Abstract
Nuclear transcription factor Nrf2, binds with antioxidant response element (ARE) in the promoter regions of cytoprotective genes leading to its increased expression and cellular protection. In the present study, we investigated the role of Nrf2 in the regulation of anti-apoptotic Bcl-xL protein and its effect on cellular apoptosis. Treatment of mouse Hepa-1 cells with the antioxidant tert-butylhydroquinone (t-BHQ) led to induction of Bcl-xL gene expression. Promoter mutagenesis, transfection, and chromatin immunoprecipitation assays identified an antioxidant-response element (ARE) between nucleotides −608 to −600 in the forward strand of the proximal Bcl-xL promoter that bound to Nrf2 and led to increased Bcl-xL gene expression. In addition, short interfering RNA inhibition and overexpression of Nrf2 led to a respective decrease and increase in Bcl-xL gene expression. These results implicated Nrf2 in the regulation of expression and induction of Bcl-xL protein. Nrf2 mediated expression of Bcl-xL protein down regulated Bax and decreased caspases 3/7 activities. Both siRNA inhibition of Nrf2 and Bcl-xL increased susceptibility of cancer cells to etoposide-mediated cell death and reduced cell survival. Moreover, dysfunctional/mutant INrf2 in human lung cancer cells failed to degrade Nrf2 resulting in increased Bcl-xL levels and increased cell survival. These data provide the first evidence of Nrf2 in control of Bcl-xL expression and apoptotic cell death with implications in antioxidant protection, survival of cancer cells, and drug resistance.
Keywords: Nrf2, INrf2 (Keap1), Bcl-xL, Anti-apoptotic proteins, Apoptosis
Introduction
The Nrf2:INrf2 complex serves as sensor of chemical and radiation induced oxidative and electrophilic stress [1,2]. Nrf2 resides predominantly in the cytoplasm where it interacts with actin-associated cytosolic protein, INrf2 (inhibitor of Nrf2) or Keap1 (Kelch-like ECH-associated protein 1). INrf2 functions as a substrate adaptor protein for a Cul3-Rbx1-dependent E3 ubiquitin ligase complex to ubiquitinate and degrade Nrf2 thus maintaining a steady-state levels of Nrf2 [1]. The mechanisms by which Nrf2 is released from INrf2 under stress have been actively investigated. One mechanism is that cysteine thiol groups of INrf2 were shown to function as sensors for oxidative stress which are modified by the chemical inducers, causing formation of disulfide bonds between cysteines of two INrf2 peptides. This results in conformational change that render INrf2 unable to bind to Nrf2 [2,3]. On the other hand, we and others have shown that antioxidant-induced phosphorylation of Nrf2Serine40 by protein kinase C delta leads to dissociation of Nrf2 from INrf2 [4,5]. Nrf2 stabilizes and translocates in the nucleus leading to induction of downstream cytoprotective proteins [4,5]. Recent studies have shown that the two mechanisms act in concert to activate Nrf2 and downstream genes in response to chemical and radiation stress [6].
Antioxidant response element (ARE) is a cis-acting regulatory element in the promoter regions of several genes encoding phase II detoxification enzymes and antioxidant proteins, such as NAD (P)H:quinine oxidoreductase-1 (NQO1), glutathione S-transferase (GST),γ-glutamylcysteine synthetase (γ-GCS), heme oxygenase 1 (HO-1), thioredoxin reductase-1, and thioredoxin [1]. Transcriptional activation through ARE is mainly regulated by Nrf2. Recently, Nrf2 and its downstream proteins have been shown to be critical regulators in protection of cells from oxidative stress- and chemical-induced damages of liver and lung tissues. It has been demonstrated that Nrf2 knockout mice are more sensitive to hyperoxic injury of lung [7]. The primary astrocyte of Nrf2−/− mice is also more susceptible to oxidative stress and inflammation than that of Nrf2+/+ mice [8,9]. Leung et al. [9] showed that deficiency of Nrf2 results in early embryonic lethality with severe oxidative stress. These observations, collectively, imply that Nrf2 is a master regulator of ARE-driven transcriptional activation for antioxidant genes in maintaining the homeostasis of redox status within cells. On contrary, evidence also suggests that persistent accumulation of Nrf2 in the nucleus is harmful. INrf2-null mice demonstrated persistent accumulation of Nrf2 in the nucleus that led to postnatal death from malnutrition resulting from hyperkeratosis in the esophagus and forestomach [10]. Reversed phenotype of INrf2 deficiency by breeding to Nrf2-null mice suggested tightly-regulated negative feedback might be essential for cell survival [11]. The systemic analysis of INrf2 genomic locus in human lung cancer patients and cell lines showed that deletion, insertion, and missense mutations in functionally important domains of INrf2 results in reduction of INrf2 affinity for Nrf2 and elevated expression of cytoprotective genes which resulted in drug resistance and cell survival in lung cancer cells [12,13]. Unrestrained activation of Nrf2 in cells increases the risk of adverse effects including survival of damaged cells, tumorigenesis and drug resistance [6]. Therefore, it appears that cells contain mechanisms that auto-regulate cellular abundance of Nrf2 [14,15]. Indeed, these evidences suggested that, Nrf2 play an important role in cell survival/protection in normal cell and drug resistance in cancer cells.
In cancer, apoptosis is a critical process that is dysregulated resulting in tumourigenesis [16]. Bcl-2-family proteins regulate cell death and survival [17,18]. Bcl-2 family proteins are the prototypic anti-apoptotic proteins, and Bcl-xL was the first protein discovered with a similar function [19]. Since then the Bcl-2 family has expanded to include more than 6 anti-apoptotic and many pro-apoptotic members [20]. Bcl-2 and Bcl-xL display 43% amino acid identity, share regions of sequence similarity, as well as a C-terminal hydrophobic region required for membrane localization [21]. Bcl-2 and Bcl-XL appear to function in the same apoptotic pathway and both confer resistance to multiple chemotherapy agents when tested in experimental systems. Over-expression of either protein is usually associated with poor prognosis in many human cancers. However, in some cancer types multiple anti-apoptotic proteins are expressed [22], and have opposing effects on prognosis indicating that there may be subtle, but clinically and biologically relevant functional differences between family members. Experiments in mice with deletion of individual anti-apoptotic genes indicate that the phenotypes are not identical [23]. However, it is generally accepted that this is due to expression in different tissues or in the same tissue but at different times rather than being a consequence of differences in the potency or mechanism of action of the different anti-apoptotic proteins. The mechanisms of action of Bcl-2 and Bcl-XL are complex, with many postulated interactions with other proteins, and the role of any single interaction in the final phenotype at the cellular level remains unknown.
Several studies have investigated the INrf2:Nrf2 regulation of anti-apoptotic proteins and its role in cellular apoptosis especially during chemical and radiation stress. H2S-mediated stabilization of Nrf2 increased the levels of anti-apoptotic proteins Bcl-2 and Bcl-xL that led to cardioprotection in mice [24]. INrf2-mediated degradation of anti-apoptotic Bcl-2, and Bcl-xL proteins contributed to increased cellular apoptosis [25,26]. Nrf2 regulated Bcl-2 expression also prevented cellular apoptosis under stress conditions that led to cell survival [27]. However, the role of Nrf2 in regulation of Bcl-xL gene expression and induction during stress remains unknown.
In the present study, we investigated the role of Nrf2 in the regulation of anti-apoptotic factor Bcl-xL and its contribution to cellular apoptosis. Deletion mutagenesis of Bcl-xL gene promoter identified antioxidant-response element in the forward strand of the proximal Bcl-xL promoter that bound to Nrf2 and activated Bcl-xL gene expression. Nrf2 mediated up-regulation of Bcl-xL down regulated pro-apoptotic protein Bax, reduced caspases 3/7 activities and protected cells from etoposide mediated apoptosis. These results led to conclusion that Nrf2-mediated induction of Bcl-xL plays an important role in decreasing cellular apoptosis and enhancing cell survival.
Materials and Methods
Plasmid construction
1.565 Kb mouse Bcl-xL gene promoter was isolated from mouse tail genomic DNA by PCR usingthe forward 5’- ATTCTTGCTAGCTAGTGTCTGGAAGCCACTGGG-3’ and reverse 5'-ACCGCCAGATCTGCCTGTGTTTAGCGATTCTCTTC-3' set of primersand high fidelity platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA).The PCR-amplified promoter fragment was cloned into pGL2-basic luciferase vector (Promega, Madison, WI) using NheI and Bgl II restriction sites. The resulting plasmid was designed as pGL2b-1.565 kb (−64 to −1565, +1 is ATG site). Two deletion plasmids of Bcl-xL promoter were generated using specific set of primers. Forward primer 5'-ATTATTGCTAGCTGGCTGGAGCCTGGAGCAGAGA-3' (for -0.650 kb plasmid) and 5'-ATTATTGCTAGCTTCGCAATTCCTCTGTCGCCTTCT-3' (for −0.588 kb plasmid),and the same reverse primer 5'-ACCGCCAGATCTGCCTGTGTTTAGCGATTCTCTTC-3' were used to generate Bcl-xL promoter deletion plasmids. Forward primer 5’-GATGGAGGAACCAGGTTGACTGGGGATAGGTTCCTAAG-3’ and reverse primer 5’-CAACCTGGTTCCTCCATCGACCAGATCGAGGGCGGC-3’ and Gene tailor site directed mutagenesis kit (Invitrogen) were used to generate mutant ARE-F1 plasmid. In addition, we generated pGL2p-ARE-F1 and mutant ARE-F1 plasmids. ARE-F1 oligonucleotides (plus strand-5'-ATTGCACCCGGGGCTAGCCAGGTTGTGAGGGGGCAGGTTCCT-3' and minus strand-5’-ATTCGGCCCGGGGCTAGCAGGAACCTGCCCCCTCACAACCTG-3’) were synthesized, annealed, digested with Sma I and Nhe I enzymes and cloned into pGL2p vector to generate pGL2p-ARE-F1 plasmid. Similarly, mutant ARE-F1 oligonucleotides plus strand 5'-ATTGCACCCGGGGCTAGCCAGGTTGAATGGGGTTAGGTTCCT-3' and minus strand 5’-ATTCGGCCCGGGGCTAGCAGGAACCTAACCCCATTCAACCTG-3’ were cloned in pGL2p to generate pGL2p-mutant ARE-F1. We replaced luciferase coding sequence in wild type pGL2b-1.565 WT and ARE-F1 mutant pGL2b-1.565 MT plasmids with Bcl-xL cDNA using Bgl II and Cla I sites. Bcl-xL coding sequence was PCR amplified using forward primer 5’ – ATTCGAAGATCTACCGCCATGTCTCAGAGCAACCGG-3’ and reverse prime 5’-TTACATATCGATCTACTTCCGACTGAAGAGTGAGCCCAG-3’. The sequence accuracy of all plasmids was confirmed by DNA sequencing using ABI3700 capillarysequencer (Applied Biosystems, Foster City, CA). The construction of luciferaseplasmid harboring human NQO1 gene ARE, pCMV-FLAG-Nrf2, pCMV-FLAG-INrf2 and pcMV-Flag-Bcl-xL plasmids were described previously [26, 28].
