Abstract
Aedes aegypti is a major vector of arthropod-borne viruses such as yellow fever virus and dengue viruses. Efforts to discern the function of genes involved in important behaviors such as vector competence and host seeking through reverse genetics would greatly benefit from the ability to generate targeted gene disruptions. Homing endonucleases are selfish elements which catalyze double-stranded DNA (dsDNA) breaks in a sequence-specific manner. In this report we demonstrate that the homing endonucleases I-PpoI, I-SceI, I-CreI and I-AniI are all able to induce dsDNA breaks in adult female Ae. aegypti chromosomes as well as catalyze the somatic excision of a transgene. These experiments provide evidence that homing endonucleases can be used to manipulate the genome of this important disease vector.
Keywords: Aedes aegypti, Homing endonuclease, Sindbis virus, gap repair
Introduction
Aedes aegypti is the primary vector responsible for the transmission of viruses which cause dengue fever, dengue hemorrhagic fever, and yellow fever, with approximately half of the world's population at risk from infection (Halstead 2007). Understanding the genetic basis for important phenotypes such as vector competence, bloodfeeding, and host seeking is a critical priority, and will likely provide insight into novel control strategies for this pest species.
Ae. aegypti has long been the subject of genetic research (Craig and Hickey 1967). In addition to a high-coverage genome sequence (Nene et al. 2007), a number of tools are available for genetic studies in this organism. These include transient expression systems such as recombinant double-subgenomic Sindbis viruses (dsSINV) (Hahn et al. 1992; Higgs et al. 1996), classical transposon-based transformation using Mos1 (Coates et al. 1998), Hermes (Jasinskiene et al. 1998) or piggyBac (Kokoza et al. 2001) transposable elements, site-specific transgene excision using cre recombinase (Jasinskiene et al. 2003), and site-specific integration using phiC31 integrase (Nimmo et al. 2006). The insertion of transgenes in the genome via transposons is a random process, and while cre recombinase and phiC31 integrase catalyze site-specific events, both of these rely on the prior insertion of docking sites via random integration. Thus there is an urgent need for tools which will promote or aid in site-specific gene inactivation or homologous recombination in this mosquito. This need is compounded by the lack of other genetic tools such as a detailed physical map and balancer chromosomes, which would facilitate the recovery of loss-of-function mutants following random mutagenesis.
Meganucleases are site-specific double-stranded DNA (dsDNA) endonucleases whose recognition sequences are rare in, or absent from, large eukaryotic genomes. Homing endonucleases (HEs) are naturally occurring meganucleases which recognize target sequences which can range from 14-40 bp (Belfort and Roberts 1997; Kowalski and Derbyshire 2002). The ability of HEs to recognize and cleave rare DNA sequences has lent them to a variety of uses in genome manipulation [reviewed in (Jasin 1996)]. Recombinant HEs have been used to study homologous recombination in Drosophila (Gong and Golic 2003; Rong and Golic 2000; Rong and Golic 2001), human cells (Saleh-Gohari and Helleday 2004) and plant cells (Gisler et al. 2002); double-stranded break repair in Drosophila (Bellaiche et al. 1999; Rong and Golic 2003) and mammalian cultured cells (Guirouilh-Barbat et al. 2004; Monnat et al. 1999); chromosomal rearrangements in Drosophila (Egli et al. 2004), and to insert transgenes into fish (Thermes et al. 2002). Combined with the ability to re-engineer HEs to recognize novel target sites (Arnould et al. 2006; Arnould et al. 2007; Chames et al. 2005; Rosen et al. 2006; Smith et al. 2006), this class of molecules represents a powerful tool for triggering targeted gene disruptions or homologous recombination without the need for random processes such as with transposable elements.
Due to the selfish manner of their propagation and maintenance in nature, homing endonuclease genes have been proposed as a mechanism to drive desirable phenotypes into vector populations (Burt 2003; Deredec et al. 2008; Sinkins and Gould 2006). Homing endonuclease genes could be used to disrupt a gene or genes necessary for pathogen transmission or to trigger homologous recombination and gene conversion in order to increase the frequency of an introduced anti-pathogen gene (Deredec et al. 2008; Sinkins and Gould 2006). Most recently, HEs have been proposed as a method of genetic sterilization or sex-ratio distortion in Anopheles gambiae (Windbichler et al. 2007; Windbichler et al. 2008). Using the HE gene I-PpoI, which recognizes a conserved sequence present in the 28S rDNA repeat region, Windbichler et al. (2008) have shown that I-PpoI induces complete embryonic lethality when expressed in the male germline. While the HE genes I-SceI and I-PpoI have now been used successfully in An. gambiae (Windbichler et al. 2007), there are as yet no reports describing the use of HEs in Culicine mosquitoes.
We sought to determine whether homing endonucleases are capable of recognizing and catalyzing double-stranded DNA breaks at their specific target sites in Ae. aegypti. We used dsSINV expression systems to express homing endonucleases in Ae. aegypti, due to their ability to achieve robust expression of exogenous gene products and rapidly infect most tissues of adult mosquitoes in a non-cytopathic fashion (Higgs et al. 1997). We found that the homing endonucleases I-PpoI, I-SceI, I-CreI and I-AniI are all capable of generating dsDNA breaks in Ae. aegypti chromosomes, and that this could result in the excision of chromosomal segments. Repair of homing-endonuclease-induced dsDNA breaks was associated with deletions of various sizes, indicating that these molecules could be used for targeted gene disruptions.
