Abstract
Mutations in ZIC3 cause human X-linked heterotaxy and isolated cardiovascular malformations. A mouse model with targeted deletion of Zic3 demonstrates an early role for Zic3 in gastrulation, CNS, cardiac and left–right axial development. The observation of multiple malformations in Zic3null mice and the relatively broad expression pattern of Zic3 suggest its important roles in multiple developmental processes. Here, we report that Zic3 is primarily required in epiblast derivatives to affect left–right patterning and its expression in epiblast is necessary for proper transcriptional control of embryonic cardiac development. However, cardiac malformations in Zic3 deficiency occur not because Zic3 is intrinsically required in the heart but rather because it functions early in the establishment of left–right body axis. In addition, we provide evidence supporting a role for Zic3 specifically in the perinodal region of the posterior lateral plate mesoderm for the establishment of laterality. These data delineate the spatial requirement of Zic3 during left–right patterning in the mammalian embryo, and provide basis for further understanding the molecular mechanisms underlying the complex interaction of Zic3 with signaling pathways involved in the early establishment of laterality.
INTRODUCTION
Zic3 (zinc finger protein of cerebellum 3), encoded on the X chromosome in both human and mouse, is a member of the broadly conserved Zic family of transcription factors. These genes were originally identified as a paralogous family encoding zinc finger proteins expressed in the mouse cerebellum (1,2). ZIC3 was the first gene unequivocally associated with human Heterotaxy Syndrome (3), a class of congenital disorders resulting from failure to establish normal left–right asymmetry during embryonic development (4). X-linked heterotaxy syndrome (HTX1) is rare and a relatively small number of loss-of-function mutations in ZIC3 have been identified in individuals with either heterotaxy or isolated looping-type congenital heart defects.
Studies of Zic3 in different model organisms reveal multiple functions during embryogenesis. In Xenopus, Zic3 plays a role in ectodermal development by inducing proneural gene expression and neural crest differentiation (5). Xenopus Zic3 is also critical for mesodermal development by acting upstream of nodal signaling during left–right axis formation (6). More recently, Zic3 is shown to regulate notochord and organizer development in Xenopus embryos through suppression of the Wnt/β-catenin pathway (7). In mouse, the naturally occurring bent tail mutant has a deletion that includes the murine Zic3 locus (8,9). Targeted deletion of Zic3 in mice disrupts nodal signaling in the lateral plate mesoderm, resulting in significant lethality and multiple complex laterality defects similar to human heterotaxy patients, including abnormal heart looping, dextrocardia, septal and outflow tract defects, as well as atrial and pulmonary isomerism (10,11). Additionally, Zic3null mice develop axial skeleton and neural tube defects, which are less consistently associated with left–right patterning defects. Craniofacial abnormalities and axial skeleton defects mimicking Goldenhar Syndrome were observed in mice bearing a targeted neo-insertion allele that resulted in mild overexpression of Zic3 (12). Recent in vitro studies reveal that maintenance of ESC pluripotency, and development of retinal progenitor cells also requires Zic3 function (13,14). These findings suggest that Zic3 activities are pleiotropic.
The broad and dynamic expression pattern of Zic3 provides further evidence for a complex role in embryonic development. In both Xenopus and mouse, Zic3 is first expressed in the pre-streak stage embryo and the newly formed mesoderm at the beginning of gastrulation. As gastrulation proceeds, Zic3 transcript is located in the mesoderm of the embryonic region, the primitive streak and ectoderm adjacent to the expressing mesoderm (15,16). Loss of Zic3 at this early stage disrupts morphogenesis of primitive streak, node and notochord, resulting in variable gastrulation defects in mouse embryos (17). These data suggest that Zic3 may play an essential role in the development of mesodermal components in mice, similar to its function in Xenopus. As mouse embryos progress to 8.5 dpc, Zic3 expression is detected in the dorsal central nervous system, posterior neural plate and somites. At later embryonic stages, its expression further extends to the developing eye, limb buds, tail bud and central nervous system (18). The expression pattern, together with the CNS defects found in Zic3null mice, suggest that Zic3 is also critical for neuroectodermal development in mice. Zic3 is weakly expressed in 10.5 dpc mouse cardiac tissues and functionally interacts with serum response factor (SRF) in vitro (19). Cardiac specific expression combined with the findings of ZIC3 mutations in patients with isolated cardiovascular malformations suggest the hypothesis that Zic3 may play a role in cardiac development independent of left–right patterning. Alternatively, it is possible that disturbances in the earlier process of left–right patterning may secondarily affect the subsequent cardiac development through disruption of signaling cascades necessary for cardiac organogenesis.
Although much evidence indicates that Zic3 plays pleiotropic roles in multiple developmental processes, it is not clear in which region(s) of the embryo Zic3 acts specifically to affect left–right patterning and cardiac development. In the present study, we used Cre-mediated conditional deletion of Zic3 to dissect its differential functions in mouse embryogenesis. We provide evidence that Zic3 is required in epiblast derivatives to affect left–right patterning and heart development; whereas its function in extraembryonic tissues may be critical for early gastrulation. We also directly address the possibility of whether Zic3 has a cell-autonomous role in heart development independent of its role in establishing the left–right axis. We show that although Zic3 plays an important part in transcriptional control of embryonic heart development, deletion of Zic3 in cardiac progenitors and outflow tract does not affect cardiac structural development. Thus, heart defects found in Zic3null embryos arise due to an extra-cardiac requirement for Zic3. These results support the hypothesis that disturbances in early establishment of left–right axis secondarily affect heart development. Finally, we further narrow the requirement of Zic3 to lateral plate mesoderm (LPM) adjacent and posterior of the node specifically for left–right patterning and cardiac development; while its function in ectoderm may be critical for CNS development. Taken together, these results directly define tissue specific functions of Zic3 in mammalian embryo development for the first time, and provide the basis for understanding how Zic3 acts to establish laterality around the node in the future.
RESULTS
Epiblast-specific deletion of Zic3 recapitulates all the cardiac, laterality and CNS defects seen in Zic3null mice
Previous studies show that Zic3 is critical for the initiation and progression of gastrulation prior to left–right axis specification and later organogenesis (10,17). To distinguish different functions of Zic3 in the embryo proper or in extraembryonic derivatives during early post-implantation development, we deleted Zic3 in epiblast cells using Sox2-cre, which induces recombination efficiently in all epiblast cells by 6.5 dpc (Supplementary Material, Fig. S2A–C) (20). Genotyping of epiblast conditional knockout (CKO) mice at weaning showed departure from the expected mutant-to-wild-type ratio, indicating significant lethality in these mice (Table 1). However, the penetrance of lethality was reduced to ∼40% in the epiblast CKO mice, whereas ∼80% of the Zic3null mice were lost by weaning. All viable epiblast CKO males had similar ‘kinked’ tails as described in the Zic3null mice (10). Other than complete penetrance of tail abnormalities, a small portion of them also developed anus and eye anomalies and axial skeleton defects leading to hind-limb paralysis (data not shown). Genotyping at 9.5 dpc showed no observable loss of the epiblast CKO and Zic3null embryos, but developmental anomalies and cardiac looping defects were frequently found (Fig. 1, Table 1). Fourteen out of 32 epiblast CKO embryos and 12 out of 18 Zic3null embryos at 9.5 dpc displayed various degrees of developmental delay or arrest. Typical heart looping anomalies described by Ware et al. (11) were seen in five out of 32 epiblast CKO embryos and four out of 18 Zic3null embryos at 9.5 dpc. Genotyping at 15.5 dpc showed that epiblast CKO embryos were still present in the expected Mendelian ratios; in contrast, 40–50% of the Zic3null mice were lost by this stage (Table 1). This suggested that epiblast CKO embryos die either in late gestation or perinatally. Although no significant lethality was found in epiblast CKO embryos at 15.5 dpc, ∼35% of these mutants exhibited multiple defects, similar to abnormalities found in ∼35% of the surviving Zic3null embryos at this stage. Four out of 38 epiblast CKO embryos and five out of 58 Zic3null embryos at 15.5 dpc had visible CNS anomalies including exencephaly, cleft palate and neural tube defect (Fig. 2). MRI analyses demonstrated that classical laterality defects were evident in two out of eight epiblast CKO embryos and three out of 15 Zic3null embryos at 15.5 dpc. These internal malformations include dextrocardia (Fig. 3D, F, G, I, J and L), right aortic arch (Fig. 3L), atrial septal defect (Fig. 3G and J), ventricular septal defect (Fig. 3D, F, J and L), double-outlet right ventricle (Fig. 3F), situs inversus and interruption of inferior vena cava (Fig. 3E, F, H and I), separate entrance of hepatic veins (Fig. 3H, I, K and L), atrium and pulmonary isomerism (Fig. 3J and Fig. 4F and H), pulmonary situs inversus (Fig. 4D) as well as midline and right-sided stomach (Fig. 4C, E and G). These anomalies are typical of situs ambiguous in humans. Taken together, these results demonstrate that Zic3 is primarily required in epiblast derivatives to affect left–right patterning and heart development.
