Abstract
We report a novel sexual-cycle-specific gene-silencing system in the genetic model Aspergillus nidulans. Duplication of the mating type matAHMG gene in this haploid organism triggers Mat-induced silencing (MatIS) of both endogenous and transgenic matA genes, eliminates function of the encoded SRY structural ortholog, and results in formation of barren fruiting bodies. MatIS is spatiotemporally restricted to the prezygotic stage of the sexual cycle and does not interfere with vegetative growth, asexual reproduction, differentiation of early sexual tissues, or fruiting body development. MatIS is reversible upon deletion of the matA transgene. In contrast to other sex-specific silencing phenomena, MatIS silencing has nearly 100% efficiency and appears to be independent of homologous duplicated DNA segments. Remarkably, transgene-derived matA RNA might be sufficient to induce MatIS. A unique feature of MatIS is that RNA-mediated silencing is RNA interference/Argonaute-independent and is restricted to the nucleus having the duplicated gene. The silencing phenomenon is recessive and does not spread between nuclei within the common cytoplasm of a multinucleate heterokaryon. Gene silencing induced by matA gene duplication emerges as a specific feature associated with matAHMG regulation during sexual development.
Keywords: Aspergillus, premeiotic gene silencing, sexual reproduction
DISCOVERY of homology-dependent gene silencing (HDGS) has opened a new dimension to our understanding of eukaryotic genome integrity, structure, and expression. HDGS as a consequence of gene duplication is a ubiquitous phenomenon that has been reported across the kingdoms in various species of fungi, plants, and animals. Transgene-mediated gene duplication often triggers simultaneous silencing of both the transgene and the homologous endogenous gene at the transcriptional and/or post-transcriptional level (Bingham 1997; Cogoni and Macino 1999b; Cogoni 2001).The basic molecular machinery for gene silencing shares common mechanistic features with plants, animals, and fungal species (Bingham 1997; Selker 1997; Cogoni and Macino 1999b; Cogoni 2001; Vaucheret and Fagard 2001). Silencing is usually induced by duplicated homologous coding sequences that trigger RNA-mediated post-transcriptional degradation of the gene-specific messenger RNA (mRNA) or RNA/DNA-mediated DNA methylation and/or chromatin modification resulting in transcriptional inhibition of gene expression (Cogoni and Macino 1999b; Moazed 2009). Gene silencing is believed to be an ancient phenomenon that evolved as a genome defense mechanism responding to virus infection or transposon invasion. It plays a major role in genome stability, maintenance, and regulation of chromatin structure and gene expression (Cogoni and Macino 1999b; Moazed 2009).
Several components of gene-silencing pathways are conserved. RNAse III (Dicer), RNA-dependent RNA polymerase (RdRP), Argonaute proteins, RNA-silencing complexes [RNA-induced silencing complexes (RISCs) and RNA-induced transcriptional silencing complexes (RITS)], and chromatin-remodeling complexes have been characterized in various eukaryotic species from protists to humans (Cerutti and Casas-Mollano 2006; Moazed 2009). However, precise molecular mechanisms and mechanistic details underlying HDGS pathways are still poorly defined and remain largely unknown (Cogoni and Macino 1999b; Vaucheret and Fagard 2001; Catalanotto et al. 2004; Chicas et al. 2004; Forrest et al. 2004; Hammond and Keller 2005; Wassenegger 2005).
