Abstract
Mathematical models of meiosis that relate offspring to parental genotypes through parameters such as meiotic recombination frequency have been difficult to develop for polyploids. Existing models have limitations with respect to their analytic potential, their compatibility with insights into mechanistic aspects of meiosis, and their treatment of model parameters in terms of parameter dependencies. In this article I put forward a computational approach to the probabilistic modeling of meiosis. A computer program enumerates all possible paths through the phases of replication, pairing, recombination, and segregation, while keeping track of the probabilities of the paths according to the various parameters involved. Probabilities for classes of genotypes or phenotypes are added, and the resulting formulas are simplified by the symbolic-computation system Mathematica. An example application to autotetraploids results in a model that remedies the limitations of previous models mentioned above. In addition to the immediate implications, the computational approach presented here can be expected to be useful through opening avenues for modeling a host of processes, including meiosis in higher-order ploidies.
Keywords: meiotic recombination frequency (MRF), tetraploidy, quadrivalent, pairing-partner switch, coefficient of double reduction
MEIOTIC recombination is the exchange of chromosomal regions between homologous chromosomes during meiosis, a specialized type of cell division in sexually reproducing eukaryotes (Lao and Hunter 2010; Lichten and De Massy 2011; Pradillo and Santos 2011). Meiotic recombination ensures the proper segregation of chromosomes through the formation of crossovers between interhomologous DNA strands (Lichten and De Massy 2011) and increases genetic diversity among offspring (Yanowitz 2010).
Polyploidy is the accumulation of additional complete sets of chromosomes and is widespread among flowering plants, including important crop species (Brownfield and Köhler 2011; Jackson et al. 2011). While evidence that polyploidy has promoted speciation or morphological diversification is lacking (Otto 2007), the possibility has been acknowledged (Comai 2005; Otto 2007), and it has recently been suggested that increased rates of meiotic recombination might have contributed to the evolutionary success of polyploid plants (Pecinka et al. 2011).
Tetraploids are generally classified as allotetraploids or autotetraploids. Allotetraploids such as Arabidopsis suecica (Pecinka et al. 2011) are derived from two different (diploid) species (Brownfield and Köhler 2011) and are traditionally considered to show pairing between homologous chromosomes only, although pairing between homeologous chromosomes may occur (Yang et al. 2013). Autotetraploid genomes are derived from a single species (Brownfield and Köhler 2011) and generally show no pairing preference among the four homologous chromosomes, although again preferential pairing might occur (R. Wu et al. 2001).
Mathematical models of meiosis that relate the genotypes of offspring to those of their parents at single- or multiple-marker locations through parameters such as the meiotic recombination frequency between a marker pair are easily formulated for diploids and allopolyploids, but have been difficult to develop for autopolyploids. The difficulties lie in the more complex meiosis in autopolyploids, in which not only bivalents are formed—pairs of homologous chromosomes, parts of which are then exchanged later through crossovers—but possibly also higher-order valents such as quadrivalents. In quadrivalents, each chromosomal region is still paired with only one other chromosome, but pairing partners can switch in a position-dependent manner. Such pairing-partner switches can lead to the phenomenon of double reduction, in which homologous segments of sister chromatids—“sister factors” (Levings and Alexander 1966)—end up in the same gamete (Mather 1935; Haynes and Douches 1993). Double reduction of a locus requires homologous chromosomes to form a quadrivalent and a recombination between the locus and the centromere (Mather 1936; Luo et al. 2004). Estimates of coefficients of double reduction can provide insights into the extent of quadrivalent formation (see, e.g., Wang and Luo 2012), and estimates of the recombination frequency r are necessary for the construction of genetic maps (see, e.g., Ripol et al. 1999).
In 1947, Fisher described 11 modes of tetraploid gamete formation for two linked loci (Fisher 1947), but it was not until over half a century later that frequencies of these gamete types were expressed in terms of recombination and double-reduction parameters under a tetrasomic model of inheritance that takes the formation of quadrivalents into account (S. S. Wu et al. 2001; Luo et al. 2004; Wu and Ma 2005; Lu et al. 2012). The resulting sets of equations, however, have at least three limitations: First, the models are descriptive rather than analytical; that is, they do not explain the probability distribution of gamete modes in terms of biological (here meiotic) mechanisms; instead, gamete modes are described in terms of observables, that is, whether a gamete is recombined or not and double reduced or not. Second, as a consequence, the resulting models might not be compatible with any given mechanistic model of meiosis, either agreed upon for a particular species or hypothesized with the aim of further testing. Third, the parameterizations of probability distributions treat recombination rates and coefficients of double reduction as independent, thus putting aside the fact that recombination is required to allow double reduction to take place.
In this article I put forward a computational approach to the probabilistic modeling of meiosis, with an example application to autotetraploids. The approach takes account of the major mechanistic aspects of autotetraploid meiosis, including mixed bivalent and quadrivalent pairings, pairing-partner switches, and recombination on a chromatid level. A computer program enumerates all possible paths through the phases of replication, pairing, recombination, and segregation, while keeping track of the probabilities of the paths according to the various parameters involved. Probabilities for classes of genotypes or phenotypes are added, and the resulting formulas are simplified by the symbolic-computation system Mathematica (Wolfram Research, Inc. 2010).
The approach is applied to the case of extreme autotetraploids with tetrasomic inheritance, the mixed formation of bivalents and quadrivalents, two linked marker loci on the same arm of a chromosome, and one partner switch per quadrivalent. Model parameters model the ratio of quadrivalent to bivalent formation (τ), the location of the partner switch (between the centromere and the centromere-proximal locus, between the two loci, or outside these two regions), the recombination frequency between the centromere and the centromere-proximal locus (q), and the recombination frequency between the two loci (r).