Table 1.
Deletion of Zic3 in epiblast results in significant perinatal lethality and multiple developmental defects similar to Zic3null mutants
|
Sox2-cre/+ X Zic3flox/flox | ||||||
|---|---|---|---|---|---|---|
| Stage | Total | Zic3flox/y | Zic3flox/+ | Zic3flox/+; Sox2-cre | Zic3flox/y; Sox2-cre | P-value |
| 9.5 dpc | 108 | 27 (7%) | 23 | 26 (27%) | 32 (59%) | 0.67 (χ2 = 1.56) |
| 15.5 dpc | 179 | 41 (2%) | 46 (2%) | 54 | 38 (35%) | 0.35 (χ2 = 3.27) |
| Post-birth 3 weeks | 299 | 87 | 83 | 74 | 55 | 0.04 (χ2 = 8.12) |
| Zic3−/y X Zic3+/− | ||||||
| Stage | Total | Zic3+/y | Zic3+/− | Zic3−/y | Zic3−/− | P-value |
| 9.5 dpc | 41 | 11 (9%) | 12 (50%) | 8 (100%) | 10 (80%) | 0.83 (χ2 = 0.9) |
| 15.5 dpc | 151 | 46 (2%) | 47 (2%) | 34 (30%) | 24 (35%) | 0.02 (χ2 = 9.39) |
| Post-birth 3 weeks | 642 | 261 | 257 | 74 | 50 | <0.0001 (χ2 = 243) |
Percentage in parentheses represents proportions of embryos that have observable abnormalities. Dead or absorbed embryos were not included in the count. P-values were calculated from the χ2 analysis.
Figure 1.
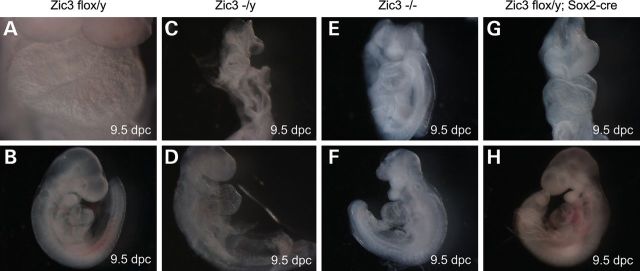
Deletion of Zic3 in epiblast results in significant developmental anomalies and cardiac looping defects, similar to Zic3null embryos. Typical morphology of Zic3flox/y control (A and B), Zic3null (C–F) and Zic3flox/y; Sox2-cre epiblast CKO (G and H) embryos at 9.5 dpc shows various degree of developmental defects, including head anomalies (C, E, F) and open neural tube (E), in the mutants compared with the controls (A and B). Hearts show normal dextral looping in Zic3flox/y controls (A and B), sinistral looping in Zic3null (F) and Zic3flox/y; Sox2-cre epiblast CKO (G and H) embryos, and ventral looping (C–E) in Zic3null embryos. Embryos are shown in a frontal view (A, C, E and G), right lateral position (B and D) and left lateral position (F and H), respectively.
Figure 2.
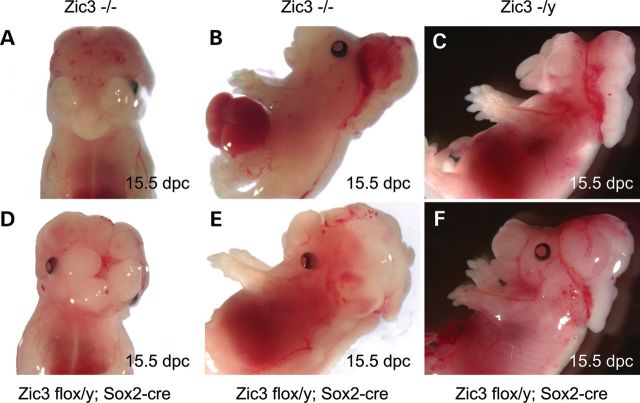
Multiple laterality and CNS anomalies are seen in Zic3null (A–C) and Zic3flox/y; Sox2-cre epiblast CKO (D–F) embryos at 15.5 dpc. These abnormalities include exencephaly (A–F), cleft palate (A, C and D), omphalocele (B), malposition or loss of eyes (C and D), and neural tube closure defect (B and E).
Figure 3.
