Summary
The Cmr complex is an RNA-guided effector complex that cleaves invader RNA in the prokaryotic immune response mediated by the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat)-Cas system. Here we report the crystal structure of a Cmr subcomplex containing Cmr2 (Cas10) and Cmr3 subunits at 2.8 Å resolution. The structure revealed a dual Ferredoxin fold and glycine-rich loops characteristic of previously known repeat-associated mysterious proteins (RAMPs) and two unique insertion elements in Cmr3 that mediate its interaction with Cmr2. Surprisingly, while mutation of both insertion elements significantly weakened Cmr3-Cmr2 interaction, they exhibit differential effects on Cmr-mediated RNA cleavage by the Cmr complex, suggesting stabilization of Cmr2-Cmr3 interactions by other subunits. Further mutational analysis of the two conserved (but non-Cmr2-binding) glycine-rich loops of Cmr3 identified a region that is likely involved in assembly or the RNA cleavage function of the Cmr complex.
Introduction
Microorganisms are challenged by viruses, plasmids and other mobile genetic elements that can threaten their survival. Therefore, bacteria and archaea have developed strategies for defending themselves against the invading nucleic acids of these molecular parasites. CRISPR-Cas systems provide host cells with a mechanism that captures inheritable memory of an infection and uses the information to disable the invasive agents (Bhaya et al., 2011; Makarova et al., 2011a; Terns and Terns, 2011; Wiedenheft et al., 2012). CRISPRs (clustered regularly interspaced short palindromic repeats) are genetic loci of identical repeats interspaced with distinct spacer or guide sequences derived from genetic elements of past invaders (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). A set of protein-encoding genes typically found immediately adjacent to the repeat-spacer array direct the synthesis of CRISPR-associated (Cas) proteins. Together, the transcribed CRISPR repeat-spacer array and Cas proteins provide an RNA-mediated defense mechanism to the microorganisms (Haft et al., 2005; Makarova et al., 2011a).
CRISPR immunity is characterized by three functional stages. In the adaptation stage, infected cells incorporate a small portion of the invader’s genetic sequence (~30 base pairs) into their genome as a “spacer” by largely unknown mechanisms (Barrangou et al., 2007; Datsenko et al., 2012; Erdmann and Garrett, 2012; Garneau et al., 2010; Horvath et al., 2008; Swarts et al., 2012; Yosef et al., 2012). In the biogenesis stage, the CRISPR repeat-spacer array is transcribed and processed to yield small CRISPR RNAs (crRNA), each of which is comprised of a spacer sequence and some repeat sequence (Brouns et al., 2008; Carte et al., 2008; Hale et al., 2008; Haurwitz et al., 2010; Sashital et al., 2011; Gesner et al., 2011). Finally, in the silencing stage, crRNAs assemble with Cas proteins and guide the ribonucleoprotein particles to degrade (or otherwise silence) invading nucleic acids (Hale et al., 2009; Jinek et al., 2012; Westra et al., 2012; Zhang et al., 2012).
There are ten or more CRISPR-Cas systems that are categorized into three superfamilies based primarily on bioinformatic analysis of cas genes. Type I CRISPR-Cas systems, exemplified by the Cse system of Escherichia coli (E. coli), use specific ribonucleases to process crRNA (Brouns et al., 2008). A multi-subunit ribonucleoprotein complex comprised of Cse proteins and crRNA recognizes invading nucleic acids by complementary base-pairing and recruits a helicase-nuclease, Cas3, to cleave the invader (Jore et al., 2011; Lintner et al., 2011; Westra et al., 2012; Wiedenheft et al., 2011). Type II CRISPR-Cas systems such as the Csn systems of Streptococcus thermophilus and Streptococcus pyogenes use a trans-activating CRISPR RNA (tracrRNA) and non-Cas protein, RNase III, to process crRNA, and the dual active site nuclease Cas9 protein for cleaving nucleic acid targets (Deltcheva et al., 2011; Garneau et al., 2010; Jinek et al., 2012). Type III CRISPR-Cas systems also use sequence-specific ribonucleases for processing crRNA but a crRNP complex containing a polymerase/nucleotide cyclase-like Cas10 protein for destruction (Hale et al., 2012; Hale et al., 2009; Zhang et al., 2012; Zhu and Ye, 2012). For both the biogenesis and silencing processes, where biochemical mechanisms have been determined, a diverse range of Cas protein types, in some cases evolutionarily unrelated, have been observed to serve the same functional roles. However, while the protein composition and mechanism of action differ significantly between type I and III effector complexes, both include members of the repeat-associated mysterious protein (RAMP) family, suggesting an evolutionary link among the seemingly diverse CRISPR-Cas systems. Structures of the RAMP proteins studied so far revealed a common dual-ferredoxin fold capable of binding (Wang et al., 2012), and in some cases (the CRISPR RNA endoribonucleases) cleaving RNA (Carte et al., 2008; Haurwitz et al., 2010; LaRonde-LeBlanc, 2012; Nam et al., 2012; Sashital et al., 2011; Sternberg et al., 2012). There is currently no data on how RAMP proteins partner with other Cas proteins in the assembly of the various CRISPR-Cas complexes. Structure and function studies of Cas proteins and their complexes are required for a complete understanding of the CRISPR-Cas immune systems.