The resulting model is profoundly different from previous ones in several aspects, including the distribution of probability mass across gamete modes, most strikingly by one of the modes having zero probability. The coefficient of double reduction at the centromere-proximal locus, α, and the coefficient of double reduction at the centromere-distal locus, β, are not independent variables, but are fully explained through the other parameters. Further symbolic elimination of the partner switch location parameters turns the five-parameter model into a four-parameter model of r, q, α, and β; the mixing parameter τ disappears, showing that the ratio of quadrivalent to bivalent formation does not influence the frequencies of gamete modes beyond α and β.
The resulting model is also applied to a recent study of recombination frequencies in tetraploids based on a seed-fluorescence assay (Pecinka et al. 2011) and in a simulation study, in which it is compared to previously published models.
In addition to the immediate implications described, the computational approach presented here can be expected to be useful through opening avenues for modeling a host of processes, among them manifestations of meiosis that contain multiple-partner switches, crossover and chromatid interference, and meiosis in higher-order ploidies, such as hexaploidy and beyond.
Materials and Methods
The definition of the 11 gamete modes is taken from Table 1 in Luo et al. (2004), but see also Table 10 in Fisher (1947), there in a different order. With A denoting the centromere-proximal locus and B the centromere-distal locus, and with 1 ≤ i, j, k, l ≤ 4 (i, j, k, l all different) denoting the four copies of the marker-carrying chromosome, the 11 gamete modes are defined as
1: AiBi / AiBi
2: AiBj / AiBj
3: AiBi / AiBj
4: AiBj / AiBk
5: AiBi / AjBi
6: AiBj / AkBj
7: AiBi / AjBj
8: AiBi / AjBk
9: AiBj / AjBi
10: AiBj / AjBk
11: AiBj / AkBl
with the two columns of A’s and B’s showing the two chromosomes of the gamete.
The computational procedure, as described in detail in the Results section, has been implemented in the functional programming language Haskell. The computer program outputs sums of probabilities for gamete modes and other classes of interest that are then simplified by the symbolic-computation system Mathematica (Wolfram Research, Inc. 2010). The program together with ASCII text versions of the formulas presented in this article are available from the author.
Results
A five-parameter model
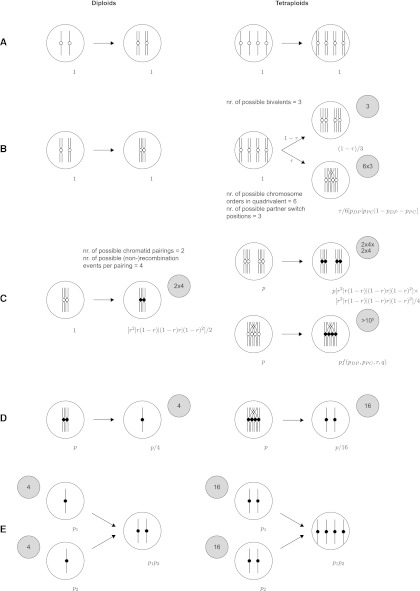
The approach put forward here is to computationally enumerate all possible paths of meiosis through the phases of replication, pairing, recombination, and segregation, while keeping track of the probabilities of the paths according to the various parameters involved. Probabilities for classes of genotypes or phenotypes are added, and the resulting formulas are simplified by the symbolic-computation system Mathematica. As an example application, I present the development of a model of meiosis in an autotetraploid with two linked marker loci on the same arm of a chromosome, taking into account mixed bivalent and quadrivalent pairings, pairing-partner switches, and recombination on a chromatid level (see Figure 1, together with a comparison to diploid meiosis).
Figure 1.
(A–E) Models of meiotic recombination in diploids and autotetraploids. The left column shows diploids and the right column tetraploids. (A) Replication of chromosomes. (B) Pairing of chromosomes into bivalents (diploids, tetraploids) and quadrivalents (tetraploids). (C) Recombination of paired chromosomes. See also Figure 2. (D) Distribution of (possibly) recombined chromosomes into gametes. (E) Crossing of gametes into seeds. Shaded circles show overall numbers of possibilities in that step. 1, p, p1, and p2 on the left sides of steps denote probabilities from previous steps. Formulas on the right sides of steps denote probabilities up to and including the current step. Alternatives are separated by vertical bars in square brackets, r is the frequency of recombination between the two marker loci, q is the recombination frequency between the centromere and the centromere-proximal marker, τ is the quadrivalent-to-bivalent ratio, and pDP and pPC are the probabilities of having a partner switch between the markers and between the centromere-proximal marker and the centromere, respectively. f(pDP, pPC, r, q) denotes a function of the parameters indicated.
Autotetraploids are modeled here as extreme autotetraploids, that is, without pairing preferences between chromosomes. Chromosomes can form either quadrivalents or bivalents, following a quadrivalence-to-bivalence ratio τ.
Consistent with a model of quadrivalent formation in which chromosome pairing starts at the ends of chromosomes, a (single) pairing-partner switch can occur such that one end of a chromosome pairs with one chromosome and the other end pairs with a different chromosome. Upon completion of pairing, this manifests itself in a partner switch, the location of which is either between the two markers (occurring with probability pDP, D for “distal” and P for “proximal”), between the centromere-proximal marker and the centromere (occurring with probability pPC), or outside these regions (occurring with probability 1 − pDP − pPC). For simplicity, in the first two cases, partner switches are modeled to be in the middle of the respective regions; the third case is treated as equivalent to the formation of two bivalents, and the location of the switch is irrelevant.
