Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is a common form of muscular dystrophy characterized by an asymmetric progressive weakness and wasting of the facial, shoulder and upper arm muscles, frequently accompanied by hearing loss and retinal vasculopathy. FSHD is an autosomal dominant disease linked to chromosome 4q35, but the causative gene remains controversial. DUX4 is a leading candidate gene as causative of FSHD. However, DUX4 expression is extremely low in FSHD muscle, and there is no DUX4 animal model that mirrors the pathology in human FSHD. Here, we show that the misexpression of very low levels of human DUX4 in zebrafish development recapitulates the phenotypes seen in human FSHD patients. Microinjection of small amounts of human full-length DUX4 (DUX4-fl) mRNA into fertilized zebrafish eggs caused asymmetric abnormalities such as less pigmentation of the eyes, altered morphology of ears, developmental abnormality of fin muscle, disorganization of facial musculature and/or degeneration of trunk muscle later in development. Moreover, DUX4-fl expression caused aberrant localization of myogenic cells marked with α-actin promoter-driven enhanced green fluorescent protein outside somite boundary, especially in head region. These abnormalities were rescued by coinjection of the short form of DUX4 (DUX4-s). Our results suggest that the misexpression of DUX4-fl, even at extremely low level, can recapitulate the phenotype observed in FSHD patients in a vertebrate model. These results strongly support the current hypothesis for a role of DUX4 in FSHD pathogenesis. We also propose that DUX4 expression during development is important for the pathogenesis of FSHD.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is the third most common muscular dystrophy affecting 1 in 20 000 individuals. FSHD patients develop progressive muscle weakness initially of facial, shoulder, upper arm muscles that often show asymmetry and later of trunk and lower limb muscles (1). In addition to muscle involvement, non-muscle tissues are also affected in FSHD. It has been reported that hearing loss and retinal vasculopathy occur in 75 and 60% of FSHD patients, respectively (2). FSHD is associated with a contraction of D4Z4 microsatellite repeats on chromosome 4q35 (3), but the causative gene remains controversial. FSHD patients carry 10 or less repeats of D4Z4 units, whereas normal individuals have 11 to 100 repeats (4). The severity of the disease is somewhat correlated with the residual repeat size, but the disease is not seen in individuals with complete loss of the repeats (5–7), suggesting the essential role of D4Z4 unit in the pathogenesis of FSHD. The disease is further complicated by the requirement of the contractions occurring on the ‘permissive’ 4qA allele that is associated with specific polymorphisms immediately distal to the last D4Z4 unit (8).
Accumulating evidence supports the hypothesis that derepression of DUX4 expression plays a major role in the pathogenesis of FSHD (9,10). DUX4 is a double homeobox retrogene encoded within each of D4Z4 units (11,12). Lemmers et al. revealed that the polymorphisms on 4qA allele create a polyadenylation signal at the end of the DUX4 locus and it stabilizes DUX4 transcripts expressed from the last D4Z4 unit (13). Two different DUX4 mRNA isoforms have been identified in the human skeletal muscle with 4qA allele: a full-length open reading frame mRNA (DUX4-fl) and a short spliced isoform (DUX4-s) that maintains the two homeobox domains, but lacks the C-terminal region unique to DUX4-fl protein (9). Several groups have reported that DUX4-fl mRNA and protein have been detected only in FSHD patients' muscle (9,13,14), whereas DUX4-s mRNA has been detected in both patients and controls (9). DUX4 is a potent transcription factor, and its overexpression affects the expression of myogenic regulators (15), induces atrophy-related genes (16) and activates germline genes and immune mediators (17) in cell culture. Moreover, DUX4-fl overexpression causes massive apoptosis both in cell culture (15,18) and in vivo (19,20). These results support the hypothesis that misexpression of DUX4 and the subsequent dysregulation of its downstream target genes are involved in the pathogenesis of FSHD.
However, this hypothesis is still controversial because the endogenous level of DUX4 expression in FSHD muscle is extremely low (9,13,14), and only a small subset of cells, estimated as approximately one cell per 1000, express DUX4 protein (9). There are to date no animal models that match this low level of DUX4 expression. In addition, we have recently found that DUX4-fl is also detectable in normal muscle-derived cells albeit at lower levels than FSHD (21), suggesting that the timing of misexpression of DUX4 may be important for disease pathology. In the present study, we introduced ≈1–2 × 105 copies of human DUX4-fl mRNA, which would be approximately 1 RNA molecule per 1000 cells before somitegenesis when diluted during development, into fertilized zebrafish eggs. The injection of the small amount of DUX4-fl mRNA caused asymmetric abnormalities of the eyes, ears, fins and/or trunk muscle in zebrafish embryos and mislocalization of myogenic cells during development. Our results suggest that the disturbance of normal development by DUX4-fl is important for FSHD pathogenesis.
RESULTS
DUX4-fl misexpression perturbs zebrafish development via its DNA-binding activity
To investigate whether the extremely low abundance of DUX4 is sufficient to cause pathology seen in FSHD in vivo, we introduced human DUX4 mRNAs into developing zebrafish embryos. First, DUX4-fl, DUX4-s cDNAs and a functionally deficient mutant of DUX4-fl (HOX1mut), which has mutations in the first homeobox domain (20), were cloned. DUX4 mRNAs were synthesized from these clones in vitro and injected into fertilized eggs at one-cell stage (Fig. 1A). Injection of 10 pg (≈1 × 107 copies) of DUX4-fl mRNA into fertilized eggs killed 96.2% of zebrafish embryos within 24 h, whereas 10 pg of DUX4-s or HOX1 mutant mRNA resulted in most of injected embryos developing identically to uninjected embryos (Fig. 1B–E). Introduction of the HOX1 mutant transcript caused abnormalities in body shape by 2 day post fertilization (dpf), but did not cause embryonic death with most embryos surviving to 4 dpf (Fig. 1F–I). The same experiment performed with 1.0 or 0.5 pg of DUX4 mRNAs had DUX4-fl killing over 90% of embryos by 4 dpf, whereas DUX4-s and HOX1mut did not cause any abnormal phenotypes (Supplementary Material, Fig. A). These results demonstrated that the expression of human DUX4-fl protein is extremely toxic, but that of DUX4-s is not toxic in zebrafish as has been shown in human cells (18,22), and in zebrafish, the DUX4-fl toxicity depends on its DNA-binding activity.