Results
Expression of homing endonucleases in mosquito cells using Sindbis virus expression systems
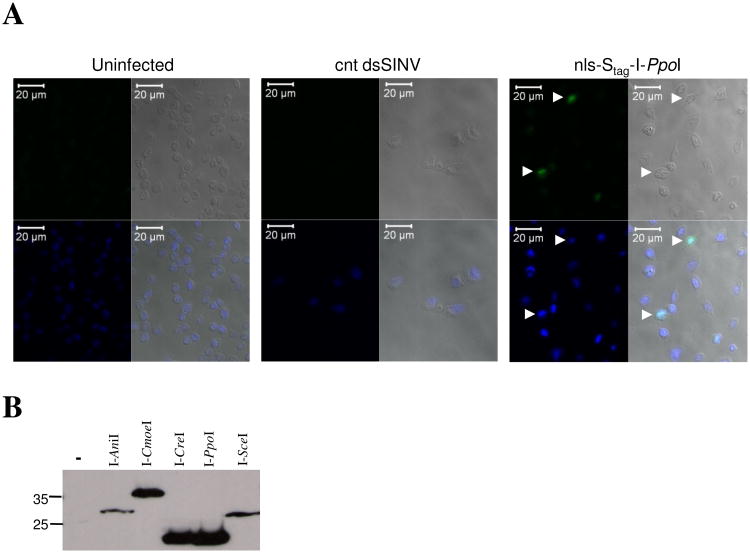
To ensure proper tracking and to simplify detection of each recombinant homing endonuclease (rHE), we inserted an in-frame nuclear localization signal (nls) and epitope tag (Stag) at the N-terminus of each homing endonuclease ORF (Fig. 1). For simplicity, we refer to these fusion proteins as nls-Stag-rHE, and the recombinant dsSIN viruses which express each of these as nls-Stag-rHE viruses. Following the successful rescue of each nls-Stag-rHE virus, we sought to validate that our recombinant homing endonuclease fusion proteins were indeed being expressed and translocated into mosquito cell nuclei. Following infection of C6/36 cells (Ae. albopictus) with each nls-Stag-rHE virus, we performed an immunofluorescence assay (IFA) using a primary antibody recognizing the Stag epitope. Figure 2A shows a typical IFA result following infection of C6/36 cells with nls-Stag-I-PpoI virus or a control dsSINV. While no FITC-fluorescence was observed in uninfected cells or cells infected with the control dsSIN virus, nls-Stag-I-PpoI protein was consistently observed in nuclei of nls-Stag-I-PpoI-infected cells, as shown by co-localization with DAPI (Fig. 2A, white arrows). Similar results were obtained with nls-Stag-I-AniI, nls-Stag-I-CreI, nls-Stag-I-CmoeI and nls-Stag-I-SceI viruses (data not shown). To verify that nls-Stag-rHE viruses were producing a single Stag-fused protein of the expected molecular weight, we performed western analysis on total cell protein extracts from nls-Stag-rHE-infected C6/36 cells at 24 hours post-infection (Fig. 2B). As expected, a single band was observed for each nls-Stag-rHE protein. We conclude from these experiments that our recombinant nls-Stag-rHE viruses are suitable for experiments involving the transient expression of homing endonucleases in whole mosquitoes.
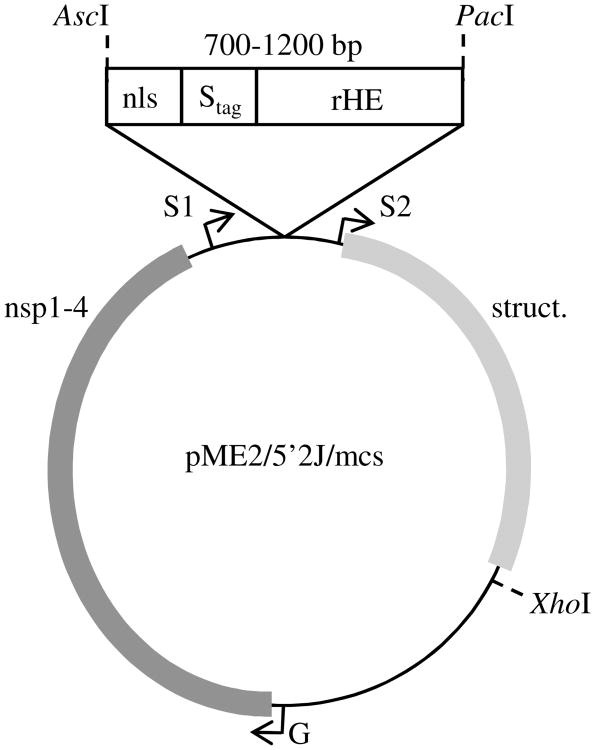
Figure 1. Construction of DNA plasmids encoding recombinant nls-Stag-rHE SIN viruses.
As described in Experimental Procedures, an AscI/PacI fragment containing the SV40 nuclear localization signal (MPKKKRKV), Stag, and rHE as a single coding region was inserted into a modified pME2/5′2J (Pierro et al. 2003) downstream of the duplicated subgenomic promoter (S1). In this construct, the genomic promoter (G) drives the production of full-length viral genomes and replication machinery (nsp1-4), and the second subgenomic promoter (S2) drives the expression of the viral structural genes (struct.).
Figure 2. Localization and expression of rHEs in C6/36 mosquito cells.
(A) nls-Stag-I-PpoI protein expressed from a dsSINV localizes to mosquito cell nuclei (white arrows). C6/36 cells (uninfected) or infected with nls-Stag-I-PpoI virus or a control (cnt) dsSINV were subject to IFA. For each group, four panels are shown: FITC (upper left); white light (upper right); DAPI (lower left); merged (lower right). (B) Western analysis of rHE expression in C6/36 cells infected with five nls-Stag-rHE viruses. Molecular weight markers are indicated to the left (kD).
Homing endonuclease somatic assay with single target site
A search of the Ae. aegypti genome using the canonical recognition sites of I-AniI, I-CreI, I-CmoeI, I-SceI and I-PpoI revealed that only I-PpoI had perfect matches (in the 28S rDNA repeats). However, as homing endonucleases tolerate degeneracy in their recognition sites, it is impossible to predict in advance whether any cryptic sites might be recognized. Thus, while endogenous target sites capable of being recognized by various homing endonuclease genes may or may not be present in the Ae. aegypti genome, we sought to simplify our analysis by introducing perfect recognition sequences for the five homing endonuclease genes under investigation into the Ae. aegypti genome via transposable element transformation. A Mos1 vector containing the recognition sites for I-PpoI, I-SceI, I-CreI, I-AniI and I-CmoeI downstream of the 3xP3-DsRED marker gene was inserted into the Ae. aegypti genome as previously described (Adelman et al. 2008; Coates et al. 1998). Two transgenic lines were obtained, and Southern analysis confirmed that these insertions were associated with single integration events (data not shown). One of these strains, referred to as UUGFP#18, was selected for use in subsequent experiments. The UUGFP#18 transgenic strain also expresses EGFP under the control of a novel promoter and this strain will be described in more detail elsewhere. To determine the capability of each recombinant homing endonuclease to recognize and catalyze site-specific dsDNA breaks, female UUGFP#18 mosquitoes were first intrathoracically inoculated with each nls-Stag-rHE virus. Following an incubation period, we performed Southern analysis on genomic DNA isolated from nls-Stag-rHE virus-infected mosquitoes. Despite several attempts, we were unable to identify any evidence of unrepaired dsDNA breaks using this method (data not shown). We reasoned that if dsDNA breaks are rapidly repaired in Ae. aegypti, then direct detection might not be possible. Therefore we shifted our approach to look for evidence of imperfect gap repair, which would be expected to occur in a subset of any homing endonuclease-induced dsDNA breaks as a result of mistakes made during the non-homologous end-joining process (NHEJ) [for a review of the NHEJ process, see (Mahaney et al. 2009)].