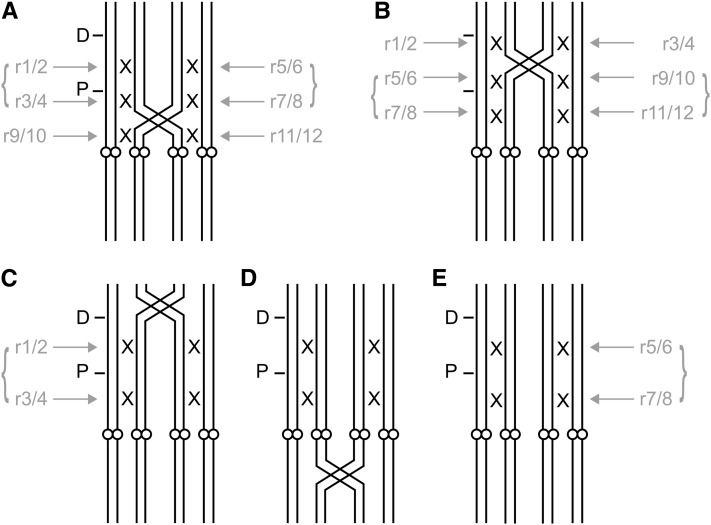
Chromatids of paired (segments of) chromosomes are modeled as pairing randomly, with sister chromatids not being allowed to pair. Depending on the valency and, in the case of a quadrivalent, on the position of the partner switch, up to three (effective) recombination events per chromatid can occur (see Figure 2). If the region between proximal and distal markers is partner switch free, recombination between these loci occurs with frequency r. If the partner switch is in this region, two recombinations can occur, one in each of the halves defined by the partner switch, with frequency . Similarly, for the region between centromere-proximal marker and centromere, either one recombination event with frequency q or two events with frequency each can occur. The half-recombination frequencies r′ and q′ are solutions of r = 2r′(1 − r′) and q = 2q′(1 − q′), respectively, expressing the fact that a recombination in the whole interval effectively occurs if and only if a recombination occurs in any of the halves but not in the other. In a partner switch free region, modeling two half-recombinations with frequency r′ would be equivalent to one recombination with frequency r and likewise for q′ and q. Every region or half-region of a chromatid can undergo recombination with the respective region of the pairing partner. Recombination events are modeled as being independent. Every region or half-region can be subject to at most one (effective) recombination event. Since recombination events in the distal-marker telomere region and the arm that does not carry the markers have no effect on marker distributions, they can be ignored here.
Figure 2.
(A–E) Chromosome pairing and recombination events. (A) Quadrivalent with partner switch between centromere and centromere-proximal locus. (B) Quadrivalent with partner switch between loci. (C and D) Quadrivalents with partner switch outside the two regions in A and B, effectively equivalent to E. (E) Two bivalents (no partner switch). D, centromere-distal locus; P, centromere-proximal locus; open circles, centromeres. X’s denote possible recombination events (two events per X). Shaded text and arrows describe the order of recombination events taken account of in the computer program. Curly brackets indicate that chromatid partner choice is not independent. Annotation of recombination events is implicitly the same in C–E.
Paired centromeres together with their attached chromatids are randomly distributed toward opposite poles (modeling anaphase I), and the two chromatids attached to a common centromere are again randomly distributed toward different poles (modeling anaphase II). This results in four gametes, each carrying two chromatids that are not sister chromatids. Note though that through recombination, parts of chromatids that were originally sister chromatids (sister factors) can now end up in the same gamete.
Sorting the resulting gametic products into gamete modes (see Materials andMethods for a definition of modes), and summing up probabilities per mode, results, after simplification with Mathematica, in the following five-parameter model, with r being the recombination frequency between the two markers, q the recombination frequency between the centromere and the centromere-proximal marker, τ the quadrivalent-to-bivalent ratio, pDP the probability of a pairing-partner switch between the two markers, pPC the probability of a pairing-partner switch between the centromere and the centromere-proximal marker, p1–p11 the probabilities of the 11 modes, and r′ the half-recombination frequency as defined above:
Coefficients of double reduction
Instead of sorting gametic products into gamete modes we can also classify them according to whether they are double reduced, in the centromere-proximal marker locus, in the centromere-distal marker locus, or in both marker loci. Summing up probabilities defines the coefficients of double reduction at the proximal locus, α, and at the distal locus, β, as
| (1) |
| (2) |
Elimination of parameters
The above probabilities of the 11 gamete modes are defined in terms of the five parameters r, q, τ, pDP, and pPC. Since the coefficients of double reduction, α and β, are also defined in terms of pDP and pPC, we can solve these two equations for pDP and pPC and substitute the solutions in the gamete mode equations, effectively eliminating pDP and pPC, and making α and β parameters. This procedure results in the following model, with – being the probabilities of the 11 modes, with parameters eliminated (indicated by the superscript e) and again r′ as defined above:
Surprisingly, the quadrivalent-to-bivalent ratio τ also disappears, indicating that it does not influence gamete mode probabilities beyond the coefficients of double reduction α and β. The resulting model is now one of four parameters, r, q, α, and β. Note, however, that α and β are not independent parameters, but depend on r and q, and that probabilities are undefined for r = 0 or q = 0. For r = 0, limits can be found by setting r = 0 in the original p1–p11and eliminating parameters again, resulting in the probabilities of all gamete modes becoming zero, except modes 1 and 7:
Note that α = β in this case. Limits for q = 0 cannot be found in the same way, since α becomes also zero, and, as a consequence, pDP and pPC cannot be both eliminated.
