Abstract
Presence of ectopic lipid droplets (LDs) in cardiac muscle is associated to lipotoxicity and tissue dysfunction. However, presence of LDs in heart is also observed in physiological conditions, such as when cellular energy needs and energy production from mitochondria fatty acid β-oxidation are high (fasting). This suggests that development of tissue lipotoxicity and dysfunction is not simply due to the presence of LDs in cardiac muscle but due at least in part to alterations in LD function. To examine the function of cardiac LDs, we obtained transgenic mice with heart-specific perilipin 5 (Plin5) overexpression (MHC-Plin5), a member of the perilipin protein family. Hearts from MHC-Plin5 mice expressed at least 4-fold higher levels of plin5 and exhibited a 3.5-fold increase in triglyceride content versus nontransgenic littermates. Chronic cardiac excess of LDs was found to result in mild heart dysfunction with decreased expression of peroxisome proliferator-activated receptor (PPAR)α target genes, decreased mitochondria function, and left ventricular concentric hypertrophia. Lack of more severe heart function complications may have been prevented by a strong increased expression of oxidative-induced genes via NF-E2-related factor 2 antioxidative pathway. Perilipin 5 regulates the formation and stabilization of cardiac LDs, and it promotes cardiac steatosis without major heart function impairment.
Keywords: lipids, hypertrophy, myocardial cardiomyopathy
Studies in patients with diabetes and/or obesity have found an association between increased cardiac triglyceride (TAG) accumulation and cardiac dysfunction, suggesting that accumulation of neutral lipid may exert a toxic effect on the myocardium (1). Additional studies in humans find that this association is not so clear, as there are many situations where an increase in myocardial TAG content is not associated with cardiac dysfunction (2). Further, recent studies in transgenic mice show that elevated TAG accumulation can be dissociated from cardiac dysfunction and suggest that the packaging of TAG into lipid droplets (LD) may be of critical importance in determining whether accumulation of excess lipid exerts a toxic effect on the myocardium (3). Finally, a novel study in humans suggests a defect in lipid droplet packaging associated with severe heart failure (4). LDs play a critical role in lipid homeostasis, mediating transient storage of excess fatty acid (FA) in TAG (5); however, their contribution to cardiomyopathy remains controversial, in part because LDs can be cardio protective by preventing accumulation of bioactive lipids or production of toxic lipids at times when FA supply is high, such as with fasting, poorly controlled diabetes, or advanced heart failure (6, 7). While cardiomyocyte ectopic fat accumulation is associated with cardiac lipotoxicity and dysfunction in transgenic mice that overexpress genes facilitating transport and use of FA (3), identical levels of cardiomyocyte fat accumulation are observed in three-month-old mice overexpressing specifically DGAT-1 but without cardiac function abnormalities (8). It is thus clear that development of tissue lipotoxicity and dysfunction is not simply due to presence or amount of ectopic fat; rather, it might be due to alterations in LD function, resulting in abnormal management of excess energy in diseased tissues.
It was recently demonstrated that LDs are highly dynamic and depend on protein activities associated with their monolayer surface (9). Depending on physiological and pathological circumstances, some proteins are associated with LDs in a transient manner, whereas others are associated in a more permanent fashion (10). The surface of LDs are coated with at least one of five perilipin proteins, with perilipin 5 (Plin5) found mostly in oxidative tissues, such as heart, whereas perilipin 1 (Plin1) and perilipin 4 (Plin4) are mostly expressed in adipose tissue, and Plin2 and Plin3 are ubiquitous. Our previous work in cell culture models, along with the work of others, has shown that Plin5 plays an important role by positively regulating LD accumulation (11–13). We postulated that Plin5 may play a role in LD hydrolysis (11). At a molecular level, Plin5 acts as a scaffolding protein for lipolytic proteins (14–17), such as its interaction with ATGL (14, 17), which is a property unique to Plin5 (14), and with CGI-58, which is a property shared with Plin1 (14, 16, 18, 19). The significance of intracellular cardiac LD hydrolysis was revealed by an unexpected phenotype of whole-body ATGL-knockout mice (20), which exhibited massive accumulation of TAG in the heart, cardiomyopathy, and shortened life expectancy. This resembles patients with defective ATGL function who suffer from cardiomyopathy, illustrating the importance of LD regulation in disease (21). Furthermore, it was recently demonstrated that ATGL-mediated LD hydrolysis can regulate cardiac mitochondrial function via peroxisome proliferator-activated receptor (PPAR)α and PGC-1 (22). Endogenous ligands for PPARα activation have low affinity, and LD hydrolysis may generate acutely high concentrations, inducing PPARα/PGC-1 activities. The ability of Plin5 to provide a scaffold for ATGL and CGI-58 at the LD surface and to regulate ATGL activity suggests that Plin5 plays an important role regulating cardiac mitochondrial function. In addition, it was recently recognized that Plin5 has a distinctive property of tethering mitochondria to the LD surface (23); we identified a Plin5 protein domain necessary to support this function (23). This past work performed mostly in cultured cells supported the view that lipid storage is characterized by a reorganization of organelles to maximize this process and is cell type specific. More importantly, it suggested that Plin5 may mediate important metabolic and physical interactions between cardiac LDs and mitochondria. The overall body Plin5 knockout phenotype was recently published, and its main features include absence of cardiac LDs, increased cardiac β-oxidation, and increased reactive oxygen species (ROS) content. These transgenic mice develop heart failure with age (24).
In the present investigation, we directly address the role of cardiac LDs by overexpressing Plin5 in heart and examine its consequences on cardiac LDs, mitochondrial function, and cardiac function. We provide evidence that by overexpressing Plin5 in cardiac muscle, we have generated a mouse model with a robust increase in cardiac LDs, indicating that Plin5 plays a critical role in formation and stabilization of cardiac LDs in vivo. Chronic cardiac excess of Plin5-coated LDs was found to result in a modest cardiac dysfunction with decreased expression of a subset of PPARα target genes, decreased mitochondria function, and left ventricular concentric hypertrophy. Lack of more severe heart function complications may have been prevented by a strong increased expression of oxidative-induced genes via the nuclear factor (erythroid-derived 2)-like 2 (Nrf2) pathway. Taken together, these findings indicate that Plin5 is an important regulator of cardiac LDs.
MATERIALS AND METHODS
Mice
All procedures that involved animal handling were approved by the Institutional Animal Care and Use Committee at the University of Maryland School of Medicine. Mice were housed under the 12 h light/dark cycles (lights on at 7:00 AM, lights off at 7:00 PM) and temperature-controlled room. Mice were kept on a regular chow diet (2018SX Teklad Global 18% Protein Rodent Diet, Harlan Laboratory, 6.2% fat) and allowed at libitum access to food and water except when stated otherwise. Only males were used in these studies. For fasting studies, fasting was started before the dark cycle.
Creation of transgenic mice and mouse breeding
Transgenic mice expressing mouse myc-tagged-Plin5 cDNA selectively in cardiac muscle were made using the cardiac-specific promoter from the gene encoding α-myosin heavy chain (α-MHC). To generate a αMHC-epitope-tagged myc Plin5 transgenic construct, we attached a myc epitope to the NH2 terminus of Plin5. The pBS II SK vector containing the α-myosin heavy α-chain (MHC) promoter region (GenBank U71441) was kindly obtained from Dr. Robbins (Department of Pediatrics, Children's Hospital Research Foundation, Cincinnati, OH) (25). The amplified Myc-Plin5 was fused to the α-MHC promoter at the Sal I/Hind III site of the vector. After agarose gel electrophoresis and isolation of the transgene with the QuickCleanII Gel extraction kit (Qiagen, Valencia, CA), the transgene was injected into the pronucleus of B6C3HF/C57BL/6J zygotes and transferred to the oviducts of Swiss Webster pseudopregnant females (University of Maryland Transgenic Core). PCR analysis with genomic DNA isolated from tail tips was performed to identify offspring carrying the Myc-Plin5 transgene. Two founder lines were established showing elevated expression of the myc-Plin5 transgene exclusively in cardiac muscle and were developed after six backcrosses with the wild-type C57BL/CJ (NCI, Frederick, MD). To maximize our ability to examine the effect of excess cardiac LDs on mitochondria and heart function, we used the line with the highest Plin5 expression in the present study.
Phenotypic characterization of MHC-Plin5 hearts
Hearts from three- to four-month-old MHC-Plin5 and wild-type (WT) mice were collected to measure mRNA and protein expression of heart perilipin proteins, TAG content, LD quantification, and mitochondria size, number, and function, as well as heart function (see description below).
Tissue processing for electron microscopy and micrograph analysis
For electron microscopy, hearts were excised, quickly cut in small pieces, fixed in situ with Trump's fixative, and postfixed in 1% osmium tetroxide as previously described (26). Briefly, tissues were then dehydrated in graded alcohols, en-bloc stained with uranyl acetate, and embedded in epoxy resin. One micron sections and ultrathin sections were cut on either a Porter-Bloom or LKB ultramicrotome, then stained with lead acetate and examined in a JEOL 1200 EX. Thirty-five (heart) fields containing longitudinally arrayed myofibrils were photographed from each section at 3,200× or 5,000× magnification. Scanned micrographs were analyzed using ImageJ software to manually generate masks of mitochondrial and LD contours, which were used for the calculation of mitochondrial and LD area maximum and total mitochondrial and LD numbers.