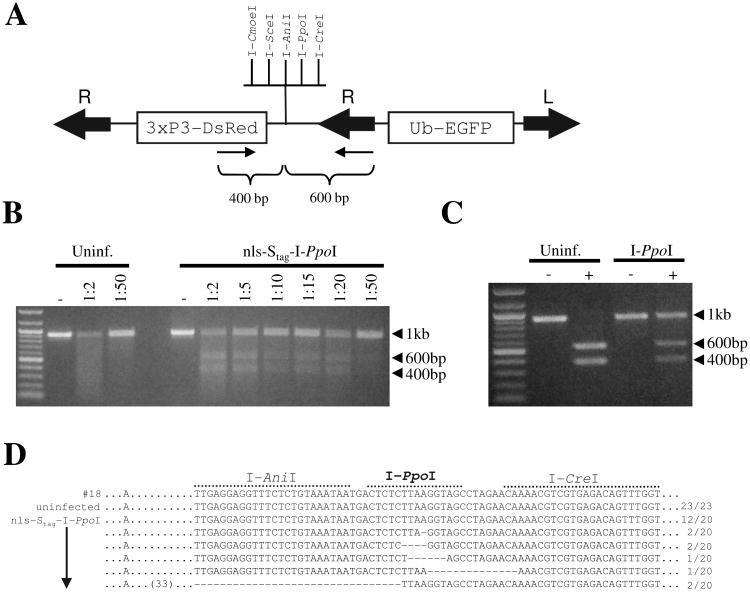
Mismatch specific DNA endonucleases are commonly found in plants (Yang et al. 2000), and the enzymes CEL I and CEL II, isolated from celery, have been used to identify mutations and mismatch repair following treatment with site-specific nucleases (Maeder et al. 2008; Santiago et al. 2008). We utilized a CEL II-based Surveyor Nuclease assay (Transgenomic, Omaha, NE) to detect evidence of mismatched bases as a result of imperfect repair at the cluster of exogenously introduced homing endonuclease recognition sites. At 10 days following injection with nls-Stag-I-PpoI virus, PCR amplicons were generated from genomic DNA from UUGFP#18 mosquitoes using primers which recognize the inserted transgene and flank the cluster of homing endonuclease sites (Fig. 3A). Amplicons were melted, reannealed, and subjected to digestion with various dilutions of Surveyor Nuclease (Fig. 3B). If I-PpoI-induced dsDNA breaks had occurred and were followed by imperfect gap repair at the I-PpoI recognition site, the melted/reannealed amplicon should contain mismatches, resulting in cleavage into approx. 400 and 600 bp fragments. This was indeed the case, as for all dilutions of Surveyor Nuclease tested we observed partial digestion of the initial 1 kb amplicon following infection with nls-Stag-I-PpoI virus (Fig. 3B). Surveyor Nuclease was unable to digest amplicons from uninfected mosquitoes (Fig. 3B), or amplicons from mosquitoes infected with a control dsSINV (data not shown). Alternatively, we digested the 1 kb amplicon obtained from nls-Stag-I-PpoI virus-infected mosquitoes with a commercial preparation of I-PpoI (Promega). Imperfect repair of the I-PpoI site in the mosquito would render the amplicon resistant to re-digestion. Consistent with the Surveyor Nuclease assay, approximately half of the amplicon DNA generated from nls-Stag-I-PpoI-infected mosquitoes was resistant to re-digestion with I-PpoI (Fig. 3C). To confirm that these results were due to imperfect repair at the I-PpoI recognition site, 1 kb amplicons from uninfected UUGFP#18 or nls-Stag-I-PpoI virus-infected mosquitoes were cloned and sequenced. Sequence results from all clones (23/23) obtained from uninfected mosquitoes revealed no alteration at the exogenous I-PpoI site or neighboring region (Fig. 3D). In contrast, 8 out of 20 clones (40%) obtained from nls-Stag-I-PpoI virus-infected mosquitoes contained small deletions in the I-PpoI recognition site. In 5 out of the 8 clones, these deletions were observed exclusively at the I-PpoI recognition site, while in the remaining 3 clones, deletions also overlapped some of the neighboring rHE sites (Fig. 3D). I-PpoI-induced deletions ranged from 1 to 65 bp, with a median deletion size between 4 and 6 bp. No base changes were observed in any other region of the amplicon. In total, these results confirm that the meganuclease I-PpoI is able to induce dsDNA breaks in Ae. aegypti.
Figure 3. Somatic footprint assay to detect imperfect gap repair at a single I-PpoI recognition site in Ae. aegypti.
(A) Schematic depiction of the UUGFP#18 transgenesis construct. The relative locations of primers used to generate amplicons are indicated (small arrows). Mos1 right (R) and left (L) inverted terminal repeats are indicated by large arrows. (B) Detection of imperfect gap repair using a mismatch-specific nuclease. PCR amplicons from uninfected or nls-Stag-I-PpoI virus-infected mosquitoes were digested with the indicated dilutions of Surveyor Nuclease, or were undigested (−). (C) PCR amplicons from uninfected or nls-Stag-I-PpoI virus-infected mosquitoes were digested (+) with I-PpoI or were undigested (−). (D) Sequence analysis of uninfected or nls-Stag-I-PpoI virus-infected cloned UUGFP#18 amplicons. The number of clones obtained for each sequence compared with the total number is shown to the right of each sequence. Dashes indicate deleted bases. The first row (#18) indicates the sequence of the parent transformation construct used to generate line UUGFP#18.
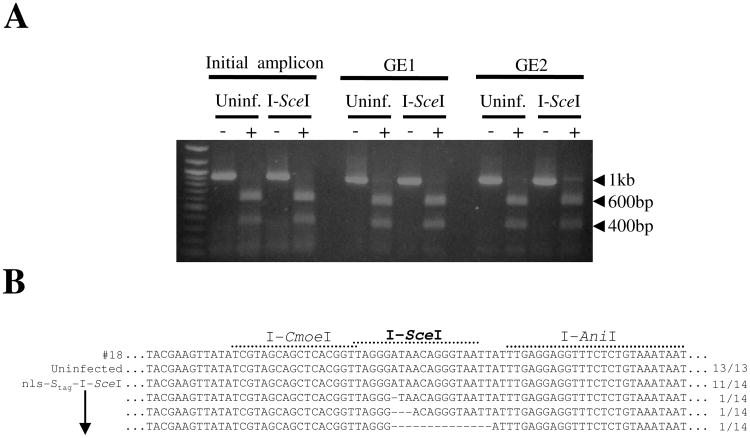
We performed similar experiments using the homing endonuclease I-SceI. Transgenic UUGFP#18 mosquitoes were infected with nls-Stag-I-SceI virus, and after an incubation period of 10 days, genomic DNA was extracted and used as a template in PCR as described for I-PpoI. Amplicons obtained from nls-Stag-I-SceI virus-infected mosquitoes were digested with Surveyor Nuclease or with a commercial preparation of I-SceI (New England Biolabs, Ipswich, MA). While no evidence of imperfect repair was observed with Surveyor Nuclease (data not shown), after 2 rounds of PCR enrichment we obtained a 1 kb amplicon that partially resisted digestion with commercial I-SceI (Fig. 4A). This could not be explained by mutations introduced during the PCR enrichment, as amplicons enriched from uninfected mosquitoes remained completely susceptible to I-SceI digestion (Fig. 4A). We cloned and sequenced both enriched amplicons. All clones obtained from uninfected mosquitoes revealed no alteration at the I-SceI site, or anywhere else in the amplicon (Fig. 4B). However, 21% (3/14) of clones exposed to I-SceI contained small deletions at the I-SceI recognition site (Fig. 4B). These deletions only occurred at the I-SceI site and ranged between 1 and 14 bp. Thus we conclude that the homing endonucleases I-PpoI and I-SceI, both previously shown to generate dsDNA breaks in An. gambiae (Windbichler et al. 2007), are also capable of generating dsDNA breaks in Ae. aegypti.