Cell culture and generation of stable Flp-In T-REx HEK293 cells expressing tetracycline-inducibleNrf2 and INrf2
Mouse hepatocarcinoma (Hepa-1) and Human hepatoblastoma (Hep-G2) cells were obtained from the American Type Culture Collection (Manassas, VA). Human embryonic kidney (HEK-293) cells were obtained from Invitrogen, Carlsbad, CA. Hepa-1 and Hek-293 cells were grown in Dulbecco’s Modified Eagle’sMedium supplemented with 10% fetal bovine serum, penicillin (40 units/ml), and streptomycin (40 µg/ml). Hep-G2 cells were grown in alpha Minimum Essential Medium (α-MEM) containing 10% fetal bovine serum, penicillin (40 units/ml), and streptomycin (40 µg/ml). INrf2 mutant lung cancer A549 cells were grown in F12/DMEM medium. We also generated wild type INrf2 expressing stable A549 cells by transfection of pcDNA-INrf2 followed by selection of clones with neomycin (G148). For generation of stable Nrf2 and INrf2 expressing cells, Flp-In T-REx HEK293 cells were purchased from Invitrogen, Carlsbad, CA and co-transfected with FLAG-Nrf2 or FLAG-INrf2 in pcDNA/FRT/TO plasmids along with pOG44 plasmid (Invitrogen, Carlsbad, CA) by Effectene method (Qiagen, Valencia, CA) and the manufacturer's instructions. Forty eighthours after transfection, the cells were grown in medium containing200 µg/ml hygromycin B (Invitrogen, Carlsbad, CA). The 293/FRT/FLAG-Nrf2 and 293/FRT/FLAG-INrf2cells stably expressing tetracycline-inducible Nterminal FLAG-taggedNrf2 and FLAG-tagged INrf2 were selected. The stably selected cells were grown andtreated with 1 to 2 µg/ml tetracycline (Sigma, St. Louis, MO) for varyingperiods of time to follow the over-expression of FLAG-taggedNrf2 protein and FLAG-tagged INrf2 protein. The cellswere grown in monolayer in an incubator at 37°C in 95%air and 5% CO2.
Preparation of cell lysates and western blotting
Hepa-1or Hep-G2 and Hek293 cells were seeded in 100-mm plates and transfected/treated as displayedin the figures. Cells were washed twice with ice-cold phosphate-bufferedsaline, trypsinized, and centrifuged at 1500 rpm for 5 min. For making whole cell lysates, the cells were lysedin RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.2 mM EDTA, 1% NonidetP-40, 0.5% Sodium deoxycholate, with 1X proteaseinhibitor mixture (Roche Applied Science, Branford, CT). The protein concentration was determinedusing the protein assay reagent (Bio-Rad, Hercules, CA). Sixty to eighty microgramsof proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 3% non-fat dry milk in TBST. For immunoblotting, the following antibodies were used: anti-Nrf2 (H-300) (1:1000), anti-Bcl-xL (H-5) (1:1000), anti-Bax (P-20) (1:1000) and anti-lamin B (1:1000), all purchased from Santa Cruz Biotechnology (CA). Anti-FLAG-HRP and anti-actin antibodies were obtained from Sigma, St. Louis, MO. Immunoreactive bands were visualized using a chemiluminescence ECL system (GE Healthcare Biosciences, Pittsburg, PA). The intensity of the protein bands after immunoblotting were quantitated byusing Quantity One 4.6.3 Image software (ChemiDoc XRS, Bio-Rad, Hercules, CA) and normalized against proper loading controls. Cytoplasmic and nuclear fractions were prepared using the Active Motif nuclear extract kit (Active Motif, Carlsbad, CA) following the manufacturer'sprotocol. To confirm the purity of nuclear-cytoplasmic fractionation, the membranes were re-probed with cytoplasm-specific, anti-lactatedehydrogenase (LDH) (Chemicon International Inc., Temecula, CA) and nuclear specific, anti-lamin-Bantibodies (Santa Cruz Biotechnology, CA). In related experiments, the cells were treated with 50 µMt-BHQ or 30 µg/ml cycloheximide or 2 µg/ml actinomycin D and DMSO as a vehicle control.
Transient transfection and luciferase assay
Hepa-1 cells were plated in 100 mmplates at a density of 1×106 cells/plate 24h prior to transfection. In the related experiments, the cells were transfected with 1 µg of the indicated plasmids using Effectene transfection reagent (Qiagen, Valencia, CA) according to the manufacturer’s instructions. After 36h of transfection, cells were harvested and cellular specific protein regulation was examined by western blotting. For luciferase reporter assay, Hepa-1 cells were grown in monolayer cultures in 12-well plates. After 12h, cells were co-transfected with 0.1 µg of indicated Bcl-xL promoter ARE-Luc reporter constructs and 10 times less quantities of fireflyRenilla luciferase encoded by plasmid pRL-TK. Renilla luciferasewas used as the internal control in each transfections. After 12h of transfection, the cells were further treated with DMSO or tBHQ for 24h. Cells were washed with 1X phosphate-buffered saline and lysed in 1X Passive lysis buffer from the Dual-Luciferase®reporter assay system kit (Promega, Madison, WI). The luciferase activity was measured and plotted.
siRNA interference assay
Mouse and human specific Nrf2 and Bcl-xL siRNA were obtained from Applied Biosystem, Foster City, CA and used to inhibit Nrf2 and Bcl-xL protein. Hepa-1 or Hep-G2 cells were transfected with 50 to 100 nM of Nrf2 siRNA or Bcl-xL siRNA or GAPDH control siRNA as indicated in different figures using Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA)according to the manufacturer’s instructions. Thirty two hours after transfection, the cells were harvested and lysates were immunoblotted for Nrf2, Bcl-xL, NQO1 and actin proteins.
Real time quantitative PCR
Hepa-1cells were treated with t-BHQ for various time periodsor transfected with siRNA against Nrf2 as indicated in the figures. The total RNAwas isolated using the RNeasy mini kit (Qiagen, Valencia, CA). Two µg of totalRNA was subjected to reverse transcription using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystem, Foster City, CA). After synthesis of cDNA at37°C for 60 min, the PCR was performed using 7500 Real Time PCR system as per manufacturer’s instructions. For Real Time PCR, Bcl-xL primer probe amplicon Mm00437783_m1, NQO1 primer and probe amplicon Mm00500821_m1 and GusB amplicon Mm00446953_m1 as an internal control (Applied Biosystems, Foster City, CA) were used. Total mix was run on 7500 Real Time System (Applied Biosystems, Foster City, CA) using relative quantitation according to the manufacturer's protocols.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using a kit from Active Motif as described previously [14]. Briefly, 70% confluent Hepa-1 cells were treated with DMSO or 50 µM t-BHQ for 4h and then fixed in 1% formaldehyde for 15 min. Cells were lysed and nuclei pelleted by centrifugation. Nuclei were resuspended and sheared using a sonicator (MisonixInc., Farmingdale, NY) with five pulses of 20sec at 25% of maximum output. Sheared chromatin was immuno precipitated with 2 µg of anti-Nrf2 or control IgG antibody. The cross-links reversed overnight at 65°C and deproteinated with 20 µg/ml proteinase K. Nrf2 bound Bcl-xL ARE-F1 was detected by PCR amplification with the primers as follows: forward primer 5'-TTCTCCTGACTCCCAGTAGGAGG-3' and reverse primer, 5'-AAATCTATCTCCGGCGACAGCAAGCG-3'. GAPDH PCR was performed as internal control. Primers used for GAPDH amplification were: forward primer 5’ACCACAGTCCATGCCATCAC-3’ and reverse primer 5’TCCACCACCCTGTTGCTGTA3’. The PCR conditions used for ChIP assay were 37 cycles of a denaturing step at 94°C for 30s, an annealing step at 65°C for30s, and an extension step at 72°C for 30s. PCR products were separated on 2% agarose gel containing ethidium bromide andimaged using Quantity One 4.6.3 Image software (ChemiDoc XRS, Bio-Rad, Hercules, CA).The relative binding of Nrf2 to the ARE-F1 region of Bcl-xL promoter was also measured by quantitative Real-Time PCR using custom made probes and primes obtained from Applied Biosystem (ID-AIPADGH). The mixture was run on 7500 Real Time System (Applied Biosystems) according to the manufacturer's protocols.
Caspases activity
The effect of Nrf2, INrf2, antioxidant tBHQ and etoposide on the caspase-3/7 activity was analysed. Hepa-1 cells were transfected with FLAG-Nrf2 or FLAG-INrf2 for 24h and cells were treated with etoposide (20 µM) for 36h. One set of cells were further treated with tBHQ for additional 24h as indicated in figures. Cells were harvested and lysed in the lysis buffer. Twenty micrograms of cell lysates were mixed with Caspase/Glo 3/7 substrate (Promega, Madison, WI) and caspases 3/7 activity was measured as manufactures instructions and plotted.
DNA fragmentation/cell death assay
Hepa-1 or Contol-293 or FRT-Nrf2-293 cells were plated at a density of 2000 cells per well in 96 well plates. After 20h, Hepa-1 cells were treated with DMSO or tBHQ (24h) or transfected with pcDNA or Flag-Bcl-xL plasmid. Nrf2-293 cells were treated with water or 1 µg/ml of tetracycline for 24h, then, both cell lines were exposed to various indicated concentrations of etoposide for 30h. Cells were harvested and a photometric enzyme immunoassay was performed for the quantitative in vitro determination of cytoplasmic histone-associated DNA fragments (mono and oligonucleosomes) using Cell Death Detection ELISA kit (Roche: as per manufacturer’s instructions). Each combination of cell line and drug concentration was set up in eight replicate wells, and the experiment was repeated thrice. Each data point represents a mean ± SD and normalized to the value of the corresponding control cells.
MTT cell survival assay
Hepa-1 or HepG2 cells were plated at a density of 2000 cells per well in 96 well plates, allowed to recover for 12h, and then transfected with Nrf2 or Bcl-xL or control siRNA for 24h. Control and tranfected cells were exposed with etoposide (25 µM) for 30h. In some experiments Bcl-xL knockdown cells further treated with DMSO or tBHQ for 24h. Cells were incubated with fresh MTT solution (200 µl/well; stock 5 mg/ml in PBS) for 2h and absorbance at 570 nm was measured. Each combination of cell line and drug concentration was set up in eight replicate wells, and the experiment was repeated thrice. Each data point represents a mean ± SD and normalized to the value of the corresponding control cells.
Clonogenic cell survival assay
A549 cells and INrf2-A549 cells were grown to 70% confluence and transfected with Bcl-xL or control siRNA and treated with either DMSO or tBHQ in presence of etoposide (15 µM) for 30h. Cells were trypsinized and reseeded for 9 days. The fresh medium was added at day 5. After 10 days of incubation, a freshly prepared 2 ml clonogenic reagent (0.25% 1,9 dimethyl-methylene blue in 50% ethanol) was added into the plates and plates were kept at room temperature for 45 min. Cells were washed with PBS twice and blue colonies were counted. Each data point represents a mean ± SD and normalized to the value of the corresponding control cells.
Statistical analyses
Data from luciferase assays, real time PCR, DNA fragmentation and cell survival assay were analyzed using a two-tailed Student's t test. Data are expressed as mean ± S.D. of three independent experiments. Significance values are represented as *, p< 0.05; **, p< 0.03; *** p<0.01 and are shown in the figures.