Western blot analysis
For whole-cell extract, individual hearts were homogenized with a glass homogenizer with RIPA buffer (Teknova Hollister, CA) containing 150 mM sodium chloride, 1% Triton X-100,1% deoxycholic acid-sodium salt, 0.1% sodium dodecyl sulfate, 50 mM Tris-HCL (pH 7.5), 2 mM EDTA and a pill of complete inhibitor as directed by the manufacturer (Roche Diagnostics). Samples were then centrifuged at 16,000 g for 30 min at 4°C. For supernatant and fat-cake fractions, five fasted mice hearts from each genotype were pooled and homogenized in Tris and EDTA sucrose buffer supplemented with a pill of complete inhibitor as directed by the manufacturer (Roche Diagnostics), and then samples were processed as previously described (26). Immunoblot analyses were performed as previously described (23). For whole extract, 2 mg equivalents of heart tissue homogenates were subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and probed with specific antibodies against Plin2–5 and Nrf2 (23, 27). GAPDH (1:1000) (Cell Signaling, Beverly, MA) and β-actin (1:5000) (Sigma, St. Louis, MO) were used as the housekeeping genes. For supernatant and fat-cake samples, the protein content of each sample was determined as described previously (15), and the precise amounts of protein loaded on gels were as indicated in the figure legends. The immunoblot signals were detected with Supersignal chemiluminescence reagents (Pierce, Rockford, IL). Mouse anti-GFP antibody (1:10,000) was from Convance (Emeryville, CA). The membranes were visualized with chemiluminescence reagent ECL (Amersham, NJ) and exposed to Kodak XAR film. Quantitative analysis was performed using Un-scan (Silk Scientific Corporation, UT).
Total lipid, total lipid composition, and glycogen content
The hearts were perfused with 3 ml of PBS from the left ventricle. Tissues were excised, weighed, and frozen. Lipids were extracted from 20–40 mg of the left ventricles. Total lipids were extracted by the Folch method (28), and the TAG content was measured using a commercially available kit (Sigma). Lipid composition was determined using thin-layer chromatography as previously described (29). The glycogen content was measured by a commercially available kit (BioVision, CA).
Gene expression analysis
Total RNA was extracted using Qiagen RNease kit (Qiagen), quantitated using a ND1000 nanodrop spectrophotometer, and then between 100 and 400 ng was reverse-transcribed using a using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Switzerland). Steady-state mRNA levels were determined by two-step quantitative real time PCR (qRT-PCR) using the LightCycler 480 (Roche) (30) and Taqman probe/primer sets (Applied Biosystems, CA). GAPDH was used as an internal control for normalization. Primers sequences are listed in supplementary Table II.
For microarray (GEO accession number GSE44192), hearts were harvested from four MCH-Plin5 mice and four control mice at the age of 12 weeks. The total of eight RNA samples was individually processed with the Ambion WT expression kit and GeneChip WT terminal labeling and controls kit (Affymetrix, CA) and hybridized to the Mouse Gene 1.0 ST array (Affymetrix). The array was then read by GCS3000 laser scanner (Affymetrix), and microarray image analysis was carried out using Partek Genomics Suite software. Subsequent analysis was carried out using BioConductor. Data from the eight samples were subjected to background subtraction, quantile normalization, log2 transformation, and probe-set summarization using the robust multichip average algorithm (31). Only the probe sets that interrogate genes were retained, and those meant for quality control and normalization purposes were excluded from further analysis. The significance analysis of microarrays procedure (32) was then used to obtain multiple test-corrected q values for differential expression of genes between the hearts from MCH-Plin5 mice and control. Genes upregulated or downregulated in αMCH-Plin5 hearts were selected with a maximum q value of 0.05. Gene ontology analysis was performed in David Informatics Resources 6.7 with GO BP-FAT (33).
Mitochondrial DNA quantification
Relative amounts of mitochondrial DNA (mtDNA) and nuclear DNA were determined by real-time PCR as described previously (34). Genomic DNA was extracted from mouse heart using the DNeasy Blood and Tissue Kit (Qiagen), according to the manufacturer's instructions. Real-time PCR amplification was performed using the LightCycler 480 (Roche, Switzerland). Levels of mtDNA were measured by normalizing the mitochondrial gene (cytochrome b) to the nuclear gene (actin). A total of 250 ng genomic DNA was used for mtDNA and nuclear DNA markers, respectively, in a 20 μl reaction containing SYBR Green I Master (Roche, IN). The following forward and reverse gene-specific primers were used. Mouse cytochrome b, sense: GGC TAC GTC CTT CCA TGA GGA C; antisense: GAA GCC CCC TCA AAT TCA TTC GAC. Mouse actin, sense: CAT CTC CTG CTC GAA GTC TAG; antisense: ATC ATG TTT GAG ACC TTC AAC ACC C.
Preparation of individual mitochondrial subpopulations
Subsarcolemmal mitochondria (SSM) and interfibrillar mitochondria (IFM) were isolated from adult mouse hearts as previously described in detail (35). Two hearts from the same genotype of mouse were pooled for each experimental data point, and all three genotypes were studied on the same day. Size and membrane potential of SSM and IFM were determined by flow cytometry as previously reported (9).
Mitochondria size and internal complexity
To index mitochondrial subpopulation size and complexity, flow cytometry analyses were performed using a FACS as previously described (35). Briefly, Mitotracker deep red 633 (Invitrogen, Carlsbad, CA), which moves into intact mitochondria due to membrane potential (ΔΨm), was used to selectively stain intact mitochondria (emission wavelength, 633 nm; fluorescence, 633 nm; red diode laser), and debris was excluded by gating for only Mitotracker deep red 633-positive events (intact mitochondria). Forward scatter (FSC) and side scatter (SSC) detectors were used to examine size (FSC) and complexity (SSC) in isolated mitochondrial subpopulations, and data were represented as histograms plotted against the number of gated events as previously described (35).
Electron transport chain respiration
Oxygen consumption in SSM and IFM was measured using a Clark-type oxygen electrode (Qubit Systems, Ontario, Canada). Mitochondria (0.25 mg protein) were maintained in 0.5 ml solution consisting of 100 mM KCl, 50 mM MOPS, 1 mM EGTA, and 0.5 mg fatty acid-free albumin at pH 7.0 and 37°C. State 3 (ADP-stimulated) and state 4 (ADP-limited) respiration were measured with pyruvate + malate (10 mM and 5 mM, respectively) (glycolytic substrates), palmitoyl-CoA (10 mM) (fatty acid substrate), and then succinate plus rotenone (10 mM and 7.5 µM, respectively) was used to assess respiration through complex II of the ETC exclusively. State 4 respiration was measured ± oligomycin. Respiratory control ratios (RCR; state 3 divided by state 4) reflect the control of oxygen consumption by phosphorylation (“coupling”). The ADP/O ratios (number of ADP molecules added for each oxygen atom consumed) are an index of the efficiency of oxidative phosphorylation. These indexes were calculated as described (36).
ROS production
ROS content was estimated by measuring malondiadehyde using a lipid peroxidation assay kit according to the manufacturer's instructions (Oxford Biomedical Research, MI).
Enzyme activity assays
The enzyme activities of citrate synthase (CS), medium-chain acyl-CoA dehydrogenase (MCAD), and aconitase were assayed in tissue homogenates and normalized to milligram of wet tissue weight. To obtain homogenized tissue, 10–20 mg of tissue was combined with 150 μl buffer containing 0.1 M Tris-HCl and 15 mM tricarballylic acid (pH 7.8). Tissue was homogenized in a Bullet blender (Next Advance, Averill Park, NY). For mitochondrial enzymatic measurements, mitochondria were isolated as described above and diluted to 1 mg of protein per milliliter of homogenate.
For aconitase activity, 10 μl of myocardial tissue homogenate or 20 μl of mitochondrial homogenate was added to cuvettes containing 490 μl of aconitase reaction mix (0.083% chloroform, 1.67 mM sodium citrate, 26.7 mM triethanolamine, 0.5 mM NADP+, 0.5 mM MgCl2, pH 7.4), and the increase in absorbance at 340 nm was measured over 5 min. Activity was then determined by multiplying the slope to the molar concentration constant for NADPH (6.7 M−1).
For CS activity, 0.5 μl of myocardial tissue homogenate or 2 μl of mitochondrial homogenate was added to cuvettes containing 500 μl of CS reaction mix (0.1 M Tris-HCl, 1.25 mM 5,5′-dithiobis[2-nitrobenzoic acid], pH 8). Then, 25 μl of 50 mM oxaloacetate and 5 mM acetyl-CoA were added to the reaction mixture, and the increase in absorbance at 412 nm was measured over 5 min. Activity was then determined by multiplying the slope to the molar concentration constant for 5,5′-dithiobis[2-nitrobenzoic acid] (13,600 M-1).
For MCAD activity, 5 μl of myocardial tissue homogenate or 10 μl of mitochondrial homogenate was added to cuvettes containing 500 μl of 100 mM KH2PO4, 1 mM EDTA, 0.5 mM sodium tetrathionate, and 200 μM ferrocenium hexafluorophosphate, pH 7.4. Then, 50 μl of 0.5 mM octanoyl-CoA was added to the reaction mixture, and the decrease in absorbance at 300 nm was measured over 5 min. Activity was then determined by multiplying the slope to the molar concentration constant for ferrocenium hexafluorophosphate (4,300 M−1).
Triglyceride hydrolase activities assays
Tissues were surgically removed and washed in ice-cold PBS and 1 mM EDTA. Tissues were homogenized on ice in lysis buffer A (0.25 M sucrose, 1 mM EDTA, 1 mM dithiothreitol, complete proteases cocktail inhibitor, pH 7.0; Roche Diagnostic) using a glass homogenizer. The infranatants were obtained after centrifugation at 20,000 g at 4°C for 90 min. Cells were harvested by scraping with a rubber policeman, and homogenization was performed on ice in lysis buffer A. The substrate for the measurement of triglyceride hydrolase activity, containing triolein and [9,10-3H]triolein (PerkinElmer Life Sciences), was emulsified with phosphatidylcholine/phosphatidylinositol (3:1) using a probe sonicator (Virsonic 475, Virtis, Gardiner, NJ), as described previously (34). Total-cell lysates (0.1 ml) composed of 10 µl of cell lysate containing ATGL and CGI-58, 80 µl of cell lysate containing either empty vector or Plin5 or Plin3 from cells expressing the constructs described in supplementary Table III were added to 0.1 ml of the substrate and then incubated in a water bath at 37°C for 30 min. The reaction was terminated by adding 3.25 ml of methanol/chloroform/heptane (10:9:7) and 1 ml of 0.1 M potassium carbonate, 0.1 M boric acid, pH 10.5. After centrifugation at 800 g for 20 min, the radioactivity in 0.5 ml of the upper phase was determined by liquid scintillation counting as described previously (15).