Figure 4. Somatic footprint assay to detect imperfect gap repair at a single I-SceI recognition site in Ae. aegypti.
(A) PCR enrichment for rare imperfect repair events in UUGFP#18 mosquitoes following infection with nls-Stag-I-SceI virus. PCR amplicons generated from uninfected or nls-Stag-I-SceI virus-infected mosquitoes were subjected to 1 (GE1) or 2 (GE2) rounds of PCR enrichment. (−) denotes undigested and (+) denotes digested with commercial I-SceI. (B) PCR enriched amplicons from GE2 were cloned and sequenced, as described in Fig. 3.
Somatic transgene excision: homing endonuclease assay with two target sites
The presence of a single target site simulates a gene mutagenesis strategy, whereby a homing endonuclease or other meganuclease is used to generate disruptions in a target gene. Meganucleases also have the potential to be used for the selective excision of genes/transgenes or possibly even large chromosomal segments. In this case, a target region would be flanked by two homing endonuclease recognition sites. To test whether homing endonucleases are capable of catalyzing the excision of genomic segments from the Ae. aegypti genome, we constructed a second transgenic strain (UUGFP#P17A) through Mos1-mediated germline transformation. The transgenic construct used to generate this strain, pictured in Figure 5A, displays two groups of homing endonuclease sites now flanking a promoter-EGFP gene cassette. Simultaneous rHE-induced dsDNA breaks on either side of the EGFP gene cassette and subsequent repair via non-homologous end-joining would result in the loss of this gene. Importantly, we arranged the homing endonuclease recognition sites asymmetrically between the two clusters so that each rHE would leave a unique and identifiable pattern of remaining sites following the excision of the EGFP gene cassette.
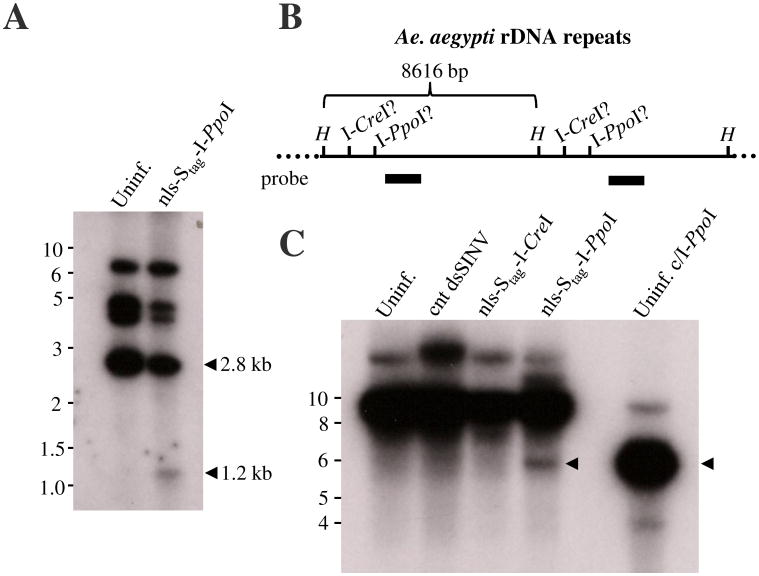
Figure 5. rHEs catalyze the somatic excision of genome segments in Ae. aegypti.
(A) Schematic depiction of the Mos1 transgenesis construct used to generate Ae. aegypti transgenic line UUGFP#P17A. Two clusters of homing endonuclease recognition sites flanking the EGFP gene cassette are indicated. Large arrows indicate the right (R) and left (L) inverted terminal repeats of Mos1, small arrows indicate the relative locations of primers used in PCR analysis. Black bars indicate sequences present in the random-primed probe used in the Southern analysis presented in Fig. 6A. (B-E) Sequence analysis of amplicons obtained from uninfected UUGFP#P17A mosquitoes (B), or mosquitoes infected with nls-Stag-I-PpoI (C), nls-Stag-I-CreI (D), or nls-Stag-AniI (E) viruses. The number of clones recovered for each sequence compared with the total number of sequenced clones per group is shown on the right. Red dashes indicate deleted bases. The top row of each dataset indicates the sequence of the parent transformation construct used to generate line UUGFP#P17A (B), or a hypothetical sequence based on the perfect excision of the EGFP gene cassette catalyzed by the respective homing endonuclease (C-E). Blue characters indicate sequences derived from the cluster of rHE recognition sites upstream of EGFP while orange characters indicate sequences from the cluster downstream of the EGFP gene cassette.
Female UUGFP#P17A mosquitoes were inoculated with nls-Stag-I-PpoI, nls-Stag-I-CreI, nls-Stag-I-AniI or nls-Stag-I-CmoeI viruses and held for 10 days prior to the extraction of genomic DNA. A single amplicon of ∼2.1 kb was obtained following PCR from uninfected UUGFP#P17A mosquitoes, and sequence analysis of this amplicon revealed that in all clones (16/16) the EGFP gene cassette was present, with no alterations at any homing endonuclease recognition site or anywhere else in the amplicon (Fig. 5B). In contrast, following PCR of genomic DNA isolated from nls-Stag-I-PpoI virus-infected mosquitoes, a second, smaller amplicon was obtained. Cloning and sequencing of this smaller amplicon revealed that in all clones (21/21), the EGFP gene cassette had been excised with upstream and downstream boundaries located precisely at each I-PpoI recognition site (Fig. 5C). In 2 out of the 21 clones (10%), the two I-PpoI recognition sites had been repaired perfectly, restoring a single intact I-PpoI site. In the remaining 19 clones (90%) we observed evidence of imperfect gap repair in the form of small deletions, with most (18/19) deletions occurring exclusively at the I-PpoI recognition site (Fig. 5C). While one larger deletion was observed (106 bp), the median deletion size following I-PpoI-induced gene excision was just 1 bp.
Similarly, we found that both I-CreI and I-AniI were able to catalyze the excision of the EGFP cassette in transgenic line UUGFP#P17A. For I-CreI, we sequenced 20 clones containing putative excision events. In 4/20 clones (20%) we observed regeneration of a single I-CreI site, indicating perfect repair. In the remainder of sequenced clones, we observed evidence of imperfect gap repair, with deletions ranging from 2-146 bp and in one case an insertion of 4 bp (Fig. 5D). Unlike what we observed with I-PpoI, repair of I-CreI-induced dsDNA breaks resulted in a consistently larger number of deleted bases, as the median deletion size was 65 bp (average deletion size was 60 bp). For I-AniI, we obtained sequence data from 19 clones containing putative EGFP excision events (Fig. 5E). In 12 clones (63%), we observed restoration of an intact I-AniI recognition site. The remaining clones contained deletions of various sizes, ranging from 1 to 135 bp with an average size deletion of 28 bp and a median size of 2 bp. We did not recover any putative EGFP excision events following infection with nls-Stag-I-CmoeI virus.