Results
Antioxidant tBHQ up-regulates Bcl-xL gene expression
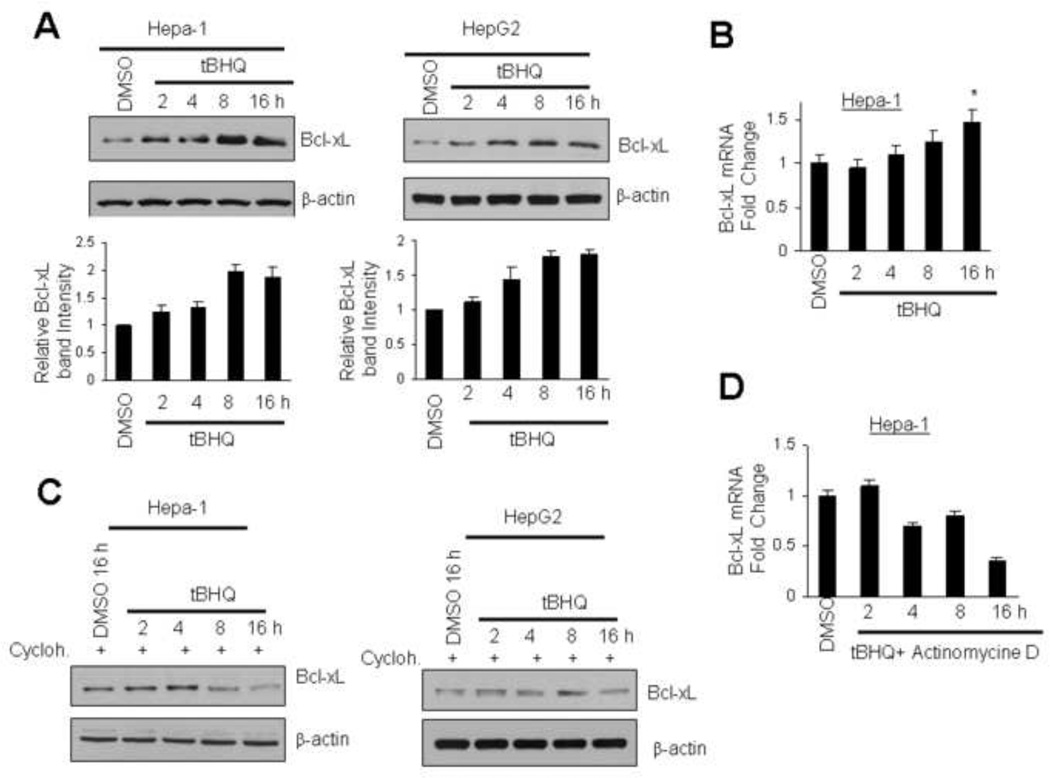
Immunoblot analysis of Hepa-1 and Hep-G2 cells treated with t-BHQ showed time dependent increase in Bcl-xL protein (Fig. 1A). Highest induction was observed around 8-16h after t-BHQ treatment in both cell lines (Fig.1A, upper and lower panels). Hepa-1 cells treated with t-BHQ were also analyzed for Bcl-xL RNA transcripts by quantitative real time PCR to test if tBHQ induced transcription of Bcl-xL gene to increase Bcl-xL protein (Fig. 1B). Treatment of Hepa-1 cells with tBHQ (8-16h) increased Bcl-xL mRNA by 1.6 fold compared with DMSO control (Fig. 1B). In related experiments, the pre-treatment of Hepa-1 and Hep-G2 cells with protein synthesis inhibitor cycloheximide failed to demonstrate tBHQ-mediated increase in Bcl-xL protein (Fig.1C). In addition, RT-PCR analysis revealed that pre-treatment of Hepa-1 cells with transcription inhibitor actinomycin D also failed to induce tBHQ-mediated Bcl-xL gene transcription (Fig. 1D). These results together suggested that the antioxidant tBHQ up-regulated Bcl-xL gene expression.
Fig. 1.
Antioxidant t-BHQ induces Bcl-xL gene expression. (A) Hepa-1 and HepG2 cells were treated with DMSO or antioxidant t-BHQ (50 µM) for different time periods and 60 µg cell extracts were immunoblotted with Bcl-xL and actin antibodies. The Bcl-xL band intensities were quantified and plotted (Fig. 1A, lower panels). (B) The effect of t-BHQ on Bcl-xL mRNA expression was analysed by real time quantitative PCR. (C) Hepa-1 and HepG2 cells were pre-treated with cycloheximide (30 µg/ml, 2h) and followed by treatment with DMSO or t-BHQ for indicated time periods in presence of cycloheximide. Cells were lysed and 60 µg lysates were immunoblotted with indicated antibodies. (D) Hepa-1 cells were pre-treated with 2 µg/ml of actinomycin D for 2h followed by 50 µM tBHQ+actinomycin D for indicated time periods. Bcl-xL mRNA expression was analysed by real time quantitative PCR. The data shown are mean ± S.D. of three independent experiments.
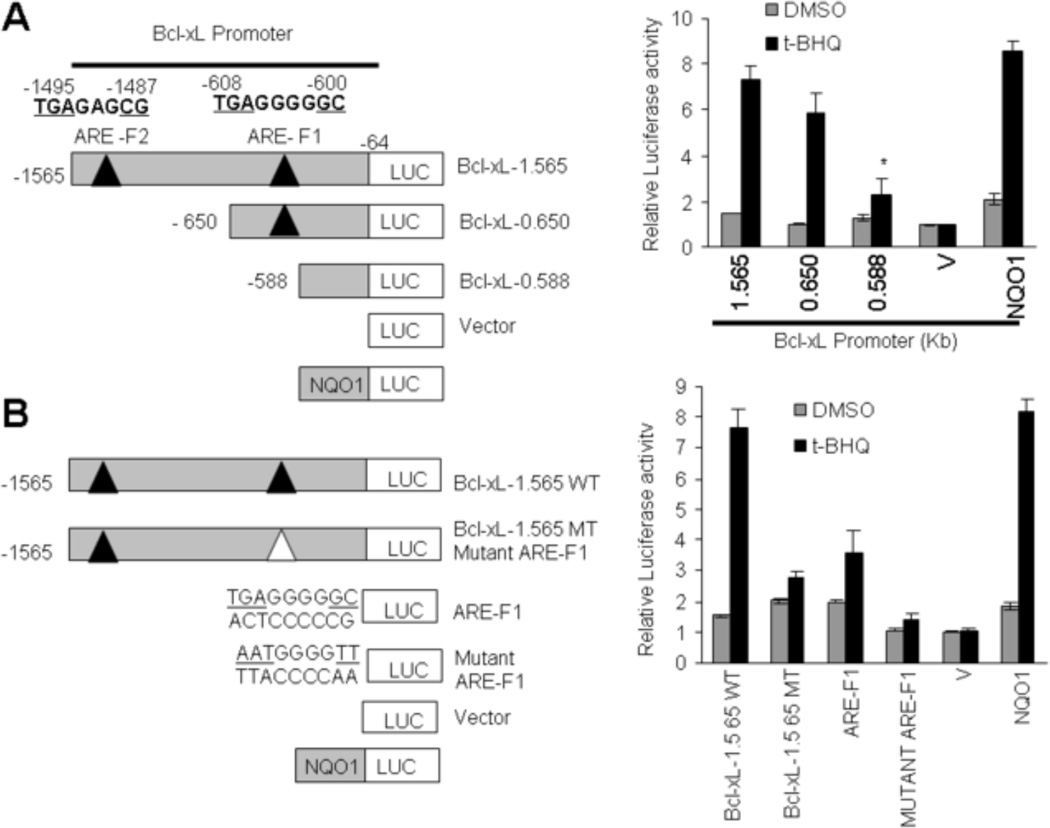
An ARE in the proximal Bcl-xL promoter on forward strand mediates expression and t-BHQ induction of Bcl-xL gene expression
Nucleotide sequence analyses of 1565 bp Bcl-xL promoter revealed the presence of two putative AREs (Fig. 2A, left panel). Both ARE elements were present on the forward strand at nucleotide positions −600 (ARE-F1)and −1487 (ARE-F2) from the start site of translation (ATG, +1). 1565 bp Bcl-xL gene promoter attached to the luciferase gene upon transfection in Hepa-1 cells produced luciferase activity that was induced in response to t-BHQ (Fig. 2A, left and right panels). The t-BHQ induction of Bcl-xL gene expression was comparable to known t-BHQ induction of NQO1-luciferase gene expression (Fig. 2A, right panel). Deletion mutagenesis in 1565 bp Bcl-xL gene promoter and transfection analysis in Hepa-1 cells revealed that promoter region between nucleotides −650 to −588 containing ARE-F1 contributed to a majority (>90%) of t-BHQ induction of Bcl-xL gene expression in response to t-BHQ (Fig. 2A, left and right panels). Deletion mutagenesis and transfection studies also showed that the contribution of nucleotide region between nucleotides −1565 to −650 containing ARE-F2 to t-BHQ induction appeared minor (<10%). Mutation of the ARE-F1 element in 1565 bp Bcl-xL promoter showed a significant reduction in t-BHQ induction as compared with the wild type 1565 bp Bcl-xL promoter (Fig. 2B, left and right panels). In related experiments, ARE-F1 but not mutated ARE-F1 cloned separately in the pGL2p promoter vector upon transfection in Hepa-1 cells demonstrated t-BHQ induction of luciferase gene expression through the heterologouspromoter (Fig. 2B, left and right panels). The mutant ARE-F1 in the same experiments showed significantly reduced basal expression and the loss of t-BHQ induction of luciferase gene expression, as compared with wild type ARE-F1. The results combined suggested that ARE-F1 between nucleotide region −608 to −600 of Bcl-xL promoter was required for t-BHQ induction of the Bcl-xL gene expression.
Fig. 2.
ARE-F1 between nucleotides −608 to −600 in the forward strand of Bcl-xL gene promoter is essential for antioxidant induction of Bcl-xL gene expression.(A). Systematic representation and cloning strategy of mouse Bcl-xL gene promoter into PGL2B or pGL2P luciferase reporter vectors. Two putative ARE sequences of the Bcl-xL promoter (ARE-F1 and ARE-F2) on the sense strand are shown (upper left panel). Mouse Bcl-xL promoter (1.565 kb) and deletions were separately cloned into PGL2B luciferase (Luc) vector and plasmids were separately transfected in Hepa-1 cells. Cells were treated with DMSO or 50 µM t-BHQ for 24h, and luciferase activity was measured (right panels). Human NQO1-ARE luciferase reporter plasmid was also transfected in Hepa-1 cells as a positive control for t-BHQ-mediated luciferase gene induction. (B). Bcl-xL 1.56-WT (wild type) and Bcl-xL-1.56 MT (mutated ARE-F1) promoter plasmids were separately transfected in Hepa-1 cells and analyzed for luciferase gene expression. In the same experiment ARE-F1 and mutant ARE-F1 sequences were attached to SV40 basal promoter hooked to luciferase reporter gene by cloning in vector pGL2P, transfected in Hepa-1 cells, treated with DMSO or t-BHQ (50 µM for 24h), and analyzed for luciferase activity (rigut panel). Human NQO1-ARE luciferase reporter plasmid was also transfected in Hepa-1 cells as a positive control for t-BHQ-mediated luciferase gene induction. The data shown are mean ± S.D. of three independent transfection experiments. V, vector control.