Cell culture
CHO-K1 cells were obtained from ATCC (Manassas, VA). Cells were grown in Ham's F-12 medium supplemented with 10% fetal calf serum, 2 mmol/liter L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C with 5% CO2 and 95% humidity. The constructs ATGL-CFP, CGI-58-YFP, Plin5-YFP, and Plin3-GFP were introduced into the CHO-K1 cells according to the manufacturer's instructions. All constructs have been validated previously (14, 15)
Echocardiography
Cardiac function was assessed by echocardiography using a Vevo 2100 high-resolution imaging system (Visual Sonics, Toronto, Canada) with an integrated rail system and a 55 MHz transducer (MS550D). Measurements were performed on mice anesthetized with 1% isoflurane. Mice were shaved and placed in a supine position on a warming pad. M-mode frames were recorded from the parasternal short axis, and Doppler measurements were recorded from the apical view. Absolute wall thickness (AWT) and relative wall thickness (RWT) were calculated as: (PWTd + AWTd) and (PWTd + AWTd) / EDD, where PWTd is the end diastolic posterior wall thickness, AWTd is the end diastolic anterior wall thickness, and EDD is the end diastolic diameter. Ejection fraction was calculated as: (EDV − ESV) / EDV × 100%, where EDV is the end diastolic volume and ESV is the end systolic volume. EDV and ESV were calculated as: ((7.0 / (2.4 + EDD)) × EDD3 and ((7.0 / (2.4 + ESD)) × ESD3, respectively, where ESD is the end systolic diameter. Myocardial performance index (MPI) was calculated as (isovolumetric contraction time + isovolumetric relaxation time) / ejection time.
Intraventricular catheterization
Animals were anesthetized with inhaled 2.5% isoflurane and ventilated at a tidal volume of 0.30 ml at 80 breaths/minute (Harvard Apparatus Inspira, Holliston, MA). Anesthesia was confirmed by toe pinch, and the heart was exposed with a midline sternotomy. A high-fidelity manometer-tipped catheter (SPR-1000 model, Millar, Houston, TX) was rapidly inserted through the apex into the left ventricle (LV), and pressure was recorded for 1–2 min, after which the terminal necropsy procedure was performed.
Tail-cuff blood pressure and heart rate measurement
Systolic blood pressure and heart rates were measured on unanesthetized mice using a Hatteras Instruments SC1000 Dual-Channel Blood Pressure Analysis System (Cary, NC). Mice were warmed to increase blood flow in the tail. Blood pressure was measured on three consecutive days, each analysis consisting of a 5 min preliminary acclimation phase followed by 10 min of data recording. Systolic blood pressure and heart rates were recorded each minute for the 10 min period. Reported values are for all values obtained over the last two consecutive days.
Statistical analysis
All values are expressed as means ± SE. Statistical significance was tested using either one-way ANOVA or a two-tailed Student t-test (GraphPad Software).
RESULTS
Myocardial TAG content is chronically increased in MHC-Plin5
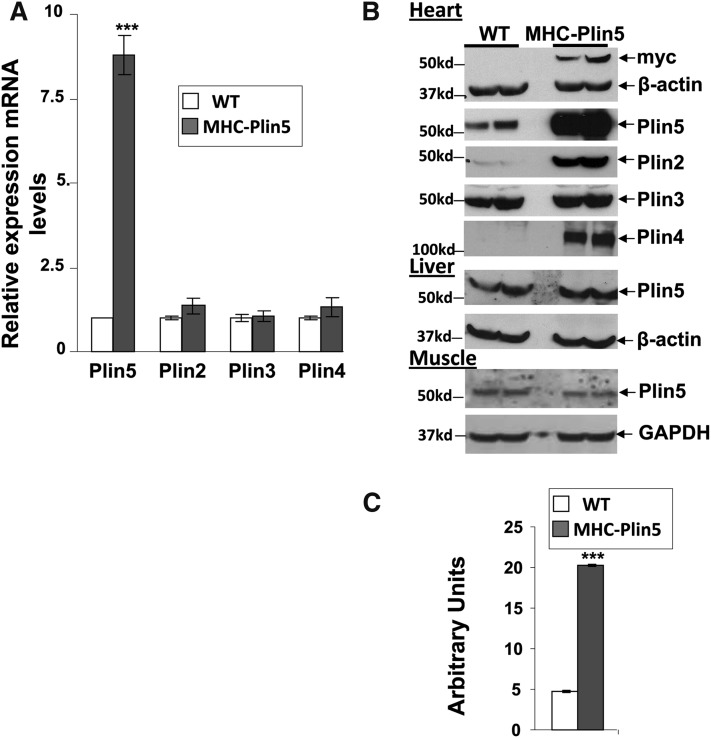
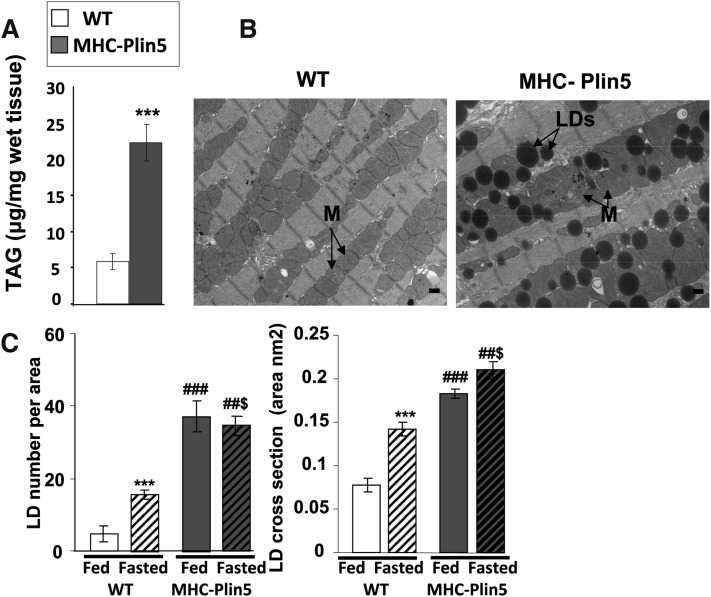
Mice with cardiac myocyte-targeted Plin5 expression (MHC-Plin5) are viable and survive to adulthood. Hearts from MHC-Plin5 mice showed a significant increase in both Plin5 mRNA levels (Fig. 1A) and Plin5 protein expression (Fig. 1B, C). Overexpression of Plin5 was myocardium specific, as Plin5 protein expression in other tissues, such as liver and skeletal, was unchanged (Fig. 1B). Plin5 overexpression did not affect Plin3 protein or Plin2–4 RNA expression, but it did markedly increase Plin2 and Plin4 protein levels in cellular extracts (Fig. 1A, B). Additionally, Plin4 was found increased in Plin4 supernatant as well as fat-cake fractions (supplementary Fig. III), suggesting a role of Plin5 to stabilize LDs in the heart. This was further confirmed by our histomorphological analysis and TAG measurements showing a marked increase in LDs and intracellular TAG in MHC-Plin5 hearts (Fig. 2A–C). TAG was present in larger and more numerous LDs (Fig. 2C). Lipid fat cake composition assayed by thin-layer chromatography showed that TAG constitutes the major lipid species in heart and confirmed that it were strikingly elevated in MHC-Plin5 mice (supplementary Fig. I). While fasting increased the number and the LD droplet size in WT mice cardiac myocytes as previously reported (37), larger and more numerous LDs were chronically present in MHC-Plin5 cardiac myocytes, irrespective of the nutritional status of the mice. We confirmed by histocytochemistry and Western blots that Plin5 was present at the lipid surface in cardiac muscles and in the fat-cake fraction from fed MHC-Plin5 mice (supplementary Fig. IIA, B).
Fig. 1.
Heart-specific Plin5 overexpression results in posttranscriptional expression of other cardiac perilipin proteins. (A) mRNA expression of Plin5 is markedly increased in fed three-month-old male MHC-Plin5 compared with age-matched WT. Means are errors bars ± SEM; n = 9 per group; ***P < 0.001. (B) Western blot analysis of cardiac perilipin protein expression profile. Left ventricles were excised, and equal milligram equivalents of wet tissue were loaded in each lane. Experiments were performed twice. Liver and gastrocnemius muscle Plin5 content were determined to control for the specificity of Plin5 overexpression. A myc antibody was used to recognize ectopic expression of Plin5. β-actin and GAPDH were used as control loading. (C) Quantification of Plin5 protein expression in WT and MHC-Plin5 hearts by densitometry of Western blots. Means are errors bars ± SEM; n = 4 for each genotype; ***P < 0.001 for WT-fasted versus MHC-Plin5-fasted mice.
Fig. 2.