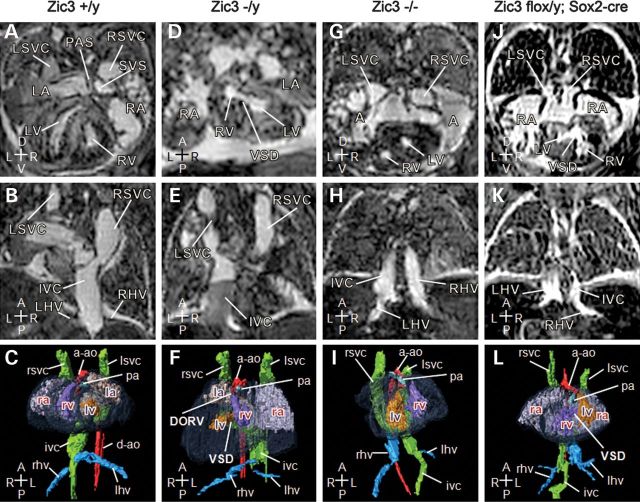
Deletion of Zic3 in epiblast recapitulates complex cardiovascular laterality defects seen in Zic3null embryos. Magnetic resonance imaging of 15.5 dpc embryos. (A–C) Transverse, coronal sections and three-dimensional reconstruction (ventral view) of a typical wild-type control heart and vasculature showing a pectinated right atrium (RA), with a systemic venous sinus (SVS), into which drains the right superior vena cava (RSVC), the left superior vena cava (LSVC) via the coronary sinus and the inferior vena cava (IVC). The left atrium (LA) is characterized by the primum atrial septum (PAS). The right ventricle (RV) is dextral to the left, and gives rise to the main pulmonary artery (pa). The left ventricle (LV), separated from the right ventricle by interventricular septum (not labeled), gives rise to the ascending aorta (a-ao), which arches to the left of the body axis. The left and right hepatic veins (LHV, RHV) join the inferior vena cava on the right of the body axis prior to entering the right atrium. (D–L) Transverse, coronal sections and 3D reconstruction (ventral view) of Zic3null and Zic3flox/y; Sox2-cre epiblast CKO embryos' heart and vasculature. (D–F) Zic3−/y embryo exhibits situs inversus with heart malpositioned to the right. Atria and ventricles are inverted with ventricular septal defect (VSD). Both aorta and pulmonary artery arise from the right ventricle creating a double-outlet right ventricle (DORV). Inferior vena cava ascends on the left of the body axis. (G–I) Zic3−/− heart showing a large atrial septal defect, resulting in a common atrium (A). The heart is malpositioned to the right (dextrocardia) with inverted ventricles. Inferior vena cava ascends on the left and receives the left hepatic veins prior to entering the atrium, while the right hepatic veins enter separately. (J–L) Zic3flox/y; Sox2-cre heart showing a large atrial septal defect, leading to a common atrium, which is pectinated on each side and drains the bilateral superior vena cava. The coronary sinus is absent. These appearances indicate right atrial isomerism. Dextrocardia is also present with large ventricular septal defect (VSD). Right-sided aortic arch is noted with malposition of the great arteries and the aorta ascending anterior to the pulmonary artery. Inferior vena cava is continuous on the right and receives the right hepatic veins before draining to the atrium, while the left hepatic veins enter separately. Axis: D, dorsal; V, ventral; R, right; L, left; A, anterior; P, posterior.
Figure 4.
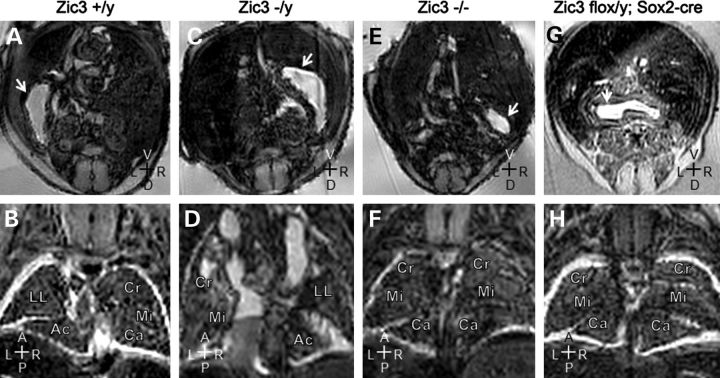
Deletion of Zic3 in epiblast also recapitulates non-cardiovascular laterality defects seen in Zic3null embryos. Magnetic resonance imaging of 15.5 dpc embryos. (A and B) Transverse and coronal sections of a wild-type embryo showing a normal left-sided stomach (arrow), and normal pulmonary topology with cranial (Cr), middle (Mi), caudal (Ca) and accessory (Ac) lobes of the right lung and the left lung (LL). (C and D) Corresponding sections through a Zic3−/y embryo showing a right-sided stomach and inverted pulmonary topology. (E and F) Corresponding sections through a Zic3−/y embryo showing that the stomach is malpositioned to the right, and both lungs have multiple lobes suggesting right pulmonary isomerism. (G and H) Zic3flox/y; Sox2-cre embryo showing a midline stomach and right pulmonary isomerism. Axis: D, dorsal; V, ventral; R, right; L, left; A, anterior; P, posterior.
Zic3 is required in epiblast for proper transcriptional control of embryonic heart development
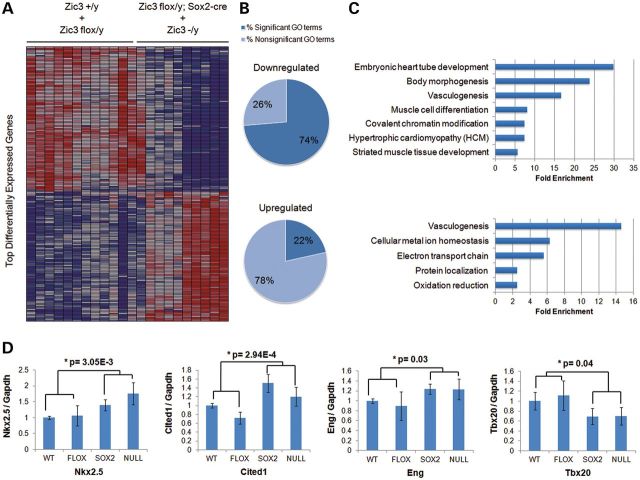
Deficiency of Zic3 is associated with decreased expression of several cardiac-specific genes in ES cells (19). However, whether this remains true during later heart development is not clear, and its direct impact on gene expression within the embryonic heart has not been addressed. In order to better understand the molecular basis for the heart defects seen in Zic3null and epiblast CKO embryos, we further investigated whether complete or epiblast-specific deletion of Zic3 would impact embryonic heart development at the transcriptional level by whole genome expression microarray. The whole heart was carefully dissected out from 15.5 dpc Zic3+/y, Zic3flox/y, Zic3flox/y; Sox2-cre, and Zic3-/y embryos, RNA was extracted, processed and hybridized onto the Illumina MouseWG-6 v2.0 Expression BeadChips. Significant expression changes were identified in 2569 transcripts after imposing an FDR (false discovery rate) cutoff of 0.05 and Q-value <0.05 between the Zic3 mutant hearts and the control hearts, whereas no significant difference within the two control groups and the two mutant groups were evident for these transcripts. One-hundred and ninety-nine of these 2569 differentially expressed transcripts showed fold change >1.5 (Fig. 5, Supplementary Material, Table S2). Due to the assay's design, some probes mapped to different regions or alternatively spliced transcripts of the same gene. Thus, the reporters corresponding to 199 differentially expressed transcripts in our data set uniquely mapped to 146 RefSeq-annotated genes and 44 non-annotated gene candidates. Out of 146 unique RefSeq-annotated genes, 66 genes were downregulated and 80 genes were upregulated in the Zic3 mutant hearts (Fig. 5A). Evaluation of the gene content by association of Gene Ontology (GO) terms in the DAVID bioinformatics database revealed that downregulated genes in the Zic3 mutants were much more represented in the significant GO categories than the upregulated genes (Fig. 5B). Multiple processes involved in heart development, such as embryonic heart tube development, vasculogenesis and muscle cell differentiation, were significantly enriched for these differentially expressed gene sets (Fig. 5C). In addition, Shh signaling may also be disturbed in the mutant hearts, as evidenced by decreased expression for key components of the signaling pathway, such as Hey2 and Ezh2 (Supplementary Material, Table S2). The top differentially expressed genes that have known functions for cardiac development, including Nkx2.5, Tbx20, Cited1 and Eng, were also validated by real-time PCR (Fig. 5D). These results demonstrate that expression of Zic3 in epiblast derivatives is necessary for proper transcriptional control of embryonic stage cardiac development, and suggest that Zic3 may be required in early development of epiblast, later heart morphogenesis or both.
Figure 5.