In fungi, a variety of different HDGS phenomena have been reported, all demonstrating conserved but also unique features. HDGS processes have been observed in Neurospora (Cogoni and Macino 1997a,b), Ascobolous (Barry et al. 1993; Malagnac et al. 1997), Schizophyllum (Schuurs et al. 1997), Coprinus (Freedman and Pukkila 1993), Phytophthora (van West et al. 1999), and Cryptococcus (Wang et al. 2010). The most studied and best characterized are premeiotic repeat-induced point mutation (RIP) in Neurospora crassa and methylation induced premeiotically (MIP) in Ascobolous immersus. Both RIP and MIP phenomena represent examples of transcriptional gene silencing (TGS). MIP and RIP occur specifically at the sexual stage and are induced in the haploid nuclei during the period between fertilization and karyogamy. In both silencing phenomena, a minimal size of 400 bp of DNA homology between repeated genes is required to trigger pairwise transcriptional gene silencing of homologous duplicated sequences. Gene inactivation by RIP is irreversible because duplicated sequences are heavily methylated and permanently mutagenized, whereas MIP involves only DNA methylation and is reversible when affected sequences are demethylated (Selker 1990; Barry et al. 1993; Cogoni and Macino 1999b; Cogoni 2001).
Other extensively studied gene-silencing phenomena are quelling and meiotic silencing by unpaired DNA in N. crassa. Both quelling and meiotic silencing result in homology-dependent gene inactivation by mRNA degradation by components of the post-transcriptional gene silencing (PTGS) pathway. PTGS in quelling functions through small interfering RNA (siRNA) molecules that are embedded in specific RISCs that recognize and degrade homologous mRNA particles in the cytoplasm. Quelling operates during the vegetative stage and requires homology between duplicated coding DNA segments (as small as 132 bp). Neither homology between promoter regions nor transgene-derived RNA or protein products are required to induce quelling (Cogoni and Macino 1997b). The silencing effect in quelling is dominant and acts in trans to inactivate homologous genes in both transformed and untransformed nuclei of heterokaryons. Quelling is reversible when transgenes are removed (Barry et al. 1993; Cogoni and Macino 1999b; Cogoni 2001). Unlike quelling, meiotic silencing operates specifically in the zygotic cell after karyogamy. Meiotic silencing is a genome surveillance mechanism that scans the pairing and alignment of homologous chromosomes in the meiotic prophase. Unpaired DNA segments with homology to the transcript are required to trigger self-silencing of unpaired genes and trans-silencing of all homologous copies of the gene, whether or not they are paired (Lee et al. 2004). Meiotic silencing appears to affect a broad array of genes coding for functions required during meiosis, such as APSES-domain transcription factor, Asm-1; β-tubulin, Bml; actin, act; histones H3 and H4-1, hH3-H4-1; plasma membrane ATPase, pma-1; and RecA/RAD51 homolog, mei-3 (Shiu et al. 2001). Recently reported SIS in the fungal pathogen C. neoformans represents a novel example of transgene-induced post-transcriptional gene silencing that is specific to the sexual stage. SIS is triggered by tandem integration of a transgene array and is mediated by RNAi (Wang et al. 2010).
Gene-silencing phenomena reported in other fungal species represent a great range of complexity and variation of mechanistic details. Transnuclear transcriptional gene silencing in Phytophthora infestans is diffusible and dominant but does not involve siRNA molecules. Rather, it has been demonstrated that DNA methylation is required for the silencing effect (van West et al. 1999, 2008).
The classic model organism Aspergillus nidulans provides a valuable and sophisticated system for the molecular dissection of the gene-silencing phenomenon. A. nidulans is a haploid, multicellular, filamentous fungus with an experimentally amenable sexual reproductive cycle. Sexual development in A. nidulans is a complex multistep process that requires special environmental conditions and is governed by the mating-type genes matA(HMG-box) and matB(α-box) that transcriptionally coordinate expression of sex-specific genes. Sexual morphogenesis in A. nidulans has been previously described (Champe et al. 1994; Sohn and Yoon 2002; Champe and Simon 2009). Sexual reproduction results in the formation of macroscopic fruiting bodies (cleistothecia) containing meiotic progeny (ascospores). Mating and sexual differentiation involves formation of the fruiting body wall (cleistothecium shell) and reproductive ascogenous tissue containing many dividing haploid nuclei. Two haploid nuclei undergo karyogamy within specialized dikaryotic cells (croziers) to form a diploid zygote (ascus mother cell). The zygotic nucleus undergoes meiosis followed by a postmeiotic mitosis, which results in the formation of eight haploid ascospores within an ascus. An individual cleistothecium contains hundreds of thousands of ascospores.