MHC-Plin5 mice show massive increase of LDs in their heart. (A) TAG content in cardiac muscle of male WT and MHC-Plin5 at three months old. Errors bars are means ± SEM; n = 6 for WT and n = 4 for MHC-Plin5; **P < 0.001 for WT versus MHC-Plin5. (B) Representative electron micrographs depict the histological appearance of left ventricles from fed WT and MHC-Plin5 mice at three months old (magnification 3,200, bar equals 500 nm). MHC-Plin5 hearts accumulated LDs (arrowheads) around clusters of mitochondria (M, arrowheads), whereas no lipid droplets were observed in similar regions of the left ventricle from WT mice. We used four mice per group and studied 35 positions per sample. (C) Morphometric analysis of LDs. Using six electron micrographs of left ventricles from three fed and overnight-fasted WT and MHC-Plin5 mice three months old, the LD number was manually counted and mitochondria area was determined using ImageJ software. Means are errors bars ± SEM; ***P < 0.001 for WT fed versus WT fasted, ###P < 0.001 for MHC-Plin5 fed versus MHC-Plin5 fasted, ##$P < 0.001 fasted MHC-Plin5 versus fasted WT.
Next, we checked whether the LD accumulation was due to the increased presence of Plin5 and not due to a decreased capacity to hydrolyze TAG in MHC- Plin5 cardiac myocytes. Measurements of total heart hydrolase activity performed on whole- tissue extracts were not affected by Plin5 overexpression (4.6 ± 0.54 nmol FA released/hour/milligram protein for WT and 5.19 ± 0.7 nmol FA released/hour/milligram protein for MHC-Plin5 mice, values are ± SEM, n = 4). In addition, RNA expression levels for ATGL and for CGI-58 were found unchanged (supplementary Fig. IVA). ATGL protein expression was not found different in whole cardiac tissue lysates between genotypes (supplementary Fig. IVB). In addition, ATGL protein expression was measured in fat-cake and supernatant fractions (supplementary Fig. IVC). In both fractions, an antibody against mouse ATGL recognized a doublet appearing at 54 kDa and at 50 kDa, respectively. The 54 kDa band was similar in the supernatant of both WT and MHC-Plin5 supernatant samples; the 50 kDa band was increased in the MHC-Plin5 supernatant and MHC-Plin5 fat-cake samples; and the 54 kDa band was found increased in the WT fat-cake samples. Differences in detergent content of the buffer used for whole extract as opposed to fractionation of fat cake might explained these differences in ATGL protein (see Material and Methods). CGI-58 protein expression was found increased in both supernatant and fat-cake fractions (supplementary Fig. IVD).
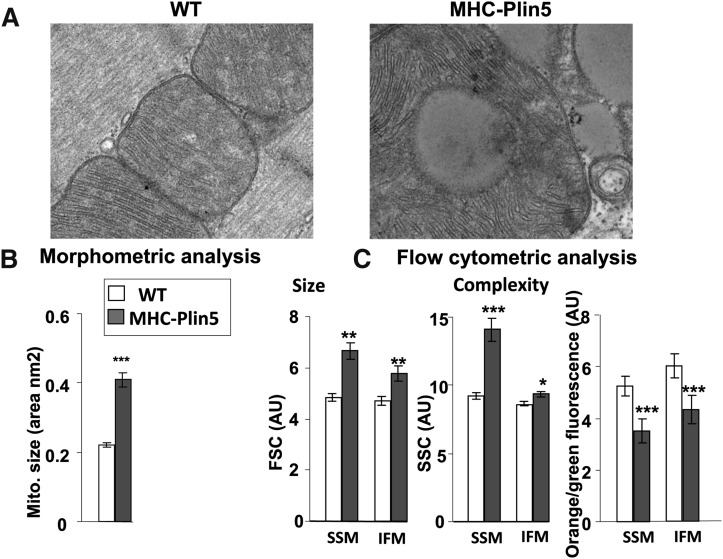
Mitochondria in MHC-Plin5 appeared to always be distributed in very tight cluster around LDs and in some cases, the enlarged mitochondria in the MHC-Plin5 tissue displayed further abnormalities in their internal structure, such as loss of cristae and formation of inner membrane vesicles (Fig. 3A and supplementary Fig. V). Morphometric analysis of mitochondria from cardiac muscles from WT and MHC-Plin5 size revealed a significant increase in mitochondrial size in cardiac muscle from the MHC-Plin5 mice (Fig. 3B). In contrast, the relative amount of mitochondrial DNA in cardiac muscle from the WT and MHC-Plin5 mice was unchanged as estimated by the determination of the ratio of α-actin/cytochrome b (32.1 ± 3.6 for WT and 28.7 ± 3.8 for MHC-Plin5 mice, values are ± SEM, n = 4), thus suggesting similar number of mitochondria in the hearts of MHC-Plin5 mice. To further characterize the consequences of Plin5 overexpression on mitochondria morphology, cardiac mitochondrial subpopulations were isolated. Mitochondrial size was found to be larger by flow cytometry for both subsarcolemma mitochondria (SSM) and interfibrillar mitochondria (IFM) in the MHC-Plin5 hearts than in WT (Fig. 3C), consistent with the electron microscopy analysis (Figs. 2B, 3A, and supplementary Fig. IV). The membrane potential of SSM and IFM was lower with high expression of Plin5 in both SSM and IFM (Fig. 3C).
Fig. 3.
Heart-specific Plin5 overexpression results in mitochondria morphology alteration. (A) Representative electron micrographs depict the histological appearance of mitochondria in left ventricles from fed WT and MHC-Plin5 mice at 3 months old (magnification 21,000, bar equals 100 nm). (B) Morphometric analysis of mitochondria. Using six electron micrographs of left ventricles from four overnight fed WT and MHC-Plin5 mice at 12 weeks old, mitochondria area were determined using ImageJ software. Means are errors bars ± SEM; ***P < 0.001. (C) Flow cytometry analysis of subpopulations of mitochondria and ΔΨm. The relative size and internal complexity of distinct mitochondrial subpopulations were determined using flow cytometric analyses. Mitochondrial subpopulations were stained with Mitotracker deep red 633, a membrane potential (ΔΨm)-dependent dye, and gated based on incorporation of the dye. Analysis of FSC and SSC were calculated per 20,000 gated events for all mitochondrial subpopulations. FSC and SSC are expressed in arbitrary units (AU). Means are errors bars ± SEM; n = 18 for WT and n = 12 for MHC-Plin5; *P < 0.05, **P < 0.01, ***P < 0.001 for WT versus MHC-Plin5. ΔΨm was assessed by staining mitochondrial subpopulations with JC-1 dye and assessing the shift from green to orange fluorescence with flow cytometry. Ψm was calculated based on orange-to-green fluorescence ratios in WT versus MHC-Plin5 cardiac mitochondrial subpopulations. Orange-to-green fluorescence ratios are expressed in AU. Means are errors bars ± SEM; n = 18 for WT and n = 12 for MHC-Plin5; ***P < 0.001 for WT versus MHC-Plin5.
Plin5 overexpression induces a gene expression pattern indicative of downregulation of FA utilization, upregulation of glucose utilization, and activation of the Nrf2 antioxidative pathway
To identify cellular pathways likely to be affected by overexpression of Plin5 in the heart, a microarray analysis of gene expression in myocardium from three-month-old mice was performed, and results are reported in Tables 1, 2, and supplementary Table I. Gene ontology analysis showed that genes downregulated in MHC-Plin5 myocardium were substantially enriched in mitochondrial bioenergetic processes and lipid metabolism (P < 0.01), whereas genes upregulated in MHC-Plin5 myocardium were enriched in oxidative stress-induced gene expression via Nrf2 (P < 0.005), especially genes involved in gluthatione metabolism (Table 2 and supplementary Table I). Further pathway analysis showed that many differentially expressed genes in the MHC-Plin5 hearts encoded enzymes that are involved in mitochondria β-oxidation, oxidative phosphorylation, and fatty acid metabolism and that most of these genes were previously reported to be PPARα target genes (38). In contrast, a shift in energetic utilization toward glucose is suggested by the regulation of key regulatory genes promoting glycolysis with decreased expression for 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (Pfkfb1) and increased expression of glucokinase (Gck), hexokinase (Hk1), and glucose transporter 1 (Slc2a1) (supplementary Table I), although measured cardiac glycogen stores remained unchanged (amount of glucose was 0.88 ± 0.45 mg/g wet tissue in MHC-Plin5 hearts versus 0.92 ± 0.98, n = 4 in control WT). Importantly, the expression of a number of genes previously involved with cytoskeleton reorganization in MHC-Plin5 hearts was found increased, as well as a small subset of genes reported to be associated with early heart failure conditions (Table 2).
TABLE 1.
Genes downregulated in knockout versus WT myocardium
| Process | Gene Symbol | Gene Description | F.C. | P |
| Acyl CoA formation/hydrolysis/binding | Acsm5 | Acyl-CoA synthetase medium-chain family member 5 | −2.4 | 1.1E-04 |
| Acsl6 | Acyl-CoA synthetase long-chain family member 6 | −1.9 | 4.6E-04 | |
| Acot1 | Acyl-CoA thioesterase 1 | −1.7 | 4.2E-04 | |
| Lpcat3 | Lysophosphatidylcholine acyltransferase 3 | −1.7 | 1.2E-05 | |
| Acss1 | Acyl-CoA synthetase short-chain family member 1 | −1.4 | 6.1E-04 | |
| Mitochondrial β-oxidation/oxidative phosphorylation | Scd4 | Stearoyl-CoA desaturase 4 | −3.0 | 9.2E-07 |
| Ucp3 | Uncoupling protein 3 (mitochondrial, proton carrier) | −2.9 | 7.6E-04 | |
| Acaa2 | Acetyl-CoA acyltransferase 2 (mitochondrial 3-oxoacyl-CoA thiolase) | −2.0 | 1.7E-04 | |
| Slc25a20 | Solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 | −1.7 | 4.2E-04 | |
| Fads3 | Fatty acid desaturase 3 | 1.6 | 3.1E-04 | |
| Acadsb | Acyl-CoA dehydrogenase, short/branched chain | −1.4 | 7.4E-04 | |
| Acss1 | Acyl-CoA synthetase short-chain family member 1 | −1.4 | 6.1E-04 | |
| Fh1 | Fumarate hydratase 1 | −1.4 | 1.1E-03 | |
| Ndufs8 | NADH dehydrogenase (ubiquinone) Fe-S protein 8 | −1.4 | 1.1E-03 | |
| Ketogenesis | Hmgcs2 | 3-Hydroxy-3-methylglutaryl-CoA synthase 2 | −1.9 | 1.4E-04 |
| Ech1 | Enoyl CoA hydratase 1, peroxisomal | −1.7 | 1.8E-04 | |
| Acat1 | Acetyl-CoA acetyltransferase 1 | −1.5 | 1.1E-03 | |
| Peroxisomal β-oxidation | Peci | Peroxisomal delta3, delta2-enoyl-CoA isomerase | −1.5 | 4.2E-04 |
| Lipases | Ces3 | Carboxylesterase 3 | −2.5 | 1.2E-04 |
| Lipe | Lipase, hormone sensitive | −1.5 | 5.8E-01 | |
| Biotransformation | Ephx2 | Epoxide hydrolase 2, cytoplasmic | −1.5 | 2.9E-04 |
Enriched biological processes of genes were differentially expressed in myocardium of MCH-Plin5 versus WT control mice. F.C., fold change.
