Abstract
Genetic variations in fat mass- and obesity (FTO)-associated gene, a well-replicated gene locus of obesity, appear to be associated also with reduced regional brain volumes in elderly. Here, we examined whether FTO is associated with total brain volume in adolescence, thus exploring possible developmental effects of FTO. We studied a population-based sample of 598 adolescents recruited from the French Canadian founder population in whom we measured brain volume by magnetic resonance imaging. Total fat mass was assessed with bioimpedance and body mass index was determined with anthropometry. Genotype–phenotype associations were tested with Merlin under an additive model. We found that the G allele of FTO (rs9930333) was associated with higher total body fat [TBF (P = 0.002) and lower brain volume (P = 0.005)]. The same allele was also associated with higher lean body mass (P = 0.03) and no difference in height (P = 0.99). Principal component analysis identified a shared inverse variance between the brain volume and TBF, which was associated with FTO at P = 5.5 × 10−6. These results were replicated in two independent samples of 413 and 718 adolescents, and in a meta-analysis of all three samples (n = 1729 adolescents), FTO was associated with this shared inverse variance at P = 1.3 × 10−9. Co-expression networks analysis supported the possibility that the underlying FTO effects may occur during embryogenesis. In conclusion, FTO is associated with shared inverse variance between body adiposity and brain volume, suggesting that this gene may exert inverse effects on adipose and brain tissues. Given the completion of the overall brain growth in early childhood, these effects may have their origins during early development.
INTRODUCTION
The fat mass and obesity-associated gene (FTO) is a well-replicated gene locus of obesity (1–4). It was originally identified as a gene locus of type-2 diabetes, and only later it was discovered that it increases risk of this disease through its primary effect on adiposity (1). However, the mechanisms through which FTO may increase adiposity are still not clear. In vitro studies suggest that FTO encodes 2-oxoglutarate-dependent nucleic-acid demethylase that may regulate gene expression (5–7). Beginning from early embryogenesis, the gene is expressed throughout the body and the brain (5,8,9). The relatively high expression of FTO in the adult hypothalamus, a brain structure regulating energy balance, suggested that FTO might influence adiposity through its impact on energy homeostasis. Conflicting results of both human and animal studies exist, however, as to whether the gene influences either food intake or energy expenditure in a way that would increase risk for obesity [reviewed in (3,4)].
Obesity is a major risk factor for cognitive decline (10). In part, this association may be mediated through the cardiometabolic sequelae of obesity that promote cerebrovascular disease and subsequent neuronal degeneration (11,12). Recently, the FTO-risk allele for obesity has been associated with lower regional volumes of (cortical) brain tissue in elderly (13). These differences were not, however, associated with differences in cholesterol levels, hypertension or white-matter hyperintensities (13), suggesting that other factors may be at play.
The aim of the present study was to investigate an association between FTO, brain and adiposity in a developmental context. For this reason, we have focused on adolescence and used total brain volume as the main dependent variable. Total brain volume reaches 83% of adult values by the end of the second year of postnatal life (14) and 95% by 6 years of age (15). Thus, values of the total brain volume obtained in adolescence are likely to reflect mainly biological processes at play in utero and during early postnatal years. We performed these studies in a sample of 598 adolescents recruited from the French Canadian founder population as part of the Saguenay Youth Study (SYS-Discovery) and replicated our findings in 413 adolescents who were recruited and phenotyped later in the same study (SYS-Replication) and in 718 adolescents of mixed European background who were recruited as part of the IMAGEN study. In all three samples, brain volumes were assessed with magnetic resonance imaging (MRI), and a variant of FTO (i.e. rs9930333) previously reported as associated with body mass index (BMI) (16) was studied.
RESULTS
FTO associations with BMI and brain volume
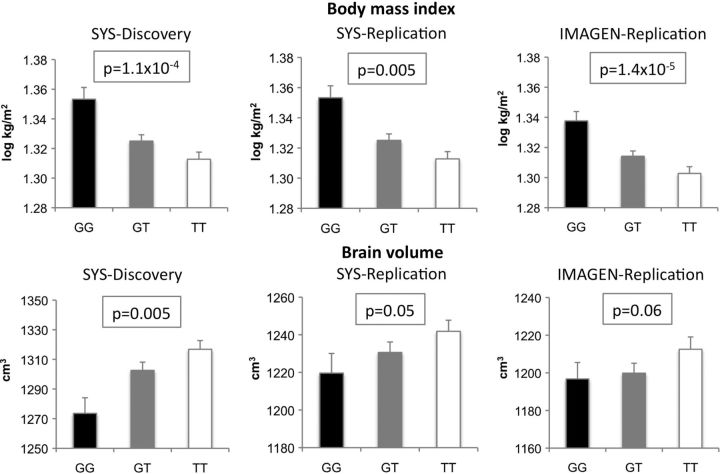
Basic characteristics of the SYS-Discovery, SYS-Replication and IMAGEN samples are provided in Table 1. The prevalence of overweight or obesity was 23.9, 32.7 and 19.0% in the SYS-Discovery, SYS-Replication and IMAGEN samples, respectively, which is similar to that of current adolescent populations (17,18). In the SYS, the G allele of rs9930333 was associated with higher BMI (P = 1.1 × 10−4) and lower brain volume (P = 0.005, Fig. 1 and Table 2). Similar associations were observed in the replication (SYS-Replication and IMAGEN) samples, and the G allele was associated with higher BMI (P = 0.005 and P = 1.4 × 10−5, respectively) and trends towards lower brain volume (P = 0.05 and P = 0.06, respectively; Fig. 1 and Table 2). In a meta-analysis involving 1729 SYS-Discovery, SYS-Replication and IMAGEN adolescents, the G allele was associated with higher BMI (P = 8.9 × 10−11) and lower brain volume (P = 1.5 × 10−4, Table 3). These results suggest that FTO may impact inversely on BMI and brain volume.