Application to a seed fluorescence assay
Melamed-Bessudo et al. (2005) introduced transgenic A. thaliana lines carrying two fluorescent seed markers that the authors used to estimate meiotic recombination frequencies (MRFs) between the two marker loci in diploid Arabidopsis plants. In Pecinka et al. (2011), the procedure was applied to tetraploidized Arabidopsis plants, with one copy of chromosome 3 carrying the markers on its top arm (green centromere-distal and red centromere-proximal), and MRFs were found to be significantly higher than in diploids. However, MRFs were estimated on the basis of a diploid model of meiosis, which is equivalent to a tetraploid model in which only bivalents form (see Wang and Luo 2012 and Rehmsmeier 2012 for an in-depth discussion). For a mixed bivalent–quadrivalent model we can specialize the five-parameter gamete model to the following equations (either by computationally sorting gametes into the various seed fluorescence classes or by manually adding probabilities from corresponding gamete modes), which describe the probabilities of seeds that contain only the red-fluorescent marker (pr, “red only”), of seeds that contain only the green-fluorescent marker (pg, “green only”), of seeds that contain both markers (py, “yellow”), and of seeds that contain none of the markers (pb, “brown”):
Eliminating pDP and pPC (and implicitly τ), as was done above for the 11 gamete modes, results in the following four-parameter model of seed fluorescences:
Note again that that probabilities are undefined for r = 0 or q = 0, but that limits for r = 0 can be found by setting r = 0 in the original pr, pg, py, and pb and eliminating parameters again, resulting in the following equations:
Note again that α = β in this case. Also again, limits for q = 0 cannot be found in the same way, since α becomes also zero, and, as a consequence, pDP and pPC cannot be both eliminated.
While the four-parameter model of seed fluorescences cannot be solved for the four parameters—since there are only three independent equations, the four equations summing up to unity—one can find the relationships
allowing estimation of the coefficients of double reduction α and β from seed count differences. Note that these same equations can also be derived from other models, such as the one in Luo et al. (2004) as used in Rehmsmeier (2012) (see Supporting Information, File S1), suggesting that this particular seed fluorescence assay is not sensitive to the underlying model of meiosis when coefficients of double reduction are to be determined. This, however, does not hold in cases where two or three copies of a chromosome carry the markers (see File S1).
Seed fluorescence probabilities for selfed plants can be defined by combining gamete probabilities, following the procedure in Wang and Luo (2012) and Rehmsmeier (2012).
Simulations
To understand the implications of the five-parameter model with respect to parameter estimation, I simulated gamete formation with fully informative markers (see, e.g., S. S. Wu et al. 2001) according to the five-parameter model and estimated parameters with the same model, with Luo et al.’s model (Luo et al. 2004) and with Wu et al.’s model (S. S. Wu et al. 2001; Wu and Ma 2005). Parameters r and q covered ranges from small to large values (0.01, 0.1, and 0.3), and τ, pDP, and pPC were fixed to 0.67, 0.21, and 0.34, respectively. The sample size was 1000, each sampling was repeated 100 times, and means and standard deviations were calculated from these 100 repetitions. Since gamete modes 7 and 9 cannot be distinguished by their genotypes, nor can modes 8 and 10 (see, e.g., S. S. Wu et al. 2001), the respective counts were added, as were the corresponding probability functions. S. S. Wu et al. (2001) and Wu and Ma (2005) give slightly different equations for the recombination frequency r (denoted θ in S. S. Wu et al. 2001), and both versions were used here. Parameters for the five-parameter model and for Luo et al.’s model were estimated with a maximum-likelihood approach, using R’s optim function in conjunction with the BFGS method and gradient functions (R Core Team 2012). For Wu et al.’s model, the recombination frequency r was directly estimated with a closed-form solution to their equations describing r (Equation 6 in S. S. Wu et al. 2001 and Equations 4, 5, and 8 in Wu and Ma 2005), and coefficients of double reduction, α and β, were calculated directly as sums of observed gamete frequencies [ (S. S. Wu et al. 2001)]. For Luo et al.’s model, β was calculated from r and α according to Equation 1 in Luo et al. (2004). True values of α and β and their estimates from the five-parameter model were calculated from the (true or estimated, respectively) r, q, τ, pDP, and pPC.
Table 1 shows mean estimates of the recombination frequency r and standard deviations of the estimates, expressed relative to the true values. Mean estimates for the five-parameter model are mainly perfect (1.0) or near perfect (0.99); for very small values of r and q, the mean estimate is slightly lower, at 0.97. The mean estimates for Luo et al.’s and Wu et al.’s models are between 0.91 and 1.03 of the true values. Mean estimates for Luo et al.’s and Wu et al.’s models are identical, and mean estimates for S. S. Wu et al. (2001) and Wu and Ma (2005) are nearly identical. Relative standard deviations are similar for all methods and always very small (≤0.025).
Table 1. Mean estimates of recombination frequency r and standard deviations of estimates (in parentheses), relative to true values.
| Relative mean estimated r |
|||||
|---|---|---|---|---|---|
| r | q | Five-parameter model | Luo et al. (2004) | S. S. Wu et al. (2001) | Wu and Ma (2005) |
| 0.01 | 0.01 | 0.97 (0.025) | 0.91 (0.023) | 0.91 (0.023) | 0.91 (0.023) |
| 0.01 | 0.10 | 0.99 (0.024) | 0.91 (0.022) | 0.91 (0.022) | 0.91 (0.022) |
| 0.01 | 0.30 | 1.00 (0.025) | 0.92 (0.022) | 0.92 (0.022) | 0.92 (0.022) |
| 0.10 | 0.01 | 0.99 (0.022) | 0.95 (0.020) | 0.95 (0.020) | 0.95 (0.020) |
| 0.10 | 0.10 | 0.99 (0.022) | 0.95 (0.020) | 0.95 (0.020) | 0.95 (0.020) |
| 0.10 | 0.30 | 1.00 (0.022) | 0.95 (0.021) | 0.95 (0.021) | 0.95 (0.021) |
| 0.30 | 0.01 | 1.00 (0.023) | 1.03 (0.023) | 1.04 (0.024) | 1.03 (0.023) |
| 0.30 | 0.10 | 1.00 (0.022) | 1.03 (0.022) | 1.04 (0.024) | 1.03 (0.022) |
| 0.30 | 0.30 | 1.00 (0.020) | 1.04 (0.022) | 1.05 (0.023) | 1.04 (0.022) |
r and q are varied as indicated; τ, pDP, and pPC are fixed to 0.67, 0.21, and 0.34, respectively.