The mating-type gene matA encodes a transcription factor with a conserved HMG high mobility group (HMG) DNA-binding domain that is typical of proteins involved in both chromatin architecture and gene transcription. MatA and other Mat-HMG proteins are members of the SOX/MATA/TCF protein family based on the ability of the HMG box to bind to specific DNA sequences (Laudet et al. 1993). The fungal Mat-HMG box domain demonstrates a high level of structural similarity with the human SRY gene (sex determining region Y) HMG box (Idnurm et al. 2008). Fungal Mat-HMG proteins are required for fine-tuning and balanced spatiotemporal expression of different sets of target genes directly involved in male and female fertility, fruiting body morphogenesis, fruiting body abundance, and ascospore formation (Debuchy and Turgeon 2006). Manipulations of mating-type gene structure and/or function affect sexual phenotype and make possible conversions between fungal reproductive lifestyles (Yun et al. 1999; Lee et al. 2003). In this study, we demonstrate evidence for premeiotic Mat-induced silencing (MatIS) that is triggered by duplication of the matA gene.
We present the first report of gene silencing associated with matA mating-type function during the premeiotic stage of the sexual cycle. Silencing is not a generalized feature of sex-specific gene duplication. Additional copies of the matB mating-type gene or the tubB meiosis-specific α-tubulin do not induce MatIS or cause infertility. MatIS is not activated during vegetative growth or early sexual development. Silencing takes place in the population of prezygotic cells (croziers) and results in the failure of karyogamy, meiosis, and, consequently, lack of meiotic progeny. MatIS is induced by the matA RNA transcript derived from the transgene; however, unlike other silencing systems it is independent of transcript abundance or homologous matA DNA segments per se and demonstrates essentially 100% silencing efficiency. MatIS is also unique among reported HGDS phenomena in fungi because it occurs in the absence of Argonaute, and RNAi involving RITS or RISC complexes is apparently not involved. One of the characteristic features of RNA-mediated silencing reported in plants and fungi is the dominance effect and spreading of silencing between multiple nuclei located within a common cytoplasm. Interestingly, MatIS in A. nidulans is recessive and does not spread between nuclei within heterokaryons.
Discovery of the unique features of HDGS associated with mating-type function in A. nidulans provides a valuable model system that can be used to unravel molecular mechanisms responsible for sexual-cycle-specific silencing phenomena. Further studies of gene-silencing systems in fungal genetic model organisms such as A. nidulans offer the opportunity to identify novel molecular components and mechanistic details of eukaryotic gene silencing.
Materials and Methods
Strains, culture conditions, and molecular techniques
A. nidulans strains used in this study are listed in Table 1. Complete and appropriately supplemented media were prepared as described by Pontecorvo et al. (1953), Kafer (1977), and Vallim et al. (2000). Standard molecular techniques, DNA and RNA extraction, Southern blot analysis, and fungal transformations were performed according to protocols previously described by Miller et al. (1985, 1987), Wu and Miller (1997), Pyrzak et al. (2008), and Yelton et al. (1983). Sexual development was induced under standard culture conditions as described previously (Miller et al. 1985; Vallim et al. 2000; Pyrzak et al. 2008). Fertility was determined by random sampling of at least 10 cleistothecia from an induction plate. Each cleistothecium was cleaned by gently rolling in 3% water agar, transferred to an Eppendorf tube containing 100 μl 0.1% Tween80, and crushed with a glass rod to release ascospores. Two ascospore counts were made for each cleistothecium using a hemacytometer.