The Cmr complexes of P. furiosus (Pf) and S. solfataricus (Ss) have been well characterized biochemically and they serve as models for studies of type III subtype B (IIIB) CRISPR-Cas systems (Hale et al., 2012; Hale et al., 2009; Zhang et al., 2012). The Pf Cmr complex contains six proteins (Cmr1-Cmr6) that co-purify with crRNAs of two sizes (39nt and 45nt) (Hale et al., 2012; Hale et al., 2009). The Cmr complex crRNAs consist of 8-nt of repeat sequence and 31-nt or 37-nt of guide sequence. While the guide sequence is used to bind target RNA through base-pairing, the 8-nt tag sequence is thought to serve as the anchor with which the Cmr proteins associate specifically. Upon recognition of a target RNA, the Cmr complex cleaves at the 14th based-paired nucleotide of the target RNA. Although the cleavage activity is metal dependent, the cleavage products contain 5′-OH and 3′- phosphate or 2′,3′-cyclic phosphate groups, suggesting a metal independent catalytic mechanism (Hale et al., 2009). The molecular details of how the Cmr complex interacts with the crRNA and destroys the target RNA remain largely unknown.
We have begun to elucidate functional roles of each Cmr protein by using in vitro reconstituted Pf Cmr complexes. Previously, we studied structure and function of the hallmark Cas10 subunit, Cmr2 (Cocozaki et al., 2012). Deletion or mutation of two previously predicted nuclease active sites of Cmr2 (the N terminal putative histidine-aspartate (HD) domain and an internal, highly conserved aspartate (GGDD) cluster), did not affect target RNA cleavage by the Cmr complex indicating that Cmr2 is not likely the catalytic component of the Cmr complex (Cocozaki et al., 2012; Makarova et al., 2002). This raises the question of where the ribonuclease activity resides among the Cmr subunits. The fact that four of the six Cmr proteins, Cmr1, Cmr3, Cmr4, and Cmr6, belong to the RAMP family of Cas proteins whose members include crRNA processing endoribonucleases, suggests that one of these four essential proteins could possess the catalytic site. In this work, we report the structure of Pf Cmr3 bound with the HD-domain-deleted Cmr2 (Cmr2dHD) and ATP at 2.8 Å resolution. We show that Cmr3 structurally resembles crRNA processing endonucleases Cas6 (Wang et al., 2011) and Cse3 (Brouns et al., 2008; Gesner et al., 2011; Sashital et al., 2011). Together with mutagenesis analysis, we have identified important functional regions in Cmr3 required for binding Cmr2 and possibly for subsequent steps in the Cmr complex function.
Results
Untagged Pf Cmr3 (Pf1128) was co-purified as a 1:1 complex with an N-terminally tagged Pf Cmr2dHD (Pf1129) lacking the N-terminal HD domain (residues 1 - 212) (Cocozaki et al., 2012). The Cmr2dHD-Cmr3 complex was crystallized in space group I222 with one protein complex in the asymmetric unit. Since Cmr2dHD had previously been shown to interact with an adenine nucleotide, the crystals were soaked in a solution containing ATP prior to diffraction. The structure of the Cmr2dHD-Cmr3-ATP complex was solved by single wavelength anomalous diffraction (SAD) phasing using a crystal containing selenomethionine-labeled Cmr2dHD. The structure was refined to an Rwork of 21.4% and an Rfree of 26.4%. The structure of the Cmr2dHD-Cmr3-ATP complex contains Cmr2 residues 213-402, 415-559, 569-608, 638-819 and 824-871 and Cmr3 residues 3-11, 32-48, 61-141 and 170-321. Data collection, phasing, and refinement statistics are presented in Table 1.
Table 1.
Data collection and refinement statistics.
| Cmr2dHD-Cmr3-ATP | |
|---|---|
| Space group | I222 |
| a | 102.5 |
| b | 135.9 |
| c | 189.6 |
| Resolution range (Å) | 50.0-2.85 (2.90-2.85) |
| No. of observed unique reflections | 32647 (1557) |
| Redundancy | 8.0 (6.6) |
| Completeness (%) | 99.9 (99.9) |
| I/σ(I) | 33.1 (2.5) |
| Rsym(%) | 8.1 (75.9) |
| Refinement statistics | |
| Resolution range (Å) | 36.833-2.804 |
| Rwork(%) | 23.2 |
| Rfree(%) | 26.8 |
| Root-mean-square deviations (rmsd) | |
| Bond length (Å) | 0.008 |
| Bond angle (°) | 1.146 |
| Ramachandran plot of protein residues | |
| Preferred regions (%) | 94.7 |
| Allowed region (%) | 5.3 |
| Disallowed region (%) | 0 |
Values in parentheses are for the highest resolution shell
Cmr3 shares structural homology with other RAMP proteins
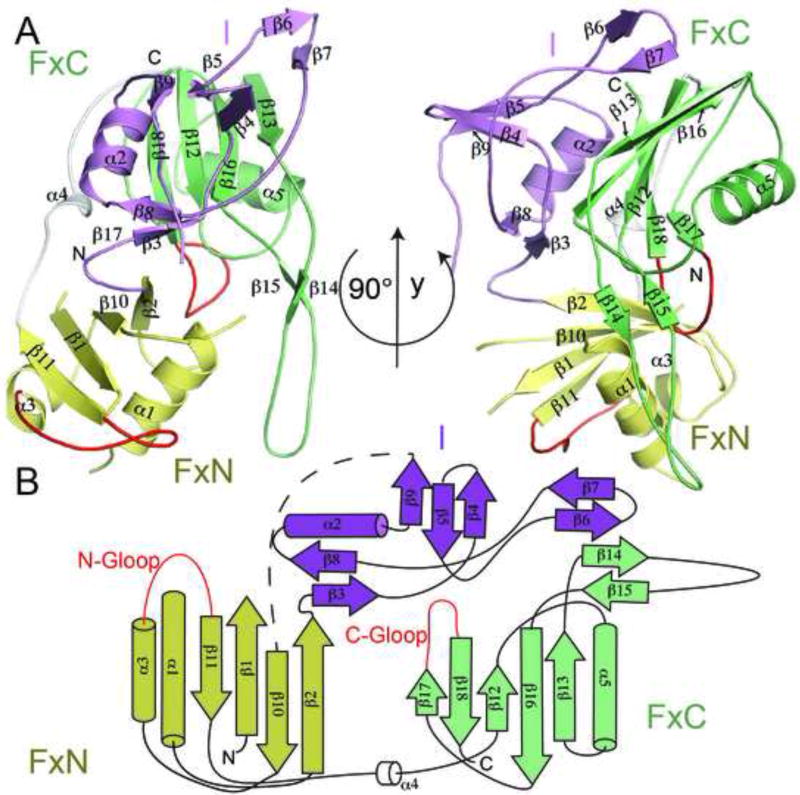
Cmr3 has three domains: two Ferredoxin fold domains (FxN and FxC) and an insertion domain (I) (Figure 1A and 1B). The two Ferredoxin domains are arranged similarly to those in the crRNA processing endoribonuclease members of the RAMP family (Przybilski et al., 2011; Sashital et al., 2011; Wang et al., 2011). Although structural disorder is observed in three regions of Cmr3 (Leu12-Ile31, Lys49-Glu60, and Glu142- Leu169), it still is evident that Cmr3’s FxN domain follows a classic βαββαβ topology (β1α1β2β10α3β11), suggesting a great stability of the Ferredoxin fold itself (Figure 1B).