Table 2 shows mean estimates of the coefficient of double reduction at the centromere-proximal locus, α, and standard deviations of the estimates, expressed relative to the true values. Mean estimates for the five-parameter model and for Luo et al.’s model are between 0.97 and 1.07 and are mainly identical. Estimates with the direct method (sums of gamete mode frequencies) are between 0.98 and 1.11. Relative standard deviations are small for all methods, although slightly larger for the direct method.
Table 2. Mean estimates of the coefficient of double reduction α and standard deviations of estimates (in parentheses), relative to true values.
| Relative mean estimated α |
|||||
|---|---|---|---|---|---|
| r | q | α | Five-parameter model | Luo et al. (2004) | S. S. Wu et al. (2001) |
| 0.01 | 0.01 | 0.00057 | 1.07 (0.029) | 1.07 (0.029) | 1.11 (0.033) |
| 0.01 | 0.10 | 0.00570 | 0.97 (0.027) | 0.97 (0.028) | 0.98 (0.031) |
| 0.01 | 0.30 | 0.01708 | 0.97 (0.028) | 0.98 (0.028) | 0.99 (0.032) |
| 0.10 | 0.01 | 0.00057 | 1.07 (0.029) | 1.07 (0.029) | 1.11 (0.033) |
| 0.10 | 0.10 | 0.00570 | 0.97 (0.028) | 0.97 (0.028) | 0.98 (0.031) |
| 0.10 | 0.30 | 0.01708 | 0.97 (0.028) | 0.98 (0.028) | 0.99 (0.032) |
| 0.30 | 0.01 | 0.00057 | 1.07 (0.029) | 1.07 (0.029) | 1.11 (0.033) |
| 0.30 | 0.10 | 0.00570 | 0.97 (0.027) | 0.97 (0.028) | 0.98 (0.031) |
| 0.30 | 0.30 | 0.01708 | 0.97 (0.028) | 0.98 (0.028) | 0.99 (0.032) |
r and q are varied as indicated; τ, pDP, and pPC are fixed to 0.67, 0.21, and 0.34, respectively; the true α is determined by q, τ, and pPC through Equation 1.
Table 3 shows mean estimates of the coefficient of double reduction at the centromere-distal locus, β, and standard deviations of the estimates, expressed relative to the true values. Mean estimates from the five-parameter model are between 0.98 and 1.00. Mean estimates from sums of gamete mode frequencies are between 0.92 and 1.02. Estimates from Luo et al.’s model are between 1.2 and 4.7. Standard deviations are small for all methods.
Table 3. Mean estimates of the coefficient of double reduction β and standard deviations of estimates (in parentheses), relative to true values.
| Relative mean estimated β |
|||||
|---|---|---|---|---|---|
| r | q | β | Five-parameter model | Luo et al. (2004) | S. S. Wu et al. (2001) |
| 0.01 | 0.01 | 0.0020 | 0.98 (0.029) | 3.3 (0.036) | 0.92 (0.032) |
| 0.01 | 0.10 | 0.0069 | 0.98 (0.025) | 1.6 (0.031) | 0.94 (0.030) |
| 0.01 | 0.30 | 0.0179 | 0.99 (0.030) | 1.2 (0.030) | 0.99 (0.032) |
| 0.10 | 0.01 | 0.0141 | 0.98 (0.028) | 4.2 (0.032) | 0.97 (0.031) |
| 0.10 | 0.10 | 0.0174 | 1.00 (0.029) | 3.7 (0.030) | 0.99 (0.031) |
| 0.10 | 0.30 | 0.0247 | 1.00 (0.029) | 2.9 (0.029) | 1.01 (0.030) |
| 0.30 | 0.01 | 0.0346 | 0.98 (0.030) | 4.7 (0.026) | 1.02 (0.032) |
| 0.30 | 0.10 | 0.0354 | 0.98 (0.030) | 4.7 (0.025) | 1.02 (0.032) |
| 0.30 | 0.30 | 0.0372 | 0.99 (0.029) | 4.6 (0.023) | 1.01 (0.033) |
r and q are varied as indicated; τ, pDP, and pPC are fixed to 0.67, 0.21, and 0.34, respectively; the true β is determined by r, q, τ, pDP, and pPC through Equation 2.
Table 4 shows mean estimates of the recombination frequency q and standard deviations of the estimates, expressed relative to the true values. Mean estimates are accurate for the majority of cases, but deviate strongly from the true values for small values of r and q.
Table 4. Mean estimates of recombination frequency q and standard deviations of estimates (in parentheses), relative to true values.
| r | q | Relative mean estimated q, five-parameter model |
|---|---|---|
| 0.01 | 0.01 | 25.6 (2.27) |
| 0.01 | 0.10 | 4.0 (0.57) |
| 0.01 | 0.30 | 1.4 (0.26) |
| 0.10 | 0.01 | 4.7 (1.24) |
| 0.10 | 0.10 | 1.8 (0.55) |
| 0.10 | 0.30 | 1.0 (0.23) |
| 0.30 | 0.01 | 1.4 (0.18) |
| 0.30 | 0.10 | 1.1 (0.19) |
| 0.30 | 0.30 | 1.1 (0.19) |
r and q are varied as indicated; τ, pDP, and pPC are fixed to 0.67, 0.21, and 0.34, respectively.
Table 5 shows mean estimates of the quadrivalent-to-bivalent ratio τ, the probability of a pairing-partner switch between the two markers pDP, and the probability of a pairing-partner switch between the centromere and the centromere-proximal marker pPC and standard deviations of the estimates, expressed relative to the true values. Mean estimates of τ are accurate, with values between 0.83 and 1.11, but have standard deviations of mainly between 0.25 and 0.3. Mean estimates of pDP and pPC are inaccurate for small values of r and q (r and q = 0.01), with values of 2.7 and 0.41. Mean estimates of pDP and pPC are accurate for larger values of r and q, with values between 0.69 and 1.6 for r and q ≥ 0.1 and values of 0.92 and 1.1 for r and q = 0.3. Standard deviations for estimates of pDP and pPC are large, with values between 0.29 and 0.82.