Whole heart transcriptional response to complete or epiblast-specfic deletion of Zic3 shows distinct gene expression patterns between Zic3 mutants and the controls at 15.5 dpc. (A) Heatmap illustrating distinct expression patterns for top differentially expressed transcripts between two groups with Q-value <0.05, false discovery rate (FDR) <0.05 and fold change larger than 1.5. Red indicates a higher expression value; blue indicates a lower expression value. (B) Proportions of significant and non-significant Gene Ontology (GO) categories for each of the two expression patterns. (C) Selection of the most important GO terms for each expression pattern. Higher fold enrichment means a GO term is more likely to be enriched in a given set of differently expressed genes. (D) Real-time PCR confirms upregulation of Nkx2.5, Cited1 and Eng, and downregulation of Tbx20 in the mutant hearts at 15.5 dpc. Four independent heart samples are used for each of the four genotype groups (WT: Zic3+/y, FLOX: Zic3flox/y, SOX2: Zic3flox/y; Sox2-cre, NULL: Zic3−/y), and each sample is assayed in triplicate and normalized to the endogenous control Gapdh. One-way ANOVA was used for statistical analyses in Qbase PLUS (Biogazelle).
Deletion of Zic3 in heart progenitors and outflow tract does not affect normal development
Previous studies have shown that Zic3 is weakly expressed in the embryonic heart and its deficiency leads to reduced trabeculation and thinner myocardium in a proportion of Zic3null embryos that do not exhibit heart looping defects and gross morphological abnormalities (19). However, it is still unclear whether the heart defects seen in Zic3null embryos occur due to an intrinsic requirement for Zic3 in the heart, or as a consequence of abnormal left–right patterning of the early embryo or both. To address this issue, we used two independent Cre lines to delete Zic3 specifically from the developing heart. Vertebrate heart arises from the primitive heart tube that gives rise to ventricles and atria, outflow tract which later divides into aorta and pulmonary trunk, as well as inflow tract (definitive horns of sinus venosus) which eventually develops into inferior and superior vena cava. Nkx2.5-cre mediates specific and efficient deletion in the first and second heart fields that form the primitive heart tube (Supplementary Material, Fig. S2D) (21); whereas Wnt1-cre induces efficient recombination in all neural crest derivatives, including cardiac neural crest that contributes to the formation of out flow tract (Supplementary Material, Fig. S2E and F) (22). Thus, use of these two Cre lines could delete Zic3 in crucial parts of the developing heart. In contrast to the Zic3null and epiblast CKO mutants, deletion of Zic3 in heart progenitors and outflow tract did not affect survival (Table 2). Cardiac or neural crest CKO mice developed well into adulthood (>1 year) without any observable anomalies and were phenotypically indistinguishable from their littermate controls. Examination of these mutants at 9.5 dpc yielded no evidence of heart looping anomalies (Table 2), and MRI analyses at 15.5 dpc showed none with heart or great vessel defects (data not shown). These results reveal that Zic3 is not required in the heart for its normal development; instead heart morphogenesis must rely on Zic3 functioning in extra-cardiac tissues early during the establishment of left–right axis.
Table 2.
Deletion of Zic3 from the heart and outflow tract does not affect viability and cardiac laterality
|
Nkx2.5-cre/+ X Zic3flox/flox | ||||||
|---|---|---|---|---|---|---|
| Stage | Total | Zic3flox/y | Zic3flox/+ | Zic3flox/+; Nkx2.5-cre | Zic3flox/y; Nkx2.5-cre | P-value |
| 9.5 dpc | 71 | 20 | 16 | 16 | 19 | 0.87 (χ2 = 0.72) |
| Post-birth 3 weeks | 154 | 42 | 44 | 34 | 34 | 0.54 (χ2 = 2.15) |
| Wnt1-cre/+ X Zic3flox/flox | ||||||
| Stage | Total | Zic3flox/y | Zic3flox/+ | Zic3flox/+; Wnt1-cre | Zic3flox/y; Wnt1-cre | P-value |
| 9.5 dpc | 93 | 26 | 23 | 21 | 23 | 0.9 (χ2 = 0.57) |
| Post-birth 3 weeks | 230 | 59 | 57 | 61 | 53 | 0.89 (χ2 = 0.62) |
No observable abnormalities were found in Zic3flox/y; Nkx2.5-cre and Zic3flox/y; Wnt1-cre mutants. P-values were calculated from the χ2 analysis.
Deletion of Zic3 in posterior LPM adjacent to the node, but not in cranial or cardiac mesoderm, results solely in laterality and cardiac defects
Zic3 is known to be expressed in the emerging mesoderm during gastrulation and its deficiency results in the randomization of Nodal and Pitx2 expression in the LPM of the Zic3null embryos (10). Nodal is a key morphogen that plays an integral role during left–right patterning by its asymmetric expression at the node and left LPM. Zic3 genetically interacts with Nodal and acts upstream of Nodal signaling in the maintenance of left–right identity (11). Thus, Zic3 may be required specifically in the LPM for left–right patterning and subsequent cardiac development. To test this hypothesis, we deleted Zic3 using two well-characterized mesoderm specific Cre-expressing lines (Supplementary Material, Fig. S2G and H). Mesp1-cre expresses Cre efficiently in the cranial, cardiac and extraembryonic mesoderm anterior of, but not adjacent to, the node during gastrulation (23,24); whereas T-cre mediates efficient deletion broadly in mesoderm along the entire primitive streak encompassing the LPM adjacent and posterior to the node soon after gastrulation initiates (25,26). Deletion of Zic3 from LPM anterior to the node using Mesp1-cre did not affect survival or normal development, these mice lived well into adulthood with no observable anomalies. Examination of mutants at 9.5 dpc showed all with normal laterality and correct heart looping (Table 3). In contrast, deletion of Zic3 from LPM adjacent and posterior to the node using T-cre led to significant lethality in ∼50% of Zic3flox/y; T-cre mice before weaning (Table 3). All viable T-cre CKO males, unlike Mesp1-cre CKO males, had similar ‘kinked’ tails as the Zic3null and epiblast CKO mice. Careful examination at 9.5 dpc revealed typical heart-looping defects in four out of 22 T-cre CKO embryos, but no discernible embryo loss or developmental anomalies were found at this stage (Fig. 6, Table 3). In addition, no significant lethality was found in T-cre CKO embryos at 15.5 dpc (Table 3), indicating these mutants die either in late gestation or perinatally, similar to the epiblast CKO mutants. However, unlike epiblast CKO embryos, CNS development in T-cre CKO embryos was normal at 15.5 dpc. MRI analyses demonstrated classical laterality defects in four out of 18 T-cre CKO embryos at 15.5 dpc (Fig. 7), these internal malformations were similar to what were seen in Zic3null and epiblast CKO embryos. Together, these results reveal that Zic3 functions specifically in the posterior LPM around the node for left–right patterning and cardiac development; while its function in ectoderm may be critical for CNS development.
Table 3.
Deletion of Zic3 by T-cre but not Mesp1-cre results in significant perinatal lethality and early laterality defects
|
T-cre/+ X Zic3flox/flox | ||||||
|---|---|---|---|---|---|---|
| Stage | Total | Zic3flox/y | Zic3flox/+ | Zic3flox/+; T-cre | Zic3flox/y; T-cre | P-value |
| 9.5 dpc | 86 | 24 (4%) | 20 (5%) | 20 (5%) | 22 (18%) | 0.91 (χ2 = 0.55) |
| 15.5 dpc | 118 | 32 | 34 | 23 | 29 (22%) | 0.51 (χ2 = 2.33) |
| Post-birth 3 weeks | 178 | 61 | 44 | 50 | 23 | 0.0007 (χ2 = 17) |
| Mesp1-cre/+ X Zic3flox/flox | ||||||
| Stage | Total | Zic3flox/y | Zic3flox/+ | Zic3flox/+; Mesp1-cre | Zic3flox/y; Mesp1-cre | P-value |
| 9.5 dpc | 97 | 32 | 22 | 21 | 22 | 0.34 (χ2 = 3.38) |
| Post-birth 3 weeks | 123 | 35 | 33 | 24 | 31 | 0.53 (χ2 = 2.23) |
Percentage in parentheses represents proportions of embryos that have observable abnormalities. Dead or absorbed embryos were not included in the count. P-values were calculated from the χ2 analysis.