Figure 1.
Structure and topology of Cmr3 when bound to Cmr2dHD. (A) Secondary and domain structures of Cmr3 in two orientations. “FxN” and “FxC” refer to the Ferredoxin folds of the N-terminal and C-terminal domains and “I” refers to the insertion domain. Cmr3 domains are shown in different colors (FxN: Yellow, FxC: Light green, I: purple) (B) Topology of the Cmr3 structure. The two glycine-rich loops are shown in red. “N-Gloop” refers to the glycine-rich loop of the N-terminal domain and “C-Gloop” refers to the glycine-rich loop of the C-terminal domain.
Both the FxN and FxC domains include secondary structure appendages to the basic βαββαβ fold (Figure 1A and 1B). The previously predicted Glycine-rich loop (G-loop) near the end of the FxN domain (Makarova et al., 2011a) is well ordered between α3 and β11 (Figure 1A and 1B). The insertion domain is the largest addition to the FxN domain (Figures 1 A and 1B). This domain (Thr74-Ile141) emanates from the second and returns to the third β-strand of the FxN fold. The insertion domain is composed of three nearly isolated β-strand units (β3β8, β9β4β5, and β6β7) and a single helix (α2). It rests on the flat β-sheet surface of the FxC domain and is also stabilized by the bound Cmr2dHD protein (Figure 3A and 3C). The isolated β-strands of the insertion domain are stabilized by their interactions with both the FxC domain and Cmr2dHD. In the absence of these interactions, these β-strands are likely to be disordered. The insertion domain is essential to the integrity of the Cmr3 structure; its removal was detrimental to the solubility of Cmr3 (data not shown).
Figure 3.
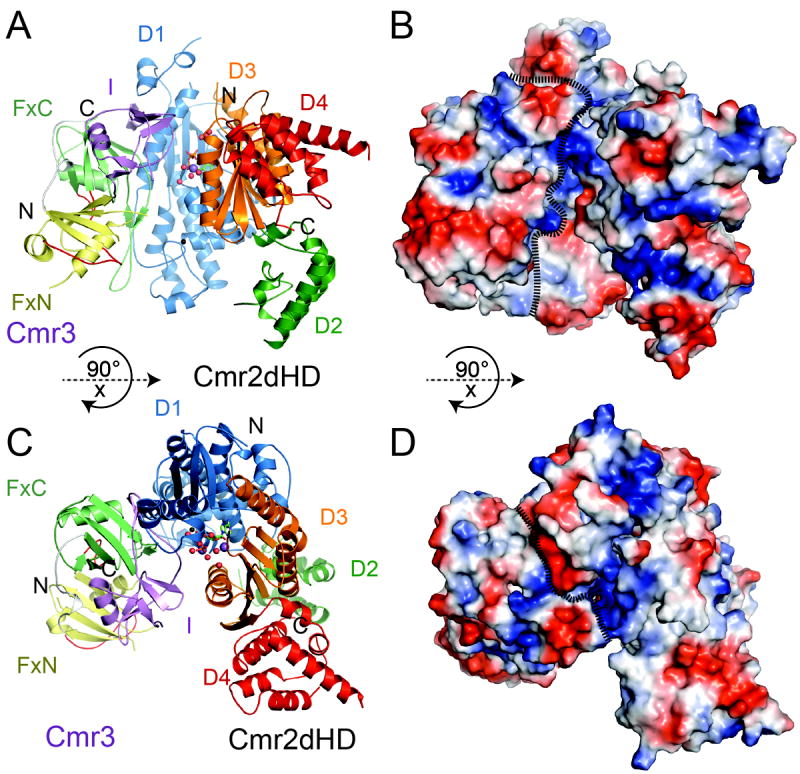
Overall structure of the Cmr2dHD-Cmr3-ATP complex of Pyrococcus furiosus. (A) Ribbons representation of the Cmr2dHD-Cmr3 complex with Cmr2dHD domains shown in different colors (D1:Blue, D2: Green, D3: Orange, D4: Red). “FxN” and “FxC” refer to the N-and C-terminal Ferredoxin folds of Cmr3, respectively, and “I” refers to the insertion domain of Cmr3. Cmr3 domains are shown in different colors (FxN: Yellow, FxC: Light green, I: purple). Ordered water molecules are shown as red spheres, Na2+ metal ions are shown as purple spheres and a Zn2+ metal ion bound to the zinc-finger of the D1 domain is shown as a black sphere. An ATP molecule is represented by ball-and-stick models. (B) Surface electrostatic potential distribution of the Cmr2dHD-Cmr3 complex with positive potential areas shown in blue and negative potential areas shown in red. The complex is in the same orientation as in (A). (C) Cmr2dHD-Cmr3 complex structure in a different orientation relative to that of (A). (D) Electrostatic potential distribution of the Cmr2dHD-Cmr3 complex in the same orientation as in (C). For (B) and (D), the dotted lines are used to define the surfaces of Cmr2 and Cmr3.