Table 5. Mean estimates of the quadrivalent-to-bivalent ratio τ, of the probability of a pairing-partner switch between the two markers pDP, and of the probability of a pairing-partner switch between the centromere and the centromere-proximal marker pPC, and standard deviations of estimates (in parentheses), relative to true values.
| Five-parameter model |
||||
|---|---|---|---|---|
| r | q | Relative mean estimated τ | Relative mean estimated pDP | Relative mean estimated pPC |
| 0.01 | 0.01 | 0.93 (0.43) | 2.68 (0.82) | 0.41 (0.40) |
| 0.01 | 0.10 | 1.05 (0.38) | 2.15 (0.79) | 0.62 (0.41) |
| 0.01 | 0.30 | 1.11 (0.30) | 1.49 (0.69) | 0.91 (0.39) |
| 0.10 | 0.01 | 0.82 (0.25) | 0.74 (0.58) | 1.52 (0.47) |
| 0.10 | 0.10 | 0.90 (0.29) | 0.95 (0.65) | 1.35 (0.51) |
| 0.10 | 0.30 | 0.83 (0.26) | 0.69 (0.51) | 1.58 (0.46) |
| 0.30 | 0.01 | 0.92 (0.25) | 1.11 (0.32) | 1.21 (0.34) |
| 0.30 | 0.10 | 0.96 (0.26) | 1.01 (0.36) | 1.20 (0.40) |
| 0.30 | 0.30 | 1.05 (0.26) | 0.92 (0.29) | 1.08 (0.36) |
r and q are varied as indicated; τ, pDP, and pPC are fixed to 0.67, 0.21, and 0.34, respectively.
Discussion
Owing to its complexities, polyploid meiosis has resisted the development of mathematical models. Indeed, Luo et al. point out that a tedious analysis was necessary to be able to express probabilities of gamete modes as functions of meiotic recombination frequency and the coefficient of double reduction (Luo et al. 2004). Any attempt to manually formulate the complex model of tetraploid meiosis presented here—one that is consistent with postulated mechanistic behaviors such as the formation of pairing-partner switches—would be doomed. The computational approach to developing mathematical models of polyploid meiosis that is presented here makes complex models feasible to begin with.
In addition to the methodological advances, the resulting model of autotetraploid meiosis gives unexpected insights into the process of double reduction and its consequences for model parameterization and gamete mode probabilities. Luo et al.’s model (Luo et al. 2004) is a model of two parameters, the frequency of recombination between the two loci, r, and the coefficient of double reduction at the centromere-proximal locus, α. For double reduction at the centromere-proximal locus to occur, a partner switch between the centromere and the locus is necessary, and the probability of such a switch is, in addition to other requirements, expressed by α. Luo et al. couple the coefficient of double reduction at the centromere-distal locus, β, to α by β = (α(3 − 4r)^2 + 2r(3 − 2r))/9 (Equation 1 in Luo et al. 2004). Double reduction at that locus, however, requires a partner switch between it and the centromere—the probability of which cannot be controlled by α and r alone (see Wu and Ma 2005 for a related discussion). In fact, letting one locus coincide with the centromere, thus forcing α to be zero, would make β entirely defined by r.
In response to this problem, Wu and Ma (2005) developed a model that is parameterized with β as well as with α and r. The model presented here contains a further parameter, the frequency of recombination between the centromere and the centromere-proximal locus, q. This additional parameter is naturally introduced when the possibility of a pairing-partner switch between the centromere and the centromere-proximal locus is taken into account. Indeed, upon setting the probability of such a pairing-partner switch, pPC above, to zero, q disappears again from the equations, which can easily be seen in the definitions of p1–p11 by noting that q occurs only with pPC as a factor.
In the model presented here, the coefficients of double reduction, α and β, are not independent parameters, unlike α in Luo et al. (2004) and α and β in Wu and Ma (2005), but are defined in terms of the quadrivalent-to-bivalent ratio τ, the probabilities of partner switches, pDP and pPC, and the recombination frequencies r and q. The coefficients of double reduction are thus decomposed into what constitutes them mechanistically (in terms of τ, pDP, pPC, r, and q), allowing for a deeper understanding of these individual aspects.