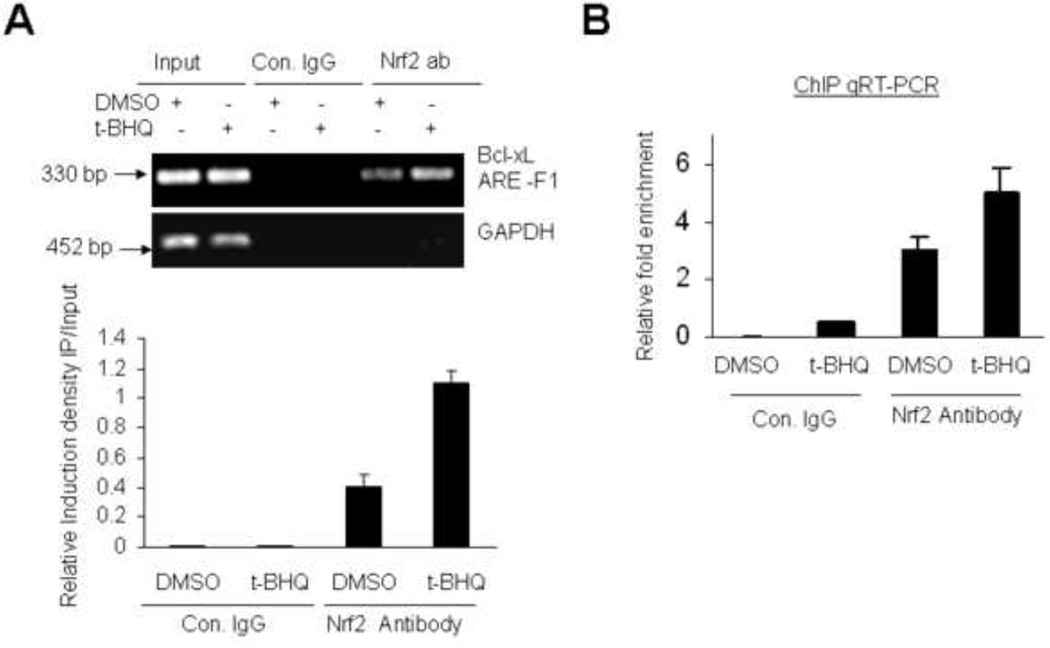
Antioxidant t-BHQ Increases in vivo binding of Nrf2 to the ARE-F1 of Bcl-xL promoter
Nuclear factor Nrf2 is known to bind to NQO1 gene ARE leading to increase in NQO1 gene expression [1]. We performed ChIP assays to test whether Nrf2 interacts with Bcl-xL gene ARE-F1 (Fig. 3). Nrf2-specificantibody and PCR primers covering the ARE-F1 region in the Bcl-xL promoter determined the binding of Nrf2 to ARE-F1 in DMSO and t-BHQ-treated Hepa-1 cells (Fig. 3A). The results demonstrated Nrf2 binding to Bcl-xL gene ARE-F1 that was enhanced 2.6 fold in response to antioxidant t-BHQ (Fig.3A, upper and lower panels). Nrf2 binding to the ARE-F1 was not detected when chromatin was immunoprecipitatied with control rabbit IgGs (Fig. 3A). ChIP assay and quantitative Real-Time PCR also determined the relative binding of Nrf2 to the Bcl-xL ARE-F1 in control and t-BHQ treated cells (Fig. 3B). These results demonstrated specific interaction of nuclear factor Nrf2 to Bcl-xL gene ARE-F1, which was enhanced upon t-BHQ treatment.
Fig. 3.
Antioxidant increases binding of Nrf2 to Bcl-xL gene ARE-F1. (A) ChIP assay. Hepa-1 cells were treated with 50 µM t-BHQ for 4h, fixed with formaldehyde, cross-linked, and sheared the chromatin. The chromatin was immunoprecipitated with anti-Nrf2 antibody or control IgG. Nrf2 binding to Bcl-xL promoter was analyzed by PCR with specific primers for region containing AREF-1 of Bcl-xL promoter. GAPDH primers were used as a control (upper panel). The relative binding of Nrf2 to the Bcl-xL-ARE-F1 promoter was quantified from the band intensities from upper panel and plotted (lower panel). (B) ChIP and qRT-PCR. The chromatin was immunoprecipitated as described in “A” from Hepa-1 cells. The binding of Nrf2 to the ARE-F1 region of Bcl-xL promoter was measured by quantitative Real-Time PCR using custom made probes and primes from Applied Biosystem. The mixture was run on 7500 Real Time System (Applied Bio systems) using relative quantitation according to the manufacturer's protocols.
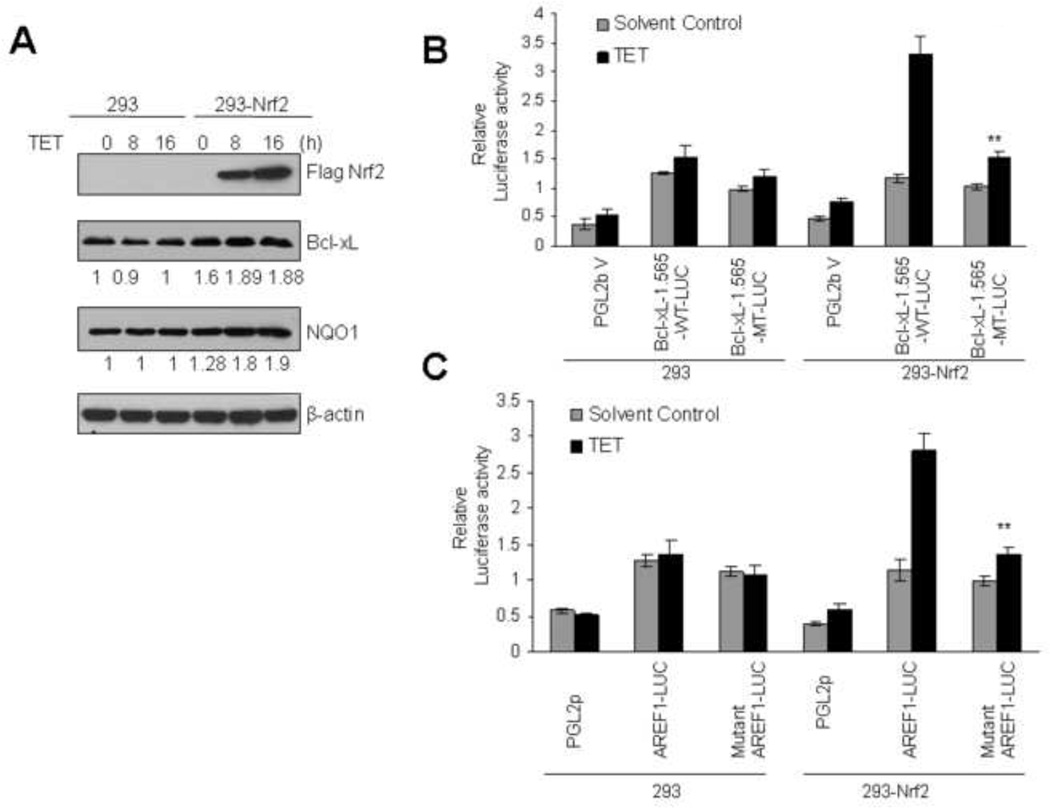
Nrf2 mediates t-BHQ induction of Bcl-xL gene expression
Nrf2 overexpression and knock down strategies were used to determine a role of Nrf2 in the regulation of Bcl-xL gene expression. We successfully generated stable Flag-Nrf2-Hek-293 (293-Nrf2) cell lines, which expressed Flag-Nrf2 protein upon exposure to tetracycline (Tet ‘On’ and ‘Off’ system) as also described previously (Fig. 4A, ref. 14). In the same experiment, 293 cells upon exposure to tetracycline failed to demonstrate overexpression of Flag-Nrf2 and served as control (Fig. 4A). Overexpression of Flag-Nrf2 in 293-Nrf2 cells by tetracycline significantly increased Bcl-xL and NQO1 protein levels (Fig. 4A). In the same experiment, control 293 cells that did not overexpress Nrf2 failed to demonstrate an increase in Bcl-xL or NQO1 gene expression (Fig. 4A). In similar experiments, the transfection of wild type Bcl-xL-1.565WT-LUC (Fig. 4B) or wild type Bcl-xL-ARE-F1-LUC (Fig. 4C) plasmids in 293-Nrf2 cells upon treatment with tetracycline showed significant increase in luciferase gene expression. The mutant plasmid Bcl-xL-1.565-MT-LUC and mutant ARE-F1-LUC in the same experiment failed to demonstrate an increase in luciferase gene expression upon stimulation with tetracycline (Fig. 4B &C). On contrary, the transfection of wild type or mutant Bcl-xL-1.565-LUC or ARE-F1 plasmids in control Hek-293 cells failed to induce tetracycline mediated luciferase activity (Fig. 4B & C).
Fig. 4.
Overexpression of Nrf2 up regulates endogenous and transfected Bcl-xL gene expression. (A). HEK-293 and HEK-293-Nrf2 cells expressing tetracycline-induced FLAG-tagged Nrf2 were treated with 2 µg/ml tetracycline for the indicated times. Sixty µg cell lysates were immunoblotted with anti-FLAG, anti-Bcl-xL, anti-NQO1 and anti-actin antibodies (upper panel). The band intensities of Bcl-xL and NQO1 from “A” were quantified and presented below the blots. (B & C). HEK293 and HEK293-Nrf2 cells were co-transfected with wild type pGL2B-Bcl-xL-1.565-WT-LUC or mutant type pGL2B-Bcl-xL-1.565-MT-LUC with the internal control Renilla luciferaseplasmid pRL-TK (B) or co-transfected with pGL2p-ARE-F1-LUC or pGL2p-mutant ARE-F1-LUC plasmids with the internal control Renilla luciferase plasmid pRL-TK (C). pGL2B and pGL2p vectors were also transfected as negative control. Twenty-four hours after transfection the cells were treated with 2µg/ml tetracycline for 16h and lysates were analyzed for luciferase activity. The data shown are mean ± S.D. of three independent transfection experiments.
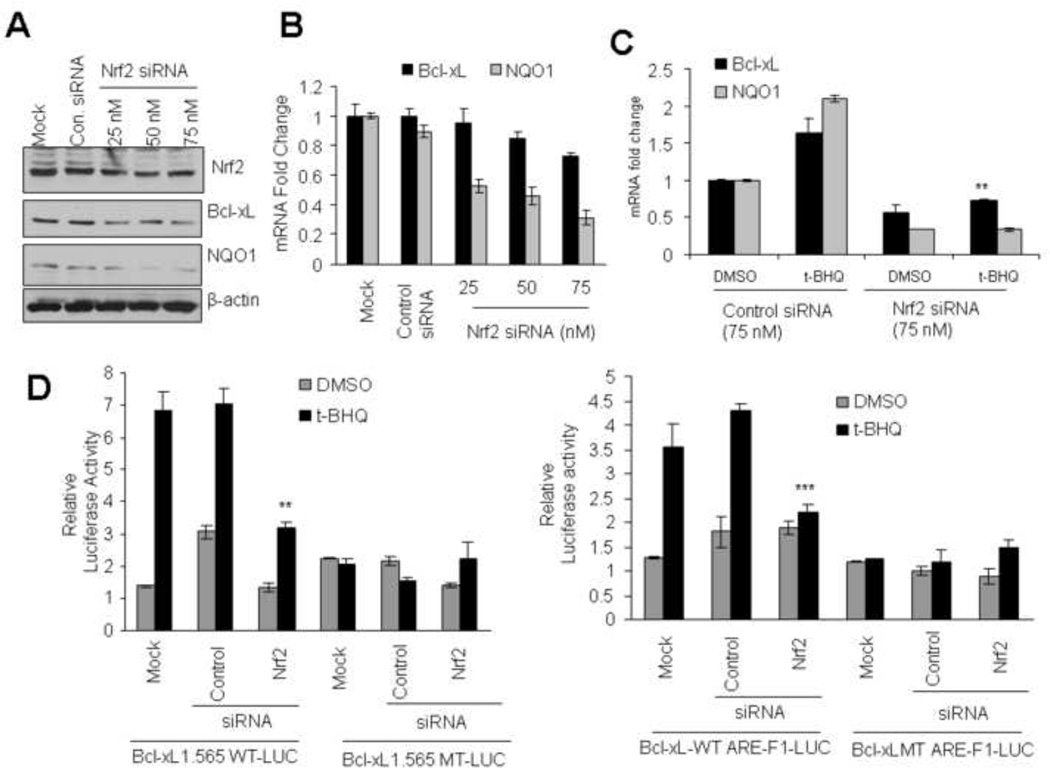
To strengthen the overexpression results, we used siRNA to inhibit Nrf2 expression in Hepa-1 cells and determined its effect on Bcl-xL and NQO1 expression (Fig. 5). Transient trasfection of Hepa-1 cells with Nrf2 siRNA led to siRNA concentration dependent decrease in Nrf2, Bcl-xL and NQO1 (Fig. 5A). RT PCR analysis also showed that siRNA-mediated inhibition of Nrf2 led to significantly deceased Bcl-xL and NQO1 transcripts in Hepa-1 cells (Fig. 5B). In addition, siRNA-mediated decrease in Nrf2 led to reduced Bcl-xL and NQO1 RNA (Fig. 5C). In similar experiments, siRNA inhibition of Nrf2 led to significant decrease in t-BHQ induction of luciferase in Bcl-xL1.565-WT-LUC (5D, left panel) and Bcl-xL-WT AREF1-LUC (Fig. 5D, right panel) transfected Hepa-1 cells. The decrease in luciferase activity was absent in Hepa-1 cells transfected with plasmid mutant Bcl-xL-MT-LUC (Fig. 5D, left panel) and Bcl-xLMT-ARE-F1-LUC (Fig. 5D, right panel). These results together with overexpression results suggested that Nrf2 through ARE-F1 controls expression and t-BHQ induction of Bcl-xL gene expression. This control was similar to NQO1 gene expression and induction (Current report and ref. 4).