The FxC domain is connected to the FxN domain through a non-conserved linker (Lys211-Lys224) (Figure 1A and Figure S1). The FxC domain has a degenerate βαββαβ topology (β12α5β13β16β18) with the second α-helix replaced by a long β-hairpin (β14-β15, residues Ser260-Pro285). The sequence of the β-hairpin is well conserved (Figure S1), which is consistent with its role in mediating Cmr2dHD-Cmr3 interactions (discussed below). The FxC domain ends with another G-loop, characteristic of RAMP proteins. As in other RAMP proteins, the FxC G-loop is positioned between the two Ferredoxin domains, suggesting its role in maintaining folding integrity of RAMP proteins (Figure 1A).
The structure of Pf Cmr3 was compared with other known protein structures using the DALI server search. The RAMP superfamily endoribonucleases Cas6 (3PKM) and Cse3 (2Y8W) emerged as the two top structural homologs. Despite low sequence homology (10%-12 % sequence identity) with Cas6 (Wang et al., 2011) and Cse3 (Sashital et al., 2011), Cmr3 has a Root-Means-Square-Deviation (RMSD) of 3.4 Å (155 aligned Ca) and 4.3 Å (155 aligned Ca) for Cas6 (3PKM) and Cse3 (2Y8W), respectively (Figure 2). Alignments of Cmr3 to other RAMP proteins were limited to the Ferredoxin folds (Figure 2).
Figure 2.
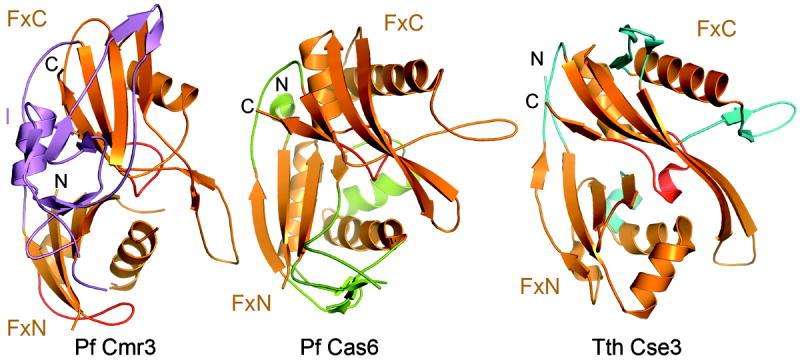
Comparison of the structure of Pyrococcus furiosus Cmr3 with those of RNA-bound Pyrococcus furiosus Cas6 (3PKM) (Wang et al, 2011) and Thermus thermophilus Cse3 (2Y8W) (Sashital et al, 2011). Superimposed protein structures are displayed in separate panels for clarity. Ferredoxin folds of the three proteins are shown in orange and their appendages are shown in violet for Cmr3, green for Cas6 and cyan for Cse3. The N-terminal and C-terminal glycine-rich loops are shown in red.
Interestingly, although the structure of Cmr3 was obtained without bound RNA, it is more similar to the RNA-bound than the isolated forms of Cas6 and Cse3. This is largely due to the β-hairpin of the FxC domain in all three structures (Figure 2). In both Cas6 and Cse3, the β-hairpin is used to bind RNA (Sashital et al., 2011; Wang et al., 2011), and in Cmr3 it interacts with Cmr2 (Figure 3). In the RNA-unbound structures of Cas6 and Cse3, the β-hairpin is unstructured (Sashital et al., 2011; Wang et al., 2011) rendering the lower similarity between those structures and that of Cmr2dHD-bound Cmr3. This comparison suggests that the FxC domain provides a basic framework for insertion elements that serve the function of protein-protein or protein-RNA interaction.
Overall structure of the Cmr2dHD-Cmr3 complex
The structural features of isolated Pf Cmr2dHD bound to ADP were previously described (Cocozaki et al., 2012). Briefly, Cmr2dHD is composed of four domains (D1-D4) that form an overall flat triangular shape. Domains D1 and D3, both of the Ferredoxin fold, form a homodimeric nucleotide-cyclase-like arrangement. In addition, D1 contains a conserved zinc-finger subdomain while D3 contains a highly conserved GGDD motif that is part of a binding site for divalent metals and ADP (Cocozaki et al., 2012). Cmr3 binds to Cmr2dHD through extensive interactions with the C-terminus of the D1 domain (Figure 3A and 3C). There are no contacts made between Cmr3 and the D2, D3, or the D4 domain of Cmr2dHD. Computed surface electrostatic potential showed that the Cmr2dHD-Cmr3 complex contains basic patches on surfaces of Cmr3 as well as that of a pocket formed between Cmr2dHD and Cmr3 (Figure 3B and 3D) suggesting potential sites of interaction with RNA.
Similar to the structure of isolated Cmr2dHD, that of the Cmr2dHD in complex with Cmr3 contains a single ATP molecule bound at its nucleotide-binding site between the D1 and D3 domains (Figure 3A and 3C). The bound Cmr3 is about 5Å away from the ATP and partially blocks its access to solvent (Figure 3A and 3C). The fact that no direct contacts are observed between Cmr3 and the bound ATP suggests that Cmr3 does not directly facilitate ATP-binding though it may stabilize it through its interaction with Cmr2dHD.