Figure 1.
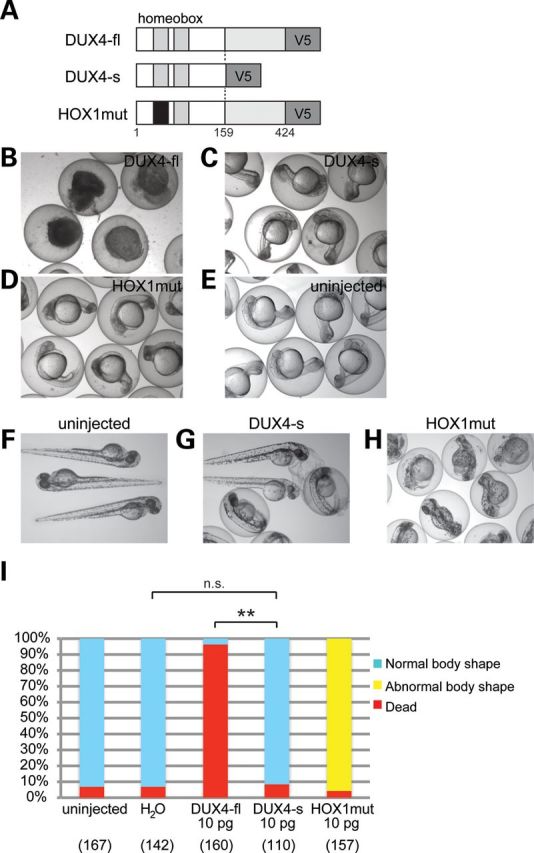
High-level expression of DUX4 in zebrafish development. (A) Illustration of DUX4 constructs. DNA-binding region in the first homeobox domain in HOX1mut is mutated (black). The numbers indicate amino acid residue of DUX4 protein. (B–E) Embryos injected with 10 pg of DUX4 variants mRNA at 24 hpf. (B) DUX4-fl-injected embryos. Lethality was evidenced by the deformed embryoid bodies in the zebrafish eggs. (C) DUX4-s-injected embryos. (D) HOX1mut-injected embryos. (E) uninjected control. (F–H) Embryos injected with 10 pg of DUX4 variants mRNA at 2 dpf. (F) uninjected control. (G) DUX4-s-injected embryos. (H) HOX1mut-injected embryos. (I) Quantitation of the phenotypes at 2 dpf is shown in the graph with the number of analyzed embryos. H2O: embryos injected with water containing 0.1% phenol red as a negative control. The number of dead embryos was significantly increased under DUX4-fl injection when compared with DUX4-s injection, but the number was not significantly different between H2O-injected embryos and DUX4-s-injected embryos. **P < 0.01, n.s. means not significant (χ2 test).
Asymmetric abnormalities caused by extremely low levels of DUX4-fl expression
To more closely resemble the expression of DUX4 in FSHD, lower levels (0.2 or 0.1 pg) of DUX4-fl mRNA (≈1–2 × 105 copies that when diluted during development would be approximately 1 RNA molecule per 1000 cells between the shield stage and bud stage; ZFIN http://zfin.org/) or DUX4-s mRNA were injected into the fertilized eggs. At 4 dpf, 67.4% of embryos had died, 14.7% showed gross body shape abnormalities upon 0.2 pg of DUX4-fl injection (Fig. 2A and D) in contrast, 95.2% appeared normal following 0.2 pg of DUX4-s injection (Fig. 2C and D). With these lower amounts of DUX4-fl injection, 17.8% or 44.4% of injected embryos (0.2 pg or 0.1 pg, respectively) exhibited asymmetric abnormalities of their eyes, ears, and/or fins (we classified these as ‘mildly abnormal’) (Fig. 2B and D). The eye abnormalities were most frequently observed (Fig. 2E and I). The severity was variable in the injected embryos. In most of the affected embryos, the structure of the eyes seemed to be normally formed, but the pigmentation of one eye was disturbed. Some severely affected embryos had a small eye on one side, but had a normal eye on the other side. Frequently, the ear phenotype as depicted in Figure 2 was observed (Fig. 2G and H). The number, size and position of otoliths in the ears were altered in DUX4-fl-injected embryos when compared with the wild type. The wild-type embryos have a larger posterior otolith on the medial wall of otic vesicle, and a smaller anterior otolith lies in a lateral position (Fig. 2G). DUX4-fl-injected embryos frequently lacked one of the otolith (Fig. 2H and I). In most affected embryos, neither otoliths were seen and the otic vesicle was small. The ear abnormality was also asymmetric and always seen on the same side as an abnormal eye. We also observed embryos of which only one fin appeared deformed or lost in DUX4-fl-injected embryos (Fig. 2F and I).
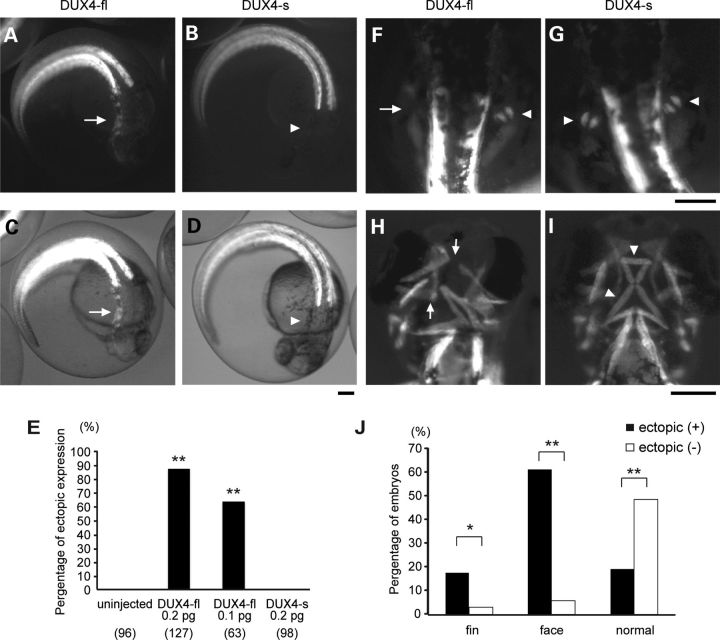
Figure 2.