Table 1.
Basic characteristics of studied participants
| Outcome | SYS-Discovery |
SYS-Replication |
IMAGEN |
|||
|---|---|---|---|---|---|---|
| Number | Mean ± SD | Number | Mean ± SD | Number | Mean ± SD | |
| Age (years) | 592 | 15.1 ± 1.9 | 416 | 14.4 ± 1.8 | 710 | 14.5 ± 0.4 |
| Sex (males/females) | 592 | 282/310 | 416 | 210/206 | 718 | 348/370 |
| Height (cm) | 590 | 163.3 ± 9.5 | 415 | 163.1 ± 9.5 | 663 | 168.4 ± 8.0 |
| Body weight (log kg) | 581 | 1.7 ± 0.1 | 412 | 1.8 ± 0.1 | 669 | 1.8 ± 0.1 |
| BMI (log kg/m2) | 582 | 1.3 ± 0.1 | 413 | 1.3 ± 0.1 | 658 | 1.3 ± 0.01 |
| Overweight/obesity (y/n) | 582 | 139/443 | 413 | 135/278 | 654 | 124/530 |
| TBF (log kg) | 559 | 1.0 ± 0.3 | 397 | 1.0 ± 0.3 | N/A | N/A |
| LBM (log kg) | 566 | 1.7 ± 0.1 | 389 | 1.7 ± 0.1 | N/A | N/A |
| Brain volume (cm3) | 562 | 1301 ± 119 | 395 | 1235 ± 112 | 712 | 1195 ± 113 |
| IC volume (cm3) | 529 | 1813.5 ± 156 | 355 | 1811.5 ± 151 | 653 | 1818.1 ± 156 |
| Ventricular volume (log cm3) | 558 | 1.0 ± 0.2 | 401 | 1.0 ± 0.2 | 708 | 1.1 ± 0.2 |
| MAF (rs99930333) | 594 | 0.31 | 422 | 0.36 | 717 | 0.34 |
Ventricular volume is defined as the sum of volumes of the left lateral, right lateral, third and fourth ventricles.
MAF, minor allele frequency.
Figure 1.
FTO associations with BMI and brain volume in the SYS-Discovery, SYS-Replication and IMAGEN samples. Associations between FTO (rs99930333) and the body and brain variables were tested with Merlin-1.1.2 under an additive model.
Table 2.
Associations between FTO (rs9930333) and body and brain outcomes
| Outcome | Effect size ± SE (G allele) |
P-value |
||||
|---|---|---|---|---|---|---|
| SYS-Discovery | SYS-Replication | IMAGEN | SYS-Discovery | SYS-Replication | IMAGEN | |
| BMI (log kg/m2) | 0.017 ± 0.004 | 0.017 ± 0.006 | 0.016 ± 0.004 | 1.1 × 10−4 | 0.005 | 1.4 × 10−5 |
| Body weight (log kg) | 0.016 ± 0.005 | 0.021 ± 0.007 | 0.013 ± 0.004 | 0.003 | 0.004 | 0.003 |
| TBF (log kg) | 0.052 ± 0.016 | 0.063 ± 0.022 | – | 0.002 | 0.004 | – |
| LBM (log kg) | 0.008 ± 0.004 | 0.008 ± 0.005 | – | 0.05 | 0.11 | – |
| Height (cm) | 0.001 ± 0.488 | 0.60 ± 0.53 | −0.001 ± 0.004 | 1.00 | 0.26 | 0.72 |
| Brain volume (cm3) | −16.8 ± 6.0 | −12.8 ± 6.6 | −9.8 ± 5.2 | 0.005 | 0.05 | 0.06 |
| IC volume (cm3) | −17.4 ± 8.0 | −3.8 ± 10.0 | −8.5 ± 7.4 | 0.03 | 0.70 | 0.25 |
| Ventricular volume (log cm3) | 0.002 ± 0.010 | 0.003 ± 0.01 | 0.006 ± 0.009 | 0.84 | 0.81 | 0.50 |
| Component 1 | 0.16 ± 0.09 | 0.15 ± 0.12 | 0.05 ± 0.07 | 0.10 | 0.21 | 0.51 |
| Component 2 | −0.29 ± 0.06 | −0.25 ± 0.07 | −0.15 ± 0.06 | 5.5 × 10−6 | 5.9 × 10−4 | 0.007 |
Associations were tested with Merlin-1.1.2. Ventricular volume is defined as the sum of volumes of the left lateral, right lateral, third and fourth ventricles. Components 1 and 2: principal components 1 and 2 from PCA of brain volume and BMI determinants (i.e. TBF, LBM and height).
