferation by retinoic acid. Carcinogenesis. 25: 2061–2066. [DOI] [PubMed] [Google Scholar]
- 45.Ohtake Y., Maruko A., Ohishi N., Kawaguchi M., Satoh T., Ohkubo Y. 2008. Effect of retinoic acid on transglutaminase and ornithine decarboxylase activities during liver regeneration. Cell Biochem. Funct. 26: 359–365. [DOI] [PubMed] [Google Scholar]
- 46.López-Valencia V., Rangel P., Rodriguez S., Hernandez-Munoz R. 2007. Involvement of alcohol and aldehyde dehydrogenase activities on hepatic retinoid metabolism and its possible participation in the progression of rat liver regeneration. Biochem. Pharmacol. 73: 586–596. [DOI] [PubMed] [Google Scholar]
- 47.Omori M., Muto Y., Nagao T. 1981. Cellular retinoid-binding proteins in reginerating rat liver: demonstration of a novel cellular retinoid-binding protein. J. Lipid Res. 22: 899–904. [PubMed] [Google Scholar]
- 48.Kwon H. S., Huang B., Ho Jeoung N., Wu P., Steussy C. N., Harris R. A. 2006. Retinoic acids and trichostatin A (TSA), a histone deacetylase inhibitor, induce human pyruvate dehydrogenase kinase 4 (PDK4) gene expression. Biochim. Biophys. Acta. 1759: 141–151. [DOI] [PubMed] [Google Scholar]
- 49.Liu M., Iavarone A., Freedman L. P. 1996. Transcriptional activation of the human p21(WAF1/CIP1) gene by retinoic acid receptor. Correlation with retinoid induction of U937 cell differentiation. J. Biol. Chem. 271: 31723–31728. [DOI] [PubMed] [Google Scholar]
- 50.Ray W. J., Bain G., Yao M., Gottlieb D. I. 1997. CYP26, a novel mammalian cytochrome P450, is induced by retinoic acid and defines a new family. J. Biol. Chem. 272: 18702–18708. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto Y., Zolfaghari R., Ross A. C. 2000. Regulation of CYP26 (cytochrome P450RAI) mRNA expression and retinoic acid metabolism by retinoids and dietary vitamin A in liver of mice and rats. FASEB J. 14: 2119–2127. [DOI] [PubMed] [Google Scholar]
- 52.Sherr C. J., Roberts J. M. 2004. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 18: 2699–2711. [DOI] [PubMed] [Google Scholar]
- 53.Weymann A., Hartman E., Gazit V., Wang C., Glauber M., Turmelle Y., Rudnick D. A. 2009. p21 is required for dextrose-mediated inhibition of mouse liver regeneration. Hepatology. 50: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albrecht J. H., Poon R. Y., Ahonen C. L., Rieland B. M., Deng C., Crary G. S. 1998. Involvement of p21 and p27 in the regulation of CDK activity and cell cycle progression in the regenerating liver. Oncogene. 16: 2141–2150. [DOI] [PubMed] [Google Scholar]
- 55.Estep J. M., O'Reilly L., Grant G., Piper J., Jonsson J., Afendy A., Chandhoke V., Younossi Z. M. 2010. Hepatic stellate cell and myofibroblast-like cell gene expression in the explanted cirrhotic livers of patients undergoing liver transplantation. Dig. Dis. Sci. 55: 496–504. [DOI] [PubMed] [Google Scholar]
- 56.Makino H., Shimizu H., Ito H., Kimura F., Ambiru S., Togawa A., Ohtsuka M., Yoshidome H., Kato A., Yoshitomi H., et al. 2006. Changes in growth factor and cytokine expression in biliary obstructed rat liver and their relationship with delayed liver regeneration after partial hepatectomy. World J. Gastroenterol. 12: 2053–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schirmacher P., Geerts A., Pietrangelo A., Dienes H. P., Rogler C. E. 1992. Hepatocyte growth factor/hepatopoietin A is expressed in fat-storing cells from rat liver but not myofibroblast-like cells derived from fat-storing cells. Hepatology. 15: 5–11. [DOI] [PubMed] [Google Scholar]
- 58.Kwon H. S., Harris R. A. 2004. Mechanisms responsible for regulation of pyruvate dehydrogenase kinase 4 gene expression. Adv. Enzyme Regul. 44: 109–121. [DOI] [PubMed] [Google Scholar]
- 59.Shaw N., Elholm M., Noy N. 2003. Retinoic acid is a high affinity selective ligand for the peroxisome proliferator-activated receptor beta/delta. J. Biol. Chem. 278: 41589–41592. [DOI] [PubMed] [Google Scholar]
- 60.Blaner W. S., Olson J. A. 1994. Retinol and retinoic acid metabolism. In The Retinoids: Biology, Chemistry, and Medicine. 2nd edition. M. B. Sporn, A. B. Roberts, and D. S. Goodman, editors. Raven Press, New York. 229–256.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.