Fig. 5.
siRNA inhibition of Nrf2 decreases t-BHQ-inducible expression of Bcl-xL.(A) Western analysis. Hepa-1 cells were transfected with control or 25, 50, and 75 nM of Nrf2 siRNA. Forty-eight hours after transfection, cells were harvested, lysed, and cell extracts were immunoblotted. (B) Real time-PCR analysis. Hepa-1 cells were transfected with control or Nrf2 siRNA. Twenty-four hours after siRNA transfection, cells were harvested, total RNA extracted and converted to cDNA. The mRNA levels of Bcl-xL and NQO1 were quantified by Real Time-PCR. (C) Real time-PCR analysis. Hepa-1 cells were transfected with control or Nrf2 siRNA (75 nM). Twenty-four hours after siRNA transfection, cells were treated with tBHQ for additional 16h, cells were harvested and total RNA was extracted. The mRNA levels of Bcl-xL and NQO1 were quantified by Real time-PCR. (D) Reporter analysis. Hepa-1 cells were transfected with control or Nrf2 siRNA. Twenty-four hours after transfection, cells were transfected with wild type or mutant Bcl-xL-1.565 (left panels) or with ARE-F1 or mutant ARE-F1 sequences in the pGL2p vector plasmids (right panels), incubated with DMSO or t-BHQ (50µM) for 24h and analyzed for luciferase activity. The data shown are mean ± S.D. of three independent transfection experiments.
Antioxidant (tBHQ) treatment stabilized Nrf2 up-regulates Bcl-xL expression leading to decrease in Bax and Caspases 3/7 activities
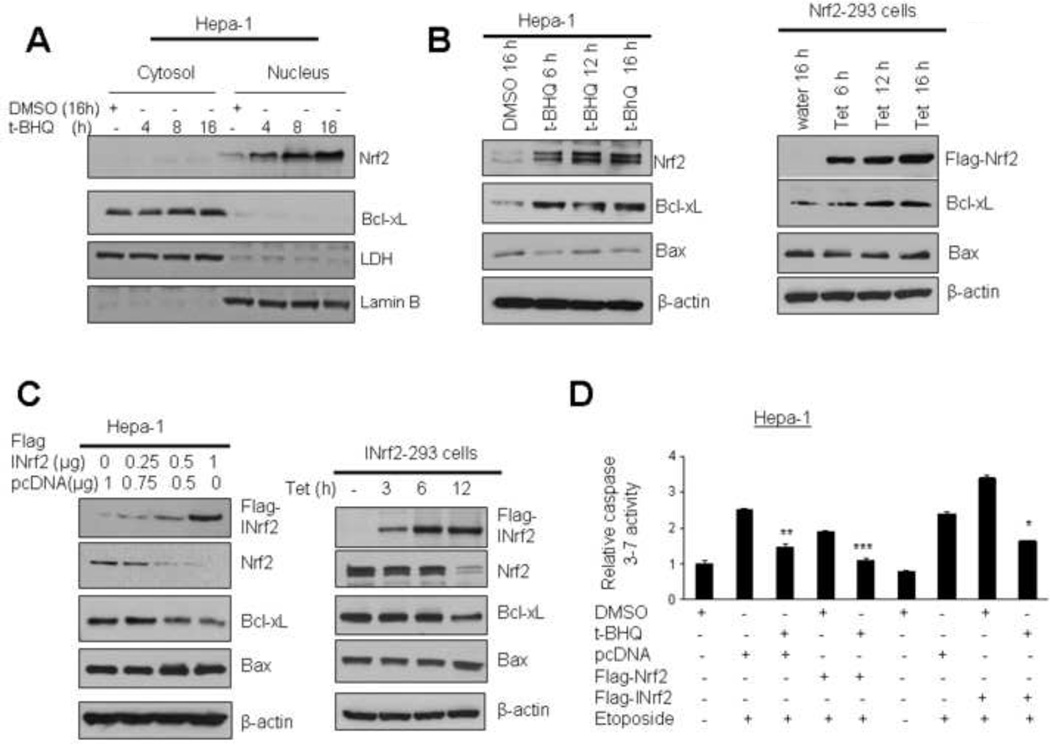
Bcl-xL is anti-apoptotic cellular factor that prevents cellular apoptosis. Results as described above suggested that antioxidant tBHQ and Nrf2 control Bcl-xL gene expression. This raised an important question whether Nrf2 mediated Bcl-xL expression contributes to control of cellular apoptosis. Hepa-1 cells were treated with t-BHQ for different time periods, cytosolic and nuclear fractions made and analyzed by immunoblotted with Nrf2, Bcl-xL, LDH and lamin B antibodies (Fig. 6A). The results demonstrated that t-BHQ stabilized Nrf2 that localized in the nucleus and led to increased expression of Bcl-xL in the cytoplasm (Fig. 6A). Next, we examined the effect of Nrf2 mediated up-regulation of Bcl-xL protein on apoptosis marker proteins Bax and caspases. Hepa-1 cells treated with tBHQ or Nrf2-293 cells overexpressing tetracycline-induced Flag-Nrf2 protein in Nrf2-Hek293 cells were immunoblotted for Nrf2, Bcl-xL, Bax and actin (Fig. 6B). t-BHQ-mediated stabilization of Nrf2 in Hepa-1 and overexpression of Flag-Nrf2 in Nrf2-Hek293 cells caused time dependent increase in Bcl-xL and decrease in Bax. In related experiments, dose dependent overexpression of INrf2 (inhibitor of Nrf2 or Keap1) in Hepa-1 cells by transient transfection or Flag-tagged-INrf2 overexpression in Hek-293 cells by tetracycline treatment led to degradation of Nrf2 and subsequent decrease in the expression of Bcl-xL and stabilization of Bax protein (Fig 6C). In related experiments, the treatment of Hepa-1 cells with etoposide enhanced endogenous caspases3/7 activity by 3.1 fold indicating increased apoptosis (Fig. 6D). Post-treatment of Hepa-1 cells with tBHQ or overexpression of Nrf2 both decreased caspases3/7 activity by 1 to 2 fold (Fig. 6D). On the other hand, overexpression of INrf2 and etoposide treatment increased caspase3/7 (~3.5 fold) that was decreased (1.7 fold) upon treatment with tBHQ. These results suggested that Nrf2 overexpression or tBHQ treatment up-regulated Bcl-xL protein and down regulated the activities of pro-apoptotic factors Bax and caspases3/7.
Fig. 6.
Antioxidant tBHQ and overexpresion of Nrf2 up regulates Bcl-xL leading to decreased Bax and caspases3/7 activity. (A) Hepa-1 cells were treated with DMSO or antioxidant tBHQ for different time periods and nuclear and cytoplasmic extracts were generated. Eighty microgram nuclear and cytoplasmic extracts were immunoblotted. (B) Hepa-1 cells were exposed to DMSO or tBHQ for different time intervals (left panel). In related experiments, HEK293-Nrf2 cells were treated with tetracycline for various time periods (right panel). The endogenous levels of Nrf2 in Hepa-1 cells, overexpressed FLAG-Nrf2 in HEK-293-Nrf2 cells, Bcl-xL, Bax and actin levels were analysed by western blotting. (C) Hepa-1 cells were transfected with pcDNA or increasing concentration of FLAG-INrf2 plasmid (left panel) or HEK-INrf2-293 cells were treated with tetracycline for different time points (right panel), the levels of Flag-INrf2 in Hepa-1 cells, overexpressed FLAG-INrf2 in HEK-293-INrf2 cells, Nrf2, Bcl-xL, Bax and actin were analysed by western blotting. (D) Hepa-1 cells were transfected with Flag-Nrf2 or Flag-INrf2 for 16h and treated with etoposide (20 µM) 30h alone or in presence of 50 µM tBHQ or DMSO as indicated. Cells were harvested and lysed in the lysis buffer. Twenty µg cell lysate were mixed with Caspase Glo 3/7 substrate (Promega) and Caspase 3/7 activity was measured and plotted. The experiment was repeated thrice. Each point represents a mean ± SD and normalized to the value of the corresponding control cells.