Cmr3 binds to Cmr2dHD using appendages to its Ferredoxin folds
Analysis of the accessible surface area of Cmr2dHD and Cmr3 alone and as a complex using the Protein Interfaces, Surfaces and Assemblies (PISA) server revealed a moderate 1672 Å2 of buried surface area comprising 57% polar and 43% nonpolar residues. Residues at the interface between Cmr2dHD and Cmr3 are only marginally conserved. Cmr3 contributes 43 residues, primarily from its insertion domain and β14-β15 hairpin, while Cmr2dHD contributes 45 residues from the C-terminus of its D1 domain to the interface.
Protein-protein interactions provide mutual stabilization to both Cmr3 and Cmr2dHD. Clear structural changes are observed for Cmr2 upon binding to Cmr3. In the unbound Cmr2dHD, residues Asp377-Ala402 and Arg436-Lys444 showed no electron density (Cocozaki et al., 2012). Upon binding Cmr3, Asp377-Ala402 of Cmr2 form well-ordered secondary structures that include α9’ and α10’ between β3 and α9, while residues Arg436-Lys444 form the C-terminal portion of α9 and a loop that connects α9 to the zinc finger motif (Figure 4). In addition, Lys606-Ile608 of the β4-α18 loop and Phe699-Gly707 of the α19-β7 loop of Cmr2dHD were also disordered in the isolated Cmr2 structure (Cocozaki et al, 2012), but become structured upon binding to Cmr3 (Figure 4). In addition, unbound Cmr3 is highly insoluble (data not shown) indicating that the structure of Cmr3 is stabilized upon association with Cmr2dHD.
Figure 4.
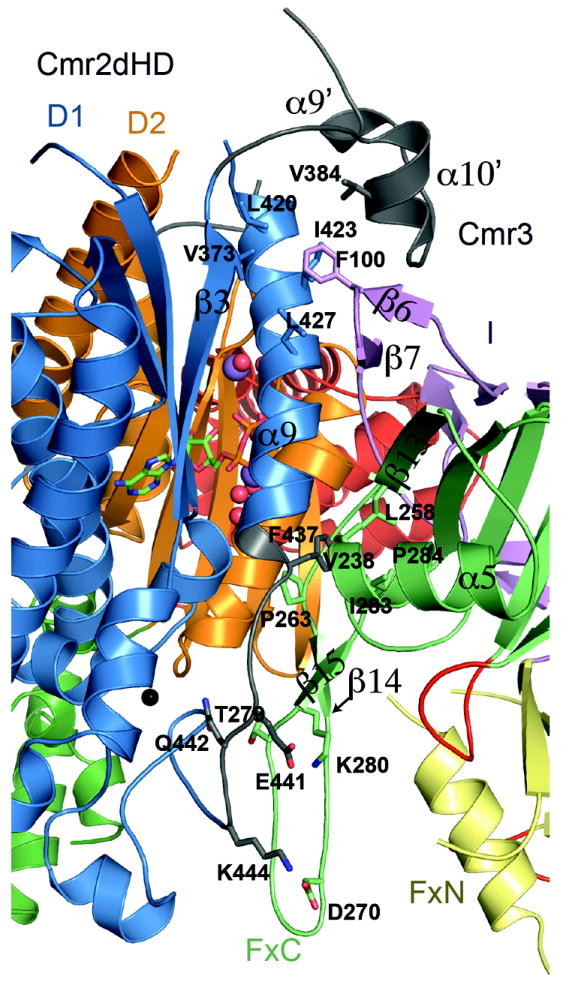
Interaction interface between Cmr2dHD and Cmr3. Parts of Cmr2dHD that become structured after binding to Cmr3 are shown in white. Conserved residues involved in protein-protein interactions are shown in ball-and-stick models. Ordered water molecules are shown as red spheres, Na2+ metal ions are shown as purple spheres and a Zn2+ metal ion bound to the zinc-finger of the D1 domain is shown as a black sphere.
Cmr3 mainly uses insertions to the Ferredoxin folds to provide a longitudinal interface for interacting with the N-terminal D1 domain Cmr2dHD. Cmr3 binds to Cmr2dHD via the β6-β7 hairpin of its insertion domain, and the first βαβ unit and the long inserted β14-β15 hairpin of the C-terminal Ferredoxin fold. The β6-β7 hairpin loop of Cmr3 wraps around the α9 helix of Cmr2dHD, and at the same time, makes contact with a newly observed α10′ α-helix of Cmr2dHD (Ser377-Glu404), leading to formation of a hydrophobic core at the interface. Phe100 of Cmr3 (Figure S1) packs into this hydrophobic pocket (Figure 4). Contacts with α9 of Cmr2dHD are also made from α5 and β13 of the first βαβ unit of the FxC domain, although these contacts are primarily nonspecific. The side chain of Phe437 of Cmr2dHD, which is unstructured in the free Cmr2dHD structure and exposed to solvent, packs against a hydrophobic patch formed by residues from the C-terminal Ferredoxin domain of Cmr3 (Figure 4). Phe437 is conserved in sequence as a bulky hydrophobic residue (Phe or Ile). However, the sequences of both Cmr3 and Cmr2dHD are mostly non-conserved in this region. The long β14-β15 hairpin of Cmr3 is mostly conserved and runs parallel to the mostly non-conserved loop following the α9 helix of Cmr2dHD, with which it makes multiple polar and non-polar interactions. In particular, three pairs of conserved salt bridges are formed by Cmr3-Lys280 and Cmr2dHD-Glu441, Cmr3-Asp270 and Cmr2dHD-Lys444, and Cmr3-Thr279 and Cmr2dHD-Glu442 (Figure 4 and Figure S1). These results suggest that RAMP proteins utilize appendages to their Ferredoxin folds for interactions with other proteins.