In addition to performing PCR-based assays, we performed Southern analyses directly on genomic DNA isolated from mosquitoes following the injection of nls-Stag-I-PpoI virus (Fig. 6A and C). I-PpoI-catalyzed excision of the EGFP gene cassette followed by non-homologous end-joining repair would be expected to reduce the size of a SalI-generated genomic fragment. Consistent with the PCR-based assays, I-PpoI was able to catalyze the excision of the EGFP gene cassette, as evidenced by the appearance of a 1.2 kb hybridization signal (Fig. 6A). Larger fragments at ∼4.5 kb and 7 kb likely represent junction fragments with mosquito genomic DNA, as the probe used was also capable of hybridizing with the transposon arms. The homing endonucleases I-PpoI and I-CreI have been found to cleave endogenous recognition sequences found within the 28S rDNA repeats of An. gambiae and D. melanogaster (Windbichler et al. 2007 (Maggert and Golic 2005). Digestion of Ae. aegypti genomic DNA with HindIII is predicted to generate an 8616 bp rDNA fragment (Fig. 6B) and (Nene et al. 2007). This fragment would be expected to be shortened into 5685 bp or 6598 bp fragments by I-PpoI or I-CreI, respectively, based on the probe sequence used (Fig. 6B). As shown in Fig. 6C, hybridization fragments were reduced in both commercial I-PpoI and nls-Stag-I-PpoI treated mosquitoes (black arrows). This indicates that I-PpoI can catalyze dsDNA breaks in the rDNA genes of Ae. aegypti mosquitoes both in vivo and in vitro. Similar dsDNA breaks were not observed in nls-Stag-I-CreI virus-infected mosquitoes.
Figure 6. Southern analysis of I-PpoI-induced somatic excision of the EGFP transgene or rDNA genes.
(A) Genomic DNA from uninfected UUGFP#P17A mosquitoes (Uninf.) or UUGFP#P17A mosquitoes infected with nls-Stag-I-PpoI virus was extracted 10 days post-infection and was subjected to SalI digestion. A 32P-dATP-labeled random-primed probe derived from HindIII-digestion of the transgene construct was used for hybridization (see Fig. 5A legend for location of probe sequence in transgene construct). Arrows denote the expected size of the internal SalI-generated hybridization signal before (2.8 kb) or following (1.2 kb) excision of the EGFP gene cassette. (B) Schematic representation of the Ae. aegypti rDNA repeats (not to scale). HindIII sites (H) and putative I-CreI and I-PpoI sites are indicated. The probe sequence used in (C) is indicated by the thick black bar. (C) Genomic DNA from uninfected UUGFP#P17A mosquitoes (Uninf.), or UUGFP#P17A mosquitoes infected with nls-Stag-I-PpoI virus, nls-Stag-I-CreI virus or a control virus (cnt dsSINV) was digested with HindIII and hybridized to a random-primed probe derived from a portion of the 28S rDNA. Genomic DNA was also digested with commercial I-PpoI as a positive control. Black arrows indicate the expected size of HindIII-I-PpoI double-digested product.
Discussion
We have demonstrated that four different homing endonucleases: I-PpoI, I-SceI, I-CreI, and I-AniI, are able to recognize and induce dsDNA breaks at their target site when present in the Ae. aegypti genome. Our results are consistent with work performed with I-PpoI and I-SceI in An. gambiae (Windbichler et al. 2007; Windbichler et al. 2008), and represent the first report of I-CreI and I-AniI inducing dsDNA breaks in any mosquito species.
I-PpoI appeared to be the most efficient rHE at introducing dsDNA breaks in Ae. aegypti, as we detected evidence of mismatches following exposure to I-PpoI using Surveyor Nuclease while we were unable to detect any evidence of imperfect dsDNA break repair following exposure to I-SceI, I-CreI, I-AniI or I-CmoeI with this assay (data not shown). However, we hesitate to draw firm conclusions relating the efficiency of one rHE to another for several reasons. First, in our experiments we were only able to detect the footprint of a rHE-induced dsDNA break based on imperfect repair or complete excision of a transgene. As little is known about the speed and efficiency of dsDNA break repair in Ae. aegypti, we are likely underestimating the total number of dsDNA breaks being induced due to the fact that such dsDNA breaks might be repaired correctly a large portion of the time, or in the case of excision events, that one dsDNA break is repaired prior to the second being induced. It is also possible that dsDNA breaks generated by some homing endonucleases may be more likely to be repaired perfectly than others, which would influence the rate at which we recover imperfect repair events. Indeed, we observed that for the homing endonuclease I-CreI most deletions were greater than 60 bp (Fig. 5D), while for I-AniI most dsDNA breaks were repaired without introducing any base changes (Fig. 5E). Lastly, while the recombinant SINV used to express each rHE is expected to infect most tissues of the adult mosquito (Olson et al. 1994), the replication kinetics of SINV have been shown to vary based on the presence of inserted sequences (Pierro et al. 2003), and without performing detailed growth curves for each recombinant virus it is not possible to say that each has the same course of infection (and thus produce the same amount of rHE).
Restriction digests of PCR amplicons containing putative imperfect repair events with commercial preparations of I-PpoI or I-SceI allowed for multiple rounds of PCR enrichment, a method used previously in the determination of sequence degeneracy for both I-PpoI and I-CreI (Argast et al. 1998). However, as commercial preparations of other homing endonucleases are not currently available, for most future experiments active protein would have to be purified by the investigating group. This is not ideal, especially if many variant meganucleases are to be tested in parallel. To overcome these obstacles, we have established a methodology to quickly assess the activity of multiple novel meganucleases in Ae. aegypti, without the need for generating individual HE-expressing transgenic strains or performing extensive embryonic injections. This is especially important for experiments involving the generation of homing endonucleases with altered target site specificity. Using a single germline transformant carrying a given pair of target sites flanking a marker gene, many candidate HE genes can be tested rapidly, decreasing the time required to recover a homing endonuclease with the required activity.
The engineering of meganucleases with novel target site specificity is a rapidly developing field, currently dominated by the re-engineering of naturally occurring homing endonuclease genes and the de novo construction of synthetic genes, such as zinc-finger nucleases. Directed evolution studies have produced variants of I-SceI which recognize new target sites with the same specificity found in wild-type I-SceI for its target site (Doyon et al. 2006). Similarly, variants of I-PpoI with amino acid substitutions in the DNA-protein interface were recovered from a yeast one-hybrid assay (Eklund et al. 2007), while modified versions of I-AniI were recovered with 100-fold greater affinity for its native target site (Takeuchi et al. 2009). However, by far the most extensive re-engineering has been performed using I-CreI (Arnould et al. 2006; Chames et al. 2005; Rosen et al. 2006; Seligman et al. 2002). As we have shown that all four of these homing endonucleases are capable of generating targeted mutations in Ae. aegypti, these molecules, and all of their variants, will likely provide a rich source of material for gene mutation and inactivation studies in disease vectors.