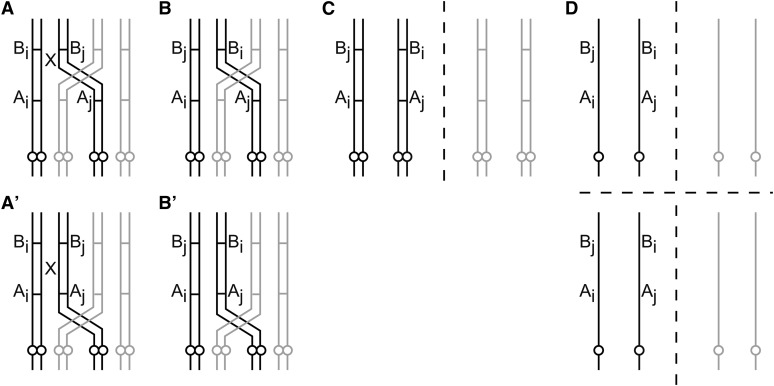
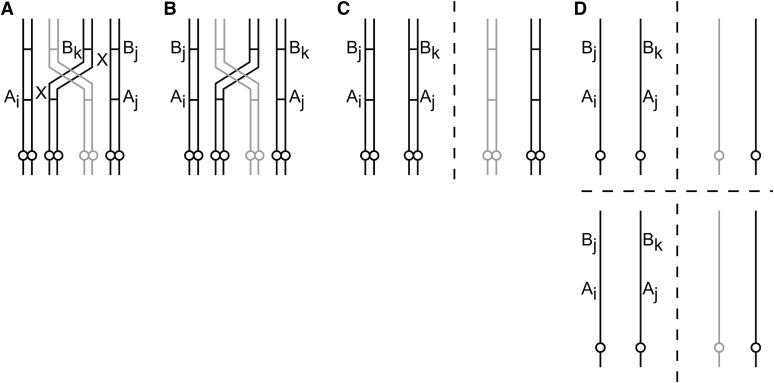
The analytic nature of the coefficients of double reduction has surprising consequences for the dependence of gamete mode probabilities on these coefficients. When setting α and β to zero in the model presented here, all modes except modes 7, 8, and 11 reduce to probability zero, in contrast to the model of Luo et al., in which, among others, modes 9 and 10 remain nonzero as well (mode 9 being described by the genotype AiBj/AjBi and mode 10 by AiBj/AjBk, with A denoting the centromere-proximal marker; B denoting the centromere-distal marker; the “/” distinguishing the two chromosomes; and i, j, and k denoting different chromosomes). The probability of mode 9 becoming zero appears counterintuitive at first glance, since this mode is not double reduced in either of the two loci. However, the manifestation of this mode requires a pairing-partner switch between the centromere and the centromere-distal locus (see Figure 3). Since here α and β are defined in terms of recombination frequencies r and q, pairing-partner switch probabilities pDP and pPC, and the quadrivalent-to-bivalent ratio τ, setting α and β to zero while leaving r and q untouched is equivalent to setting either pDP and pPC or τ to zero, thus prohibiting the necessary formation of a pairing-partner switch. This becomes also apparent from the definition of p9 in the five-parameter model. The requirement of a partner switch holds similarly for mode 10 (see Figure 4).
Figure 3.
(A–D) Gamete mode 9 through a partner switch (but without double reduction). Partner switch is between the two loci, A and B. Circles denote centromeres. X denotes two recombinations. (A) A quadrivalent forms, and the two chromosomes (solid lines) recombine twice. (B) The recombined chromosomes. The distal markers, B, have been swapped. (C) Chromosomes move apart in anaphase I. (D) Chromatids move apart in anaphase II. Two gametes now have configuration AiBj/AjBi (gamete mode 9). A single recombination leads to one gamete of mode 9. A similar process is shown (with one or two recombinations) for the partner switch being between the centromere and the centromere-proximal locus (A′ and B′, continuing with C and D).
Figure 4.
(A–D) Gamete mode 10 through a partner switch (but without double reduction). Partner switch is between the two loci, A and B. Circles denote centromeres. X’s denote two recombinations each. (A) A quadrivalent forms, and the chromosomes (solid lines) recombine twice in each of the positions indicated by X. (B) The recombined chromosomes. The distal markers, B, have been swapped. (C) Chromosomes move apart in anaphase I. (D) Chromatids move apart in anaphase II. Two gametes have now configuration AiBj/AjBk (gamete mode 10). A single recombination leads to one gamete of mode 10.
A further striking result of the model presented here is not only the generally different distribution of probability mass among the gamete modes, but also the fact that gamete mode 4 (AiBj/AiBk) has probability zero, even in the general case of mixed quadrivalent and bivalent formation. Since Ai is double reduced, there must be a partner switch between the centromere and the centromere-proximal locus. On the other hand, the two sister segments containing Bi have to cross over with two different chromosomes, j and k, which would require a second partner switch, between the two loci. Indeed, in a model that allows two partner switches, gamete mode 4 has nonzero probability (see File S2).
When the probabilities of pairing-partner switches, pDP and pPC, are eliminated and α and β are introduced, the quadrivalent-to-bivalent ratio parameter τ also disappears, indicating that τ does not contribute to the probabilities of gamete modes beyond its contribution to the coefficients of double reduction. In other words, gamete mode probabilities are fully defined in terms of recombination frequencies and coefficients of double reduction. Interestingly, this does not hold for a model in which a second partner switch can be formed (see File S3).
Double-reduction parameters in Luo et al. (2004) and Wu and Ma (2005) are conditioned on complete quadrivalency, thus being inconsistent with a definition of double reduction that does not make assumptions in this respect. Estimates of double reduction can thus be expected to be biased when the formation of bivalents is not negligible. In contrast, in the model presented here, coefficients of double reduction explicitly depend on the quadrivalent-to-bivalent ratio τ and are thus not conditioned on any particular valency.
The results of the simulations show that estimates under the five-parameter model of the recombination frequency r are very accurate and more so than estimates under existing models. Estimates of the coefficient of double reduction α are similarly accurate for all methods. Interestingly, this is also the case for the five-parameter model, even though α is not estimated directly, but calculated from the estimates of the five parameters. Estimates of the coefficient of double reduction β are very accurate for the five-parameter model and similar to the direct method of S. S. Wu et al. (2001). Estimates of β under Luo et al.’s model, by using their Equation 1, deviate considerably from the true values, demonstrating that β cannot be accurately expressed through Luo et al.’s Equation 1 by α and r alone when gamete mode frequencies follow the five-parameter model presented here. Estimates of q, the recombination frequency between the centromere and the centromere-proximal locus, deviate strongly from the true value for very small values of r and q, such as r and q = 0.01. Closer inspection shows that for these values, the estimated q is frequently close to 1/2 (see Figure S1), explaining the relative mean estimate of ∼0.25. For such small values of r and q, several gamete modes have exceedingly small probabilities. For example, for r = 0.01 and q = 0.01 (and τ = 0.67, pDP = 0.21, pPC = 0.34), the probability of gamete mode 2 is <5.7e-8. As a consequence, extremely large sample sizes would be necessary to generate gamete counts for all modes and to allow for an accurate estimation of q. Nevertheless, for larger values of r and q, mean estimates of q are accurate. Assuming that recombination frequency grows monotonously with distance from the centromere, estimates of q could assist in genetic-map construction by directly suggesting a linear order of markers on a chromosome arm.