SE, standard error.
Table 3.
Meta-analysis of FTO associations in the SYS (discovery and replication) and IMAGEN samples
| Outcome | Effect size ± SE (G allele) | P-value | HetISq | Heterogeneity P-value |
|---|---|---|---|---|
| BMI (log kg/m2) | −0.020 ± 0.003 | 8.9 × 10−11 | 0.0 | 0.98 |
| Body weight (log kg) | −0.015 ± 0.003 | 8.1 × 10−8 | 0.0 | 0.60 |
| Brain volume (cm3) | 12.8 ± 3.4 | 1.5 × 10−4 | 0.0 | 0.68 |
| IC volume (cm3) | 10.6 ± 4.8 | 0.03 | 0.0 | 0.53 |
| Component 2 | 0.22 ± 0.04 | 1.3 × 10−9 | 32.1 | 0.23 |
Meta-analysis was performed with METAL. Component 2: principal component 2 from PCA of brain volume and BMI determinants (i.e. TBF, LBM and height).
HetISq: I2 statistic that measures heterogeneity on scale of 0–100%. SE, standard error.
FTO associations with BMI determinants and intracranial (IC) and cerebrospinal volumes
Next, we tested whether the FTO association with BMI is mediated mainly through its association with body adiposity. In the SYS, the G allele was associated with higher total body fat (TBF, P = 0.002) and lean body mass (LBM, P = 0.03) and no difference in height (P = 0.99, Table 2). Similar results were observed in the replication samples. In SYS-Replication, the G allele was associated with higher TBF (P = 0.004), a trend towards higher LBM (P = 0.11) and no difference in height (P = 0.26, Table 2). In the IMAGEN sample, the G allele was associated with higher body weight (P = 0.003) and no difference in height (P = 0.72, Table 2). These results suggest that the FTO association with BMI is mediated mainly through its association with body adiposity (both SYS samples) and not with longitudinal growth (all three samples).
Furthermore, we tested whether the FTO association with brain volume is similar to that with intracranial (IC) volume (an index of cranium/head size) and ventricular volume. The G allele (associated with lower brain volume) was associated with lower IC volume in the SYS-Discovery (P = 0.03). Similar, but non-significant trends were observed in both replication samples (Table 2). In a meta-analysis involving all three samples, the G allele was associated with lower IC volume at P = 0.03 (Table 3). FTO was not associated with ventricular volume in any of the three samples (Table 2). These results suggest that FTO association with brain volume is likely mediated through processes influencing the growth of the brain and, to some degree, that of the cranium.
FTO association with shared variance between BMI determinants and brain volume
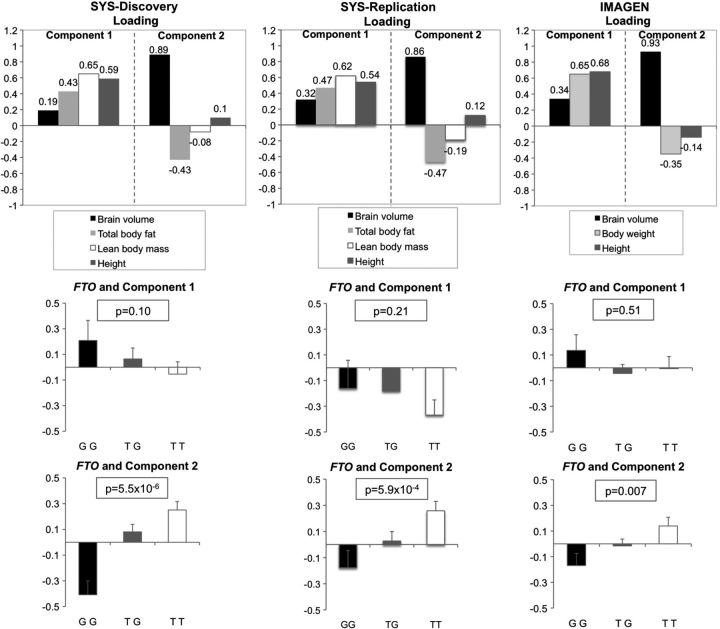
Because our results suggest that FTO may influence both body adiposity and brain size, we performed principal component analysis (PCA) to examine whether shared variance exists between body adiposity and brain volume that is independent of LBM and height. In the SYS sample, this analysis identified two significant components capturing a total of 75.6% of variance shared among brain volume, TBF, LBM and height. Component 1, which captured 50.6% of variance, was loaded positively by TBF, LBM and height, whereas Component 2, which captured 25.0% of variance, was loaded positively by brain volume and negatively by TBF (Fig. 2). Consistent with the opposite effects of the G allele on TBF and brain volume, this allele was strongly associated with Component 2 that captures the shared inverse variance between TBF and brain volume (P = 5.5 × 10−6), but not with Component 1 (P = 0.10; see Fig. 2). We obtained similar results in both replication samples (SYS-Replication: Component 1: P = 0.21, Component 2: P = 5.9 × 10−4; IMAGEN: Component 1: P = 0.51, Component 2: P = 0.007; Fig. 2). In a meta-analysis involving all three samples, FTO was associated with Component 2 at P = 1.3 × 10−9 (Table 3).