Functional importance of Cmr3 structural elements
Cmr3 and four of the other five Cmr proteins (Cmr1, Cmr2, Cmr4, and Cmr6) are required for RNA-guided RNA cleavage (Hale et al, 2009). To assess the role of Cmr3 in assembly of functional complexes and target RNA cleavage, we performed mutagenesis analysis guided by the Cmr3-Cmr2dHD complex structure (Figure 5A). We first assessed the role of the Cmr2-interacting elements in Cmr3 (Figure 5A). We deleted part of the β6-β7 (Asn96-Ile105) (dβ6-β7) or β14-β15 (Ser267-Lys277) (dβ14-β15) hairpins and tested for interaction with Cmr2dHD in co-purification assays. As expected, both mutations severely disrupted interaction with Cmr2 (Figure 5B). Interestingly however, when tested in cleavage assays with all five other Cmr proteins, while the dβ14-β15 mutant does not support RNA cleavage, complexes assembled with the dβ6-β7 mutant are functional (Figure 5C). Consistent with the lack of conservation at the β6-β7 loop and the sequence conservation of the β14-β15 loop, this result indicates that the interaction mediated by the β14-β15 hairpin of Cmr3 is essential for the function of the complex and that other Cmr subunits have some stabilization effects on the dβ6-β7 mutant.
Figure 5.
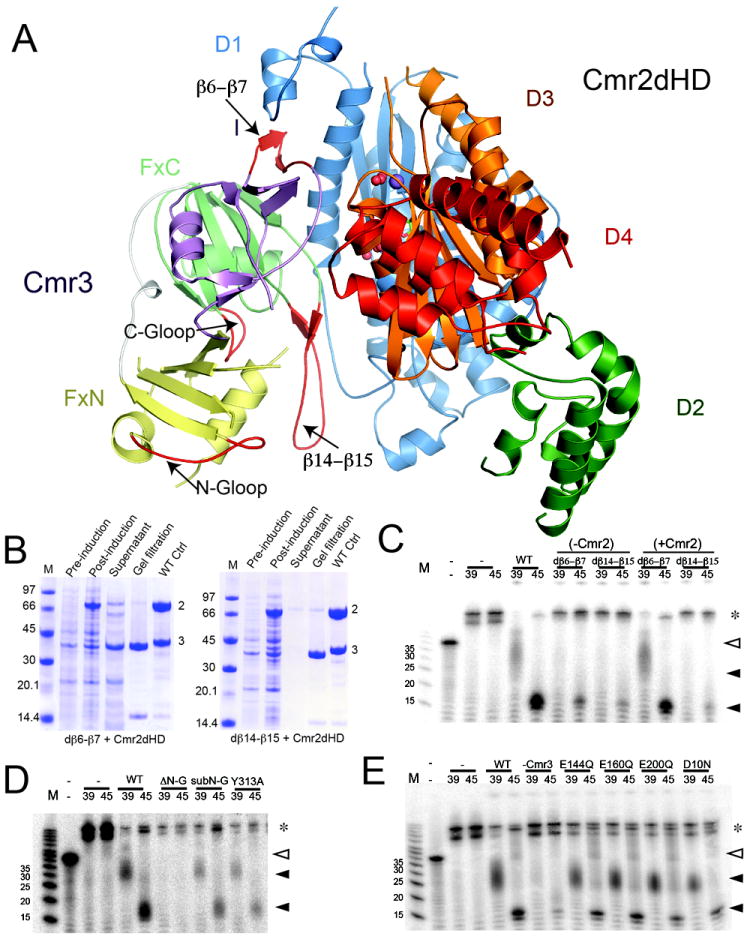
Functional analysis of Cmr3. (A) Structure of the Cmr2dHD-Cmr3 complex mapped with mutated regions shown in red. (B) Co-purification of Cmr3 loop mutants with Cmr2dHD. Untagged Cmr2dHD and tagged Cmr3 mutant were coexpressed and copurified by NiNTA affinity purification. For both panels, lane 1 is a molecular weight marker. Lanes 2, 3 and 4 represent steps prior to loading on the NiNTA column and lane 5 represents the elution products from the NiNTA column. Lane 6 is the elution from a similar co-purification experiment run in parallel using the wild-type Cmr3. (Left) Copurification of the dβ6-β7 mutant of Cmr3 with Cmr2dHD. (Right) Copurification of the dβ14-β15 mutant of Cmr3 with Cmr2dHD. (C) RNA cleavage assay results using wild-type (WT) and Cmr3 deletion mutants (dβ6-β7 and dβ14-β15). Co-purified Cmr3-Cmr2dHD samples were combined with Cmr1, Cmr4, Cmr5, and Cmr6 in the absence (-Cmr2) or presence (+Cmr2) of separately purified Cmr2dHD and incubated with crRNA (of size 39nt or 45nt) and radiolabeled target RNA. “-” denotes the control without addition of the Cmr complex. Full triangles indicate cleavage products. Empty triangles indicate uncleaved target RNA. Asterisks indicate crRNA-target duplexes not separated on denaturing polyacrylamide gel. A single cleavage product is obtained with the 39mer crRNA. The smeariness of the band is the result of the mildly denaturing conditions of the acrylamide gel (D) RNA cleavage assay results using the N-terminal deletion (ΔN-G) and N-terminal G-A substitution (subN-G) G-rich loop mutants as well as the Y313A mutant of the C-terminal G-rich loop. (E) RNA cleavage assay results using single mutations of Cmr3. Particle reconstituion and cleavage procedures are described in (C).