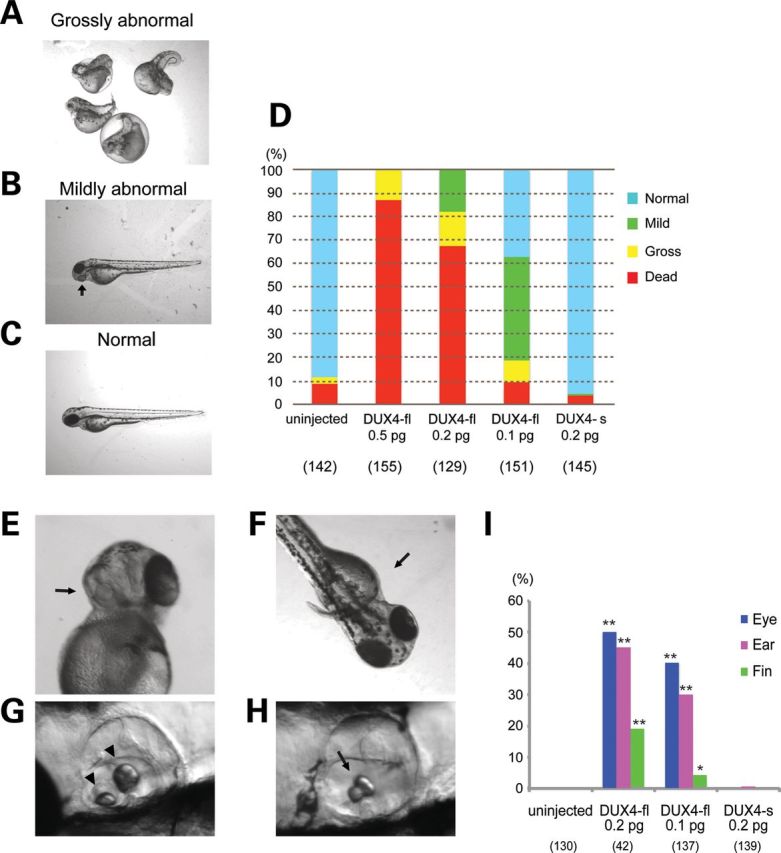
Asymmetric abnormalities of development following the injection of a smaller amount of DUX4-fl. (A–D) Classification and quantification of phenotypes of DUX4 mRNA-injected embryos at 4 dpf. (A) Representative image of ‘grossly abnormal’ embryos. The embryos show gross abnormality of their body shape. (B) Representative image of a ‘mildly abnormal’ embryo. Their body shape looks normal but their eyes, fins and/or ears are affected. An arrow indicates abnormal eye. (C) A phenotypically normal embryo. (D) Quantification of the phenotypes. The number of analyzed embryos is shown below the graph. (E–H) Asymmetric abnormalities in low levels of DUX4-fl-injected embryos. Arrows indicate abnormalities. (E) Asymmetrically disturbed eye pigmentation at 2 dpf. (F) Asymmetrical deformities of their fin at 3 dpf. (G) Normal ear in wild-type embryos at 4 dpf. Arrowheads indicate normal otoliths. (H) Ear abnormality in DUX4-fl-injected embryos at 4 dpf. An arrow indicates an abnormal otolith. (I) Quantification of the mild abnormalities at 4 dpf. The number of analyzed embryos that survived at 4 dpf is shown below the graph. Each phenotype was significantly increased under DUX4-fl injection when compared with DUX4-s injection. **P < 0.01, *P < 0.05 (χ2 test).
Abnormalities in the skeletal muscle function and structure in DUX4-fl-injected embryos
By 4 dpf, uninjected embryos and DUX4-s-injected embryos get up from the bottom of a Petri dish by continuous swimming, but in 0.2 pg or 0.1 pg of mRNA injection, many of DUX4-fl-injected embryos lay still on the bottom of the dish at 4 dpf (Fig. 3A–C). Motor activity was tested by touch-evoked escape response assay at 48 hpf (Fig. 3D). All of the analyzed uninjected and DUX4-s-injected embryos showed normal escape response (normal, 20/20), whereas 0.2 pg of DUX4-fl-injected embryos showed a flicker of movement but no swimming (flicker, 15/20), or swimming in a very short distance (short swim, 4/20). Even at 5 dpf, DUX4-fl-injected fish showed slow and short durations of swimming in response to touch, and most of them could not swim straight, often showing circular swimming (Supplementary Material, Fig. B). In zebrafish embryos, disorganization of the skeletal muscle structure can be detected by birefringence under polarizing light (23). DUX4-fl-injected embryos frequently showed misalignment of myosepta (Supplementary Material, Fig. C). Muscle birefringence appeared mildly affected, but was not remarkable. Immunostaining with anti-myosin heavy chain antibody (F59) clearly revealed muscle disorganization in DUX4-fl-injected embryos at 24 hpf (Fig. 3E–H). DUX4-fl-injected embryos showed large lesions, wide spaces between muscle fibers and split thin muscle fibers. These structural abnormalities appeared asymmetric because one side was more severely affected when compared with the other side (for example, the right side is more affected than the left side in Fig. 3E and F). DUX4-s-injected embryos showed well-organized muscle fibers without any lesions on both sides of the trunk muscle (Fig. 3G and H). Immunostaining of β-dystroglycan that is known to accumulate at myosepta in zebrafish embryos (24) revealed asymmetric arrangement of myosepta in DUX4-fl-injected embryos (Fig. 3I), whereas DUX4-s-injected embryos showed left–right symmetry of myosepta (Fig. 3J). The asymmetry of myosepta in DUX4-fl-injected embryos was confirmed by histologic analysis (Fig. 3K, black arrowheads) when compared with DUX4-s-injected embryos (Fig. 3L). Most anterior somites showing the asymmetric arrangement of myosepta were frequently associated with muscle degeneration (Fig. 3K and M, arrows). Muscle degeneration was observed only in a small area at 4 dpf, consistent with the mild phenotype of birefringence. Electron microscopic analysis showed less content of myofibrils in superficial slow fibers in DUX4-fl-injected embryos, whereas medial fast fibers seemed relatively intact (Fig. 3O and Q). Sarcomere structure was disorganized with blurry Z-disks in DUX4-fl-injected embryos when compared with the control (Fig. 3P and R).