In addition to targeted mutagenesis, homing endonucleases can be used to promote site-specific recombination. Several site-specific recombination systems have been used successfully in vector mosquitoes, such as the cre-loxP system, which has been shown to be capable of robust transgene excision in Ae. aegypti (Jasinskiene et al. 2003), though not integration (Nimmo et al. 2006), and the attP/attB system, catalyzed by phiC31 integrase which has been used to insert transgenes into Ae. aegypti (Nimmo et al. 2006). While these systems represent significant advances in vector genetics, they suffer from the requirement of an initial random integration of one or more docking/recognition sites, and so do not aid in gene tagging or gene replacement studies. Re-engineering homing endonucleases to recognize target sequences present in the mosquito genome might bring homologous recombination into the toolboxes of mosquito geneticists. All of the dsDNA break-repair events we observed appeared to be the result of non-homologous end-joining, not homologous recombination. This is not surprising, however, as all of our experiments were conducted with hemizygous individuals. Thus, no transgene sequences would be present on the homologous chromosome. Future work with distinct transgenes in homologous positions will aim to determine the efficiency of homologous recombination in dsDNA break repair in Ae. aegypti.
Homing endonucleases have been proposed as a means to genetically sterilize males or to distort sex ratios prior to their use in sterile insect programs (Windbichler et al. 2008). This is based on the observation that the recognition site for the homing endonuclease I-PpoI is present in the An. gambiae X-linked rDNA genes (Windbichler et al. 2007; Windbichler et al. 2008). Similar to An. gambiae, I-PpoI recognition sites are present in the 28S rDNA of Ae. aegypti (Fig 6C and our observations). Thus we would expect that dsSINV-mediated expression of I-PpoI should lead to shredding of the rDNA subunits, as is the case with An. gambiae (Windbichler et al. 2008). This was likely the case, as I-PpoI was able to induce dsDNA breaks in the 28S rDNA repeats, and Ae. aegypti exposed to I-PpoI had shortened lifespans compared to mosquitoes infected with control dsSINV (our observations). Whether or not conditional I-PpoI expression could be used to generate sterile-male phenotypes in Ae. aegypti is unknown, as the rDNA genes in this species (located on genomic supercont1.836) have not as yet been mapped to a specific chromosome (Nene et al. 2007). We did not obtain any evidence that I-CreI was able to induce dsDNA breaks in rDNA genes through Southern analysis, and mosquitoes infected with nls-Stag-rHE viruses appeared to have normal lifespans (our observations). This was unexpected, given that the endogenous I-CreI site described in D. melanogaster is perfectly conserved in Ae. aegypti. These results may be due to the relative insensitivity of the assays used, or to intrinsic differences in the speed of dsDNA break repair between these organisms, and as such, further work will be necessary to determine if I-CreI can generate dsDNA breaks in the rDNA of Ae. aegypti.
In summary, homing endonucleases have the potential to be used in experiments involving targeted mutagenesis or the excision of transgenes/chromosomal segments. This is especially helpful in studies of vector biology and genetics, where fewer genetic tools are typically available. Homing endonucleases might also be used in applied genetic control strategies through genetic sterilization or the inactivation of genes essential to pathogen transmission (Burt 2003; Sinkins and Gould 2006; Windbichler et al. 2008). We have developed and validated a genomic footprint assay to test the ability of any meganuclease to induce site-specific dsDNA breaks in Aedes aegypti, thus opening the door to these investigations. As the experiments described here are all based on somatic dsDNA breaks, additional experiments will be required to determine the abilities of these homing endonucleases to catalyze dsDNA breaks in germ cells.
Experimental Procedures
Generation of recombinant Sindbis viruses
To generate recombinant Sindbis viruses expressing homing endonuclease genes, an XbaI fragment containing a multiple cloning site with AscI and PacI sites was first ligated into the XbaI site of pME2/5′2J (Pierro et al. 2003) to generate pME2/5′2J/mcs. Homing endonuclease genes were subcloned into a modified pKhsp82 (Coates et al. 1996), which resulted in the addition of an N-terminal SV40-derived nuclear localization signal (nls) and Stag epitope (Novagen, Gibbstown, NJ). Homing endonuclease genes I-AniI, I-CreI and I-CmoeI were codon optimized for expression in Ae. aegypti (Morlais and Severson 2003) by de novo synthesis (Top Gene Technologies, Quebec, Canada) prior to subcloning. A fragment containing each nls-Stag-rHE was amplified using a proofreading DNA polymerase Pfx (Invitrogen, Carlsbad, CA) and primers 5′-ttttggcgcgccTTAAATTAAAACACGGATCCATGC-3′ and 5′-ttttttaattaaTGATCTTGATCTTCATGGTCGACGG-3′ (94°C, 2 min; 94°C, 30 sec; 54°C, 1 min; 68°C, 2 min; 35 cycles; 68°C, 10 min). Primer sequences contained AscI and PacI restriction sites, as indicated by underlined bases. Following restriction enzyme digestion, nls-Stag-rHE amplicons were ligated into the AscI/PacI sites of pME2/5′2J/mcs (Fig. 1) or pTE/3′2J/mcs (Adelman et al. 2008). To generate recombinant Sindbis viruses from each clone, plasmid DNAs were linearized with XhoI, and in vitro transcription reactions were performed using SP6 polymerase and electroporated into BHK-21 cells as previously described (Myles et al. 2006). The supernatant containing virus was harvested, titered by plaque assay in Vero cells, and stored at −80°C. TE/3′2J/mcs-based viruses were used for SDS-PAGE analyses as described below, while ME2/5′2J/mcs-based viruses were used for all immunofluoresence assays and all in vivo experiments involving mosquitoes.
Immunofluorescence assay
To determine whether recombinant homing endonucleases were successfully translocated to the nuclei of mosquito cells, Aedes albopictus C6/36 cells were first infected with each recombinant nls-Stag-rHE virus at a multiplicity of infection (MOI) of 1 for one hour at room temperature when cells had achieved 60% confluency. Infected cells were scraped and seeded on glass coverslips at 4 days post-infection and were fixed for 2 min with ice cold acetone:PBS (75:25) 24 hours later. Fixed cells were permeabilized in 0.3% Triton-X/PBS (10 min, RT), blocked in a solution of 2% bovine serum albumin (BSA)/1% horse serum (1 h, RT) and incubated in a humidified 37°C chamber for 1 hour with S-protein antibody (Novagen, Gibbstown, NJ) at a dilution of 1:400 in 0.1% Triton-X/0.2% BSA/PBS. Following primary antibody incubation, cells were washed with 0.1% Triton-X/0.2% BSA/PBS and incubated with a goat anti-mouse FITC conjugated antibody (Calbiochem, Gibbstown, NJ) at a dilution of 1:400 in 0.1% Triton-X/0.2% BSA/PBS before a final set of washes with PBS and a counterstain of 0.025% Evan's Blue. Coverslips were mounted on glass slides with ProLong Gold anti-fade reagent with DAPI mounting solution (Molecular Probes, Carlsbad, CA), and cells were examined using a Zeiss confocal LSM510 Meta microscope.
