Abstract
An article by Nissen et al. in the November 2012 issue of GENETICS emphasizes the importance of alternative splicing in the sex determination cascade of the honeybee Apis mellifera. This study demonstrates the application of reverse transcriptase PCR and RNA interference screens as genetic tools to better understand the regulation of transcription and splicing. It also provides the opportunity to explore the evolutionary origins of genes by considering the functions of orthologs and paralogs in different species. This Primer article provides background information and explanations of the concepts and findings of Nissen et al. and offers discussion questions for use in the classroom.
Related article in GENETICS: Nissen, I., M. Müller, and M. Beye, 2012 The Am-tra2 gene is an essential regulator of female splice regulation at two levels of the sex determination hierarchy of the honeybee. Genetics 192: 1015–1026.
Keywords: Primer, honeybee, Am-tra2
Background
THE origins of beekeeping and honey collecting in human civilizations can be traced back thousands of years. Honeybee colony management has thrived because of the important role played by honeybees in crop pollination and because human cultures worldwide consume hive products (honey, beeswax, and propolis). Importantly, bees also shape natural ecosystems by facilitating gene flow among plants.
Honeybees are eusocial insects that live in colonies of tens of thousands of individuals. A colony usually consists of three types of bees: queens, drones, and workers. The female queen (usually one per colony) is selected early in larval development and fed a diet of royal jelly. As an adult, the queen lays unfertilized and fertilized eggs. Male drones, whose main role is to mate with the queen, develop from unfertilized eggs. Female workers (the majority of bees in a colony) are the laborers that perform tasks required for colony growth and maintenance (e.g., cleaning, defense, foraging for food). Workers will produce eggs only if the colony is queen-less. Because of this social structure, honeybees are important models for studying behavior, learning, and memory as well as the genetics of sex determination.
The signal transduction cascade that directs sex determination in honeybees (i.e., development as a male drone or female queen/worker) directs developmental fate and, in turn, has a significant influence on the social structure of a colony. Over 150 years ago Johannes Dzierson hypothesized that male bees develop from unfertilized eggs and females from fertilized eggs (reviewed by Page et al. 2012). However, research to understand the genetics that controls male vs. female developmental fate is ongoing. The publication of the DNA sequence of the European honeybee (Apis mellifera) genome in 2006 (Honeybee Genome Sequencing Consortium 2006) provided important resources for studying bee genetics, including the molecular basis of sex determination. Martin Beye and his team of researchers at Heinrich Heine University in Dusseldorf, Germany, have made significant contributions in this field. In their most recent article, Beye and colleagues Inga Nissen and Miriam Müller examine the expression pattern of the gene transformer-2 (tra-2) in A. mellifera and how it regulates the splicing and expression of other genes in the sex determination cascade. Their results enhance our understanding of the mechanisms of gene regulation that lead to either female or male development in the honeybee A. mellifera.
Sex determination in honeybees: the complementary sex determiner locus
What determines whether an animal develops as a male or female? There is no universal answer, but in many animals, sex determination is triggered very early in development by chromosomal differences, such as a specific complement of sex chromosomes. For example, in humans (and many animals) males are heterogametic, having two different sex chromosomes, X and Y, while females are homogametic, having two copies of X. The total number of sex chromosomes can also determine sex: female grasshoppers have two X chromosomes (X/X) while males have only one (X/O). In some animals, hermaphrodites (individuals with both male and female characteristics) are common; in the nematode Caenorhabditis elegans, males are X/O while hermaphrodites are X/X.
The haplodiploid sex determination system is the dominant mode in Hymenoptera, the insect order that includes bees, wasps, ants, and sawflies. Sex chromosomes are completely absent in Hymenoptera; instead, ploidy (the number of chromosome sets in an individual) seems to be linked to sex determination. Fertilized eggs are diploid (two chromosome sets) and develop as females, while unfertilized eggs are haploid (one chromosome set) and develop as males. In honeybees, however, it is the allelic state of the complementary sex determiner (csd) gene, rather than ploidy, that is the primary genetic signal of sex determination.