TABLE 2.
Genes upregulated in knockout versus WT myocardium
| Process | Gene Symbol | Gene Description | F.C. | P |
| Oxidative stress via Nrf2 pathway | Gsta1 | Glutathione S-transferase, α 1 (Ya) | 12.18 | 3.5E-09 |
| Gsta2 | Glutathione S-transferase, α 2 (Yc2) | 7.00 | 6.9E-08 | |
| Atf4 | Activating transcription factor 4 | 2.83 | 9.7E-07 | |
| Esd | Esterase D/formylglutathione hydrolase | 2.20 | 5.0E-06 | |
| Txnrd1 | Thioredoxin reductase 1 | 1.93 | 6.0E-05 | |
| Hmox1 | Heme oxygenase (decycling) 1 | 1.93 | 2.3E-04 | |
| NQO1 | NAD(P)H dehydrogenase, quinone 1 | 1.80 | 3.8E-04 | |
| Gss | Glutathione synthetase | 1.75 | 4.8E-06 | |
| Por | P450 (cytochrome) oxidoreductase | 1.53 | 3.6E-04 | |
| Gclc | Glutamate-cysteine ligase, catalytic subunit | 1.53 | 9.0E-04 | |
| Mafk | V-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) | 1.50 | 6.8E-04 | |
| Gpx4 | glutathione peroxidase 4 | 1.39 | 3.3E-04 | |
| Early heart failure markers | Asns | asparagine synthetase | 22.07 | 2.7E-11 |
| Mthfd2 | Methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase | 20.65 | 1.4E-10 | |
| Maoa | Monoamine oxidase A | 6.11 | 3.0E-04 | |
| Gdf15 | Growth differentiation factor 15 | 5.90 | 4.5E-04 | |
| Myc | Myelocytomatosis oncogene | 4.93 | 3.5E-07 | |
| Uchl1 | Proteasome | 4.62 | 1.2E-03 | |
| Chac1 | ChaC, cation transport regulator-like 1 (E. coli) | 4.46 | 3.1E-07 | |
| Atf5 | Activating transcription factor 5 | 3.31 | 2.2E-06 | |
| Trib3 | Tribbles homolog 3 (Drosophila) | 2.92 | 2.3E-06 | |
| Grk5 | G protein-coupled receptor kinase 5 | 1.88 | 7.3E-04 | |
| Cytoskeleton organization | Acta1 | Actin, α 1, skeletal muscle | 3.72 | 2.1E-06 |
| Mtap1a | Microtubule-associated protein 1A | 3.38 | 2.8E-06 | |
| Ttll7 | Tubulin tyrosine ligase-like family, member 7 | 2.72 | 4.3E-05 | |
| Mtap1b | Microtubule-associated protein 1B | 2.56 | 4.0E-04 | |
| Tubgcp4 | Tubulin, γ complex-associated protein 4 | 2.07 | 3.9E-05 | |
| Flnc | Filamin C, γ | 2.05 | 1.2E-05 |
Enriched biological processes of genes were differentially expressed in myocardium of MCH-Plin5 versus WT control mice. F.C., fold change.
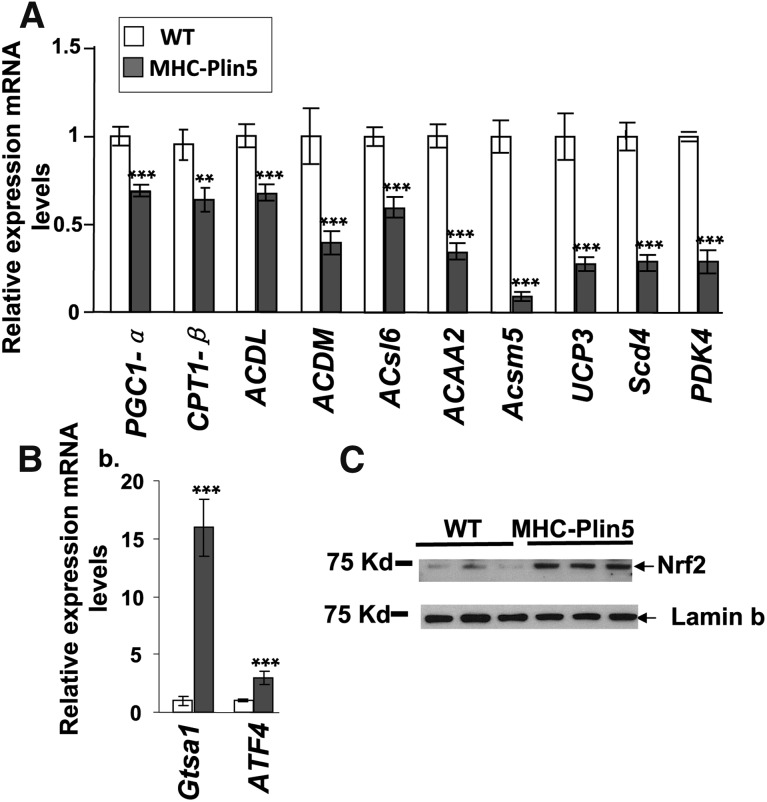
Our follow-up analysis of mRNA expression on a smaller subset of genes by quantitative real-time PCR analysis revealed a reduced expression of PPARα target genes in MHC-Plin5 hearts, including PGC1-α (PGC1-α, −30%), long-chain acyl-CoA dehydrogenase (Acadl, −40%), medium-chain acyl-CoA dehydrogenase (Acadm, −60%), acyl-CoA synthetase long-chain family member 6 (ACSL6, −40%), acetyl-CoA acyltransferase 2 (ACAA2, −65%), acyl-CoA synthetase medium-chain family member 5 (Acsm5, −65%), uncoupling protein 3 (UCP3, −70%), stearoyl-CoA desaturase 4 (scd4, −70%), and pyruvate dehydrogenase kinase 4 (Pdhk4, −70%), compared with the levels found in WT mice (Fig. 4A). We confirmed the upregulation of the oxidative stress pathway via Nrf2 by measuring increased expression of two Nrf2-targeted genes, glutathione S-transferase A1 (Gsta1, +1,600%) and activating transcription factor 4 (ATF4, +298%), by RT-PCR and the increased presence of Nrf2 protein in MHC-Plin5 nuclear extracts by Western blot (Fig. 4B). Intracellular accumulation of ROS, estimated by measuring malondiadehyde content, was found increased in MHC-Plin5 cardiac tissue homogenates compared with WT (13.2 ± 0.92 μmol/mg protein for WT and 19.2 ± 2 μmol/mg protein for MHC-Plin5 mice, values are ± SEM, n = 8).
Fig. 4.
Plin5 overexpression decreases selected PPARα target genes and increases selected Nrf2 target genes. (A) mRNA expression levels for selected PPARα target genes were determined by RT-qPCR analysis. mRNA expression of PPARα decreased in fed three-month-old MHC Plin5 hearts. Means are errors bars ± SEM; n = 9 for each group; **P < 0.01, ***P < 0.001 for WT versus MHC-Plin5. (B) mRNA levels of genes encoding Gtsa1 and ATF4 mRNA were increased in cardiac muscle. Means are errors bars ± SEM; n = 9 for each group; **P < 0.01, ***P < 0.001 for WT versus MHC-Plin5. (C) Nrf2 protein expression was found increased in MHC-Plin5 heart nuclear extracts. Lamin b was used as control loading.
Mild impaired cardiac mitochondrial respiration in MHC-Plin5
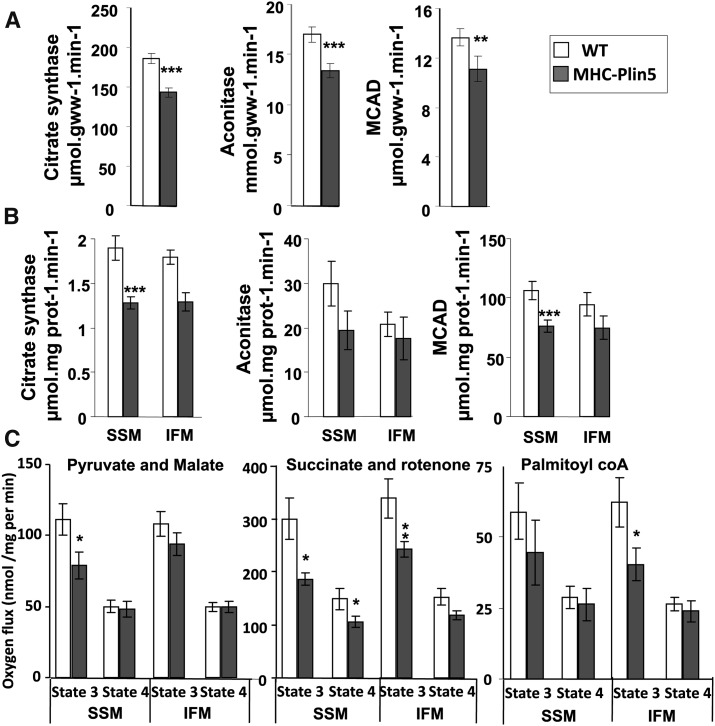
We found that heart Plin5 overexpression prompted impairment of cardiac mitochondrial function. We prepared heart homogenates from 3-month-old mice and found a small but significant decline in the activity of the mitochondrial enzymes medium chain acyl-CoA dehydrogenase (MCAD) (Fatty acid β-oxidation) of -18% and aconitase of -21% or citrate synthase (tricarboxylic acid cycle) of -23% in MHC-Plin5 hearts (Fig. 5A). Results from measurements of mitochondrial enzyme activities in isolated mitochondria subpopulations confirmed the decrease of citrate synthase, aconitase and MCAD previously found in whole cell extracts from MHC Plin5 hearts (Fig. 5B).