Figure 6.
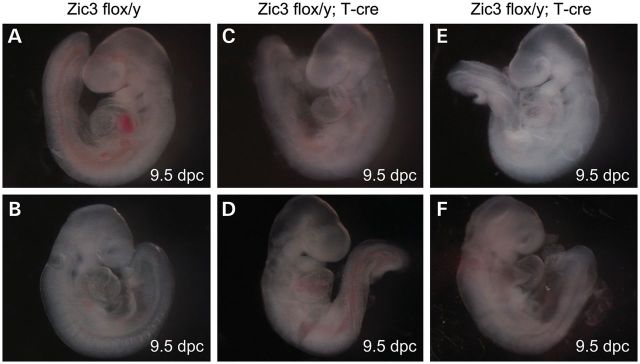
Cardiac looping abnormalities in Zic3flox/y; T-cre embryos. Gross morphology of the Zic3flox/y control (A and B) and Zic3flox/y; T-cre embryos (C–F) embryos at 9.5 dpc. Hearts show normal dextral looping (A and B), sinistral looping (C and E) and ventral looping (D and F). Embryos are shown in left lateral position (A, C and E) and right lateral position (B, D and F), respectively.
Figure 7.
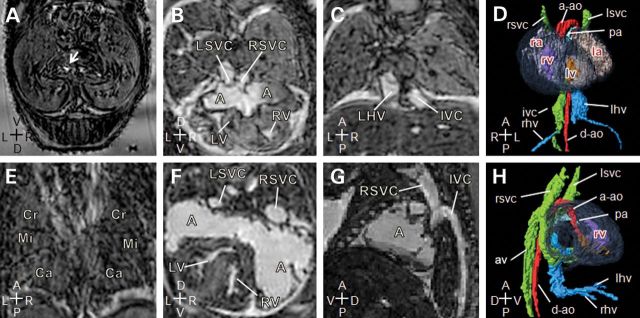
Laterality and complex cardiovascular abnormalities in Zic3flox/y; T-cre embryos. Magnetic resonance imaging of 15.5 dpc mutant embryos. (A and E) Transverse and coronal sections of a Zic3flox/y; T-cre embryo showing a midline stomach (arrow) and right pulmonary isomerism. (B–D) Transverse, coronal sections and three-dimensional reconstruction (ventral view) of a Zic3flox/y; T-cre embryo showing a large atrial septal defect, resulting in a common atrium (A), into which drains the left and right superior vena cava (LSVC, RSVC). The heart is malpositioned to the right (dextrocardia). Inferior vena cava (IVC) is continuous on the right and receives the right hepatic veins (rhv) prior to entering the atrium, while the left hepatic veins (LHV) enter separately. (F–H) Transverse, sagittal sections and three-dimensional reconstruction (right view) of a Zic3flox/y; T-cre embryo showing a normal positioned heart with a dilated common atrium (A), into which drains the left and right superior vena cava (LSVC, RSVC). Inferior vena cava (IVC) is interrupted and drains via the azygous vein (AV) to the right superior vena cava (RSVC). The hepatic veins come together and enter the atria without joining the IVC. Axis: D, dorsal; V, ventral; R, right; L, left; A, anterior; P, posterior.
DISCUSSION
Disturbances in early left–right patterning caused by the loss of Zic3 secondarily affect heart development through transcriptional cascades
The heart is the first organ to exhibit left–right asymmetry during embryonic development and human cardiovascular malformations are frequently associated with left–right patterning defects (4). Although Zic3 has been established as an important regulator of left–right patterning and heart development during embryogenesis (10,27), whether it has a cell-autonomous function in cardiac development independent of left–right patterning remains controversial (19) and its tissue specific requirement for establishing laterality has not been defined. With development of the Zic3 conditional allele in mice, these questions were directly addressed. Deletion of Zic3 in the epiblast with Sox2-cre led to significant lethality, complex cardiac and laterality defects, as well as other congenital anomalies not associated with lef–tright pattering, all of which are consistent with and fully recapitulate those previously reported with the global deletion of Zic3 (10,11). They indicate that Zic3 is primarily required in the epiblast for establishing normal laterality and subsequent heart development. Interestingly, we found that epiblast CKO mutants most likely die perinatally, whereas Zic3null mutants have an additional 40–50% embryo loss before 15.5 dpc, resulting in a much higher overall lethality rate in Zic3null mutants. We reason that it is unlikely caused by residual functions of Zic3 in the epiblast due to incomplete recombination, since Sox2-cre is very efficient (Supplementary Material, Fig. S2A–C). Additionally, we observe that Zic3null embryos exhibit more developmental anomalies as early as 9.5 dpc. This suggests that Zic3 may have additional function in extra-embryonic derivatives, most likely anterior visceral endoderm , for early gastrulation (17). In fact, there are quite a few instances in which the extra-embryonic expression of a gene is essential for early morphogenesis. For example, Smad2-deficient mouse embryos die around 7.5 dpc due to failure to gastrulate and to establish an anterior–posterior (A–P) axis, but introduction of wild-type extra-embryonic tissues enabled these embryos to develop beyond 7.5 and up to 10.5 dpc, demonstrating a requirement for SMAD2 in extra-embryonic tissues for the generation of an A–P axis and gastrulation (28). It has also been reported recently that Atp1a1 expression in the extra-embryonic yolk syncytial layer is required for heart tube elongation in zebrafish (29). In order to determine whether Zic3 has an additional role in cardiac development independent of left–right patterning, we deleted Zic3 from heart progenitors as well as the outflow tract with Nkx2.5-cre and Wnt1-cre, respectively. No cardiac structural anomalies were found in these mice, indicating normal cardiac development relies on extra-cardiac functions of Zic3 in epiblast derivatives. Microarray analyses of Zic3 epiblast conditional and null embryos' hearts demonstrate that multiple processes involved in heart development are significantly enriched for the top differentially expressed genes between mutants and controls. These data provide additional molecular evidence to support our cascade hypothesis that heart defects occur as a secondary effect of disturbances in early left–right patterning, an effect that is modulated by transcriptional control of Zic3 in the epiblast.