SDS-PAGE and western analysis
For western analysis, cells were seeded in 25 cm2 flasks and infected with recombinant nls-Stag-rHE viruses at high MOI to ensure uniform infection (MOI >5). At the indicated times, cells were scraped, washed in PBS and pelleted by centrifugation. Cell pellets were washed three times with PBS, lysed with 2X SDS Loading buffer (Novagen) and boiled at 100°C for 5 minutes. Boiled lysates were centrifuged for 1 minute at 13,000 rpm prior to loading on a 4% stacking/10% resolving SDS polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane (0.45-um pore size; Biorad, Hercules, CA), and the S-Tag HRP LumiBlot Kit (Novagen) was used for detection of nls-Stag-rHE per the manufacturer's instructions. Proteins were detected using X-ray film (Kodak, Rochester, NY) and developed with a Konica SRX-101A processor.
DNA footprint assays and Southern analyses
Aedes aegypti (khw, Liverpool, and transgenic UUGFP#18, #P17A strains) were maintained as previously described (Adelman et al. 2008). Transgenic lines were screened using a fluorescent Leica MZ16F microscope as either larvae or pupae for DsRed+ eyes. Approximately 2 day old adult female transgenic mosquitoes were intrathoracically injected with 0.4-0.5 μl of recombinant nls-Stag-rHE virus [103-104 plaque forming units (pfu)]. Injected mosquitoes were held at 28°C, 80% relative humidity until the indicated times post-infection. Mosquitoes were snap frozen in nitrogen and stored at −80°C. Genomic DNA was isolated as described previously (Adelman et al. 2008). For Southern analysis, genomic DNA was digested with SalI or HindIII prior to electrophoresis and capillary transfer to a nylon membrane. A 1.2 kb HindIII fragment derived from the MosRH/DsRED/SV40 plasmid sequence or a 448 bp 28S rDNA amplicon (F 5′-AGAGACTCTAAACCTTGGAGACCTGCTGC-3′, R 5′- AACACGAGTTAGCCAATCCTAAGCTCTATGG-3′) was labeled with [α-32P] dATP (Amersham Megaprime DNA Labeling System, GE Healthcare, Buckinghamshire, UK) and purified using illustra NICK columns (GE Healthcare). Following hybridization overnight at 65°C, membranes were washed and exposed to Kodak BioMax maximum sensitivity film at -80°C.
For footprint assay using transgenic line UUGFP#18, genomic DNA was amplified using the proofreading DNA polymerases Pfx (Invitrogen) or Phusion (New England Biolabs) and primers 5′-CGAAACGGTGAATACGGCACGCTA-3′ and 5′-CGCCACCACCTGTTCCTGTA-3′. PCR conditions were 94°C, 2 min; 94°C, 30 sec; 58°C, 1 min; 68°C 30 sec; 35 cycles; 68°C, 10 min for Pfx and 98°C, 1 min; 98°C, 15 sec; 58°C, 30 sec; 72°C, 1:30 min; 35 cycles; 72°C, 10 min for Phusion. For footprint assays using transgenic line UUGFP#P17A, genomic DNA was amplified using Pfx (Invitrogen) and primers 5′-CGCCACCACCTGTTCCTGTA-3′ and 5′-AACGTGTGAACGGTGGTTTCAACGCTTC-3′. PCR conditions were 94°C, 2 min; 94°C, 30 sec; 58°C, 1 min; 68°C 3 min; 35 cycles; 68°C, 10 min. Amplicons were digested with Surveyor Nuclease according to the manufacturer's protocol (Transgenomic, Omaha, NE); were digested with commercial preparations of homing endonuclease enzymes (I-PpoI, Promega, Madison, WI; and I-SceI, New England BioLabs, Ipswich, MA); or were directly cloned with a Zero Blunt TOPO PCR Cloning kit (Invitrogen) prior to the sequencing of individual clones.
Acknowledgments
We thank Richard B. Waring (Temple University); Monique Turmel (Université Laval); David L. Herrin (University of Texas-Austin) and Donna Muscarella (Cornell University) for providing I-AniI, I-CmoeI, I-CreI and I-PpoI expression vectors, respectively. Furthermore, we thank Collin Edler, Anthony A. James and Emir Tinaztepe, as well as members of the Adelman laboratory, for technical assistance. This work was supported by the National Institute of Allergy and Infectious Diseases [1R21AI071208-01A1 to Z.N.A.] and by Virginia Tech start up funds to Z.N.A.