Nrf2-mediated up-regulation of Bcl-xL prevents etoposide induced DNA fragmentation
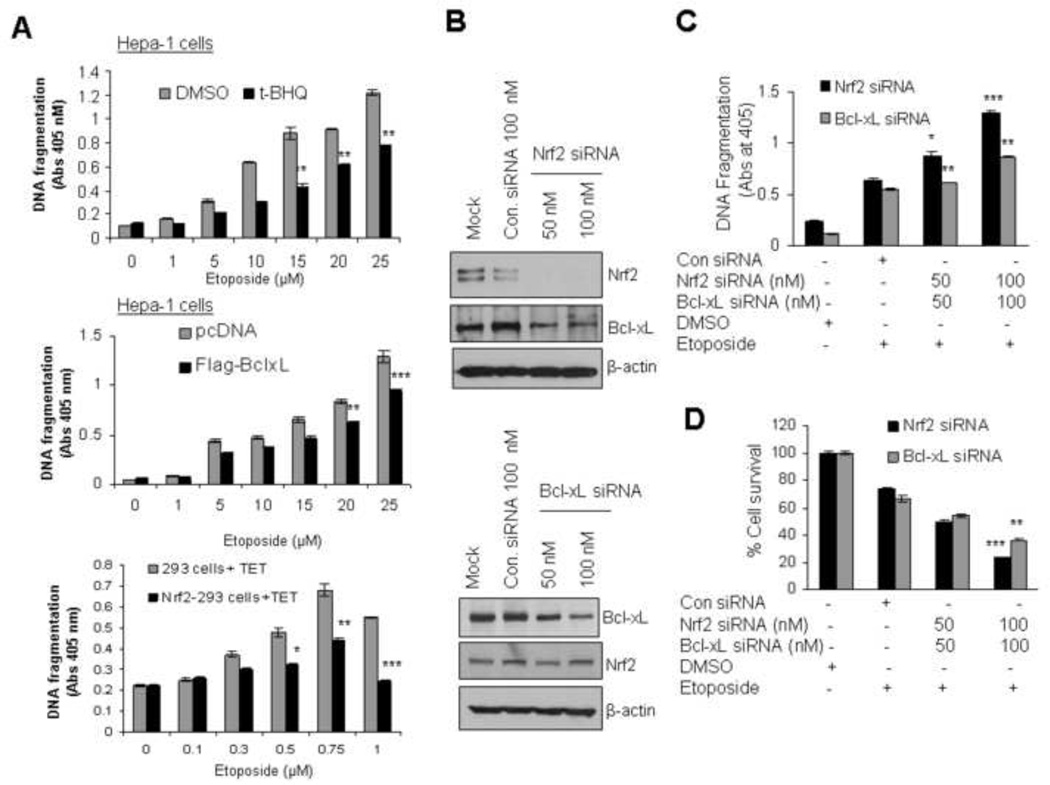
To examine the role of Nrf2 and Bcl-xL in apoptosis, drug resistance and cell survival, we quantified the etoposide mediated cytoplasmic histone-associated DNA fragmentation in Hepa-1 cells. Cells were treated with increasing concentrations of etoposide for 48h followed by treatment with either DMSO or tBHQ for an additional 24h and analyzed for DNA fragmentation (Fig. 7A, upper panel). In another experiment, Hepa-1 cells were transfected with pcDNA or Flag-Bcl-xL plasmids for 24h and treated with increasing concentrations of etoposide for 48h and analyzed for DNA fragmentation (Fig 7A, middle panel). The results demonstrated that histone-associated DNA fragmentation significantly decreased (40%) in Hepa-1 cells treated with etoposide plus tBHQ compared with etoposide plus DMSO and ~25% in Flag-Bcl-xL transfected cells compared with pcDNA transfected Heap-1 cells. In similar experiments, overexpression of tetracycline-induced Flag-Nrf2 in Nrf2-293 cells decreased ~38% of etoposide induced DNA fragmentation compared with control 293 cells treated with tetracycline (Fig. 7A, lower panel). The results again suggested that increasing the cellular levels of Nrf2 or Bcl-xL down regulated etoposide mediated DNA fragmentation. Additional experiments were performed with siRNA inhibition of either Nrf2 or Bcl-xL to strengthen the overexpression results (Fig. 7B, C & D). Hepa-1 cells transfected with Nrf2 siRNA or Bcl-xL siRNA specifically reduced more than 90% and 65% of cellular Nrf2 and Bcl-xL proteins cells, respectively (Fig.7B upper and lower panels). Nrf2 and Bcl-xL siRNA transfected cells upon treatment with etoposide led to 30 to 35% increase in DNA fragmentation and decrease 50 to 55% of cell survival compared with control siRNA transfected cells (Fig. 7C & D). The results also suggested that Nrf2 inhibition as compared with Bcl-xL inhibition had greater effect on etoposide induced apoptosis. Hepa-1 cells with inhibited Nrf2 demonstrated higher DNA fragmentation and lower cell survival, as compared with Bcl-xL inhibited cells (Fig. 7C). These results combined suggested that both Nrf2 and Bcl-xL contributed to control of apoptosis. However, Nrf2 played a bigger role than Bcl-xL presumably because of the contribution of other Nrf2 downstream proteins in control of apoptosis.
Fig. 7.
Antioxidant-mediated stabilization and overexpression of Nrf2 or Bcl-xL reduced etoposide-mediated DNA fragmentation leading to cell survival. (A) Stabilization/overexpression of Nrf2 or Bcl-xL decreased DNA fragmentation. Hepa-1 cells were treated with increasing concentrations of etoposide for 30h followed by treatment with DMSO or tBHQ for additional 24h (upper panel). Similarly Hepa-1 cells were transfected with pcDNA of Flag-Bcl-xL plasnid for 24h and treated with increasing concentrations of etoposide for 30h (middle panel). Control HEK293 and HEK293-Nrf2 cells were treated with increasing concentrations of etoposide for 24h followed by treatment with 2µg/ml of tetracycline for 24 h (lower panel). The cytoplasmic histone-associated DNA fragments [mono and oligonucleosomes) were quantified using Cell Death Detection ELISA kit (Roche) and plotted]. The experiment was repeated thrice. Each point represents a mean ± SD and normalized to the value of the corresponding control cells.(B) Nrf2 or Bcl-xL knockdown increased DNA fragmentation and decreased cell survival. Hepa-1 cells were transfected with control siRNA 100 nM or two different concentration of Nrf2 siRNA (50 nM and 100 nM) (upper panel) or Bcl-xL siRNA (lower panel). After 24 hours of transfection, eighty micrograms of cell extracts were immunoblotted with anti-Nrf2, Bcl-xL and anti β-actin antibodies.(C)Hepa-1 cells were plated at a density of 5000 cells per well in 24 well plates and cells were transfected with control siRNA 100 nM or two different concentration of Nrf2 siRNA and Bcl-xL siRNA (50 nM and 100 nM) separately for 24h. This was followed by treatment with etoposide (20 µM) for additional 30 h and etoposide mediated histone associated DNA fragmentation was analysed (upper panel). (D) Cell survival assay. Hepa-1 cells were plated at a density of 5000 cells per well in 24 well plates, and transfected with Nrf2 or Bcl-xL siRNA and treated with DMSO or etoposide 30h as indicated. Cells were incubated with fresh MTT solution for 2h at 37°C and absorbance at 570 nm was measured. The experiment was repeated thrice. Each point represents a mean ± SD and normalized to the value of the corresponding control cells.
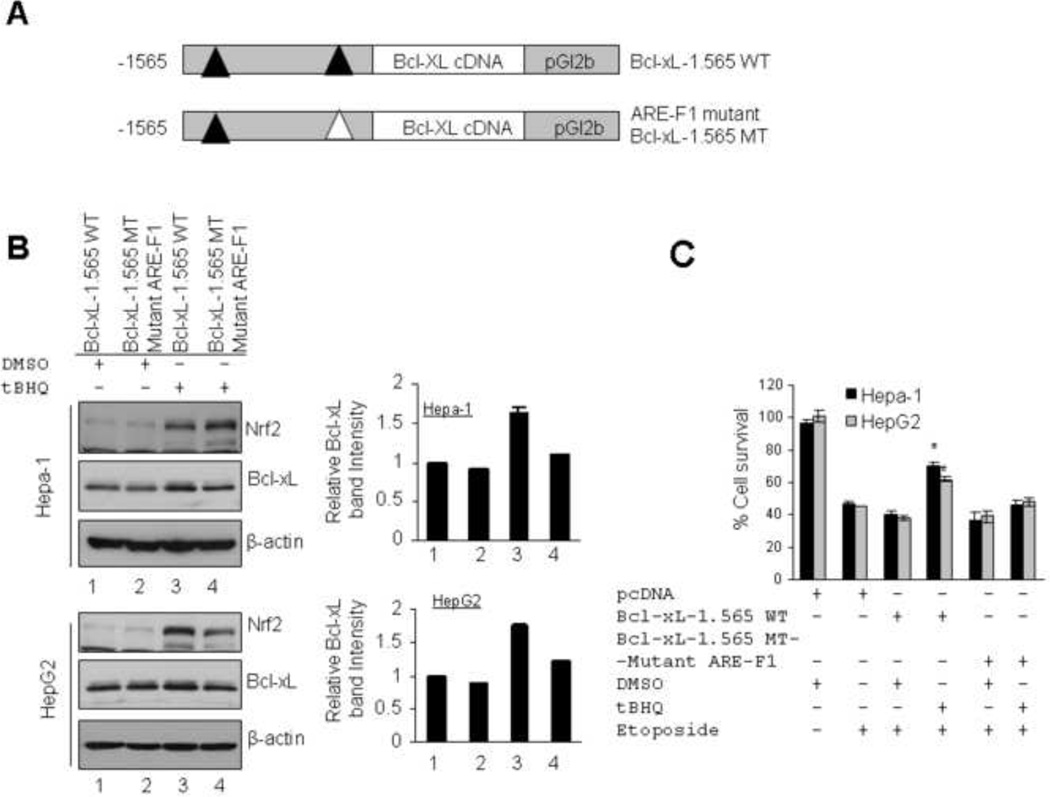
A second strategy was used to gain further support for a role of Nrf2-induced Bcl-xL in increased cell survival. We replaced luciferase coding sequence with Bcl-xL cDNA in wild type pGL2b-1.565 WT and ARE-F1 mutant pGL2b-1.565 MT plasmids to generate Bcl-xL-1.565 WT and ARE-F1 mutant Bcl-xL-1.565 MT (Fig. 8A). These plasmids were separately transfected in Hepa-1 and HepG2 cells. The transfected cells were treated with DMSO or tBHQ and lysates were immunoblotted (Fig. 8B, left and right panels). The results from both cell lines demonstrated t-BHQ-mediated increase in Nrf2 and Bcl-xL in cells transfected with plasmid Bcl-xL-1.565 WT but not in cells transfected with ARE-F1 mutant Bcl-xL-1.565 MT plasmid (Fig. 8B). These data provided additional support to the earlier observation that Nrf2 binds with ARE-F1 of Bcl-xL gene promoter leading to increased Bcl-xL gene expression. To test whether ARE-F1 mediated induction of Bcl-xL protein along with endogenous Bcl-xL contribute in cell survival, we transfected Hepa-1 and HepG2 cells with pcDNA or wild type pGL2b-1.565 WT or AREF1 mutant pGL2b-1.565 MT in separate experiments. The transfected cells were treated with etoposide alone or etoposide plus DMSO or tBHQ as indicated and relative cell survival was measured by MTT assay (Fig. 8C). The results revealed that cells transfected with wild type pGL2b-1.565 WT plasmid upon treatment with tBHQ showed significant increase in cell survival (30 to 35 %) compared with DMSO treated cells. Interestingly, the data also demonstrated that cells transfected with ARE-F1 mutant pGL2b-1.565 MT plasmid upon treatment with tBHQ showed only slight increase in cell survival (5 to 7%) compared with DMSO treated cells or same cell survival compared with etoposide alone treated cells (Fig. 8C). Indeed, these data provided the direct evidence that, under stress conditions, Nrf2 binds with ARE-F1 of Bcl-xL promoter and induced Bcl-xL expression which contributes to cell survival.
Fig. 8.
Antioxidant (tBHQ) increased Nrf2/Bcl-xL and etoposide-induced cell survival in cells transfected with wild type Bcl-xL-1.565 WT but not in cells transfected with ARE-F1 mutant BclxL-1.565 MT plasmid. (A) Luciferase cDNA was replaced with Bcl-xL cDNA in wild type pGL2b-1.565 WT and ARE-F1 mutant pGL2b-1.565 MT plasmids to generate Bcl-xL-1.565 WT and ARE-F1 mutant Bcl-xL-1.565 MT plasmids. (B) Hepa-1 and HepG2 cells were transfected with wild type pGL2b-1.565 WT and ARE-F1 mutant pGL2b-1.565 MT plasmids, treated with DMSO or tBHQ and immunoblotted (left panels). The Bcl-xL band intensities were quantified and plotted (right panels). (C) MTT assay. Hepa-1 and HepG2 cells were transfected with pcDNA or wild type pGL2b-1.565 WT or AREF1 mutant pGL2b-1.565 MT plasmids. Cells were treated with etoposide alone (Hepa-1; 30 µM, HepG2; 25 µM) or etoposide plus DMSO or tBHQ as indicated and relative cell survival was measured by MTT assay as described in methods section.