Zic3 functions in the perinodal region of the posterior LPM to establish left–right identity
The precise tissue specific functions of Zic3 have not been defined and their effect on Nodal expression in the left LPM remains unclear (10,11). The present evidence suggests that Zic3 is required in the posterior LPM around the node for proper left–right patterning and heart development. We found that deletion of Zic3 from anterior mesoderm using Mesp1-cre results in normal embryo laterality, and no defect nor significant lethality was observed in these mice. Since reporter assays demonstrate widespread Cre recombinase activity in the LPM (Supplementary Material, Fig. S2G), this suggests that Zic3 may not be required for propagation of Nodal expression in the left LPM. It is reported that Mesp1-cre mediates recombination in the LPM and paraxial mesoderm anterior of, but not adjacent to, the node (24). Importantly, Nodal ASE (asymmetric enhancer element) is required for the initiation of Nodal expression adjacent to the node, which is then propagated throughout the left LPM (30). In addition, previous studies found that Nodal ASE is responsive to Zic3 in both Xenopus and mouse (11). Therefore, it is likely that Zic3 functions in the initiation of Nodal expression in the LPM adjacent to the node. To test this hypothesis, we used T-cre, which has been shown to be active in LPM adjacent to the node and posterior of it (26), where Mesp1-cre lacks activity. Our results show that deletion of Zic3 by T-cre led to significant perinatal lethality similar to the epiblast CKO mice. More interestingly, only laterality and cardiac defects were found in these mutant embryos, whereas CNS or other early developmental anomalies frequently found in Zic3null and epiblast CKO embryos were not observed. This provides direct evidence to support the idea that Zic3 is specifically required in the posterior LPM adjacent to the node, most likely by regulating initiation of Nodal expression in this region, for proper control of left–right patterning and subsequent cardiac development. The absence of CNS defects in the T-cre conditional embryos suggests that Zic3 has additional functions outside of LPM, most likely in the neuro-ectoderm, for CNS development.
One of the most challenging questions right now is how symmetric expression of Zic3 in the LPM could modulate left–right asymmetry during embryogenesis. Previous studies show that Zic3 can physically and functionally interact with Gli3, which is an important mediator of Shh signaling (31–34). Importantly, Shh is incorporated into Nodal vesicular parcels, which are swept left-ward by the unidirectional rotation of the motile cilia (35). Shh signaling in the LPM is required for the asymmetric activation of Gdf1 (36). Gdf1 converts Nodal into a long-range signal through heterodimerization, which leads to increased Nodal activity in the perinodal region for subsequent initiation of asymmetric Nodal expression in the left LPM (37). The observation that Shh signaling may be disturbed in the Zic3null and epiblast CKO embryonic hearts, along with our data showing Zic3’s critical role in the perinodal region of the LPM for establishing laterality, strongly suggest a possible mechanism for Zic3 function in left–right patterning. Zic3 may be modulated by Shh or via interactions with Gli3 downstream of Shh signaling in the perinodal region of the LPM. Alternatively, Zic3 may function in conjunction with Notch signaling during the establishment of left–right identity. Notch signaling, activated by Wnt signaling in the node (38), is required for asymmetric transcription of Nodal in the node via Nodal enhancer (39). Recently, Zic3 was shown to control notochord and organizer development in Xenopus through suppression of the Wnt/beta-catenin signaling, providing a possible link between Zic3 and Notch signaling.
In summary, we show that Zic3 is required in epiblast derivatives to affect left–right patterning and heart development; whereas its function in extraembryonic tissues may be critical for early gastrulation. Interestingly, cardiac malformations in Zic3 deficiency arise not due to the requirement of Zic3 in the heart but rather because it functions early in the establishment of left–right body axis, and failure of this early function would disrupt transcriptional cascade that ultimately leads to complex heart defects. We further narrow the primary requirement of Zic3 to the perinodal region of the posterior LPM, possibly via regulating initiation of Nodal expression, to affect establishment of laterality. Thus, this study establishes the spatial basis for further understanding the molecular mechanisms underlying the complex interaction of Zic3 with signaling pathways involved in the early left–right patterning.
MATERIALS AND METHODS
Generation of Zic3flox mice
The targeting construct design and generation of correctly targeted embryonic stem cells with a conditional Zic3 allele (Zic3flox) is shown in Supplementary Material, Figure S1. Methods are also provided in detail in the Supplementary data. All the mice were maintained on a C57BL/6J and 129 mixed genetic background. Mice and embryos for Zic3flox-neo, Zic3flox and Zic3del were genotyped by PCR using the following primers:
P1: 5′-TGAACAGCCAGAGAGGGAGA-3′ and 5′-AGCAAGCCACAGAAACCAGT-3′
P2: 5′-GGAACGGCAACGACAAGTA-3′ and 5′-AGCAAGCCACAGAAACCAGT-3′
Conditional deletion of Zic3
Zic3flox/flox females were crossed with Sox2-cre (Tg(Sox2-cre)1Amc, imported from The Jackson Laboratory), Nkx2.5-cre (Nkx2-5IRESCre, gift from Dr James Martin, Baylor College of Medicine), Wnt1-cre (Tg(Wnt1-cre)11Rth, gift from Dr Monica Justice, Baylor College of Medicine), Mesp1-cre (Mesp1tm2(cre)Ysa, gift from Dr James Martin, Baylor College of Medicine) or T-cre (gift from Dr Mark Lewandoski, National Cancer Institute, NCI-Frederick) males to generate conditional knockout males with Zic3flox/y; cre genotypes that lack Zic3 expression in cells expressing Cre recombinase; whereas Zic3flox/+; cre females and Zic3flox mice continue to express Zic3 and serve as littermate controls. Then Zic3flox/y; Sox2-cre males were crossed to Zic3flox/+; Sox2-cre females to generate Zic3null mice, since Sox2-cre induces recombination in all epiblast cells including the germline. The specificity and efficiency of all the Cre-expressing lines were confirmed by X-gal staining of embryos derived from the intercross between Cre-expressing mice and R26R reporter mice (Gt(ROSA)26Sortm1Sor, gift from Dr Brendan Lee, Baylor College of Medicine), shown in Supplementary Material, Figure S2. However, due to lack of Zic3 specific antibodies, our ability is limited to directly demonstrate lack of Zic3 protein expression in the expected tissues for our CKO mice. Embryos were harvested at the indicated timepoints after detection of a vaginal plug, with noon of the day of appearance of the vaginal plug designated as 0.5 dpc. They were genotyped by PCR using P1, P2 (mentioned earlier, Supplementary Material, Fig. S1), general Cre specific primers: 5′-GCCACCAGCCAGCTATCAA-3′ and 5′-GCTAATCGCCATCTTCCAG-3′, as well as Sry specific primers for sex determination: 5′-CCCAGCATGCAAAATACAGA-3′ and 5′-AACAGGCTGCCAATAAAAGC-3′. All mouse experiments were performed following the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and with the approval of the Institutional Animal Care and Use Committee, at Baylor College of Medicine (Animal Welfare Assurance #A3832-01).
Morphology, X-gal staining and histology
Early heart looping morphology and other gross developmental anomalies were determined at dissection. To confirm Cre specificity and efficiency, embryos were harvested at the indicated timepoints, fixed in 4% paraformaldehyde in 1× phosphate-buffered saline (PBS) at 4°C and stained in X-gal mix overnight in the dark at 37°C. For histology studies, embryos were fixed in 4% paraformaldehyde at 4°C overnight, dehydrated in ethanol, embedded in paraffin and sections were stained with hematoxylin and eosin.
Magnetic resonance imaging and 3D reconstruction
Embryos were harvested at 15.5 dpc, fixed at 4°C for 2–4 weeks in 4% paraformaldehyde in 1× PBS, doped with 8 mm gadolinium-diethylenetriamine pentaacetic anhydride (Gd-DTPA), then embedded in 1% agarose containing 8 mm Gd-DTPA in NMR tubes and imaged on a 9.4T Bruker Avance Biospec Spectrometer, 21-cm bore horizontal scanner with a 35-mm volume resonator (Bruker BioSpin, Billerica, MA, USA). Embryos were oriented to align their A–P axis perpendicular to the long axis of the tube, with 2–3 layers per tube and 2–3 embryos per layer to achieve high-throughput imaging of up to nine embryos simultaneously. The left forelimb was removed from each embryo to facilitate the identification of the left side. Additionally, embryos had other limbs and/or tails removed before embedding to unequivocally identify each embryo in a given layer. A Fast Low Angle Shot (FLASH) pulse sequence was used with a repetition time (TR) = 15 ms and an echo time (TE) = 6.8 ms. The number of averages was 74. A matrix size of 512 × 512 × 512 at a field of view of 26 × 26 × 26 mm was achieved after imaging for ∼24 h, which yielded an experimental resolution of 51 × 51 × 51 µm when imaging up to nine embryos. 3D reconstructions were performed using Image Segmentation Editor, with SurfaceGen and TetraGen surface rendering modules in Amira 4.3 (Visage Imaging, San Diego, CA, USA).