Figure 3.

Abnormalities in the trunk muscle and motor function in DUX4-fl-injected embryos. Images of (A) uninjected or (B) DUX4-fl-injected embryos without anesthesia at 4 dpf. (C) The percentage of embryos lying on the bottom of a dish is shown in the graph with the number of analyzed embryos. The phenotype was significantly increased under DUX4-fl injection when compared with DUX4-s injection. **P < 0.01 (χ2 test). (D) Touch-evoked escape response at 48 hpf. Twenty embryos were assayed. DUX4-fl-injected embryos showed abnormal response with statistically significant difference (P < 0.01) from DUX4-s-injected embryos (Fisher's exact test). (E–H) Whole-mount immunostaining of trunk muscle with anti-myosin heavy chain antibody (F59) at 24 hpf. (E and F) Right or left side of the trunk muscle of a DUX4-fl-injected embryo, respectively. An arrow indicates a large lesion. (G and H) Right or left side of the trunk muscle of a DUX4-s-injected embryo, respectively. Lateral view. Rostral is to the right. (I and J) Whole-mount immunostaining of DUX4-fl-injected embryos (I) and DUX4-s-injected embryos (J) with anti-β-dystroglycan antibody at 4 dpf. Dorsal view. Rostral is to the top. (K and L) Semithin sections stained with toluidine blue of DUX4-fl-injected embryos (K) and DUX4-s-injected embryos (L). Dorsal view. Rostral is to the top. Black arrowheads indicate asymmetric arrangement of myosepta. White arrowheads indicate symmetric arrangement of myosepta. (M and N) Muscle degeneration in DUX4-fl-injected embryos (M) when compared with DUX4-s-injected embryos (N) at 4 dpf. Dorsal view. Rostral is to the left. Bar, 50 μm in (E–N). Arrows in (K) and (M) indicate muscle degeneration. (O–R) Ultrastructural analysis of the trunk muscle. (O and P) DUX4-fl-injected embryo. (Q and R) uninjected control. (P and R) Magnified images of (O) and (Q), respectively. SF, superficial slow fibers; FF, fast fibers. Dorsal view. Rostral is to the top. Bar, 2 μm in (O and Q), 0.5 μm in (P and R).
DUX4-fl misexpression caused aberrant localization of myogenic cells and disturbance of muscle development in the head and fin
To further characterize the effect of DUX4 misexpression on muscle development, we took advantage of the transgenic zebrafish with a enhanced green fluorescent protein (EGFP) reporter gene driven by α-actin promoter (Tg; α-actin:EGFP) (25). All skeletal muscles including myogenic precursors such as adaxial cells were labeled by EGFP in this transgenic line (26,27), enabling us to live monitor the muscle development. The injection of 0.2 or 0.1 pg of DUX4 mRNA into the fertilized eggs of this transgenic line revealed that DUX4-fl disturbs normal muscle development in an asymmetrical manner. At 1 dpf, uninjected embryos and DUX4-s-injected ones showed EGFP expression restricted to the somite (Fig. 4B and D). However, DUX4-fl-injected embryos showed asymmetrical ectopic expression of EGFP in cells outside somite boundaries: in the head region (Fig. 4A, C and E). Furthermore, the expression of EGFP clearly revealed asymmetrical muscle abnormalities at fin bud (Fig. 4F and G) and facial muscle disorganization (Fig. 4H and I) in DUX4-fl-injected embryos at 2 dpf, 3 dpf, respectively. The asymmetrical ectopic expression of EGFP occurred at random on the either side of the affected fish, but this abnormal expression and eye and/or fin abnormalities were always detected on the same side. The embryos that showed ectopic EGFP expression at 1 dpf (ectopic+) developed facial and/or fin muscle abnormalities at significantly higher rate at 3 dpf than the embryos without ectopic EGFP (ectopic–) (Fig. 4J), suggesting that the asymmetric aberrant localization of EGFP positive cells may be the cause of these abnormalities later in development.
Figure 4.
DUX4-fl expression perturbs zebrafish facial and fin muscle development. Transgenic fish line, Tg[α-actin:enhanced green fluorescent protein (EGFP)], was injected with DUX4 mRNAs. (A, C, F and H) DUX4-fl mRNA-injected embryos. (B, D, G and I) DUX4-s mRNA-injected embryos. Arrows indicate abnormal, arrowheads indicate normal. (A–D) Ectopic expression of α-actin:EGFP outside somite boundary at 24 hpf. (A and B) EGFP fluorescence (C and D) EGFP fluorescence merged with bright field image. Dorsal view. Rostral is to the bottom. (E) Quantification of embryos with ectopic expression of α-actin:EGFP at 1 dpf. The number of analyzed embryos is shown below the graph. The phenotype was significantly increased under DUX4-fl injection when compared with DUX4-s injection. **P < 0.01 (χ2 test). (F and G) Asymmetric abnormality of fin muscle at 2 dpf. Dorsal view. Rostral is to the top. (H and I) Facial muscle abnormality at 3 dpf. Ventral view. Rostral is to the top. Bar, 200 μm. (J) Correlation of ectopic EGFP expression at 1 dpf with the facial or fin bud muscle abnormalities at 3 dpf. 0.2 pg of DUX4-fl mRNA was injected. ectopic+: ectopic EGFP positive embryos (n = 183), ectopic–: ectopic EGFP negative embryos (n = 35). **P < 0.01, *P = 0.05 (χ2 test).