Fig. 5.
α-MHC-Plin5 mice have impaired mitochondria function. (A) Myocardial activity of the mitochondrial oxidative enzymes citrate synthase, aconitase, and medium-chain acyl-CoA dehydrogenase in whole-heart extract from WT and MHC-Plin5. Means are errors bars ± SEM; n = 17 for WT and n = 12 for MHC-Plin5; **P < 0.01, ***P < 0.001 for WT versus MHC-Plin5. (B) SSM and IFM from WT and MHC-Plin5 hearts were isolated, and mitochondrial oxidative enzymes citrate synthase, aconitase, and medium-chain acyl-CoA dehydrogenase were measured. Means are errors bars ± SEM; n = 17 for WT and n = 11 for MHC-Plin5; *P < 0.05 for WT versus MHC-Plin5. (C) Respiration of isolated mitochondria from cardiac tissues of three-month-old MHC-Plin5 and WT mice. ADP-driven (state 3) and state 4 were measured with pyruvate (20 mM) and malate (10 μM) or succinate (10 mM) and rotenone (7.5 μM) or palmitoyl CoA (10 mM) in SSM and IFM preparations. Means are errors bars ± SEM; n = 17; *P < 0.05, **P < 0.05.
Studies in isolated mitochondria revealed a lower yield of SSM and IFM from MHC-Plin5 hearts than from WT hearts (20.9 ± 1.13 versus 10.2 ± 1.4 mg mitochondria protein per gram wet mass for WT and MHC-Plin5 SSM, respectively, P < 0.001; 16.2 ± 1 versus 12.1 ± 1 mg mitochondria protein per gram wet mass for WT and MHC-Plin5 IFM, respectively, P < 0.001). It is likely that this lower yield of mitochondria obtained from MHC-Plin5 hearts resulted from an increased number of mitochondria still associated to the LD fraction that remained in the supernatant during mitochondria isolation. Indeed, we found increased presence of a mitochondria marker voltage-dependent anion channels (VDAC) in MHC-Plin5 supernatant (supplementary Fig. VI).
Mitochondrial respiration was significantly altered in MHC-Plin5 SSM and IFM mitochondria (Fig. 5C and supplementary Table II). State 3 respiration was decreased in MHC-Plin5 SSM mitochondria with pyruvate + malate as substrate (−30%), but no statistical difference was observed in IFM mitochondria. State 3 respiration was also decreased with palmitoylcarnitine used as a substrate in SSM mitochondria but was not statistically different, whereas in IFM mitochondria it decreased significantly (−25%). Independent of the substrate used for the experiment, levels of state 4 respiration were similar among all three groups except in presence of succinate and rotenone. RCR (state 3/state 4 ratio) values were found significantly different in MHC-Plin5 SSM mitochondria in presence of malate + pyruvate, indicating some defect in mitochondria coupling (2.4 ± 0.42 for WT and 1.64 ± 0.12 for MHC-Plin5 mice, values are ± SEM, n = 12). ADP/O ratio was normal (data not shown), indicating that overexpression of Plin5 did not affect the efficiency of phosphorylation.
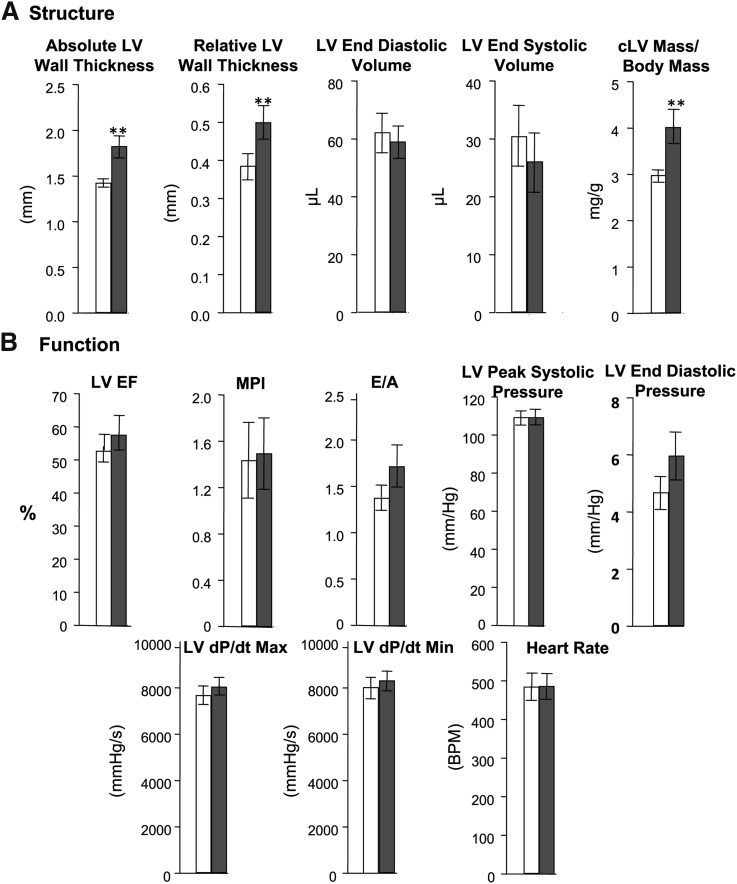
Overexpression of Plin5 leads to increased left ventricular mass and remodeling but not to heart failure
We next characterized the cardiac function of four-month-old mice. Hearts from MHC-Plin5 were enlarged with significantly elevated heart-weight to tibia-length ratios compared with WT (5.14 ± 0.15 for MHC-Plin5 versus 4.79 ± 0.1 for WT, P < 0.05). Echocardiography demonstrated a LV hypertrophy with thickening of ventricular walls (Fig. 6A), but none of the parameters related to heart function were affected in MHC-Plin5 hearts (Fig. 6B). Direct measurement of LV pressure found no differences between WT and MHC-Plin5 mice for peak LV pressure, peak positive or negative dP/dt, or end diastolic pressure, providing further support for no effect on cardiac systolic or diastolic function (Fig. 6B).
Fig. 6.
Echocardiography analysis of cardiac structure and function on four-month-old WT and MHC-Plin5 mice. (A) Cardiac structure data. Means are errors bars ± SEM; n =7 for WT and for MHC-Plin5; **P < 0.01. LV, left ventricular; cLV, concentric LV. (B) Contractile function measurements. For LV peak systolic pressure, LV end diastolic pressure, LV dP/dt max, LV dP/dt min, and heart rate, n =33 for WT and n = 22 for MHC-Plin5 mice. For measurements of LV, EF, MPI, and E/A, n = 7 for WT and for MHC-Plin5 mice.
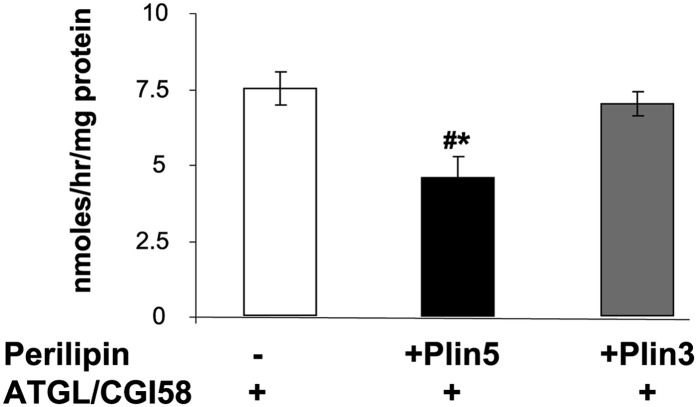
Perilipin 5 inhibits TAG hydrolase activity in a reconstituted in vitro system
To test for the ability for Plin5 to inhibit TAG hydrolyase activity, we used a reconstituted in vitro system in which we measured the ability of Plin5 to prevent the hydrolytic activity of ATGL in the presence of its colipase CGI-58 toward an exogenous substrate containing phospholipids and triolein. As control, we used Plin3, another exchangeable perilipin protein. Presence of Plin5, but not Plin3, statistically decreased the ability of ATGL and CGI-58 to hydrolyze TAG (−40%) (Fig. 7).
Fig. 7.
Plin5 inhibits TAG hydrolase activity in an in vitro reconstituted system. CHO-K1 cells were transfected with ATGL-CFP and CGI-YFP plasmids, with Plin5 alone, or with Plin3 alone. Cellular extracts (20 μl) overexpressing ATGL and CGI-58 were incubated with 80 μl of cellular extracts overexpressing either Plin5 or Plin3 and 100 μl of a radioactive exogenous substrate. After 1 h of incubation, released radiolabeled NEFA were extracted and dpm was measured with a liquid scintillation counter. Means are errors bars ± SEM; n = 3 for empty GFP vector, n = 3 for Plin5-YFP, n = 3 for Plin3-GFP; *P < 0.05 for Plin5-YFP versus empty GFP vector or versus Plin3-GFP.