Bcl-xL specifically contributes to Nrf2-mediated reduced apoptosis and increased cell survival/drug resistance
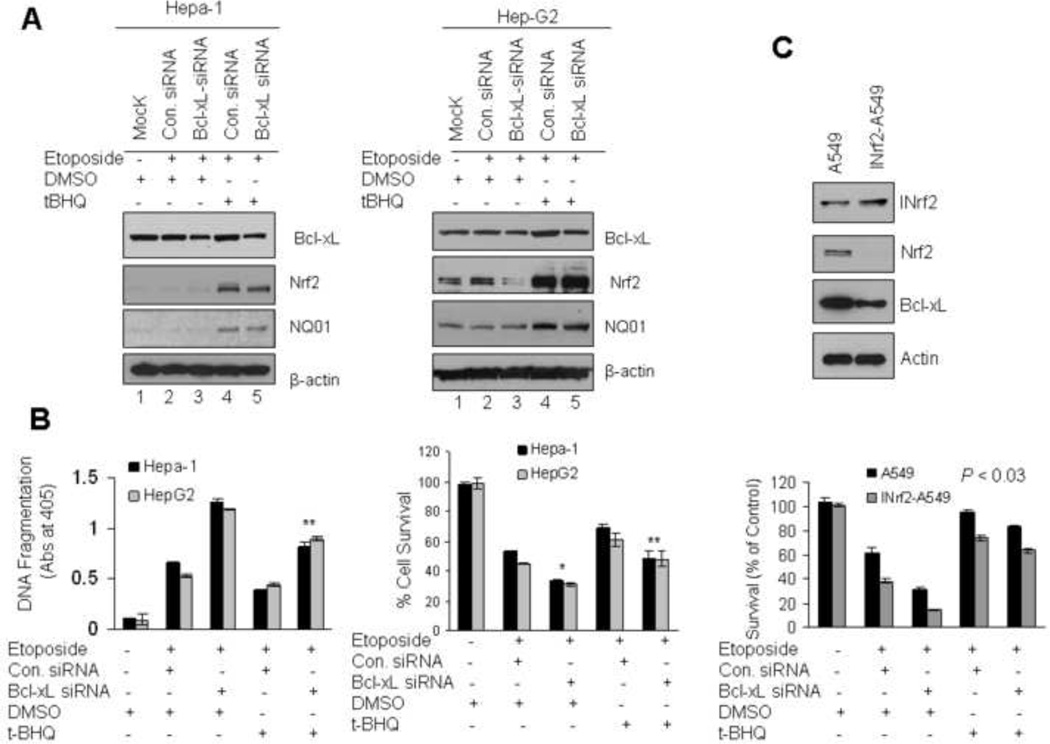
To investigate the specific role of Nrf2-mediated Bcl-xL expression in control of cellular apoptosis/survival/drug resistance, we analyzed cell survival in three different types of cancer cells exposed to etoposide. In the first experiment, Hepa-1 and Hep-G2 cells were either untransfected (Mock) or transfected with control or Bcl-xL siRNA (50 nM) followed by treatment with DMSO or t-BHQ and etopside and analyzed by immunobloting (Fig. 9A) and DNA fragmentation/cell survival (Fig. 9B). It is noteworthy that we titrated for Bcl-xL siRNA concentration in t-BHQ+Etoposide treated cells (lane 5) so that these cells express induced levels of all Nrf2 downstream gene expression except Bcl-xL expression (compare lane 5 with 2 and also lane 5 with 4).Bcl-xL expression was kept close to control siRNA transfected uninduced cells (compare Bcl-xL in lane 5 and 2). Immunoblotting results demonstrated that siRNA reduced more than 70% of Bcl-xL protein in Hepa-1 and HepG2 cells (Fig. 9A, lanes 3 & 5). Bcl-xL knock down cells when treated with tBHQ stabilized/increased Nrf2 and induced Nrf2 downstream protein NQO1 which is involved in the detoxification/cytoprotection (Fig. 9A, lanes 4 & 5). The mock and siRNA transfected Hepa-1 and Hep-G2 cells were analyzed for etoposide-induced histone associated DNA fragmentation and cell survival (Fig. 9B). The results revealed that Bcl-xL knock down induced 1.4 to 1.6 fold of etoposide mediated DNA fragmentation and decreased up to 20 to 25% of cell survival compared with control siRNA transfected cells (Fig 9B, left and right panels, compare lanes 2 and 3). The results also demonstrated approximately two-fold increase in DNA fragmentation because of siRNA-mediated specific inhibition of Bcl-xL in cells treated with t-BHQ+Etoposide (Nrf2 induced genes minus Bcl-xL induction) as compared with control siRNA transfected and t-BHQ+Etoposide treated cells (Nrf2 induced genes including Bcl-xL induction) (Fig. 9B, left panel, compare lanes 5 with 4). This led to 25–30% decrease in cell survival in cells transfected with Bcl-xL siRNA and treated with t-BHQ+Etoposide as compared with control siRNA transfected and t-BHQ+Etoposide treated cells (Fig. 9B, right panel, compare lane 5 with 4). These results collectively suggested that Nrf2-induced Bcl-xL plays a significant role in reducing apoptosis and increasing cell survival. In addition to above, we used human lung tumor derived A549 cells containing inactive INrf2 due to a point mutation at Glycine333 to Cysteine (G333C) in INrf2 protein [13]. This mutant INrf2 was unable to repress Nrf2 activity which resulted in an increase in drug resistance and cell survival [13]. By stable transfection of wild type pcDNA-INrf2 followed by selection with G148 we generated INrf2-A549 cells which express wild type INrf2 protein along with endogenous mutant INrf2. The role of mutant INrf2 in A549 cells and wild type INrf2/mutant INrf2 in INrf2-A549 cells in the regulation of Nrf2 and Bcl-xL were examined in the same isogenic cancer cells. Immunoblotting results demonstrated that, after stable transfection, the level of INrf2 in INrf2-A549 cells was increased by 1.7 fold compared with A549 parent cells (Fig. 9C, upper panel). The INrf2 expression in INrf2-A549 cells led to significant decrease in Nrf2 and Bcl-xL (Fig. 9C, upper panel). A549 and INrf2-A549 cells were transfected with control or Bcl-xL siRNA and exposed to the etoposide or etoposide plus tBHQ as shown in figure 9C and cell survival was examined by clonogenic cell survival assay. The results from clonogenic cell survival assay demonstrated that siRNA knockdown of Bcl-xL and INrf2-mediated Nrf2 degradation in INrf2-A549 cells both showed significant reduction (80%, p<0.03) in cell survival compared with control siRNA transfected cells and ~15 to 20% as compared to A549 cells (Fig. 9C, lower panel). The results also demonstrated that Bcl-xL knockdown and tBHQ treatment increased DNA fragmentation and decreased cell survival by 15 to 20% compared with control siRNA transfected cells and treated with tBHQ. These results supported the above conclusions and suggested that A459 cells carrying mutant INrf2 and unable to degrade Nrf2 led to survival of lung cancer cells.
Fig. 9.
Nrf2-mediated up-regulation of Bcl-xL enhanced cell survival and drug resistance. (A) Western analysis. Hepa-1 and HepG2 cells were untransfected (Mock) or transfected with control siRNA (50 nM) or Bcl-xL siRNA (50 nM) for 24h and treated with DMSO or tBHQ in presence of etoposide (Hepa-1; 30 µM, HepG2; 25µM) for additional 30h as indicated in figures. Eighty micrograms of cell extracts were immunoblotted with anti-Nrf2, Bcl-xL, anti-NQO1 and anti β-actin antibodies. (B) Cell death/DNA fragmentation assay. Five thousand Hepa-1 and same number of HepG2 cells were plated in 24 well plates and transfected/treated as indicated in “A”. The cytoplasmic histone-associated DNA fragments (mono and oligonucleosomes) were quantified using Cell Death Detection ELISA kit (Roche) and plotted (left panel). Similarly, after transfections/treatments Hepa-1 and Hep-G2 cells were incubated with fresh MTT solution for 2h at 37°C and absorbance at 570 nm was measured. The experiment was repeated thrice. Each point represents a mean ± SD and normalized to the value of the corresponding control cells.(C) Western blotting. Lysates from A549 cells expressing mutant INrf2 and cDNA derived wild type INrf2 (INrf2-A549) were immunoblotted with anti-INrf2, Nrf2, Bcl-xL and actin antibodies (upper panel). Clonogenic cell survival assay. In related experiments, A549 and INrf2-A549 cells also transfected with Bcl-xL siRNA or control siRNA, treated with etoposide (15 µM) in presence of DMSO or tBHQ for 30 h (lower panel). Cell were trypsinized, 1000 cells were reseeded in 100 mm tissue culture dishes (in triplicate) and incubated for 9 days and clonogenic cell survival assay was performed as described in methods section. Each data point represents a mean ± SD and normalized to the value of the corresponding control cells.
Discussion
Previous studies have shown a role of Nrf2:INrf2 in control of anti-apoptotic factor Bcl2 [25, 27] and INrf2 control of cytoprotective nuclear factor Nrf2 [1]. INrf2 is known to mediate ubiquitination and degradation of both Bcl-2 and Nrf2 thus maintaining homeostatic levels of these proteins within the cells [25]. In response to stress, both Bcl-2 and Nrf2 are released from INrf2 leading to stabilization of these factors [25, 27, 1]. Bcl-2 stabilization contributes to reduced apoptosis [27]. Nrf2 upon stabilization translocates in the nucleus leading to activation of a battery of cytoprotective proteins that also includes anti-apoptotic protein Bcl-2 resulting in cytoprotection and decreased apoptotic cell death [1, 27]. Therefore, both stabilization and Nrf2-mediated transcriptional activation of Bcl-2 contribute to decreased apoptotic cell death to ensure cell survival during stress [25, 27]. The studies have also shown that INrf2 through PGAM5 also regulates ubiquitination and degradation of anti-apoptotic protein Bcl-xL protein to control apoptosis [26]. This raised an intriguing question that if Nrf2 like Bcl-2 also regulates transcriptional activation of Bcl-xL gene expression and what is its specific contribution to apoptotic cell death.