In honeybee populations, the csd locus is highly polymorphic; there are at least 15 different csd alleles among bee populations, so diploid individuals tend to be heterozygous for csd. Recall that a diploid organism inherits two alleles of each autosomal gene, one from each parent; an organism is homozygous for a particular gene if both alleles are identical and heterozygous if there are DNA sequence differences between the two alleles. Thus, in bees, it is zygosity (the degree of similarity between two alleles) of the csd locus, rather than ploidy, that is the primary determinant of sexual fate. Individuals heterozygous for csd are female (either a queen or a worker) while haploids develop as males (they are hemizygous for csd, having one copy) (Beye et al. 2003). Diploid bees that are homozygous for csd also develop as males, although workers eat these individuals soon after they emerge from the egg.
Transcription, alternative splicing, and sex determination
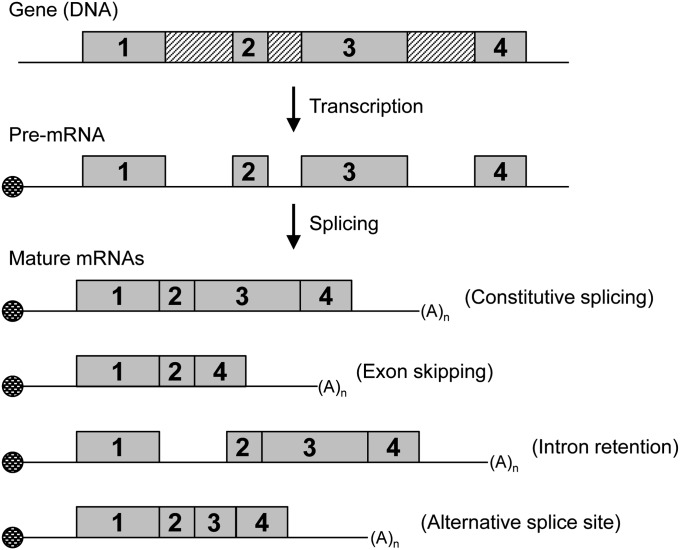
Is it that simple? Does a single gene determine honeybee sex? While csd is the primary signal of sex determination, the process actually involves a cascade of interactions among several other genes. In the honeybee, many genes involved in the sex determination pathway undergo sex-specific alternative splicing (i.e., production of a variety of messenger RNA (mRNA) transcripts from a single gene; Figure 1), producing proteins that are specific to bees of each sex. During splicing, the spliceosome catalyzes the removal of introns and joins together exons to produce a mature mRNA that contains 5′ and 3′ untranslated regions (UTRs) and exons that code for an open reading frame. SR proteins are also involved in regulating splicing. Members of this family of RNA-binding proteins contain an RS domain rich in repeated arginine–serine dipeptides that regulates constitutive splicing and, importantly, alternative splicing. During alternative splicing, RNA recognition motifs in SR proteins bind to specific splice sites (e.g., exonic splicing enhancers) within pre-mRNA.
Figure 1.
Alternative splicing can produce different mRNA transcripts from the same gene. In this figure, gray boxes with numbers are exons, and boxes with diagonal lines are introns. The circle represents the mRNA 5′ cap, and (A)n represents the poly(A) tail. DNA is transcribed into pre-mRNA that can then be spliced differently to produce transcripts of different lengths and sequence composition. Alternative splicing is indicated next to the different splice variants.
In female honeybees, heteroallelic Csd proteins direct female-specific splicing of the feminizer (fem) gene, which is then translated into a functional Fem (SR-type) protein. Fem proteins direct female-specific splicing of the doublesex gene (dsx), a transcription factor that mediates female development. Fem proteins are also necessary to direct female-specific splicing of their own pre-mRNA, thus creating an autoregulatory feedback loop (Gempe et al. 2009). In contrast, in male honeybees, homo- or hemiallelic Csd proteins lead to male-specific splicing of fem. However, this male-specific splice variant encodes a truncated nonfunctional Fem protein. Therefore, dsx is transcribed (without regulation by Fem proteins) and undergoes male-specific splicing to yield a Dsx protein that controls male development.