Figure 2.
FTO and shared variance between brain volume and BMI determinants in the SYS-Discovery, SYS-Replication and IMAGEN samples. (A) Two significant components (and their respective loadings) identified with PCA involving four variables (i.e. brain volume, TBF, LBM and height) in the SYS-Discovery and SYS-Replication samples and three variables (i.e. brain volume, body weight and height) in the IMAGEN sample are presented. Component 1 explained 50.6, 54.9 and 54.5% of variance in the SYS-Discovery, SYS-Replication and IMAGEN samples, respectively. Component 2 explained 25.0, 22.6 and 31.0% of variance in the SYS-Discovery, SYS-Replication and IMAGEN samples, respectively. (B) Association between FTO (rs9930333) and each of the two significant components were tested with Merlin-1.1.2.
In addition, we performed a conditional analysis in the discovery sample in which we tested for association between FTO and Component 2, now including TBF as a covariate. This analysis continued to show a significant association between FTO and Component 2 (effect size = −0.20, P = 5 × 10−4), confirming that this association is not driven entirely by TBF alone.
Meta-analysis of FTO co-expression networks during development
In the present study, FTO is associated with a shared (inverse) variance between body adiposity and total brain volume, suggesting that this gene may exert opposing effects on the growth of adipose and brain tissues. As pointed out in the Discussion section, we speculate that such effects may occur during early embryogenesis when a single factor may influence developmentally distant tissues. FTO being expressed during early embryogenesis (5,8,9) and encoding a nucleic acid demethylase (5–7) may modulate expression of such a factor. Based on the existing literature, we identified bone morphogenetic protein 4 (BMP4) as a possible candidate for this role. BMP4 is critical for embryonic development (19) and shows opposing effects on adipose and brain tissues (20,21) (Supplementary Material, Fig. S1). To explore the possibility that FTO may exert at least some opposing effects on body adiposity and brain volume via its interaction with BMP4 during embryogenesis, we performed a meta-analysis of FTO co-expression networks during development. We found that FTO and BMP4 exhibited the most significant positive or negative co-expressions (P < 0.01) in datasets annotated as ‘embryonic’, suggesting that the two genes may interact in embryonic tissues. Next, we searched for genes whose correlation strength with FTO was similar to that of FTO with BMP4. Ranking such ‘FTO co-expressed’ genes in this way produced a list that was enriched for functions related to the nervous system (e.g. synaptic transmission; corrected P < 0.01). The 165 top-ranking genes (r > 0.25, P < 0.01) showed significant modularity (P < 0.01) in a network constructed from the brain-derived expression data, but not in a network constructed from the non-brain-derived expression data (size and platform matched). We noted that FTO exhibited particularly strong interaction with the protocadherin α-gene cluster (Supplementary Material, Fig. S2) that is involved in the regulation of neural circuit assembly and neuronal survival (22,23).
FTO association with body fat, brain volume and shared variance between body fat and brain volume: potentially confounding factors
All statistical analyses presented above were carried out while adjusting for age and sex. But, several other potentially confounding factors might influence adiposity and brain development. In all discovery and replication samples, the participants were high-school students—therefore, the level of education did not vary among them. Data on nutrition, exercise, maternal education and family income were collected only in the SYS. Additional adjusting for these potentially confounding factors did not change the FTO associations with TBF, brain volume and the shared variance between the two variables captured by Component 2 (Supplementary Material, Table S2).
DISCUSSION
The results of the present study show that FTO is associated with shared inverse variance between body adiposity and total brain volume, suggesting that this gene may exert inverse effects on adipose and brain tissues. Given the completion of the overall brain growth in early childhood, it is plausible that these effects occur during development.
Early embryogenesis is a period of human development when such opposing effects of FTO on adipose and brain tissues may take place. As described below, a single factor may influence developmentally distant tissues, such as adipose tissue (derived from mesoderm) and brain tissue (derived from ectoderm), and this influence may be of the opposite direction. FTO, as a gene encoding nucleic acid demethylase (5–7) and expressed during early embryogenesis (5,8,9), may act as a modulator of such a factor. For example, BMP4 is a protein involved in the regulation of cell proliferation and differentiation during embryonic development [reviewed in (19)], and its inhibition in ectoderm is critical for neural induction and the development of the brain (20), whereas its inhibition in mesoderm may attenuate the development of adipose tissue (21). We speculate that a moderate increase in BMP4 expression, because of greater demethylating activity of FTO, would lead to a decrease in allocating stem cells towards brain cell lineages and an increase in allocating stem cells towards adipose tissue lineages (Supplementary Material, Fig. S1). Such relative differences in stem cell allocation, and thus life-long potential for growth of the respective tissues, would then result in a lower brain volume and a higher body fat mass postnatally. The former would be realized during major periods of brain development (prenatal and early postnatal period), whereas the latter would become apparent gradually during postnatal life as a result of a chronic positive energy balance (life-long risk for obesity) (24,25). Our meta-analysis of co-expression networks provided some support for (i) the possible existence of such FTO–BMP4 interactions during embryogenesis and (ii) the possibility that these interactions are involved in the processes of brain development. It also suggested that the latter might include the protocadherin α-gene cluster that is critical for the regulation of neural circuit assembly and neuronal survival (22,23). Further experimental studies are required to confirm these findings.