Coinjection of DUX4-s with DUX4-fl rescued abnormal phenotypes
The results shown in Figure 1 suggested that DUX4-fl toxicity depends on its DNA-binding ability. However, DUX4-s contains intact double homeobox domains, even though it did not show any effect on zebrafish development. We hypothesized that DUX4-s should bind to the same targets as DUX4-fl in zebrafish and might compete with DUX4-fl for DNA binding. Actually, it was previously shown that DUX4-s binds to the same core binding site as DUX4-fl in vitro, but does not activate transcription of the same genes (17). To test whether DUX4-s inhibits the toxicity of DUX4-fl in vivo, DUX4-s mRNA was coinjected with DUX4-fl mRNA into the fertilized eggs from Tg; α-actin:EGFP transgenic line. Coinjection of 20-fold greater amount of DUX4-s (0.2 pg DUX4-fl with 4 pg DUX4-s) dramatically suppressed ectopic expression of EGFP outside the somite boundary (Fig. 5A–E). The ratio of dead embryos, gross and mild abnormalities also decreased, and approximately 70% of embryos showed normal phenotype under the coinjection of DUX4-s (Fig. 5F). The frequency of embryos with eye, fin or facial abnormalities was also decreased (Fig. 5G–I). Coinjected embryos showed much milder muscle phenotype than single DUX4-fl-injected embryos (Fig. 5J and K).
Figure 5.
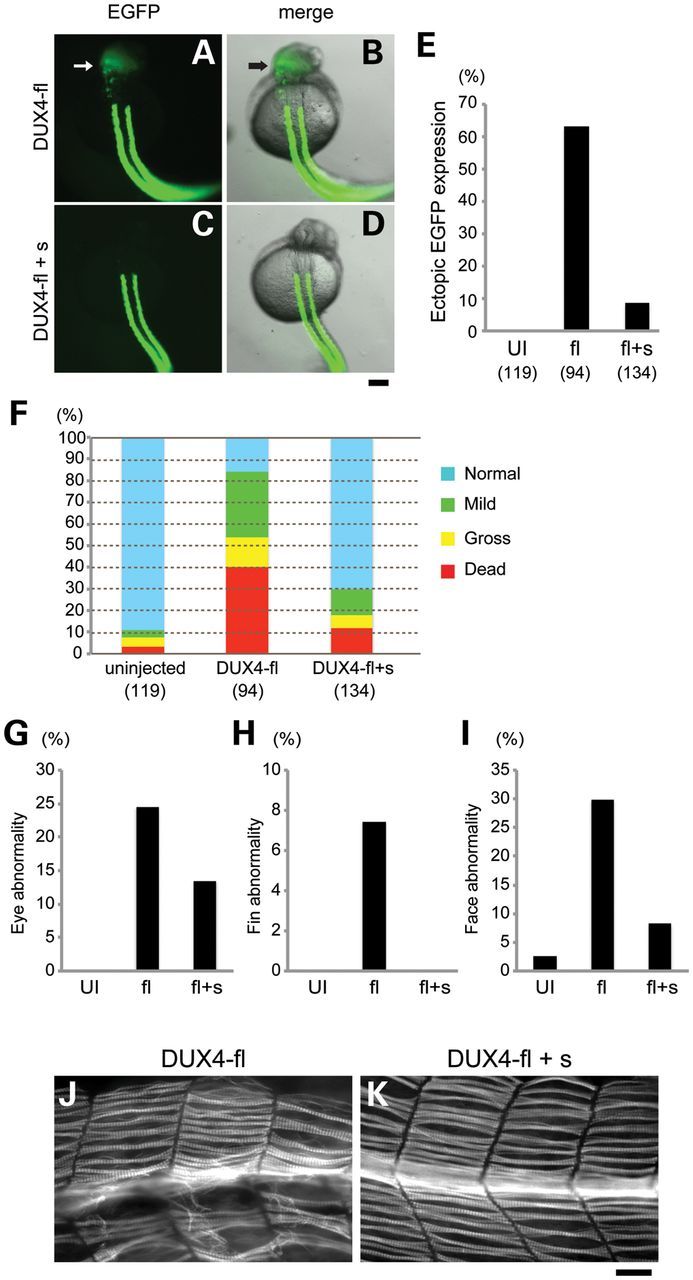
DUX4-s competes with DUX4-fl and rescues the abnormal phenotypes. Representative images of α-actin:EGFP transgenic fish with DUX4-fl injection (A and B) and coinjection of DUX4-fl and DUX4-s (DUX4-fl + s) (C and D) at 1 dpf. Arrows indicate ectopic EGFP expression in the head. (A and C) Fluorescence of EGFP (B and D) Merged image of EGFP fluorescence with bright field. Dorsal view. Rostral is to the top. Bar, 200 μm. (E) Percentage of embryos with ectopic EGFP expression is shown in the graph with the number of analyzed embryos. UI, uninjected. fl, DUX4-fl injected. fl + s, DUX4-fl and DUX4-s coinjected. (F) Quantification of the phenotypes in DUX4 mRNA-injected embryos at 4 dpf. The number of analyzed embryos is shown below the graph. (G–I) Quantification of the mild abnormalities at 4 dpf. (G) Eye abnormality. (H) Fin abnormality. (I) Facial muscle abnormality. The number of analyzed embryos is the same as (F). (J and K) Representative images of trunk muscle of 1 dpf zebrafish embryos injected with DUX4-fl or coinjected with DUX4-fl and DUX4-s. Muscle fibers were visualized with F59 antibody. Lateral view. Rostral is to the left. Bar, 50 μm.
DISCUSSION
Clinical features of FSHD were first described in 1884 by Landouzy and Dejerine, and the genetic linkage at 4q35 was identified in 1992 (3). Despite extensive research, the exact gene causing the human clinical symptoms has remained elusive. DUX4 is the most promising candidate gene, but a faithful animal model of the human disease has been lacking. Misexpresssion of DUX4 during zebrafish development represents an animal model that mirrors pathology in human FSHD, as emphasized by asymmetrical muscle and non-muscle tissue abnormalities. Human DUX4-fl perturbs zebrafish development, despite the absence of a DUX4 ortholog in zebrafish, which is analogous to the situation in humans, where DUX4-fl is also not normally expressed outside the testis (9).