DISCUSSION
The relationship between cardiac LDs and heart function is yet to be fully understood. A major finding of this investigation is that Plin5 stabilizes cardiac LDs, and we confirmed that Plin5 is a major player in cardiac LDs. Plin5 overexpression resulted in cardiac steatosis but did not lead to a major heart dysfunction phenotype despite differences in gene expression, suggesting a shift in energy substrate utilization, noticeable structural mitochondrial abnormalities, modest decrease in mitochondrial function, and left concentric ventricular hypertrophy. An interesting feature of our results is that Plin5 overexpression elicits an induction of the Nrf2 antioxidative pathway, suggesting a possible compensation by this pathway against the maladaptive cardiac response to increased cardiac Plin5-coated LDs.
Members of the perilipin protein family are the proteome signature of LDs. Four out of the five perilipin members (Plin2–5) have been found expressed in mammalian cardiac muscle (15). Among them, Plin5 must play a distinctive function as it is selectively expressed in oxidative tissues. Plin5, along with Plin3 and Plin4, has been termed an exchangeable perilipin because it can be found in the cytosol at discrete locations in the absence of FA. However, upon initiation of LD biogenesis by FA uptake, Plin5 is found bound to LD surface (39, 40). Most importantly, Plin5 has been shown to increase FA-mediated TAG accumulation in several cell culture models, and we suggested that Plin5, like adipocyte-specific Plin1, inhibits LD hydrolysis in cultured cells (23). We demonstrate here that Plin5 exerts the same property in vivo when constitutively expressed in heart. Interestingly, by overexpressing Plin5, protein levels of Plin2 and Plin4 are also found increased in absence of noticeable change in mRNA levels. Reciprocally, absence of Plin5 was reported to lead to decreased Plin2 protein in the heart (24). The positive relationship between Plin2 and Plin5 is unexpected and differs from the one observed between adipose Plin1 and Plin2: presence of Plin1 was reported to decrease Plin2 protein expression, whereas absence of Plin1 promoted increased presence of Plin2 in the LD (29, 41). A possible explanation for this “off-target effect” is that Plin5, by stabilizing LDs, increases Plin2 and Plin4 protein expression via a posttranslational mechanism. Indeed, Plin2 has been previously found to have posttranslation regulation by the ubiquitin/proteasome pathway at times when Plin2 is unable to bind to LD surface (42). Existence for such a regulation is yet to be demonstrated for Plin4. Hence, these experiments demonstrate that Plin5 plays an important role in promoting and stabilizing cardiac LDs by increasing the levels of other perilipin proteins.
Cardiac Plin5 overexpression leads to increased LDs without alteration in total heart hydrolase activity or in ATGL gene and total protein expression. In our hands, ATGL protein content in cardiac fat-cake fraction is difficult to quantify because of proteolysis occurring during the fat-cake preparation and possibly due to a lack of detergents in the fractionation buffer. In addition, it was shown that heart depletion in cardiac LDs in Plin5 knockout mice could be overcome by the use of a lipase inhibitor (24). The combined results from these transgenic mice models and from past studies performed in vitro (11, 14–17) suggest that Plin5 is a negative regulator of cardiac LD hydrolysis. The exact mechanism(s) by which Plin5 inhibits LD hydrolysis remain to be determined. Two mechanisms can be hypothesized. The coordinated increase of perilipin proteins at LD surface may be sufficient to impair access of ATGL to its TAG substrate. Indeed, presence of Plin2 was shown to inhibit ATGL binding to LD surface in cell culture systems (43), whereas absence of Plin2 in mice was associated with decreased LDs (44). Alternatively, Plin5 may act as a unique ATGL negative regulator by providing either scaffolding for ATGL or CGI-58, as suggested in vitro (14, 16). ATGL- or CGI-58-Plin5 binding may then alter the interactions between ATGL and CGI-58, an established positive regulator of ATGL (45). In support of the latter, we demonstrated here that Plin5 inhibits TAG hydrolase in a reconstituted in vitro system and that this property is not shared by Plin3, another exchangeable perilipin protein. In addition, the presence of CGI-58 is increased in the MHC-Plin5 fat-cake fraction. It was shown that Plin1 regulated ATGL activity via its binding to CGI-58 in adipose cells (46); by analogy, a similar regulation can be hypothesized for Plin5 regulation of ATGL. Although strong evidence from the existing literature and the data presented here support a role for Plin5 to regulate ATGL activity in vitro and in vivo (11, 14, 23), we cannot definitively attribute all observed consequences of Plin5 overexpression to the regulation of ATGL activity by Plin5, as overexpression of a protein might induces unexpected off-target effects. More specific to Plin5, we and others have previously found that Plin5 elicits a close physical proximity between LDs and mitochondria. Therefore, it remains a possibility that Plin5 overexpression may also directly alter mitochondria function, which in turn would result in cardiac LD accumulation. To precisely dissect the individual role of Plin5 on these organelles is difficult as both of these organelles share common regulatory pathways. Future work is needed to investigate the relationships among LDs, mitochondria, and Plin5.
Due to the ability of Plin5 overexpression to inhibit LD hydrolysis in vivo and in vitro, we predicted that the phenotype of MHC-Plin5 hearts may resemble the one recently described for the ATGL-knockout mice, leading to severe cardiac steatosis, decreased PPARα gene-targeted activities, and decreased number of mitochondria and mitochondria, function followed by heart failure and early death (22). MHC-Plin5 hearts show some adaptive features to LD overload but do not experience heart failure. Unexpectedly, chronic Plin5 expression led to oxidative stress and triggered an upregulation of the Nrf2 antioxidative pathway with specific increases in gene expression involved in gluthatione metabolism. Nrf2 is a pleiotropic protein that binds a cis-acting enhancer sequence known as the antioxidant response element (ARE) to control the basal and inducible expression of a battery of antioxidant genes and other cryoprotective phase II detoxifying enzymes (47). The cardioprotective role of Nrf2 in heart was recently investigated in a mouse model of heart failure (48). Nrf2-targeted gene expression was induced following the first two weeks of transverse aortic constriction (TAC) in control mice, whereas Nrf2 absence led to faster induction of heart failure after surgery, an indication that the transient activation of the Nrf2 antioxidative pathway is a compensatory mechanism against heart failure (47). Therefore, MHC-Plin5 hearts may be protected against progression toward heart failure due to the activation of the Nrf2 antioxidative pathway. Interestingly, mice lacking Plin5 were found to have increased cardiac ROS but developed heart dysfunction that can be preventable with an antioxidant treatment (24). Results from both transgenic mice models suggest a novel and intriguing relationship between Plin5-coated LDs and Nrf2 signaling that remains to be investigated. ROS are known to be implicated in the induction of cardiac hypertrophy in various pathologic states, and cardiac hypertrophy is observed in transgenic mice with Plin5 overexpression or loss of function. It is hypothesized that cardiac ROS production in Plin5 knockout mice might result from a surplus flux of FA into the mitochondria; in contrast, it may be hypothesized that ROS production observed in cardiac plin5 overexpression might originate from endoplasmic reticulum stress due to excess formation of LDs or from mitochondria structural alteration due to LD close proximity. Further studies are needed to confirm ROS cellular origin in these two transgenic models.
The combination of coexisting risk factors, e.g., obesity, insulin resistance, hyperlipidemia, hypertension, and type 2 diabetes, makes it difficult to attribute a direct cause and effect relationship to myocardial lipid deposition. Here, the direct manipulation of the LD compartment with overexpression of Plin5, a structural LD protein, led to an increase in cardiac LD content but failed to induce a heart failure phenotype. In contrast, chronic absence of LDs in the recently characterized Plin5 overall knockout mice led to heart dysfunction with age (15). These combined results indicate that Plin5-coated LDs play an important protective role in regulating cardiac lipid homeostasis. By its ability to regulate ATGL, Plin5 has the potential to be a key gene regulating cardiac LD hydrolysis and, therefore, tuning mitochondrial function.
Supplementary Material
Acknowledgments
The authors thank Dr. Andrew Saladino for his generous assistance with electron microscopy; Kathy Ryan who graciously helped us with the statistical analysis; and Dr. Martin Woodle for careful review and helpful editing of the manuscript.
Footnotes
Abbreviations:
- IFM
- interfibrillar mitochondria
- LD
- lipid droplet
- LV
- left ventricular
- MCAD
- medium-chain acyl-CoA dehydrogenase
- MHC-Plin5
- α-myosin heavy-chain perilipin 5
- Nrf2
- NF-E2-related factor 2
- Plin1–5
- perilipin 1–5
- PPARα
- peroxisome proliferator-activated receptor α
- ROS
- reactive oxygen species
- SSM
- subsarcolemma mitochondria
- TAG
- triglyceride
- WT
- wild-type
This work was supported by National Institutes of Health Grants 1-RO1-DK-075017 (to C.S.), DK-072488, and intramural research programs; American Heart Association Grant-in-Aid 11GRNT7600027 (to C.S.); and the Geriatric Research, Education, and Clinical Center, Baltimore Veterans Affairs Health Care Center.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of seven figures and two tables.