Microarray and pathway enrichment analysis
The whole heart was carefully dissected out for each embryo at 15.5 dpc, total RNAs were extracted and purified using RNeasy Mini Kit (QIAGEN). Spectrophotometry (NanoDrop-1000 Spectrophotometer, Thermo Fisher Scientific) and microfluidic electrophoresis (Experion Automated Electrophoresis System, Bio-Rad Laboratories) were used for RNA quality control. In vitro transcription was performed using Illumina TotalPrep RNA Amplification Kit (Applied Biosystems/Ambion). cRNAs were hybridized onto Illumina MouseWG-6 v2.0 Expression BeadChips (Illumina) as per manufacturer's instructions. Initial quality control of the microarray signal intensity data was performed using the lumi Bioconductor package in the R program as described previously (40), and two outliers out of 24 samples were excluded from further analyses. The data are MIAME compliant and have been deposited in NCBI's Gene Expression Omnibus (41), where it is accessible through GEO Series accession number GSE41674. Quantile normalization of the expression data was performed using the IlluminaNormalizer module from GenePattern (http://www.broadinstitute.org/cancer/software/genepattern/), differential expression analyses were performed using two-sided t-test with 1000 permutations in the ComparativeMarkerSelection and HierarchicalClustering modules from GenePattern. Significantly differentially expressed gene lists were analyzed using DAVID Ontology (http://david.abcc.ncifcrf.gov/) to uncover enriched pathways.
Quantitative real-time PCR
Total RNAs were isolated and purified from the whole heart of each embryo (15.5 dpc) using RNeasy Mini Kit (QIAGEN), and were converted to cDNA using High Capacity cDNA Reverse Transcription Kit (Ambion). Quantitative real-time PCR reactions were carried out using Applied Biosystems 7900HT cycler (Applied Biosystems) and pre-optimized TaqMan primer-probe sets from Applied Biosystems for selected genes. Expression levels were normalized to the endogenous control Gapdh. All reactions were performed in triplicates, and for quality control, only samples for which the threshold cycle (Ct) replicate values were within 1 Ct value of each other were used for subsequent analysis. Mean values for each reaction triplicate were analyzed. One-way ANOVA was used for statistical analyses in Qbase PLUS (Biogazelle).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the March of Dimes (1-FY08-444 to J.W.B.).
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to James Martin, Mark Lewandoski and Brendan Lee for their generous provision of Cre transgenic and R26R reporter mice and Xueqing Wang and Luis Franco for technical help with Illumina expression microarray.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Aruga J., Nagai T., Tokuyama T., Hayashizaki Y., Okazaki Y., Chapman V.M., Mikoshiba K. The mouse zic gene family. Homologues of the Drosophila pair-rule gene odd-paired. J. Biol. Chem. 1996;271:1043–1047. doi: 10.1074/jbc.271.2.1043. doi:10.1074/jbc.271.2.1043. [DOI] [PubMed] [Google Scholar]
- 2.Aruga J., Yokota N., Hashimoto M., Furuichi T., Fukuda M., Mikoshiba K. A novel zinc finger protein, zic, is involved in neurogenesis, especially in the cell lineage of cerebellar granule cells. J. Neurochem. 1994;63:1880–1890. doi: 10.1046/j.1471-4159.1994.63051880.x. doi:10.1046/j.1471-4159.1994.63051880.x. [DOI] [PubMed] [Google Scholar]
- 3.Gebbia M., Ferrero G.B., Pilia G., Bassi M.T., Aylsworth A., Penman-Splitt M., Bird L.M., Bamforth J.S., Burn J., Schlessinger D., et al. X-linked situs abnormalities result from mutations in ZIC3. Nat. Genet. 1997;17:305–308. doi: 10.1038/ng1197-305. doi:10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- 4.Zhu L., Belmont J.W., Ware S.M. Genetics of human heterotaxias. Eur. J. Hum. Genet. 2006;14:17–25. doi: 10.1038/sj.ejhg.5201506. [DOI] [PubMed] [Google Scholar]
- 5.Nakata K., Nagai T., Aruga J., Mikoshiba K. Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc. Natl Acad. Sci. U.S.A. 1997;94:11980–11985. doi: 10.1073/pnas.94.22.11980. doi:10.1073/pnas.94.22.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitaguchi T., Nagai T., Nakata K., Aruga J., Mikoshiba K. Zic3 is involved in the left-right specification of the Xenopus embryo. Development. 2000;127:4787–4795. doi: 10.1242/dev.127.22.4787. [DOI] [PubMed] [Google Scholar]
- 7.Fujimi T.J., Hatayama M., Aruga J. Xenopus Zic3 controls notochord and organizer development through suppression of the Wnt/beta-catenin signaling pathway. Dev. Biol. 2011;361:220–231. doi: 10.1016/j.ydbio.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 8.Klootwijk R., Franke B., van der Zee C.E., de Boer R.T., Wilms W., Hol F.A., Mariman E.C. A deletion encompassing Zic3 in bent tail, a mouse model for X-linked neural tube defects. Hum. Mol. Genet. 2000;9:1615–1622. doi: 10.1093/hmg/9.11.1615. doi:10.1093/hmg/9.11.1615. [DOI] [PubMed] [Google Scholar]
- 9.Carrel T., Purandare S.M., Harrison W., Elder F., Fox T., Casey B., Herman G.E. The X-linked mouse mutation Bent tail is associated with a deletion of the Zic3 locus. Hum. Mol. Genet. 2000;9:1937–1942. doi: 10.1093/hmg/9.13.1937. doi:10.1093/hmg/9.13.1937. [DOI] [PubMed] [Google Scholar]
- 10.Purandare S.M., Ware S.M., Kwan K.M., Gebbia M., Bassi M.T., Deng J.M., Vogel H., Behringer R.R., Belmont J.W., Casey B. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development. 2002;129:2293–2302. doi: 10.1242/dev.129.9.2293. [DOI] [PubMed] [Google Scholar]
- 11.Ware S.M., Harutyunyan K.G., Belmont J.W. Heart defects in X-linked heterotaxy: evidence for a genetic interaction of Zic3 with the nodal signaling pathway. Dev. Dyn. 2006;235:1631–1637. doi: 10.1002/dvdy.20719. doi:10.1002/dvdy.20719. [DOI] [PubMed] [Google Scholar]
- 12.Zhu L., Peng J.L., Harutyunyan K.G., Garcia M.D., Justice M.J., Belmont J.W. Craniofacial, skeletal, and cardiac defects associated with altered embryonic murine Zic3 expression following targeted insertion of a PGK-NEO cassette. Front. Biosci. 2007;12:1680–1690. doi: 10.2741/2180. doi:10.2741/2180. [DOI] [PubMed] [Google Scholar]
- 13.Lim L.S., Hong F.H., Kunarso G., Stanton L.W. The pluripotency regulator Zic3 is a direct activator of the Nanog promoter in ESCs. Stem Cells. 2010;28:1961–1969. doi: 10.1002/stem.527. doi:10.1002/stem.527. [DOI] [PubMed] [Google Scholar]