Although most of typical FSHD patients show clinical symptoms in the second decade, the presence of infantile form of FSHD (28) as well as marked asymmetry and unique selectivity of muscle involvement suggests that the abnormalities may arise during development. In this study, we showed that the expression of extremely small amount of DUX4-fl during zebrafish development asymmetrically disturbs the development of tissues affected in human FSHD patients such as eyes, ears, fins (comparable with arms) and facial muscles. This indicates that the DUX4 expression during development is important for FSHD pathology. Actually, it was reported that DUX4 is expressed in human embryonic stem cells even in normal, (29) and it gets silenced during differentiation. However, induced pluripotent stem (iPS) cells derived from FSHD patients fail to silence DUX4-fl, resulting in prolonged expression of DUX4-fl in the embryonic body (9). It was reported that Polycomb group of epigenetic repressors bind to D4Z4 repeats in healthy individuals and repress the expressions of 4q35 genes, including DUX4 (30). Taken together, it is speculated that D4Z4 repeats contraction reduces the repression of DUX4, allowing leaky expression of DUX4-fl protein during development, which might disturb normal development of eyes, ears and muscles.
Recently, we have reported that DUX4-fl is detectable even in muscle biopsies from some of healthy individuals, suggesting that DUX4-fl expression itself is not sufficient for FSHD muscle pathology (21). However, in our zebrafish model, the abnormal phenotypes caused by DUX4-fl injection were seen in a dose-dependent manner (Figs 2D, 2I, 3C and 4E). We hypothesize that the severity of the FSHD phenotypes may depend on the level of DUX4-fl expression during development, rather than the level at the time of biopsy. DUX4 contains homeobox domains that are found in proteins that regulate early development. We propose that in FSHD, misexpression of DUX4-fl during early embryogenesis affects certain types of progenitor cell population, which may render specific muscle groups more susceptible to disease later in life. This is consistent with the slowly progressive feature of FSHD.
Asymmetry of muscle involvement is a characteristic feature of FSHD. We observed asymmetric ectopic expression of α-actin promoter-driven EGFP in transgenic zebrafish embryos with the injection of DUX4-fl mRNA (Fig. 4). Because this asymmetric EGFP expression was followed by facial and fin muscle abnormalities, we believe that the asymmetric EGFP expression outside the somite boundary is the cause of abnormalities later in development. There are two possibilities to explain this ectopic EGFP expression: (i) DUX4 activates muscle genes in non-muscle cell lineages or (ii) DUX4 causes aberrant migration of myogenic cells to the region, where they are not supposed to go. DUX4 homeobox domains are known to be most similar to those of Pax3 and Pax7 (15). Pax3 is a key regulator of myogenesis and plays an essential role in the migration of myogenic progenitor cells from dermomyotome to the sites of myogenesis such as limb bud during development (31). Thus, it is most likely that the ectopic EGFP expression outside the somite boundary might result from altered migration of myogenic progenitor cells. We hypothesize that DUX4-fl may compete with some of the target genes with Pax3 and disturb its downstream transcriptional network related to the migration of muscle progenitor cells. Further studies are needed to understand the molecular mechanism of the ectopic EGFP expression.
We also showed that DUX4-s does not have the toxic effect on zebrafish development (Fig. 1), and coexpression of DUX4-s can rescue the abnormal phenotypes caused by DUX4-fl in the vertebrate model (Fig. 5). This is consistent with the previous report that coexpression of DUX4-s suppresses the induction of the target gene expressions by DUX4-fl in vitro (17). Together with the data that HOX1 mutant, of which first DNA-binding domain is mutated, showed remarkably reduced toxicity (Fig. 1), it is expected that the dysregulation of the DUX4-fl targets may lead to FSHD-like phenotypes. DUX4-fl shares amino acid sequences from 1 to 159, including two homeobox domains, with DUX4-s (Fig. 1A). Thus, it is most likely that the C-terminal domain unique to DUX4-fl (160–424 amino acid residues) possesses the activity that causes toxic effect. Actually, it was reported that the C-terminal domain of DUX4-fl enhances the activity of transcriptional factor. The chimeric transcript of CIC, a high mobility group box transcriptional factor, fused with DUX4 C-terminus that resulted from chromosomal translocation was detected in the cases of soft tissue sarcoma, and the CIC-DUX4 fusion protein demonstrates enhanced transcriptional activity, transforming cells into the ones with tumor-like features (32). In addition, the study using iPS cells demonstrated that DUX4-fl is expressed in undifferentiated iPS cells, and it switches to DUX4-s in the differentiated embryoid bodies, but this switching is compromised in the iPS cells derived from FSHD fibroblasts (9). This suggests that the misregulation of isoform switching may allow the misexpression of DUX4-fl later in development and disturb transcriptional network that is important for normal development via its strong transcriptional activity. Our results shown in Figure 5 imply that it might be possible to improve FSHD phenotypes by modifying alternative splicing of DUX4 gene, if DUX4-fl really plays a major role in FSHD pathogenesis. Shifting the alternative splicing from DUX4-fl to DUX4-s is expected to decrease the toxic DUX4-fl expression and to increase the protective DUX4-s expression, which may have synergistic effect to suppress the DUX4-fl toxicity. Modulation of alternative splicing is one of the most promising therapeutic approaches for other types of muscular dystrophies such as Duchenne muscular dystrophy (33–35), myotonic dystrophy (36) and Fukuyama muscular dystrophy (37). This strategy might be applied to FSHD in the future.
In summary, our DUX4 zebrafish model is the first animal model that mirrors phenotypes seen in human FSHD patients. Using this zebrafish model, further studies to understand the precise mechanism of how DUX4-fl perturbs normal development should provide further insights into the pathogenesis of FSHD. It also represents an animal model on which to test rational approaches to therapies.