We then tested the functional roles of the two G-rich loops in Cmr3, which are not observed at the Cmr2-Cmr3 interface. G-rich loops of the RAMP superfamily protein play significant roles in RNA binding and catalysis in the crRNA processing endoribonucleases (Gesner et al., 2011; Sashital et al., 2011; Wang et al., 2011). We deleted residues Gly198-Arg202 from the N-ferreodoxin G-loop to create Cmr3 ΔN-G (Figure 5A) and substituted Gly198 and Gly199 with Ala to create Cmr3 subN-G. We also attempted deleting the C-terminal G-loop or substituting its glycines into alanines but the resulting mutants were unstable in our hands (data not shown). To test the importance of the C-terminal G-loop of Cmr3, we mutated its highly conserved Tyr313 residue to create Cmr3-Y313A. All three G-loop mutants reconstituted fully assembled Cmr complexes. However, only the Cmr3 ΔN-G mutation disrupted the RNA-cleavage activity of the complex (Figure 5D) without affecting the assembly of the Cmr RNP (Figure S2). This may be explained by the possibility that substitutions of G-loop residues are less disruptive than deletion of its residues. Nonetheless, these results identified an important function in binding/cleaving RNA of the FxN domain G-loop. Strikingly, the FxN G-loop is found near the β14β15 hairpin that is also critical to Cmr complex function (Figure 5A).
Lastly, we identified four conserved residues in Cmr3 that are possible candidates for involvement in catalytic function: Asp10, Glu144, Glu160, and Glu200 (Figure 5A and Figure S1). These acidic residues have the potential for divalent metal coordination as was observed with Cmr2dHD (Cocozaki et al., 2012). However, we found that substitution of the residues did not disrupt in vitro RNA cleavage activity (Figure 5E) thus ruling out a direct role of these acidic residues in the catalytic process.
Discussion
The Cmr complex is the effector complex of the type IIIB CRISPR-Cas system and has been previously shown to possess crRNA-guided RNA degradation activity (Hale et al., 2012; Hale et al., 2009; Zhang et al., 2012). Biochemical studies with the P. furiosus Cmr complex showed that the Cmr complex is comprised of six Cmr proteins (Cmr1-Cmr6) and a crRNA, which can be one of two size forms. Depending on the length of the crRNA, the Cmr complex cleaves at a different site on the target RNA 14 nucleotides downstream of the first paired target RNA nucleotide (Hale et al., 2009). All six Cmr proteins are required for assembly and cleavage of the target RNA (Hale et al., 2009). Although divalent metals are required for RNA cleavage, the end groups of the cleaved products are consistent with a metal-independent catalytic mechanism (Hale et al., 2009). Five of the six Cmr proteins contain features that are suggestive of direct involvement in RNA binding and catalysis. However, functional roles of the subunits remain to be identified.
Previously, we studied the structure and function of the hallmark Cas10 protein family subunit, Cmr2 (Cocozaki et al., 2012). Our results suggest that Cmr2 may not be the catalytic component of the Cmr complex since removal or deletion of two conserved domains previously predicted to harbor nuclease active sites (the HD domain and polymerase/palm-like domain that binds both ATP and divalent ions) do not affect target RNA cleavage (Cocozaki et al., 2012). Here, we demonstrated a direct interaction between Cmr2 and Cmr3. Cmr3 belongs to the RAMP superfamily of proteins whose members include endoribonucleases. We determined a 2.8Å resolution structure of the Cmr2dHD-Cmr3-ATP complex and performed mutagenesis in order to probe the observed structural features of Cmr3 and its function. We showed that Cmr3 resembles the endoribonuclease RAMP proteins structurally, but has evolved unique insertion elements that are critical for the function of the Cmr complex. Interestingly, the interaction interface contains mostly non-conserved elements of Cmr3 and Cmr2dHD with the exception of the β14-β15 hairpin of Cmr3. This suggests evolutionary plasticity at the interaction interface between the two proteins. The two structural elements of Cmr3 where we found that mutation disrupts the function of the complex, the β14-β15 hairpin and FxN G-loop, are located near each other in the structure. The β14-β15 hairpin mediates the interaction of Cmr3 with Cmr2. Our findings suggest that the β14-β15 hairpin / FxN G-loop region of Cmr3 may be involved in multiple important interactions.
Four of the six Cmr proteins belong to the RAMP superfamily of proteins (Hale et al., 2009). RAMP proteins are ubiquitous in CRISPR systems and participate in both the biogenesis and silencing phases of the immune response (Hale et al., 2009; Jore et al., 2011; Makarova et al., 2011a; Przybilski et al., 2011). RAMP proteins are found in eight of the ten subtypes of CRISPR systems including all subtypes of the type I and type III CRISPR systems (Makarova et al., 2011b). Similarly to RAMP proteins, Cmr2 also contains two RRM-like folds (Cocozaki et al., 2012). The fact that the structure of Cmr3 resembles the crRNA processing endoribonucleases supports the notion that all RAMP proteins have a similar structural fold. Interestingly, unique insertions to the basic βαββαβ topology in Cmr3 mediate protein-protein (and perhaps RNA-binding) functions. The Cmr2dHD-Cmr3 structure provides a working model to understand the assembly of CRISPR-Cas complexes containing RAMP superfamily proteins.