Consistent with the above hypothesis, previous research in experimental animals and humans suggests that FTO effects observed in childhood and adulthood may stem from its effects during early development. Fto deletion in mice results in growth retardation present already at birth and a significant reduction in fat mass and, less so, in LBM (26,27). Similarly, a loss-of-function mutation in the human FTO results in severe growth retardation and development of multiple malformations, including those affecting the brain (8). In addition, duplication of a chromosomal region, including FTO in humans leads to obesity and mental retardation (28).
In the present study, FTO was associated with brain and cranium volumes, but not with height (an index of axial growth). These results are consistent with the fact that (i) the brain and cranium versus axial skeleton exhibit different developmental growth patterns: while most of the brain and cranium growth is completed during the first few years of life, the axial skeleton shows major growth during adolescence; and (ii) most of the cranium, but not the axial skeleton is of the same ectodermal origin as the brain (29). In agreement with this closer developmental relationship of the cranium with the brain than of the cranium with the axial skeleton, we show here that IC volume correlates much closer with brain volume (r2 = 0.75) than it does with height (r2 = 0.12), and note also that height is only weakly associated with brain volume (r2 = 0.04; Supplementary Material, Fig. S3). Taken together, there are likely a number of factors that influence the growth and development of the brain and cranium, but not that of the axial skeleton (e.g. FTO), and vice versa. Also supporting our results, it has been shown previously that FTO is not associated with height, despite being strongly associated with BMI (24). Whether FTO impacts primarily growth of the skull and only secondarily growth of the brain is also a possibility that requires further developmental studies.
In summary, our results suggest that FTO may exert inverse effects on adipose and brain tissues. Given the fact that these effects are detected already in adolescence, they may occur during early development.
MATERIALS AND METHODS
Adolescent samples
Discovery sample (SYS Discovery) and Replication sample 1 (SYS Replication)
Discovery sample (SYS-Discovery) included 598 SYS adolescents recruited between November 2003 and June 2009, and Replication sample 1 (SYS-Replication) included 413 SYS adolescents recruited between September 2009 and February 2012. Both SYS samples were white Caucasian adolescents (aged 12–18 years) drawn from the French Canadian genetic founder population living in the Saguenay-Lac St. Jean region of Quebec, Canada (30). The SYS is a population-based, cross-sectional study of cardiovascular, metabolic and mental health in adolescence, and recruitment and selection criteria have been described previously (30). Written consent of the parents and assent of the adolescents were obtained before the commencement of data collection. The Research Ethics Committee of the Chicoutimi Hospital approved the study protocol.
Replication sample 2 (IMAGEN)
Replication sample 2 was the IMAGEN Study involving white Caucasian adolescents (n = 718; aged 13–15 years) of mixed European background (German, English, Irish and French). The IMAGEN Study is a European multicentre study on impulsivity, reinforcement sensitivity and emotional reactivity in adolescents. The IMAGEN sample comprises 718 healthy adolescents recruited between 2008 and 2010 in local schools from 8 participating sites in Germany (Berlin, Dresden, Hamburg and Manheim), UK (London and Nottingham), France (Paris) and Ireland (Dublin). All participants and their parents provided informed written assent and consent, respectively. The local ethics committees approved the study protocol (31).
Basic characteristics of all three samples are provided in Table 1. Adolescents were defined as obese or overweight, if their BMI was ≥85th age- and sex-specific percentile of the Centers for Disease Control and Prevention BMI curves (http://www.cdc.gov/growthcharts). The prevalence of overweight or obesity was 23.9, 32.7 and 19.0% in the SYS-Discovery, SYS-Replication and IMAGEN sample (Table 1), which is similar to that in the Canadian (2004 Canadian Community Health Survey) and European adolescent populations (17,18).
Phenotyping
Anthropometry and bioimpedance
In the SYS-Discovery, SYS-Replication and IMAGEN samples, weight (0.1 kg precision) and height (1 mm precision) were measured, and the BMI was calculated as weight in kilograms divided by squared height in metres. In both SYS samples, but not in the IMAGEN sample, TBF and LBM were assessed using multi-frequency bioimpedance analysis (Xitron Technologies, Inc., San Diego, CA, USA).