MATERIALS AND METHODS
Fish strains and maintenance
The zebrafish wild-type AB strain and the transgenic line Tg(α-actin:EGFP) were maintained in the Boston Children's Hospital zebrafish facility under approved use protocols. The transgenic fish line was kindly provided by Dr Huai-Jen Tsai (National Taiwan University) (25). Zebrafish embryos were raised at 28.5°C according to standard procedures (38). EGFP fluorescence was observed under a fluorescent stereomicroscope (Nikon SMZ1500).
Construction of expression cassette for DUX4
The V5-tag sequence was inserted into XhoI- and XbaI-sites in pCS2(+) to obtain pCS2(+)-V5 vector. The human DUX4-fl cDNA and human DUX4-s cDNA were amplified by PCR, according to the conditions previously reported (9), using DUX4_pCI-neo construct (12,18) as a template and subsequently were cloned into pENTR/D vector (Invitrogen). The mutant DUX4 construct (HOX1mut) with alanine substitutions in the first homeobox domain was generated by site-directed mutagenesis to produce a functionally deficient protein (20). The cDNAs were digested with EcoRI and XhoI and subcloned into pCS2(+)-V5 vector. The primers used are listed in Supplementary Materials.
mRNA injection
The DUX4 mRNAs were synthesized by in vitro transcription with mMessage mMachine SP6 kit (Ambion) using NotI-digested DUX4 constructs. The quality of synthesized mRNAs was checked by gel electrophoresis under the denatured condition. mRNA was injected with 0.1% phenol red into the yolk of fertilized zebrafish embryos at one-cell stage by a microinjector. In the coinjection experiment, 4 pg of DUX4-s mRNA was injected into the yolk immediately followed by the injection of 0.2 pg of DUX4-fl mRNA.
Muscle birefringence and motor function
Muscle birefringence was analyzed at 4 dpf as previously described (39). Touch-evoked escape response was evaluated at 48 hpf. Behavior was categorized as flicker of movement but no swimming, swimming in a very short distance and normal swimming. Time-lapse images of embryos were acquired using Nikon SMZ1500 stereomicroscope with Openlab software version 3.1.5 (Improvision).
Whole-mount immunostaining
Immunostaining was performed as previously described (40) with minor modifications. In brief, embryos were fixed in 4% paraformaldehyde (PFA) in PBS at 4°C overnight and dehydrated in 100% methanol. After rehydration, 4 dpf embryos were incubated in 0.1% collagenase (Sigma) in PBS for 60 min. Blocking solution containing 0.2% saponin was used for 4 dpf embryos. Anti-slow muscle myosin heavy chain antibody (F59, Developmental Studies Hybridoma Bank; 1:50) and anti-β-dystroglycan antibody (Novocastra NCL-b-DG; 1:50) were used. The embryos were placed in 3% methyl cellulose or mounted on a glass slide and observed with fluorescent microscopes (Nikon Eclipse E1000 and Zeiss Axioplan2).
Histologic analysis and transmission electron microscopy
Embryos were fixed at 4 dpf in 100 mm sodium cacodylate buffer (pH 7.4) containing 2% glutaraldehyde and 2.5% PFA at 4°C overnight. Samples were postfixed in 1% Osmium tetroxide/1.5% Potassium ferrocyanide, incubated in 1% Uranyl Acetate for 30 min, dehydrated by passage through a graded series of ethanols and embedded in Epon 812. Semithin sections were cut from the dorsal to the ventral side of zebrafish embryos and stained with toluidine blue. Ultrathin sections were viewed with a Tecnai Spirit transmission electron microscope (FEI).
Statistical analysis
Results were analyzed with the χ2 test or Fisher's exact test (touch-evoked escape response) using R software version 2.15.1 (http://www.r-project.org/).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by The Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center (1U54HD060848-04) and FSH Society (FSHS-22012-02 to H.M.). Molecular Genetics Core facility was supported by Children's Hospital Boston Intellectual and Developmental Disabilities Research Center (2P30HD018655-30).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Leonard Zon and Emanuela Gussoni for comments; Fedik Rahimov for manuscript review; Chris Lawrence and Jason Best who managed our fish facility; Matthew Alexander for the maintenance of transgenic fish and his helpful advice; Jennifer Myers for technical support; Huai-Jen Tsai for providing transgenic line Tg(α-actin:EGFP); Alexandra Belayew for providing DUX4_pCI-neo construct; Louise Trakimas from Harvard Medical School Electron Microscope Facility for Epon embedding; Boston Children's Hospital Molecular Genetics Core facility for DNA sequencing.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Tawil R., Van Der Maarel S.M. Facioscapulohumeral muscular dystrophy. Muscle Nerve. 2006;34:1–15. doi: 10.1002/mus.20522. [DOI] [PubMed] [Google Scholar]
- 2.Padberg G.W., Brouwer O.F., de Keizer R.J., Dijkman G., Wijmenga C., Grote J.J., Frants R.R. On the significance of retinal vascular disease and hearing loss in facioscapulohumeral muscular dystrophy. Muscle Nerve. 1995;2:S73–S80. [PubMed] [Google Scholar]
- 3.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 4.Tawil R., Figlewicz D.A., Griggs R.C., Weiffenbach B. Facioscapulohumeral dystrophy: a distinct regional myopathy with a novel molecular pathogenesis. FSH Consortium. Ann. Neurol. 1998;43: 279–282. doi: 10.1002/ana.410430303. [DOI] [PubMed] [Google Scholar]
- 5.Goto K., Lee J.H., Matsuda C., Hirabayashi K., Kojo T., Nakamura A., Mitsunaga Y., Furukawa T., Sahashi K., Arahata K. DNA rearrangements in Japanese facioscapulohumeral muscular dystrophy patients: clinical correlations. Neuromuscul. Disord. 1995;5:201–208. doi: 10.1016/0960-8966(94)00055-e. [DOI] [PubMed] [Google Scholar]
- 6.Tupler R., Berardinelli A., Barbierato L., Frants R., Hewitt J.E., Lanzi G., Maraschio P., Tiepolo L. Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J. Med. Genet. 1996;33:366–370. doi: 10.1136/jmg.33.5.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi M., Ricci E., Colantoni L., Galluzzi G., Frusciante R., Tonali P.A., Felicetti L. The facioscapulohumeral muscular dystrophy region on 4qter and the homologous locus on 10qter evolved independently under different evolutionary pressure. BMC Med. Genet. 2007;8:8. doi: 10.1186/1471-2350-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemmers R.J., Wohlgemuth M., van der Gaag K.J., van der Vliet P.J., van Teijlingen C.M., de Knijff P., Padberg G.W., Frants R.R., van der Maarel S.M. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards M., Coppee F., Thomas N., Belayew A., Upadhyaya M. Facioscapulohumeral muscular dystrophy (FSHD): an enigma unravelled? Hum. Genet. 2012;131:325–340. doi: 10.1007/s00439-011-1100-z. [DOI] [PubMed] [Google Scholar]