- 14.Watabe Y., Baba Y., Nakauchi H., Mizota A., Watanabe S. The role of Zic family zinc finger transcription factors in the proliferation and differentiation of retinal progenitor cells. Biochem. Biophys. Res. Commun. 2011;415:42–47. doi: 10.1016/j.bbrc.2011.10.007. doi:10.1016/j.bbrc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Elms P., Scurry A., Davies J., Willoughby C., Hacker T., Bogani D., Arkell R. Overlapping and distinct expression domains of Zic2 and Zic3 during mouse gastrulation. Gene Expr. Patterns. 2004;4:505–511. doi: 10.1016/j.modgep.2004.03.003. doi:10.1016/j.modgep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Kitaguchi T., Mizugishi K., Hatayama M., Aruga J., Mikoshiba K. Xenopus Brachyury regulates mesodermal expression of Zic3, a gene controlling left-right asymmetry. Dev. Growth Differ. 2002;44:55–61. doi: 10.1046/j.1440-169x.2002.00624.x. doi:10.1046/j.1440-169x.2002.00624.x. [DOI] [PubMed] [Google Scholar]
- 17.Ware S.M., Harutyunyan K.G., Belmont J.W. Zic3 is critical for early embryonic patterning during gastrulation. Dev. Dyn. 2006;235:776–785. doi: 10.1002/dvdy.20668. doi:10.1002/dvdy.20668. [DOI] [PubMed] [Google Scholar]
- 18.Nagai T., Aruga J., Takada S., Gunther T., Sporle R., Schughart K., Mikoshiba K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev. Biol. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. doi:10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- 19.Zhu L., Harutyunyan K.G., Peng J.L., Wang J., Schwartz R.J., Belmont J.W. Identification of a novel role of ZIC3 in regulating cardiac development. Hum. Mol. Genet. 2007;16:1649–1660. doi: 10.1093/hmg/ddm106. doi:10.1093/hmg/ddm106. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi S., Lewis P., Pevny L., McMahon A.P. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 2002;119(Suppl. 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. doi:10.1016/S0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 21.Stanley E.G., Biben C., Elefanty A., Barnett L., Koentgen F., Robb L., Harvey R.P. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int. J. Dev. Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- 22.Jiang X., Rowitch D.H., Soriano P., McMahon A.P., Sucov H.M. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 23.Saga Y., Miyagawa-Tomita S., Takagi A., Kitajima S., Miyazaki J., Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 24.Lopes Floro K., Artap S.T., Preis J.I., Fatkin D., Chapman G., Furtado M.B., Harvey R.P., Hamada H., Sparrow D.B., Dunwoodie S.L. Loss of Cited2 causes congenital heart disease by perturbing left-right patterning of the body axis. Hum. Mol. Genet. 2011;20:1097–1110. doi: 10.1093/hmg/ddq554. doi:10.1093/hmg/ddq554. [DOI] [PubMed] [Google Scholar]
- 25.Perantoni A.O., Timofeeva O., Naillat F., Richman C., Pajni-Underwood S., Wilson C., Vainio S., Dove L.F., Lewandoski M. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. doi:10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- 26.Kumar A., Lualdi M., Lewandoski M., Kuehn M.R. Broad mesodermal and endodermal deletion of Nodal at postgastrulation stages results solely in left/right axial defects. Dev. Dyn. 2008;237:3591–3601. doi: 10.1002/dvdy.21665. doi:10.1002/dvdy.21665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ware S.M., Peng J., Zhu L., Fernbach S., Colicos S., Casey B., Towbin J., Belmont J.W. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. Am. J. Hum. Genet. 2004;74:93–105. doi: 10.1086/380998. doi:10.1086/380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heyer J., Escalante-Alcalde D., Lia M., Boettinger E., Edelmann W., Stewart C.L., Kucherlapati R. Postgastrulation Smad2-deficient embryos show defects in embryo turning and anterior morphogenesis. Proc. Natl Acad. Sci. U.S.A. 1999;96:12595–12600. doi: 10.1073/pnas.96.22.12595. doi:10.1073/pnas.96.22.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langenbacher A.D., Huang J., Chen Y., Chen J.N. Sodium pump activity in the yolk syncytial layer regulates zebrafish heart tube morphogenesis. Dev. Biol. 2012;362:263–270. doi: 10.1016/j.ydbio.2011.12.004. doi:10.1016/j.ydbio.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura T., Mine N., Nakaguchi E., Mochizuki A., Yamamoto M., Yashiro K., Meno C., Hamada H. Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Dev. Cell. 2006;11:495–504. doi: 10.1016/j.devcel.2006.08.002. doi:10.1016/j.devcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Koyabu Y., Nakata K., Mizugishi K., Aruga J., Mikoshiba K. Physical and functional interactions between Zic and Gli proteins. J. Biol. Chem. 2001;276:6889–6892. doi: 10.1074/jbc.C000773200. doi:10.1074/jbc.C000773200. [DOI] [PubMed] [Google Scholar]
- 32.Zhu L., Zhou G., Poole S., Belmont J.W. Characterization of the interactions of human ZIC3 mutants with GLI3. Hum. Mutat. 2008;29:99–105. doi: 10.1002/humu.20606. doi:10.1002/humu.20606. [DOI] [PubMed] [Google Scholar]
- 33.Stamataki D., Ulloa F., Tsoni S.V., Mynett A., Briscoe J. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 2005;19:626–641. doi: 10.1101/gad.325905. doi:10.1101/gad.325905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinn M.E., Haaning A., Ware S.M. Preaxial polydactyly caused by Gli3 haploinsufficiency is rescued by Zic3 loss of function in mice. Hum. Mol. Genet. 2012;21:1888–1896. doi: 10.1093/hmg/dds002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka Y., Okada Y., Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435:172–177. doi: 10.1038/nature03494. doi:10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- 36.Tsiairis C.D., McMahon A.P. An Hh-dependent pathway in lateral plate mesoderm enables the generation of left/right asymmetry. Curr. Biol. 2009;19:1912–1917. doi: 10.1016/j.cub.2009.09.057. doi:10.1016/j.cub.2009.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka C., Sakuma R., Nakamura T., Hamada H., Saijoh Y. Long-range action of Nodal requires interaction with GDF1. Genes Dev. 2007;21:3272–3282. doi: 10.1101/gad.1623907. doi:10.1101/gad.1623907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakaya M.A., Biris K., Tsukiyama T., Jaime S., Rawls J.A., Yamaguchi T.P. Wnt3a links left-right determination with segmentation and anteroposterior axis elongation. Development. 2005;132:5425–5436. doi: 10.1242/dev.02149. doi:10.1242/dev.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krebs L.T., Iwai N., Nonaka S., Welsh I.C., Lan Y., Jiang R., Saijoh Y., O'Brien T.P., Hamada H., Gridley T. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 2003;17:1207–1212. doi: 10.1101/gad.1084703. doi:10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bucasas K.L., Franco L.M., Shaw C.A., Bray M.S., Wells J.M., Nino D., Arden N., Quarles J.M., Couch R.B., Belmont J.W. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J. Infect. Dis. 2011;203:921–929. doi: 10.1093/infdis/jiq156. doi:10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. doi:10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.