References
- Adelman ZN, Anderson MA, Morazzani EM, Myles KM. A transgenic sensor strain for monitoring the RNAi pathway in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2008;38:705–13. doi: 10.1016/j.ibmb.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argast GM, Stephens KM, Emond MJ, Monnat RJ., Jr I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J Mol Biol. 1998;280:345–53. doi: 10.1006/jmbi.1998.1886. [DOI] [PubMed] [Google Scholar]
- Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, Rolland S, Prieto J, Blanco FJ, Bravo J, Montoya G, Serrano L, Duchateau P, Paques F. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J Mol Biol. 2006;355:443–58. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, Epinat JC, Duclert A, Duchateau P, Paques F. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J Mol Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- Belfort M, Roberts RJ. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–88. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaiche Y, Mogila V, Perrimon N. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics. 1999;152:1037–44. doi: 10.1093/genetics/152.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc R Soc Lond B Biol Sci. 2003;270:921–8. doi: 10.1098/rspb.2002.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chames P, Epinat JC, Guillier S, Patin A, Lacroix E, Paques F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates CJ, Howells AJ, O'Brochta DA, Atkinson PW. The 5' regulatory region from the Drosophila pseudoobscura hsp82 gene results in a high level of reporter gene expression in Lucilia cuprina embryos. Gene. 1996;175:199–201. doi: 10.1016/0378-1119(96)00149-7. [DOI] [PubMed] [Google Scholar]
- Coates CJ, Jasinskiene N, Miyashiro L, James AA. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A. 1998;95:3748–51. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig GB, Jr, Hickey WA. Genetics of Aedes aegypti. In: Wright JW, P R, editors. Genetics of Insect Vectors of Disease. Elsevier; New York: 1967. pp. 67–131. [Google Scholar]
- Deredec A, Burt A, Godfray HCJ. The Population Genetics of Using Homing Endonuclease Genes in Vector and Pest Management. Genetics. 2008;179:2013–2026. doi: 10.1534/genetics.108.089037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J Am Chem Soc. 2006;128:2477–84. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- Egli D, Hafen E, Schaffner W. An efficient method to generate chromosomal rearrangements by targeted DNA double-strand breaks in Drosophila melanogaster. Genome Res. 2004;14:1382–93. doi: 10.1101/gr.2279804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund JL, Ulge UY, Eastberg J, Monnat RJ., Jr Altered target site specificity variants of the I-PpoI His-Cys box homing endonuclease. Nucl Acids Res. 2007;35:5839–5850. doi: 10.1093/nar/gkm624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisler B, Salomon S, Puchta H. The role of double-strand break-induced allelic homologous recombination in somatic plant cells. Plant J. 2002;32:277–84. doi: 10.1046/j.1365-313x.2002.01421.x. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci U S A. 2003;100:2556–61. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–23. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Braciale TJ, Rice CM. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc Natl Acad Sci U S A. 1992;89:2679–83. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB. Dengue. Lancet. 2007;370:1644–52. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- Higgs S, Olson KE, Kamrud KI, Powers AM, Beaty BJ. Viral expression systems and viral infections in insects. In: Crampton JM, Beard CB, Louis C, editors. The Molecular Biology of Disease Vectors: a Methods Manual. Chapman & Hall; UK: 1997. [Google Scholar]
- Higgs S, Traul D, Davis BS, Kamrud KI, Wilcox CL, Beaty BJ. Green fluorescent protein expressed in living mosquitoes without the requirement of transformation. Biotechniques. 1996;21:660–4. doi: 10.2144/96214st03. [DOI] [PubMed] [Google Scholar]
- Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–8. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- Jasinskiene N, Coates CJ, Ashikyan A, James AA. High efficiency, site-specific excision of a marker gene by the phage P1 cre-loxP system in the yellow fever mosquito, Aedes aegypti. Nucleic Acids Res. 2003;31:e147. doi: 10.1093/nar/gng148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinskiene N, Coates CJ, Benedict MQ, Cornel AJ, Rafferty CS, James AA, Collins FH. Stable transformation of the yellow fever mosquito, Aedes aegypti, with the Hermes element from the housefly. Proc Natl Acad Sci U S A. 1998;95:3743–7. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoza V, Ahmed A, Wimmer EA, Raikhel AS. Efficient transformation of the yellow fever mosquito Aedes aegypti using the piggyBac transposable element vector pBac[3xP3-EGFP afm] Insect Biochem Mol Biol. 2001;31:1137–43. doi: 10.1016/s0965-1748(01)00120-5. [DOI] [PubMed] [Google Scholar]
- Kowalski JC, Derbyshire V. Characterization of homing endonucleases. Methods. 2002;28:365–73. doi: 10.1016/s1046-2023(02)00243-8. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Müller-Lerch F, Fu F, Pearlberg J, Göbel C, Dassie Justin P, Pruett-Miller SM, Porteus MH, Sgroi DC, Iafrate AJ, Dobbs D, McCray PB, Jr, Cathomen T, Voytas DF, Joung JK. Rapid “Open-Source” Engineering of Customized Zinc-Finger Nucleases for Highly Efficient Gene Modification. Molecular Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert KA, Golic KG. Highly efficient sex chromosome interchanges produced by I-CreI expression in Drosophila. Genetics. 2005;171:1103–14. doi: 10.1534/genetics.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–50. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnat RJ, Jr, Hackmann AF, Cantrell MA. Generation of highly site-specific DNA double-strand breaks in human cells by the homing endonucleases I-PpoI and I-CreI. Biochem Biophys Res Commun. 1999;255:88–93. doi: 10.1006/bbrc.1999.0152. [DOI] [PubMed] [Google Scholar]
- Morlais I, Severson DW. Intraspecific DNA variation in nuclear genes of the mosquito Aedes aegypti. Insect Mol Biol. 2003;12:631–9. doi: 10.1046/j.1365-2583.2003.00449.x. [DOI] [PubMed] [Google Scholar]
- Myles KM, Kelly CLH, Ledermann JP, Powers AM. Effects of an Opal Termination Codon Preceding the nsP4 Gene Sequence in the O'Nyong-Nyong Virus Genome on Anopheles gambiae Infectivity. J Virol. 2006;80:4992–4997. doi: 10.1128/JVI.80.10.4992-4997.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O'Leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–23. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo DD, Alphey L, Meredith JM, Eggleston P. High efficiency site-specific genetic engineering of the mosquito genome. Insect Mol Biol. 2006;15:129–36. doi: 10.1111/j.1365-2583.2006.00615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson KE, Higgs S, Hahn CS, Rice CM, Carlson JO, Beaty BJ. The expression of chloramphenicol acetyltransferase in Aedes albopictus (C6/36) cells and Aedes triseriatus mosquitoes using a double subgenomic recombinant Sindbis virus. Insect Biochem Mol Biol. 1994;24:39–48. doi: 10.1016/0965-1748(94)90121-x. [DOI] [PubMed] [Google Scholar]
- Pierro DJ, Myles KM, Foy BD, Beaty BJ, Olson KE. Development of an orally infectious Sindbis virus transducing system that efficiently disseminates and expresses green fluorescent protein in Aedes aegypti. Insect Mol Biol. 2003;12:107–16. doi: 10.1046/j.1365-2583.2003.00392.x. [DOI] [PubMed] [Google Scholar]
- Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–8. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- Rong YS, Golic KG. A targeted gene knockout in Drosophila. Genetics. 2001;157:1307–12. doi: 10.1093/genetics/157.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Golic KG. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics. 2003;165:1831–42. doi: 10.1093/genetics/165.4.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucl Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–8. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:5809–14. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002;30:3870–9. doi: 10.1093/nar/gkf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat Rev Genet. 2006;7:427–35. doi: 10.1038/nrg1870. [DOI] [PubMed] [Google Scholar]
- Smith J, Grizot S, Arnould S, Duclert A, Epinat J-C, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, Montoya G, Paques F, Duchateau P. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucl Acids Res. 2006;34:149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi R, Certo M, Caprara MG, Scharenberg AM, Stoddard BL. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 2009;37:877–90. doi: 10.1093/nar/gkn1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118:91–8. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- Windbichler N, Papathanos PA, Catteruccia F, Ranson H, Burt A, Crisanti A. Homing endonuclease mediated gene targeting in Anopheles gambiae cells and embryos. Nucleic Acids Res. 2007;35:5922–33. doi: 10.1093/nar/gkm632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler N, Papathanos PA, Crisanti A. Targeting the X chromosome during spermatogenesis induces Y chromosome transmission ratio distortion and early dominant embryo lethality in Anopheles gambiae. PLoS Genet. 2008;4(12):e1000291. doi: 10.1371/journal.pgen.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wen X, Kodali NS, Oleykowski CA, Miller CG, Kulinski J, Besack D, Yeung JA, Kowalski D, Yeung AT. Purification, Cloning, and Characterization of the CEL I Nuclease. Biochemistry. 2000;39:3533–3541. doi: 10.1021/bi992376z. [DOI] [PubMed] [Google Scholar]