The buzz on important gene terminology
When a putative gene sequence is identified, understanding the evolutionary history of that gene can be informative for developing hypotheses about its function. Genes are homologs if they are descended from a common ancestral sequence. Among types of homologs, orthologs are genes that evolved by speciation (i.e., the same gene in different species) while paralogs are genes within the same genome that evolved by duplication. Many genes (i.e., orthologs) in the Apis sex determination pathway are found in other insects. For example, despite their different names, the fem gene in Apis is an ortholog of the sex-determining gene transformer (tra) in the fruit fly Drosophila melanogaster (i.e., fem and tra evolved from a common ancestral gene millions of years ago). However, csd and fem are paralogs specific to the honeybee lineage (Hasselmann et al. 2008) that have evolved and acquired distinct roles in the sex determination pathway. To alleviate confusion when discussing orthologs from different species, genes from the honeybee will be designated with an “Am-” prefix (for A. mellifera, as in Nissen et al. 2012). Gene names are written in lowercase italics (e.g., Am-dsx) and protein names in non-italics with the first letter of the protein capitalized (e.g., Am-Dsx).
The Objective of the Study
In the honeybee, expression of csd and fem paralogs, alternative splicing of their gene products, and the function of the Csd and Fem proteins in sex determination have previously been demonstrated (Hasselmann et al. 2008). In D. melanogaster, Tra works together with another protein called Transformer-2 (encoded by the gene tra-2) to promote female-specific splicing of dsx. A sequence homology search of the honeybee genome (Dearden et al. 2006) previously identified a tra-2 ortholog; however, whether this gene is expressed or produces functional transcripts has not been demonstrated. Therefore, since tra-2 and Am-tra-2 are orthologs, the objective addressed by Nissen et al. (2012) was to determine whether Tra-2 and Am-Tra2 share the same function (i.e., regulating sex-specific splicing of dsx) in the fruit fly and in the honeybee.
Using RACE and RT-PCR to characterize transcripts
To understand the function of Am-Tra2 in the honeybee, sequences of full-length Am-tra2 transcripts (including the 5′ and 3′ UTRs) from male and female honeybees needed to be determined. RNA is single stranded and cannot be used as a template for polymerase chain reaction (PCR). Therefore, to characterize RNA sequences, mRNA is converted into complementary DNA (cDNA) using the enzyme reverse transcriptase. Together with cDNA synthesis, the RACE technique was used by Nissen et al. (2012) to determine the 5′ and 3′ sequences of a mature mRNA.
Recall that, during pre-mRNA processing, a 7-methylguanosine cap (5′ cap) is added to the 5′-end, the 3′-end is cleaved at the polyadenylation site, and a 3′ poly(A) tail (∼100–250 adenine residues) is added. Since the process of RNA isolation often partially degrades transcripts, it is necessary to select sequences that have the intact 5′ cap and 3′ poly(A) tail to characterize full-length sequences. In RACE, short oligonucleotide adapters (that contain RACE-specific primers) are added to the 5′ and 3′ transcript ends. For 5′ RACE, an adapter replaces the 5′ cap, and the transcript is then used for cDNA synthesis. In 3′ RACE, the adapter anneals to the poly(A) tail via a complementary oligo(dT) sequence that is extended by reverse transcriptase. The cDNA pools can then be used for PCR using a combination of gene-specific and RACE adapter-specific primers, and these PCR products can be isolated and sequenced to identify the first and last exons and the 5′ and 3′ UTRs for Am-tra2 transcripts.