In this article I have put forward a computational approach to the probabilistic modeling of meiosis, with an example application to autotetraploids. The relative simplicity of automatically deriving such a model from a computational description of its underlying mechanisms and the multitude of aspects in which the resulting model distinguishes itself from previously published ones demonstrate the usefulness of the approach. Possibilities now arise for the development of more complex models, such as a tetraploid three-locus model with 107 gamete modes (Fisher 1947), hexaploid two-locus models with two mixing ratios (one hexavalent vs. one quadrivalent and one bivalent vs. three bivalents) and 40 gamete modes (Fisher 1947), or models of ploidies of even higher order.
It is worth noting that the approach presented here relieves the developer of mathematical models from the burden of having to argue about properties of gamete modes in terms of model parameters. Instead, the modeler can concentrate on a computational description of the process and leave the construction and simplification of expressions to the computer. The resulting mathematical models simply emerge automatically. Not only does this approach allow one to build mathematical models that would be impossible to build manually, but also it frees creative energy that can be invested in the definition of the structure of the model and in the interpretation of the resulting mathematical formulas.
Acknowledgments
The author thanks Leonie Ringrose for advice on the writing of this article and the Editor and two anonymous reviewers for helpful comments.
Footnotes
Communicating editor: J. B. Holland
Literature Cited
- Brownfield L., Köhler C., 2011. Unreduced gamete formation in plants: mechanisms and prospects. J. Exp. Bot. 62(5): 1659–1668. [DOI] [PubMed] [Google Scholar]
- Comai L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6(11): 836–846. [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1947. The theory of linkage in polysomic inheritance. Philos. Trans. R. Soc. Lond. B Biol. Sci. 233(594): 55–87. [Google Scholar]
- Haynes K. G., Douches D. S., 1993. Estimation of the coefficient of double reduction in the cultivated tetraploid potato. Theor. Appl. Genet. 85(6): 857–862. [DOI] [PubMed] [Google Scholar]
- Jackson S. A., Iwata A., Lee S. H., Schmutz J., Shoemaker R., 2011. Sequencing crop genomes: approaches and applications. New Phytol. 191(4): 915–925. [DOI] [PubMed] [Google Scholar]
- Lao J. P., Hunter N., 2010. Trying to avoid your sister. PLoS Biol. 8(10): e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings C. S., Alexander D. E., 1966. Double reduction in autotetraploid maize. Genetics 54(6): 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., de Massy B., 2011. The impressionistic landscape of meiotic recombination. Cell 147(2): 267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Yang X., Tong C., Li X., Feng S., et al. , 2012. A multivalent three-point linkage analysis model of autotetraploids. Brief. Bioinform. (in press). [DOI] [PubMed] [Google Scholar]
- Luo Z. W., Zhang R. M., Kearsey M. J., 2004. Theoretical basis for genetic linkage analysis in autotetraploid species. Proc. Natl. Acad. Sci. USA 101(18): 7040–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K., 1935. Reductional and equational separation of the chromosomes in bivalents and multivalents. J. Genet. 30(1): 53–78. [Google Scholar]
- Mather K., 1936. Segregation and linkage in autotetraploids. J. Genet. 32(2): 287–314. [Google Scholar]
- Melamed-Bessudo C., Yehuda E., Stuitje A. R., Levy A. A., 2005. A new seed-based assay for meiotic recombination in Arabidopsis thaliana. Plant J. 43(3): 458–466. [DOI] [PubMed] [Google Scholar]
- Otto S. P., 2007. The evolutionary consequences of polyploidy. Cell 131(3): 452–462. [DOI] [PubMed] [Google Scholar]
- Pecinka A., Fang W., Rehmsmeier M., Levy A. A., Mittelsten Scheid O., 2011. Polyploidization increases meiotic recombination frequency in Arabidopsis. BMC Biol. 9: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradillo M., Santos J. L., 2011. The template choice decision in meiosis: Is the sister important? Chromosoma 120(5): 447–454. [DOI] [PubMed] [Google Scholar]
- R Core Team, 2012 R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. ISBN 3-900051-07-0, http://www.R-project.org. [Google Scholar]
- Rehmsmeier M., 2012. Response to Wang and Luo. BMC Biol. 10: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripol M. I., Churchill G. A., da Silva J. A., Sorrells M., 1999. Statistical aspects of genetic mapping in autopolyploids. Gene 235(1–2): 31–41. [DOI] [PubMed] [Google Scholar]
- Wang L., Luo Z., 2012. Polyploidization increases meiotic recombination frequency in Arabidopsis: a close look at statistical modeling and data analysis. BMC Biol. 10: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram Research, Inc., 2010 Mathematica, Version 8.0. Champaign, IL. [Google Scholar]
- Wu R., Ma C. X., 2005. A general framework for statistical linkage analysis in multivalent tetraploids. Genetics 170: 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Gallo-Meagher M., Littell R. C., Zeng Z. B., 2001. A general polyploid model for analyzing gene segregation in outcrossing tetraploid species. Genetics 159: 869–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. S., Wu R., Ma C. X., Zeng Z. B., Yang M. C. K., et al. , 2001. A multivalent pairing model of linkage analysis in autotetraploids. Genetics 159: 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Lv Y., Pang X., Tong C., Wang Z., et al. , 2013. A unifying framework for bivalent multilocus linkage analysis of allotetraploids. Brief. Bioinform. 14(1): 96–108. [DOI] [PubMed] [Google Scholar]
- Yanowitz J., 2010. Meiosis: making a break for it. Curr. Opin. Cell Biol. 22(6): 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]