REFERENCES
- 1.Wende A. R., Abel E. D. 2010. Lipotoxicity in the heart. Biochim. Biophys. Acta. 1801: 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reingold J. S., McGavock J. M., Kaka S., Tillery T., Victor R. G., Szczepaniak L. S. 2005. Determination of triglyceride in the human myocardium by magnetic resonance spectroscopy: reproducibility and sensitivity of the method. Am. J. Physiol. Endocrinol. Metab. 289: E935–E939. [DOI] [PubMed] [Google Scholar]
- 3.Khan R. S., Drosatos K., Goldberg I. J. 2010. Creating and curing fatty hearts. Curr. Opin. Clin. Nutr. Metab. Care. 13: 145–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chokshi A., Drosatos K., Cheema F. H., Ji R., Khawaja T., Yu S., Kato T., Khan R., Takayama H., Knöll R., et al. 2012. Ventricular assist device implantation corrects myocardial lipotoxicity, reverses insulin resistance, and normalizes cardiac metabolism in patients with advanced heart failure. Circulation. 125: 2844–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Listenberger L. L., Han X., Lewis S. E., Cases S., Jr.Farese R. V., Ory D. S., Schaffer J. E. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA. 100: 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park T. S., Yamashita H., Blaner W. S., Goldberg I. J. 2007. Lipids in the heart: a source of fuel and a source of toxins. Curr. Opin. Lipidol. 18: 277–282. [DOI] [PubMed] [Google Scholar]
- 7.Brindley D. N., Kok B. P., Kienesberger P. C., Lehner R., Dyck J. R. 2010. Shedding light on the enigma of myocardial lipotoxicity: the involvement of known and putative regulators of fatty acid storage and mobilization. Am. J. Physiol. Endocrinol. Metab. 298: E897–E908. [DOI] [PubMed] [Google Scholar]
- 8.Liu L., Shi X., Bharadwaj K. G., Ikeda S., Yamashita H., Yagyu H., Schaffer J. E., Yu Y. H., Goldberg I. J. 2009. DGAT1 expression increases heart triglyceride content but ameliorates lipotoxicity. J. Biol. Chem. 284: 36312–36323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beller M., Sztalryd C., Southall N., Bell M., Jäckle H., Auld D. S., Oliver B. 2008. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6: e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bickel P. E., Tansey J. T., Welte M. A. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta. 1791: 419–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalen K. T., Dahl T., Holter E., Arntsen B., Londos C., Sztalryd C., Nebb H. I. 2007. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim. Biophys. Acta. 1771: 210–227. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi T., Matsushita S., Motojima K., Hirose F., Osumi T. 2006. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 281: 14232–14240. [DOI] [PubMed] [Google Scholar]
- 13.Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Croce M. A., Gropler M. C., Varma V., Yao-Borengasser A., Rasouli N., Kern P. A., et al. 2006. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. 55: 3418–3428. [DOI] [PubMed] [Google Scholar]
- 14.Wang H., Bell M., Sreenevasan U., Hu H., Liu J., Dalen K., Londos C., Yamaguchi T., Rizzo M. A., Coleman R., et al. 2011. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J. Biol. Chem. 286: 15707–15715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H., Hu L., Dalen K., Dorward H., Marcinkiewicz A., Russel D., Gong D., Londos C., Yamaguchi T., Holm C., et al. 2009. Activation of hormone-sensitive lipase requires two steps: protein phosphorylation and binding to the PAT-1 domain of lipid droplet coat proteins. J. Biol. Chem. 284: 32116–32125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z. 2009. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J. Biol. Chem. 284: 3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z., Zhou L. 2011. Interactions of perilipin-5 (Plin5) with adipose trigylceride lipase. J. Biol. Chem. 286: 5126–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaguchi T., Omatsu N., Matsushita S., Osumi T. 2004. CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J. Biol. Chem. 279: 30490–30497. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian V., Rothenberg A., Gomez C., Cohen A. W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M. P., et al. 2004. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J. Biol. Chem. 279: 42062–42071. [DOI] [PubMed] [Google Scholar]
- 20.Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., et al. 2004. Defective lipolysis and altered energy metabolism in mice lacking ATGL. Science. 306: 1383–1386. [DOI] [PubMed] [Google Scholar]
- 21.Schweiger M., Lass A., Zimmermann R., Eichmann T. O., Zechner R. 2009. Neutral lipid storage disease: genetic disorders caused by mutations in ATGL/PNPLA2 or CGI-58/ABHD5. Am. J. Physiol. Endocrinol. Metab. 297: E289–E296. [DOI] [PubMed] [Google Scholar]
- 22.Haemmerle G., Moustafa T., Woelkart G., Büttner S., Schmidt T., van de Weijer M., Hesselink D., Jaeger P. C., Kienesberger K., Zierler R., et al. 2011. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nat. Med. 17: 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H., Sreenevasan U., Hu H., Saladino A., Polster B. M., Lund L. M., Gong D. W., Stanley W. C., Sztalryd C. 2011. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52: 2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuramoto K., Okamura T., Yamaguchi T., Nakamura T. Y., Wakabayashi S., Morinaga H., Nomura M., Yanase T., Otsu K., Usuda N., et al. 2012. Perilipin 5, a lipid droplet-binding protein, protects the heart from oxidative burden by sequestering fatty acid from excessive oxidation. J. Biol. Chem. 287: 23852–23863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robbins J., Palermo J., Rindt H. 1995. In vivo definition of a cardiac specific promoter and its potential utility in remodeling the heart. Ann. N. Y. Acad. Sci. 752: 492–505. [DOI] [PubMed] [Google Scholar]
- 26.Wolins N. E., Rubin B., Brasaemle D. L. 2001. Tip47 associates with lipid droplets. J. Biol. Chem. 276: 5101–5108. [DOI] [PubMed] [Google Scholar]
- 27.Kaspar J. W., Jaiswal A. K. 2010. An autoregulatory loop between Nrf2 and Cul3-Rbx1 controls their cellular abundance. J. Biol. Chem. 285: 21349–21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Folch J., Lees M., Sloane Stanley G. H. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 29.Tansey J. T., Huml A. M., Vogt R., Davis K. E., Jones J. M., Fraser K. A., Brasaemle D. L., Kimmel A. R., Londos C. 2003. Functional studies on native and mutated forms of perilipins. A role in protein kinase A-mediated lipolysis of triacylglycerols. J. Biol. Chem. 278: 8401–8406. [DOI] [PubMed] [Google Scholar]
- 30.de Souza Batista C. M., Yang R. Z., Lee M. J., Glynn N. M., Yu D. Z., Pray J., Ndubuizu K., Patil S., Schwartz A., Kligman M., et al. 2007. Omentin plasma levels and gene expression are decreased in obesity. Diabetes. 56: 1655–1661. [DOI] [PubMed] [Google Scholar]
- 31.Smyth G. K. 2005. Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions using R and Bioconductor. R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber, editors. Springer, New York. 397–420. [Google Scholar]
- 32.Huang da W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 33.Huang da W., Sherman B. T., Lempicki R. A. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lentz S. I., Edwards J. L., Backus C., McLean L. L., Haines K. M., Feldman E. L. 2010. Mitochondrial DNA (mtDNA) biogenesis: visualization and duel incorporation of BrdU and EdU into newly synthesized mtDNA in vitro. J. Histochem. Cytochem. 58: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baseler W. A., Dabkowski E. R., Williamson C. L., Croston T. L., Thapa D., Powell M. J., Razunguzwa T. T., Hollander J. M. 2011. Proteomic alterations of distinct mitochondrial subpopulations in the type 1 diabetic heart: contribution of protein import dysfunction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 300: R186–R200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hecker P. A., Galvao T. F., O'Shea K. M., Brown B. H., Henderson R., Jr, Riggle H., Gupte S. A., Stanley W. C. 2012. High-sugar intake does not exacerbate metabolic abnormalities or cardiac dysfunction in genetic cardiomyopathy. Nutrition. 28: 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki J., Shen W. J., Nelson B. D., Selwood S. P., Murphy G. M., Jr, Kanehara H., Takahashi S., Oida K., Miyamori I., Kraemer F. B. 2002. Cardiac gene expression profile and lipid accumulation in response to starvation. Am. J. Physiol. Endocrinol. Metab. 283: E94–E102. [DOI] [PubMed] [Google Scholar]
- 38.Mandard S., Müller M., Kersten S. Peroxisome proliferator-activated receptor alpha target genes. 2007. Cell Mol. Life Sci. 61: 393–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolins N. E., Brasaemle D. L., Bickel P. E. 2006. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 580: 5484–5491. [DOI] [PubMed] [Google Scholar]
- 40.Wang H., Sztalryd C. 2011. Oxidative tissue: perilipin 5 links storage with the furnace. Trends Endocrinol. Metab. 22: 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tansey J. T., Sztalryd C., Gruia-Gray J., Roush D. L., Zee J. V., Gavrilova O., Reitman M. L., Deng C. X., Li C., Kimmel A. R., et al. 2001. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. USA. 98: 6494–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu G., Sztalryd C., Lu X., Tansey J. T., Gan J., Dorward H., Kimmel A. R., Londos C. 2005. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J. Biol. Chem. 280: 42841–42847. [DOI] [PubMed] [Google Scholar]
- 43.Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., Brown D. A. 2007. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J. Lipid Res. 48: 2751–2761. [DOI] [PubMed] [Google Scholar]
- 44.Varela G. M., Antwi D. A., Dhir R., Yin X., Singhal N. S., Graham M. J., Crooke R. M., Ahima R. S. 2008. Inhibition of ADRP prevents diet-induced insulin resistance. Am. J. Physiol. Gastrointest. Liver Physiol. 295: G621–G628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3: 309–319. [DOI] [PubMed] [Google Scholar]
- 46.Granneman J. G., Moore H. P., Krishnamoorthy R., Rathod M. 2009. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl). J. Biol. Chem. 284: 34538–34544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baird L., Dinkova-Kostova A. T. 2011. The cytoprotective role of the Keap1-Nrf2 pathway. Arch. Toxicol. 85: 241–272. [DOI] [PubMed] [Google Scholar]
- 48.Li J., Ichikawa T., Villacorta L., Janicki J. S., Brower G. L., Yamamoto M., Cui T. Nrf2 protects against maladaptive cardiac responses to hemodynamic stress. 2009. Arterioscler. Thromb. Vasc. Biol. 29: 1843–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.