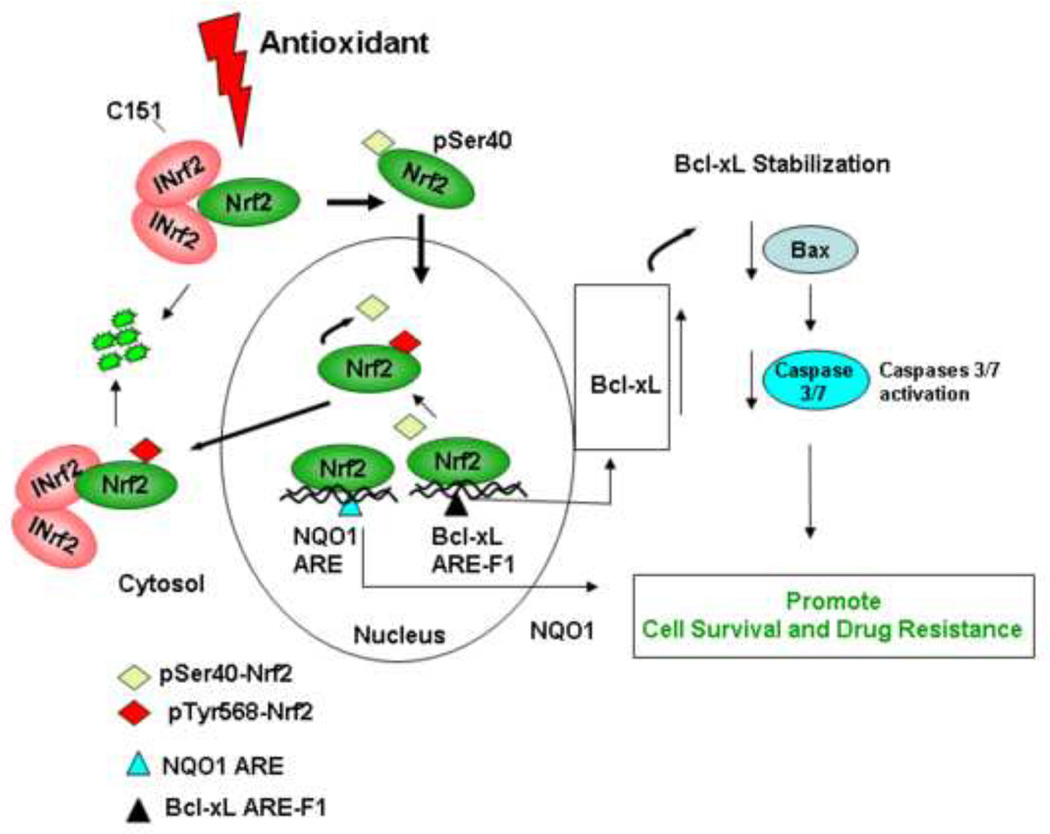
In the present report, we investigated the Nrf2 regulation of Bcl-xL gene expression and its role in cellular apoptosis, cell survival and drug resistance. Results demonstrated Nrf2 control of transcriptional activation of Bcl-xL as evident from antioxidant induction of Bcl-xL and parallel alterations in Bcl-xL with alterations in Nrf2. Deletion mutagenesis and reporter assays identified an ARE between nucleotide positions −608 to −600 that bound to Nrf2 and increased expression and induction of Bcl-xL gene expression in response to antioxidant. Further studies using several different cell lines and two different assays including apoptotic cell death/cell survival and clonogenic assays showed that siRNA mediated inhibition of Bcl-xL in t-BHQ/Nrf2 activated cells increased cellular apoptosis and decreased cell survival compared with t-BHQ/Nrf2/Bcl-xL activated cells. These results suggested a specific contribution of Nrf2 induced Bcl-xL in regulation of cellular apoptosis. The current studies together with previously published reports [26] suggested that both stress induced Nrf2-mediated transcriptional activation and stabilization of Bcl-xL contribute to reduce apoptosis and promote cell survival. A model showing a role of Nrf2-mediated increased expression of Bcl-xL gene expression and its role in decreasing apoptosis leading to increased cell survival and drug resistance is depicted in Figure 10.
Fig. 10.
Model showing antioxidant/Nrf2 mediated regulation of Bcl-xL and cellular apoptosis.
Variations and alterations in transcription/expression of Bcl-xL gene is often observed among the various tissues and in response to many signals [29–32]. In addition to ARE and Nrf2 as described above, several other transcription factors including GATA1, AP1, AP4, NF-E2A and SP1 are known to contribute to basal transcription/expression and variations in expression in the various tissues [29]. Many more factors that include hypoxia-inducible factor-1α (HIF-1α), NF-kB and Hepatocyte Growth Factor (HGF) are known to bind with promoter region of BclxL gene and positively regulate Bcl-xL expression in response to a variety of signals [30–32]. Therefore, alterations in factors as mentioned above could lead to alterations in Bcl-xL gene expression with substantial effect on the fate of cell to live or die.
INrf2:Nrf2 mediated control of anti-apoptotic proteins Bcl-2 and Bcl-xL appears to be an important physiological mechanism that along with activation of Nrf2 downstream cytoprotective proteins provides day-to-day cellular protection and ensures cell survival by reducing apoptotic cell death. The dark side of this mechanism is that significantly damaged cells escape apoptosis and might promote oncogenesis and lead to drug resistance. This especially applies to cells that contains mutated INrf2 and Nrf2 that abolish its interaction and degradation of Nrf2 leading to persistent nuclear accumulation of Nrf2, increased expression of cytoprotective and anti-apoptotic proteins, possibily of oncogenic transformation and drug resistance [12–13,33–35]. Studies have reported increased stabilization/accumulation of Nrf2 due to mutations in INrf2 resulting in loss of function in lung and many other tissue tumors [12–13, 33]. Lung cancer cell line A549 used in the current report contains INrf2G333C mutant protein that has lost its capacity to bind/degrade Nrf2 leading to accumulation of Nrf2 and induction of anti-apoptotic and cytoprotective proteins and resistance to etoposide.
In conclusion, Nrf2 functions as a master regulator of anti-apoptotic factor Bcl-xL. Antioxidant treatment activated/stabilized Bcl-xL and stabilization of Bcl-xL, decreased apoptotic cell death and increased cell survival. Nrf2 mediated up-regulation of Bcl-xl is a mechanism that protects against apoptotic cell death presumably against acute stress due to exposure to antioxidants, xenobiotics, drugs and radiation. Loss of function in INrf2 in lung cancer cells activates Nrf2 constitutively led to accumulation of Bcl-xL, resulted in decreased apoptosis and survival of cancer cells.
Nrf2 and ARE control expression of anti-apoptotic gene Bcl-xL and cellular apoptosis.
Nrf2 up-regulation of Bcl-xL prevents apoptosis leading to increased drug resistance.
Nrf2 is a potential target for reducing drug resistance.
Acknowledgement
This work was supported by NIH grants RO1 GM047466 and RO1 ES012265.
Abbreviations
- Nrf2
NF-E2 Related Factor 2
- INrf2 (Keap1)
Inhibitor of Nrf2
- t-BHQ
tert-Butyl hydroquinone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kasper JW, Niture SK, Jaiswal AK. Nrf2/INrf2 (keap1) signaling in oxidative stress. Free Radical Biol. Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD (P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 2003;278:44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- 5.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 6.Niture SK, Jain AK, Jaiswal AK. Antioxidant-induced modification of INrf2C151 and PKC delta-mediated phosphorylation of Nrf2S40 both required for stabilization and nuclear translocation of Nrf2 and increased drug resistance. J. Cell Sci. 2009;122:4452–4464. doi: 10.1242/jcs.058537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Cho HY, Jedlicka AE, Reddy SPM, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J Respir. Cell. Mol. Biol. 2002;26:175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 8.Lee JM, Shih AY, Murphy TH, Johnson JA. NF-E2-related factor-2 mediates neuroprotection against mitochondrial complex I inhibitors and increased concentrations of intracellular calcium in primary cortical neurons. J. Biol. Chem. 2003;278:37948–37956. doi: 10.1074/jbc.M305204200. [DOI] [PubMed] [Google Scholar]
- 9.Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 10.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 11.Kwak M, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1 Nrf2 pathway: identification of novel gene clusters for cell survival. J. Biol. Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 12.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provided by its point mutations in lung cancer. Mol. Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional Keap1 Nrf2 interaction in non-small-cell lung cancer. PLos Med. 2006;3:1865–1876. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee O, Jain AK, Papusha V, Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J. Biol. Chem. 2007;282:36412–36420. doi: 10.1074/jbc.M706517200. [DOI] [PubMed] [Google Scholar]
- 15.Kaspar JW, Jaiswal AK. An autoregulatory loop between Nrf2 and Cul3-Rbx1 controls their cellular abundance. J. Biol. Chem. 2010;285:21349–21358. doi: 10.1074/jbc.M110.121863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 17.Cory S, Adams JM. The Bcl-2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 18.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 19.Boise LH, González-García M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nuñez G, Thompson CB. Bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 20.Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–1284. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- 21.Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, Nettesheim D, Chang BS, Thompson CB, Wong SL, Ng SL, Fesik SW. X-ray and NMR structure of human Bcl-XL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 22.Chao DT, Linette GP, Boise LH, White LS, Thompson CB, Korsmeyer SJ. Bcl-XL and Bcl-2 repress a common pathway of cell death. J. Exp. Med. 1995;182:821–828. doi: 10.1084/jem.182.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranger AM, Zha J, Harada H, Datta SR, Danial NN, Gilmore AP, Kutok JL, Le Beau MM, Greenberg ME, Korsmeyer SJ. Bad-deficient mice develop diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA. 2003;100:9324–9329. doi: 10.1073/pnas.1533446100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvert JW, Jha S, Gundewar S, Elrod JW, Ramachandran A, Pattillo CB, Kevil CG, Lefer DJ. Hydrogen sulfide mediates cardioprotection through Nrf2 signaling. Circulation. Res. 2009;105:365–374. doi: 10.1161/CIRCRESAHA.109.199919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niture SK, Jaiswal AK. INrf2 (keap1) targets Bcl-2 degradation and controls cellular apoptosis. Cell Death and Differ. 2011;18:439–451. doi: 10.1038/cdd.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Niture SK, Jaiswal AK. Inhibitor of Nrf2 (INrf2 or Keap1) protein degrades Bcl-xL via Phosphoglycerate mutase 5 and controls cellular apoptosis. J. Biol. Chem. 2011;286:44542–44556. doi: 10.1074/jbc.M111.275073. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Niture SK, Jaiswal AK. Nrf2 protein up-regulates anti-apoptotic protein Bcl-2 and prevents cellular apoptosis. J. Biol. Chem. 2012;287:9873–9886. doi: 10.1074/jbc.M111.312694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niture SK, Jaiswal AK. Prothymosin-α mediates nuclear import of INrf2:Cul3.Rbx1 complex to degrade nuclear Nrf2. J. Biol. Chem. 2009;284:13856–13868. doi: 10.1074/jbc.M808084200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Grillot D, Gonzalez-Garcia M, Ekhterae D, Duan L, Inohara N, Ohta S, Seldin MF, Nuñez G. Genomic organization, promoter region analysis, and chromosome localization of the mouse bcl-x gene. J. Immunol. 1997;158:4750–4757. [PubMed] [Google Scholar]
- 30.Chen N, Chen X, Huang R, Zeng H, Gong J, Meng W, Lu Y, Zhao F, Wang L, Zhou Q. BCL-xL is a target gene regulated by hypoxia-inducible factor-1α. J. Biol. Chem. 2009;284:10004–10012. doi: 10.1074/jbc.M805997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glasgow J, Qiu J, Rassin D, Grafe M, Wood T, Perez-Pol J. Transcriptional regulation of the BCL-X gene by NF-kappaB is an element of hypoxic responses in the rat brain. Neurochem. Res. 2001;26:647–659. doi: 10.1023/a:1010987220034. [DOI] [PubMed] [Google Scholar]
- 32.Cao X, Littlejohn J, Rodarte C, Zhang L, Martino B, Rascoe P, Hamid K, Jupiter D, Smythe WR. Up-regulation of Bcl-xl by hepatocyte growth factor in human mesothelioma cells involves ETS transcription factors. Am. J. Pathol. 2009;175:2207–2216. doi: 10.2353/ajpath.2009.090070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kweon MH, Adhami VM, Lee JS, Mukhtar H. Constitutive overexpression of Nrf2-dependent heme oxygenase-1 in A549 cells contributes to resistance to apoptosis induced by epigallocatechin 3-gallate. J. Biol. Chem. 2006;281:33761–3377. doi: 10.1074/jbc.M604748200. [DOI] [PubMed] [Google Scholar]
- 34.Yoo NJ, Kim HR, Kim YR, An CH, Lee SH. Somatic mutations of Keap1 gene in common solid cancers. Histopathology. 2012;60:943–952. doi: 10.1111/j.1365-2559.2012.04178.x. [DOI] [PubMed] [Google Scholar]
- 35.Shibata T, Ohta T, Tong K, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in Nrf2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]