MRI of the brain
In the SYS-Discovery and SYS-Replication samples, structural magnetic resonance (MR) images of the brain were collected on a Phillips 1.0-T superconducting magnet. High-resolution anatomical T1-weighted images were acquired using the following parameters: 3D radiofrequency-spoiled gradient echo scan with 140–160 sagittal slices, 1-mm isotropic voxel size, time repetition (TR) = 25 ms, echo time (TE) = 5 ms and flip angle = 30°. In the IMAGEN study, structural MR images were obtained using 3.0-T scanners (Siemens, Philips and General Electric). The details of the entire MR protocol are described elsewhere (31). High-resolution T1-weighted anatomical images were acquired using a 3D Magnetization Prepared Rapid Acquisition Gradient Echo sequence with 160 or 170 sagittal slices (depending on the manufacturer), 1-mm isotropic voxel size, TR = 2300 ms, TE = 2.8 ms, inversion time (TI) = 900 ms and flip angle = 9°. In all three samples, total brain and ventricular volumes were derived using FreeSurfer (32), and IC volumes were estimated using the whole cranium/face model as described previously (33). For the latter, brains were removed from all MR images using the brain extraction tool algorithm, and all individual images were normalized in the same space to the average linear dimensions of the entire group. The first population average was then created via voxel-by-voxel averaging. A hierarchical iterative model-building step was then initiated, where each image was non-linearly registered to the populating average and subsequently to the model created in the previous non-linear registration step. At each iteration, finer resolution non-linear transformations were estimated. One of the outputs at the end of the model-building process is a non-linear transformation that maps each individual to the average of the group. All transformations were estimated using the minc suite of software tools (http://www.bic.mni.mcgill.ca/ServicesSoftware/HomePage) (34,35). The IC space was manually defined on the resulting population model by one of the authors (M.M.C.) and then transformed to fit each individual using the inverse transformation previously described (34,35). An example of IC mask is shown in the Supplementary Material, Fig. S4.
FTO genotyping
Both the SYS-Discovery and IMAGEN samples were genotyped with the Illumina Human610-Quad BeadChip (Illumina, San Diego, CA, USA). The genotyping was conducted at the Centre National de Génotypage (Paris, France). The SYS-Replication sample was genotyped with the HumanOmniExpress BeadChip (Illumina). The genotyping was conducted at the Genome Analysis Centre of Helmholtz Zentrum München (Munich, Germany). In all three samples, single nucleotide polymorphisms (SNPs) with call rate <95% and minor allele frequency <0.01 and SNPs that were not in Hardy–Weinberg equilibrium (P < 1 × 10−4) were excluded. A variant of FTO that showed the most significant association with BMI in our previously reported genome-wide association study (i.e. rs9930333) (16) was then tested for association with other outcomes in the SYS-Discovery sample and with all outcomes in the replication samples (SYS-Replication and IMAGEN). This variant was in linkage disequilibrium and showed the same direction of effect on BMI, as the previously reported FTO SNPs (1,2,36).
Statistical methods
Genotype–phenotype association tests
Genotype–phenotype association tests were conducted with Merlin (version 1.1.2) under an additive model (37,38). With Merlin, a simple regression model is fitted to each studied outcome, and a variance component approach is used to account for correlation between different observed phenotypes within each family (if required). For all primary and derived traits, values outside mean ± 3 SDs were excluded. Variables were log transformed, if they did not follow normal distribution. Age and sex were included as potential confounders for all variables, except for principal components that were already adjusted for age and sex prior to PCA. To combine the evidence for association from the SYS-Discovery, SYS-Replication and IMAGEN samples, we used METAL (39), which converts the direction of effect and P-value observed in each study into a signed Z-score combined across samples in a weighted sum, with weights proportional to the square root of the sample size for each study (39).
Principal component analysis
PCA was used to identify components of shared variance between individual BMI determinants and brain volume. In the SYS-Discovery and SYS-Replication samples, the BMI determinants were TBF, LBM and height. In the IMAGEN sample, only body weight and height were available. PCA is a multivariate statistical technique used to extract shared variance from correlated data (40). It transforms a number of possibly correlated variables into a number of uncorrelated variables, the so-called principal components. Each principal component represents a different linear combination of the original correlated variables. The original variables are first normalized to their respective means and then used to generate a correlation matrix. PCA is then performed by eigenvalue decomposition of the correlation matrix. Loadings of significant principal components are described in terms of eigenvectors. The significant principal components were tested for association with the studied FTO SNP (rs9930333) as described above.
Meta-analysis of FTO co-expression networks
To identify genes frequently co-expressed with FTO, we assembled 721 diverse, publicly available expression datasets containing a total of 34 019 microarrays (http://www.chibi.ubc.ca/Gemma). Using these data, we conducted co-expression analysis as previously described (41). In brief, correlations between FTO-expression profile and expression profiles of 14 184 genes (identified as all genes present in at least 500 datasets) were determined for each dataset. These correlations were then ranked to give a score of similarity between FTO-expression profile and expression profiles of all other genes in each dataset. Next, each dataset was annotated from a set of 128 terms (describing factors such as the tissue, treatment, developmental stage and/or disease); an annotation was given, if the term was present in at least 5 (and up to 25) datasets. Finally, analysis of gene function enrichment was applied to test for enrichment across dataset annotations (42). Further details on co-expression methods and results are available in Supplementary Information and at http://www.chibi.ubc.ca/FTO.