With these sequences in hand, Nissen et al. designed oligonucleotide primers to anneal to the 5′ and 3′ ends of Am-tra2 transcripts. These primers were then used in RT-PCR to amplify full-length Am-tra2 transcripts from male and female honeybee embryos and pupae. RT-PCR creates a pool of cDNA to use as a template for PCR, rather than genomic DNA. Six splice variants with minor sequence differences were identified in the pool of cDNAs created by Nissen et al. Despite the variation in the corresponding Am-Tra2 protein lengths, each isoform had the same domain structure as Tra2 orthologs in other species: a central RNA-binding domain (RBD) flanked by two RS domains. Within the RBD, two ribonucleoprotein consensus peptide elements (RNP-1 and -2) were identified. However, an amino acid substitution in RNP-1 in Apis at a position essential for female dsx splicing in D. melanogaster led Nissen et al. to ask whether Am-Tra2 has evolved a relatively different function.
Investigating the function of Am-Tra2
To investigate the role of Am-Tra2, Nissen et al. used semiquantitative PCR to measure Am-tra2 expression levels at different developmental stages in males and females. This method is similar to RT-PCR, except that it uses multiplex PCR to amplify transcripts of two different genes in the same reaction, providing an internal control in every reaction. Nissen et al. used primer pairs to amplify both Am-tra2 and elongation factor-1α (ef-1α) mRNAs in each PCR reaction. ef-1α was included because it is a housekeeping gene (one that is required for translation), performs a basic cellular function, and is expected to be constitutively expressed at all stages of development. When the PCR products for each multiplex reaction are analyzed by gel electrophoresis, gel-imaging software can compare band intensities and calculate the fold change in expression between the gene of interest (Am-tra2) and the housekeeping gene (ef-1α). In this way, relative expression of Am-tra2 is determined and used to measure relative transcription levels at different developmental stages. Nissen et al. found that Am-tra2 transcripts in both males and females were most abundant during embryonic stages (before and after csd is transcribed) but that transcription significantly decreased at the pupal stage. Sex-specific splicing patterns were nonexistent.
What have we learned so far?
Consider the findings by the authors up to this point (refer to figures 1–3 in the article by Nissen et al.). Referring to figure 1 in Nissen et al., what did the RT-PCR data reveal about Am-tra2 transcription and splicing patterns? How many splice variants are generated from the Am-tra2 gene? What is the difference between the major and minor splice variants? In Nissen et al.’s figure 2, examine the amino acid alignments of the arthropod RBD domain and identify the amino acid substitution that led the authors to hypothesize that Am-Tra2 (and perhaps Tra2 in Nasonia vitripennis, another hymenopteran) might have a function distinct from that of Tra2 in Drosophila and other holometabolous (winged) insects. In figure 3 of Nissen et al., are the splice variants equally abundant in embryos and pupae? Are they found in males and females? What does the figure demonstrate about transcription levels during embryonic and pupal development? Once the significance of the Am-tra2 gene expression data is clear, the next question to consider is: what is the cellular function of Am-Tra2 protein?
Functional studies: RNA interference knockdowns
If Tra-2 in Drosophila and Am-Tra2 are indeed homologous proteins, one might assume that Am-Tra2 possesses the function of regulating male- and female-specific splicing of Am-dsx and fem. To test this hypothesis, Nissen et al. used a technique called RNA interference (RNAi) to determine whether sex-specific splicing patterns for fem and Am-dsx were affected when expression of Am-tra2 was “silenced.”
How does RNAi work? RNAi exploits the fact that double-stranded RNA (dsRNA) can be used to repress translation of a gene of interest. In many eukaryotes, small noncoding RNAs (such as small interfering RNA) can regulate gene expression post-transcriptionally. The enzyme Dicer cleaves these small dsRNAs into shorter pieces that associate with the multi-protein RNA-induced silencing complex (RISC). This RISC–RNA complex then binds to, and stimulates, the degradation of complementary cellular mRNA. RNAi can therefore be used to repress or “silence” a specific gene to examine the phenotypic effects. This is referred to as a “gene knockdown” since mRNA degradation is likely incomplete.