- 11.Hewitt J.E., Lyle R., Clark L.N., Valleley E.M., Wright T.J., Wijmenga C., van Deutekom J.C., Francis F., Sharpe P.T., Hofker M., et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 12.Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 13.Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W., et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Matteotti C., van Acker A.M., Leo O., et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppee F., et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanderplanck C., Ansseau E., Charron S., Stricwant N., Tassin A., Laoudj-Chenivesse D., Wilton S.D., Coppee F., Belayew A. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One. 2011;6:e26820. doi: 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Matteotti C., Arias C., Corona E.D., Nunez N.G., Leo O., et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Wuebbles R.D., Long S.W., Hanel M.L., Jones P.L. Testing the effects of FSHD candidate gene expression in vertebrate muscle development. Int. J. Clin. Exp. Pathol. 2010;3:386–400. [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J., Harper S.Q. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 2011;69:540–552. doi: 10.1002/ana.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones T.I., Chen J.C.J., Rahimov F., Homma S., Arashiro P., Beermann M.L., King O.D., Miller J.B., Kunkel L.M., Charles P., et al. Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 2012;21:4419–4430. doi: 10.1093/hmg/dds284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geng L.N., Tyler A.E., Tapscott S.J. Immunodetection of human double homeobox 4. Hybridoma (Larchmt) 2011;30:125–130. doi: 10.1089/hyb.2010.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Granato M., van Eeden F.J., Schach U., Trowe T., Brand M., Furutani-Seiki M., Haffter P., Hammerschmidt M., Heisenberg C.P., Jiang Y.J., et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- 24.Parsons M.J., Campos I., Hirst E.M., Stemple D.L. Removal of dystroglycan causes severe muscular dystrophy in zebrafish embryos. Development. 2002;129:3505–3512. doi: 10.1242/dev.129.14.3505. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao C.D., Hsieh F.J., Tsai H.J. Enhanced expression and stable transmission of transgenes flanked by inverted terminal repeats from adeno-associated virus in zebrafish. Dev. Dyn. 2001;220:323–336. doi: 10.1002/dvdy.1113. [DOI] [PubMed] [Google Scholar]
- 26.Devoto S.H., Melancon E., Eisen J.S., Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- 27.Higashijima S., Okamoto H., Ueno N., Hotta Y., Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev. Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- 28.Brouwer O.F., Padberg G.W., Bakker E., Wijmenga C., Frants R.R. Early onset facioscapulohumeral muscular dystrophy. Muscle Nerve. 1995;2:S67–S72. [PubMed] [Google Scholar]
- 29.Snider L., Asawachaicharn A., Tyler A.E., Geng L.N., Petek L.M., Maves L., Miller D.G., Lemmers R.J., Winokur S.T., Tawil R., et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet. 2009;18:2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabianca D.S., Casa V., Bodega B., Xynos A., Ginelli E., Tanaka Y., Gabellini D. A long ncRNA links copy number variation to a Polycomb/Trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daston G., Lamar E., Olivier M., Goulding M. Pax-3 is necessary for migration but not differentiation of limb muscle precursors in the mouse. Development. 1996;122:1017–1027. doi: 10.1242/dev.122.3.1017. [DOI] [PubMed] [Google Scholar]
- 32.Kawamura-Saito M., Yamazaki Y., Kaneko K., Kawaguchi N., Kanda H., Mukai H., Gotoh T., Motoi T., Fukayama M., Aburatani H., et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 33.Pramono Z.A., Takeshima Y., Alimsardjono H., Ishii A., Takeda S., Matsuo M. Induction of exon skipping of the dystrophin transcript in lymphoblastoid cells by transfecting an antisense oligodeoxynucleotide complementary to an exon recognition sequence. Biochem. Biophys. Res. Commun. 1996;226:445–449. doi: 10.1006/bbrc.1996.1375. [DOI] [PubMed] [Google Scholar]
- 34.Lu Q.L., Mann C.J., Lou F., Bou-Gharios G., Morris G.E., Xue S.A., Fletcher S., Partridge T.A., Wilton S.D. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat. Med. 2003;9:1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- 35.Goemans N.M., Tulinius M., van den Akker J.T., Burm B.E., Ekhart P.F., Heuvelmans N., Holling T., Janson A.A., Platenburg G.J., Sipkens J.A., et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N. Engl. J. Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 36.Wheeler T.M., Lueck J.D., Swanson M.S., Dirksen R.T., Thornton C.A. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniguchi-Ikeda M., Kobayashi K., Kanagawa M., Yu C.C., Mori K., Oda T., Kuga A., Kurahashi H., Akman H.O., DiMauro S., et al. Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature. 2011;478:127–131. doi: 10.1038/nature10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nüsslein-Volhard C., Dahm R. Zebrafish: A Practical Approach. New York, USA: Oxford University press; 2002. [Google Scholar]
- 39.Kawahara G., Guyon J.R., Nakamura Y., Kunkel L.M. Zebrafish models for human FKRP muscular dystrophies. Hum. Mol. Genet. 2010;19:623–633. doi: 10.1093/hmg/ddp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita M., Mitsuhashi H., Isogai S., Nakata T., Kawakami A., Nonaka I., Noguchi S., Hayashi Y.K., Nishino I., Kudo A. Filamin C plays an essential role in the maintenance of the structural integrity of cardiac and skeletal muscles, revealed by the medaka mutant zacro. Dev. Biol. 2012;361:79–89. doi: 10.1016/j.ydbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.