SUPPLEMENTARY MATERIAL
FUNDING
P.P. was supported by NIH Grant GM076990 and salary awards from the Michael Smith Foundation for Health Research and the Canadian Institutes for Health Research. J.G. was supported by postdoctoral fellowships from the Canadian Institutes for Health Research, Michael Smith Foundation for Health Research and the MIND Foundation of British Columbia. IMAGEN received research funding from the European Community's Sixth Framework Programme (LSHM-CT-2007-037286).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the following individuals for their contributions in acquiring data in the SYS: Jacynthe Tremblay and her team of research nurses (Saguenay Hospital), Helene Simard and her team of research assistants (Cégep de Jonquière), Rosanne Aleong (program manager, Rotman Research Institute) and Nick Qiu (MR image processing, Rotman Research Institute). The Canadian Institutes of Health Research (Z.P., T.P.), Heart and Stroke Foundation of Quebec (Z.P.) and the Canadian Foundation for Innovation (Z.P.) supported the SYS.
Conflict of Interest statement. M.A. is a James McGill Professor of Biostatistics at McGill University. T.P. is the Tanenbaum Chair in Population Neuroscience at the Rotman Research Institute, University of Toronto. This paper reflects only the authors' views and the Community is not liable for any use that may be made of the information contained therein.
REFERENCES
- 1.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Allen H.L., Lindgren C.M., Luan J., Magi R., et al. Association analyses of 249 796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fawcett K.A., Barroso I. The genetics of obesity: FTO leads the way. Trends Genet. 2010;26:266–274. doi: 10.1016/j.tig.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larder R., Cheung M.K., Tung Y.C., Yeo G.S., Coll A.P. Where to go with FTO? Trends Endocrinol. Metab. 2011;22:53–59. doi: 10.1016/j.tem.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Gerken T., Girard C.A., Tung Y.C., Webby C.J., Saudek V., Hewitson K.S., Yeo G.S., McDonough M.A., Cunliffe S., McNeill L.A., et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia G., Yang C.G., Yang S., Jian X., Yi C., Zhou Z., He C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han Z., Niu T., Chang J., Lei X., Zhao M., Wang Q., Cheng W., Wang J., Feng Y., Chai J. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464:1205–1209. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- 8.Boissel S., Reish O., Proulx K., Kawagoe-Takaki H., Sedgwick B., Yeo G.S., Meyre D., Golzio C., Molinari F., Kadhom N., et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 2009;85:106–111. doi: 10.1016/j.ajhg.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stratigopoulos G., Padilla S.L., LeDuc C.A., Watson E., Hattersley A.T., McCarthy M.I., Zeltser L.M., Chung W.K., Leibel R.L. Regulation of Fto/Ftm gene expression in mice and humans. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;294:R1185–R1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gustafson D., Rothenberg E., Blennow K., Steen B., Skoog I. An 18-year follow-up of overweight and risk of Alzheimer disease. Arch. Intern. Med. 2003;163:1524–1528. doi: 10.1001/archinte.163.13.1524. [DOI] [PubMed] [Google Scholar]
- 11.Gustafson D., Lissner L., Bengtsson C., Bjorkelund C., Skoog I. A 24-year follow-up of body mass index and cerebral atrophy. Neurology. 2004;63:1876–1881. doi: 10.1212/01.wnl.0000141850.47773.5f. [DOI] [PubMed] [Google Scholar]
- 12.Debette S., Beiser A., Hoffmann U., Decarli C., O'Donnell C.J., Massaro J.M., Au R., Himali J.J., Wolf P.A., Fox C.S., et al. Visceral fat is associated with lower brain volume in healthy middle-aged adults. Ann. Neurol. 2010;68:136–144. doi: 10.1002/ana.22062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho A.J., Stein J.L., Hua X., Lee S., Hibar D.P., Leow A.D., Dinov I.D., Toga A.W., Saykin A.J., Shen L., et al. A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc. Natl. Acad. Sci. USA. 2010;107:8404–8409. doi: 10.1073/pnas.0910878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knickmeyer R.C., Gouttard S., Kang C., Evans D., Wilber K., Smith J.K., Hamer R.M., Lin W., Gerig G., Gilmore J.H. A structural MRI study of human brain development from birth to 2 years. J. Neurosci. 2008;28:12176–12182. doi: 10.1523/JNEUROSCI.3479-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenroot R.K., Giedd J.N. Brain development in children and adolescents: insights from anatomical magnetic resonance imaging. Neurosci. Biobehav. Rev. 2006;30:718–729. doi: 10.1016/j.neubiorev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Melka M.G., Bernard M., Mahboubi A., Abrahamowicz M., Paterson A.D., Syme C., Lourdusamy A., Schumann G., Leonard G.T., Perron M., et al. Genome-wide scan for loci of adolescent obesity and their relationship with blood pressure. J. Clin. Endocrinol. Metab. 2012;97:E145–E150. doi: 10.1210/jc.2011-1801. [DOI] [PubMed] [Google Scholar]