Nissen et al. used RNAi to knock down expression of Am-tra2 by injecting honeybee embryos with dsRNA representing a segment of Am-tra2. This induced the RNAi response and subsequently targeted complementary Am-tra2 transcripts for degradation. However, while the objective was to use RNAi to assess the role of Am-tra2 in splicing regulation, the authors encountered an unexpected result. Injecting dsRNA copies of Am-tra2 was actually lethal: male and female embryos failed to develop into larvae. And injecting lower concentrations of dsRNA only resulted in a very low frequency of larval development. If Am-tra2 was specifically involved in regulating male- and female-specific splicing of Am-tra2 and dsx, would this be the expected result (i.e., lethality for both male and female larvae)? Is this result consistent with a specific role for Am-Tra2 in the sex determination pathway? Or does it suggest that Am-Tra2 might have broader functions in the honeybee?
To address their hypothesis about the relationship of Am-tra2 and sex-specific splicing of Am-dsx and fem, Nissen et al. repeated the embryonic injections with Am-tra2 dsRNA and isolated RNA from embryos (regardless of whether larvae hatched) after ∼80 hr of development. The RNA was used in RT-PCR to amplify Am-dsx and fem transcripts and distinguish male and female splice variants.
What were their expectations? If Am-Tra2 promotes female-specific splicing of Am-dsx and fem, would male-specific or female-specific Am-dsx splice variants be expected in the Am-tra2 knockdowns? Look at the data presented in figure 4 of Nissen et al. to learn what the authors found. For example, in the Am-tra2 knockdown, only male-specific Am-dsx splice variants were produced while higher concentrations of injected Am-tra2 dsRNA compromised female-specific splicing of fem. Based on these results, what do you conclude is the role of Am-tra2? What do the authors conclude? When Am-tra2 was knocked down, male-specific fem transcripts were also absent. Does this mean that Am-Tra2 is also necessary for male-specific fem splicing? All together, these data suggest that Am-Tra2 is necessary to promote female-specific splicing of Am-dsx and fem, but it only regulates male-specific splicing for fem. Am-Tra2 does not appear to regulate male-specific splicing of dsx since male transcripts were produced in the Am-tra2 knockdown.
Approach to Classroom Use
On the surface, the concept of haplodiploid sex determination appears quite straightforward: males are haploid and females are diploid. However, recent articles have started to unravel the genetics of sex determination in the honeybee. Reading a set of sequential articles from the Beye lab allows students to experience how research on this topic has progressed over several years. This teaching method, called CREATE (consider, read, elucidate hypotheses, analyze and interpret the data and think of the next experiment), allows students to read and analyze primary literature and focus on a single line of scientific research (Hoskins et al. 2007). To apply this method, a suggested sequence of articles is the following: Beye et al. (2003) (the finding that csd heterozygosity is the primary sex-determining signal) followed by Hasselmann et al. (2008) (which characterizes the Am-fem and csd paralogs) and Gempe et al. (2009) (which explores the regulation and function of Am-fem and csd). After reading these articles, the methods and questions of Nissen et al. (2012) might seem more rational and logical to students. In addition, review articles on sex determination in Hymenoptera (Heimpel and de Boer 2008) and other insects (Gempe and Beye 2010; Verhulst et al. 2010a) are valuable reading. For a comparative approach, sex determination pathways in another Hymenopteran, the parasitoid wasp N. vitripennis, can be explored. While both Apis and Nasonia have haplodiploid sex determination, the genetics that regulates this mechanism in Nasonia is quite different (it is dependent upon the contribution of maternal transcripts in the embryo and some form of maternal imprinting) (Verhulst et al. 2010b).
The article by Nissen et al. (2012) can be used as a teaching aid to discuss laboratory techniques including RACE, RT-PCR, and RNAi as well as the concept and significance of alternative splicing in regulating gene expression. One approach might be to ask students to summarize the data that were produced using each molecular method: RACE, RT-PCR, semiquantitative PCR, and RNAi. For classroom or group discussion, considering the strengths and weaknesses of the authors’ conclusions and contemplating follow-up experiments will provide a stimulating experience and a greater understanding of the material. Additional suggested questions and problems are summarized below:
Nissen et al. used semiquantitative PCR to quantify gene expression, but what other methods could have been employed? Does having a completed genome sequence for A. mellifera provide an advantage? What are some benefits and drawbacks to these other approaches?