Methods
Protein preparation and crystallization
The Cm2dHD protein was constructed as described (Cocozaki et al., 2012). The Cmr2dHD (N-terminal poly-histidine tagged) and Cmr3 (untagged) proteins were expressed separately in Escherichia coli BL21 RIPL cells (Agilent technologies, Santa Clara, CA). Cell pellets were combined and resuspended in a lysis buffer (25 mM sodium phosphate pH 7.5, 1.0 M NaCl, 10 % (v/v) glycerol, 5.0 mM β-mercaptoethanol, and 0.2 mM phenylmethylsulfonyl fluoride). The cells were lysed by sonication and their debris was cleared by centrifugation. The supernatant was loaded onto a Ni-NTA column equilibrated with the lysis buffer supplemented with 5 mM imidazole. The column was washed with the lysis buffer containing 25 mM imidazole and the protein was eluted by increasing imidazole to 350 mM. Fractions containing protein were pooled and loaded onto a Superdex 200 (Hiload 26/60, GE Healthcare) size-exclusion chromatography column that had been equilibrated with 20 mM Tris-HCl pH 7.4, 500 mM NaCl, 5 % (v/v) glycerol, and 5 mM β-mercaptoethanol. Fractions corresponding to the Cmr3-Cmr2dHD protein complex were pooled and concentrated. The L-selenomethionine (SeMet) labeled protein complex was prepared by a similar procedure.
Native or SeMet-labeled complex was crystallized at 30°C by using the vapor diffusion hanging-drop method. Equal volumes of protein complex and a 15-22% polyethylene glycol (PEG1500) solution were combined into 2.4 μl droplets on cover slips that were then sealed with a 21-25% PEG1500 solution. Crystals formed in 6-7 days and had a hexagonal prism shape with typical dimensions of 0.4 mm × 1 mm × 0.4 mm.
Data collection and structure determination
Crystals were cryo-protected in a buffer containing 1.5 M NaCl, 20% PEG1500, and 2mM adenosine triphosphate (ATP) prior to being mounted on a goniometer head. ATP (2 mM) was included in the cryo-solution since Cmr2dHD was previously shown to bind ADP at its nucleotide binding site. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamline 22ID or 22BM. Data were indexed, integrated, and scaled using the HKL2000 software package. The space group of the crystals was determined to be I222 with one Cmr2dHD-Cmr3 complex in the asymmetric unit and the cell dimensions are listed in Table 1. Phases were determined from a highly redundant single-wavelength anomalous dispersion (SAD) data set at the anomalous peak of selenium from a SeMet-labeled crystal. Structure determination, iterative model building and structure refinement were done using the PHENIX (Adams et al., 2010) and COOT programs (Emsley and Cowtan, 2004). The refinement statistics are shown in Table 1. The atomic coordinates and structure factors were submitted to the Protein Data Bank (PDB ID: 4H4K).
RNA-cleavage reactions
In vitro RNA cleavage assay was carried out similarly as previously described (Hale et al., 2009). Briefly, purified Cmr proteins were combined in equal molar amounts to a final concentration of 50 μM in 20 mM Tris-HCl (pH 7.4), 500 mM NaCl, 5% (v/v) glycerol, and 5 mM β-mercaptoethanol. The reconstituted complex was diluted to 1 μM before being incubated with either the 39-mer or the 45-mer Pf7.01 crRNA (Hale et al., 2009) at 0.1 μM at 70°C in 20 mM sodium cacodylate (pH 6.2), 20 mM MgCl2 in the presence of 1 unit of SUPERase-In RNAse inhibitor. The RNA cleavage reaction was initiated by adding 0.016 μM 5’-radiolabeled target RNA and the reaction was incubated for 1 hour at 70°C. The reaction was quenched by the addition of 96% formamide dye. The cleavage products were resolved on a 15% polyacrylamide, 7 M urea gel and visualized by phosphorimaging.
Supplementary Material
Highlights.
Cmr3 has two Ferredoxin fold domains and a large insertion domain.
Cmr3 resembles Cas6 and Cse3.
Cmr3 uses insertions to its Ferredoxin folds to interact with Cmr2.
Cmr3 plays an important role in the assembly of the Cmr RNP complex.
Acknowledgments
This work was supported by National Science Foundation grant MCB0817638 to H.L., American Heart Association predoctoral fellowship 11PRE7090000 to A.I.C., and National Institutes of Health Grant R01 GM54682 to M.T. and R.T. We thank Caryn Hale (UGA) for preliminary data indicating an interaction between Cmr2 and Cmr3. X-ray diffraction data were collected from the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions for APS beamlines may be found at http://necat.chem.cornell.edu/ and http://www.sercat.org/members.html. Use of the Advanced Photon Source was supported by the UnitedStates Department of Energy Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
The authors declare they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocozaki AI, Ramia NF, Shao Y, Hale CR, Terns RM, Terns MP, Li H. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure. 2012;20:545–553. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Erdmann S, Garrett RA. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol. 2012;85:1044–1056. doi: 10.1111/j.1365-2958.2012.08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis M, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, 3rd, Graveley BR, Terns RM, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc Nicole A. Defense Systems Up: Structure of Subtype I-C/Dvulg CRISPR/Cas. Structure. 2012;20:1450–1452. doi: 10.1016/j.str.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011a;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011b;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Nam KH, Haitjema C, Liu X, Ding F, Wang H, Delisa MP, Ke A. Cas5d Protein Processes Pre-crRNA and Assembles into a Cascade-like Interference Complex in Subtype I-C/Dvulg CRISPR-Cas System. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Przybilski R, Richter C, Gristwood T, Clulow JS, Vercoe RB, Fineran PC. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol. 2011;8:517–528. doi: 10.4161/rna.8.3.15190. [DOI] [PubMed] [Google Scholar]
- Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zheng H, Preamplume G, Shao Y, Li H. The impact of CRISPR repeat sequence on structures of a Cas6 protein-RNA complex. Protein Sci. 2012;21:405–417. doi: 10.1002/pro.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR Immunity Relies on the Consecutive Binding and Degradation of Negatively Supercoiled Invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477(7365):486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Ye K. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR-Cas systems. FEBS Lett. 2012;586:939–945. doi: 10.1016/j.febslet.2012.02.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.