- 17.Shields M. Overweight and obesity among children and youth. Health Rep. 2006;17:27–42. [PubMed] [Google Scholar]
- 18.Lobstein T., Frelut M.L. Prevalence of overweight among children in Europe. Obes. Rev. 2003;4:195–200. doi: 10.1046/j.1467-789x.2003.00116.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen D., Zhao M., Mundy G.R. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 20.Finley M.F., Devata S., Huettner J.E. BMP-4 inhibits neural differentiation of murine embryonic stem cells. J. Neurobiol. 1999;40:271–287. [PubMed] [Google Scholar]
- 21.Bowers R.R., Kim J.W., Otto T.C., Lane M.D. Stable stem cell commitment to the adipocyte lineage by inhibition of DNA methylation: role of the BMP-4 gene. Proc. Natl. Acad. Sci. USA. 2006;103:13022–13027. doi: 10.1073/pnas.0605789103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zipursky S.L., Sanes J.R. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell. 2010;143:343–353. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Kehayova P., Monahan K., Chen W., Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc. Natl. Acad. Sci. USA. 2011;108:17195–17200. doi: 10.1073/pnas.1114357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardy R., Wills A.K., Wong A., Elks C.E., Wareham N.J., Loos R.J., Kuh D., Ong K.K. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum. Mol. Genet. 2010;19:545–552. doi: 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sovio U., Mook-Kanamori D.O., Warrington N.M., Lawrence R., Briollais L., Palmer C.N., Cecil J., Sandling J.K., Syvanen A.C., Kaakinen M., et al. Association between common variation at the FTO locus and changes in body mass index from infancy to late childhood: the complex nature of genetic association through growth and development. PLoS Genet. 2011;7:e1001307. doi: 10.1371/journal.pgen.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer J., Koch L., Emmerling C., Vierkotten J., Peters T., Bruning J.C., Ruther U. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 27.Gao X., Shin Y.H., Li M., Wang F., Tong Q., Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One. 2010;5:e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stratakis C.A., Lafferty A., Taymans S.E., Gafni R.I., Meck J.M., Blancato J. Anisomastia associated with interstitial duplication of chromosome 16, mental retardation, obesity, dysmorphic facies, and digital anomalies: molecular mapping of a new syndrome by fluorescent in situ hybridization and microsatellites to 16q13 (D16S419-D16S503) J. Clin. Endocrinol. Metab. 2000;85:3396–3401. doi: 10.1210/jcem.85.9.6776. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman D.E. The Evolution of the Human Head. Cambridge, MA: Harward University Press; 2011. pp. 56–95. [Google Scholar]
- 30.Pausova Z., Paus T., Abrahamowicz M., Almerigi J., Arbour N., Bernard M., Gaudet D., Hanzalek P., Hamet P., Evans A.C., et al. Genes, maternal smoking, and the offspring brain and body during adolescence: design of the Saguenay Youth Study. Hum. Brain Mapp. 2007;28:502–518. doi: 10.1002/hbm.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schumann G., Loth E., Banaschewski T., Barbot A., Barker G., Buchel C., Conrod P.J., Dalley J.W., Flor H., Gallinat J., et al. The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol. Psychiatry. 2010;15:1128–1139. doi: 10.1038/mp.2010.4. [DOI] [PubMed] [Google Scholar]
- 32.Dale A.M., Fischl B., Sereno M.I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 33.Chakravarty M.M., Aleong R., Leonard G., Perron M., Pike G.B., Richer L., Veillette S., Pausova Z., Paus T. Automated analysis of craniofacial morphology using magnetic resonance images. PLoS One. 2011;6:e20241. doi: 10.1371/journal.pone.0020241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins D.L., Holmes C.J., Peters T.M., Evans A.C. Automatic 3-D model-based neuroanatomical segmentation. Hum. Brain Mapp. 1995;3:190–208. [Google Scholar]
- 35.Collins D.L., Evans A.C. ANIMAL: validation and applications of non-linear registration-based segmentation. Int. J. Pattern Recogn. 1997;11:1271–1294. [Google Scholar]
- 36.Chanock S.J., Manolio T., Boehnke M., Boerwinkle E., Hunter D.J., Thomas G., Hirschhorn J.N., Abecasis G., Altshuler D., Bailey-Wilson J.E., et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 37.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 38.Abecasis G.R., Wigginton J.E. Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am. J. Hum. Genet. 2005;77:754–767. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jolliffe I.T. Principal Component Analysis. New York: Springer; 2002. [Google Scholar]
- 41.Gillis J., Pavlidis P. The role of indirect connections in gene networks in predicting function. Bioinformatics. 2011;27:1860–1866. doi: 10.1093/bioinformatics/btr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gillis J., Mistry M., Pavlidis P. Gene function analysis in complex data sets using ErmineJ. Nat. Protoc. 2010;5:1148–1159. doi: 10.1038/nprot.2010.78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.