Nissen et al. proposed a model for the role of Am-Tra2 in sex determination that implies interactions between Csd proteins with Am-Tra2 and/or Fem. What are some experiments that could be done to determine whether these protein–protein interactions take place in vitro? In vivo?
The concept of RNAi is explained in all modern genetics textbooks. However, delivery methods for dsRNA can vary between organisms. Discuss how dsRNA copies of Am-tra2 were delivered into honeybee embryos. Are there other commonly used methods of dsRNA delivery?
The finding that Am-tra2 appears to be essential for embryogenesis was intriguing and merits further investigation. What are some follow-up experiments that could determine the stages at which Am-tra2 is required for development? Are there ways to visualize the production of Am-Tra2 proteins? To characterize the localization of these proteins or transcripts in embryos?
To understand how Nissen et al. designed PCR primers, find the Am-tra2 gene sequence in an online database (e.g., http://hymenopteragenome.org/beebase/). How many exons and introns are present? How long is the coding region?
The concept of gene homologs, orthologs, and paralogs is challenging. The Nissen et al. article provides examples of orthologs in A. mellifera and D. melanogaster and paralogs within A. mellifera. To reinforce these concepts, use a BLAST search with the gene sequences identified in the Nissen et al. article to determine the phylogenetic distribution of these genes in insects (and beyond) and find evidence for gene loss or gain by duplication. Nissen et al. provide sequence accession numbers that can be used to retrieve the Am-tra2 mRNA sequence and the amino acid sequences for the six isoforms identified in their article from online sequence databases at National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). For example, amino acid queries can be used to search for homologs within the protein databases (using the BLASTP program) or the nucleotide databases [such as Whole Genome Shotgun (WGS) or Expressed Sequence Tag (EST) databases] using the TBLASTN program.
Footnotes
Communicating editor: Elizabeth A. De Stasio
Literature Cited
- Beye M., Hasselmann M., Fondrk M. K., Page R. E., Omholt S. W., 2003. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114: 419–429. [DOI] [PubMed] [Google Scholar]
- Dearden P. K., Wilson M. J., Sablan L., Osborne P. W., Havler M., et al. , 2006. Patterns of conservation and change in honey bee developmental genetics. Genome Res. 16: 1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gempe T., Beye M., 2010. Function and evolution of sex determination mechanisms, genes and pathways in insects. Bioessays 33: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gempe T., Hasselmann M., Schiøtt M., Hause G., Otte M., et al. , 2009. Sex determination in honeybees: two separate mechanisms induce and maintain the female pathway. PLoS Biol. 7(10): e1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann M., Gempe T., Schiøtt M., Nunes-Silva C. G., Otte M., et al. , 2008. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454: 519–523. [DOI] [PubMed] [Google Scholar]
- Heimpel G. E., de Boer J. G., 2008. Sex determination in the Hymenoptera. Annu. Rev. Entomol. 53: 209–230. [DOI] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium , 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443: 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins S. G., Stevens L. M., Nehm R. H., 2007. Selective use of the primary literature transforms the classroom into a virtual laboratory. Genetics 176: 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen I., Müller M., Beye M., 2012. The Am-tra2 gene is an essential regulator of female splice regulation at two levels of the sex determination hierarchy of the honeybee. Genetics 192: 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R. E., Rueppell O., Amdam G. V., 2012. Genetics of reproduction and regulation of honeybee (Apis mellifera L.) social behavior. Annu. Rev. Genet. 46: 97–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst E. C., van de Zande L., Beukeboom L. W., 2010a Insect sex determination: it all revolves around transformer. Curr. Opin. Genet. Dev. 20: 376–383. [DOI] [PubMed] [Google Scholar]
- Verhulst E. C., Beukeboom L. W., van de Zande L., 2010b Maternal control of haplodiploid sex determination in the wasp Nasonia. Science 328: 620–623. [DOI] [PubMed] [Google Scholar]