Table 1. A. nidulans strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| GR5 | pyrG89; wA3; pyroA4 | G. S. May (MD Anderson Cancer Center, Houston TX) |
| RTMH 200.10 | rsdAΔArgo::pyrG; pyroA4 | N. Keller (University of Wisconsin, Madison) |
| RTMH 202.11 | pabaA1, yA2, ΔrrpB::pyrG, ΔrrpC::metG,VeA | N. Keller (University of Wisconsin, Madison) |
| UI350 | ya2, biA1, argB2, pyrG89, riboB2 | B. L. Miller (University of Idaho) |
| UI412 | pyrG89:pWP3:pyrG, biA1, pabaA1; argB2; matA(0)::argB | This study |
| UI420-2 | pyrG89, alcA(p):medA; pyroA4; wA1 | This study |
| UI432 | pyrG89:pWP3:pyrG, alcA(p):medA, biA1, pabaA1; wA3; pyroA4 | This study |
| UI433 | pyrG89, alcA(p):medA, biA1, pabaA1; wA3; pyroA4 | This study |
| UI464 | pyrG89:pWP3:pyrG, pabaA1; wA3; argB2; matA(0)::argB | This study |
| UI465 | pyrG89, yA2, pabaA1;matA(0)::AfargB | This study |
| UI470 | pyrG89:pWP3(frameshift mutation):pyrG; wA3; pyroA4 | This study |
| UI471 | pyrG89:pWP3:pyrG; wA3; pyroA4 | This study |
| UI480 | pyrG89, wA3; pyroA4:matA:pyroA | This study |
| UI481 | pyrG89, wA3; matA:pyroA:matA; pyroA4 | This study |
| UI482 | pyrG89, wA3; ΔrsdAArgo; pyroA4:matA:pyroA | This study |
| UI483 | pyrG89; wA3; pyroA4; matB:pyroA | This study |
| UI484 | pyrG89; wA3; pyroA4, tubB:pyroA | This study |
Construction of the A. nidulans strains carrying duplication of the matA gene
Two different A. nidulans strains carrying duplications of wild-type matA gene (resident + ectopic) were constructed and used in this study. The ectopic copy of the matA gene in both strains was introduced by transformation with pWP3 and was integrated by homology at the pyrG89 locus of the recipient strain. Plasmid pWP3 carries the coding region of matA flanked by 1-kb upstream and 1.8-kb downstream genomic sequences plus pyrG as a prototrophy marker. pWP3 was constructed by cloning the AnmatA genomic region (primers AnMatAF11: P-tgggagtgtatcagcttcatg and AnmatAR11: P-tgccgtatgctacctgag) into the ppyrG plasmid (Pyrzak et al. 2008). The UI432 strain is the progeny of a cross between parental strains UI420-2 and UI412 (see Table 1). The wild-type matA gene at chromosome III was inherited from the UI420 parent, and the ectopic matA transgene (pyrG89:matA:pyrG) was inherited with chromosome I from the UI412 parent. The second transgenic strain, UI471, was created via DNA-mediated transformation of pWP3 into the GR5 recipient strain. The UI471 strain carries a wild-type matA gene at the endogenous locus and an ectopic matA transgene (pyrG89:matA:pyrG) as a result of pWP3 integration at the pyrG89 locus. The genotypes of both strains were confirmed by Southern blot analysis. A Gateway cassette (Invitrogen) was added to plasmid pAVT21 (S. Harris, University of Nebraska) containing the pyroA gene as a selectable marker. The matA genomic sequences used in pWP3 were recombined into this plasmid using clonase (Invitrogen). This resulting plasmid was used to transform strains GR5 and RTMH200.10 to pyroA prototrophy. Copy number and integration at pyroA were confirmed by quantitative RT-PCR (-PCR) and PCR, respectively. Strains carrying duplications of matB or tubB were created by cloning genomic sequences that included the coding region plus 1 kb of upstream and 1 kb of downstream sequences into pAVT21. These constructs were transformed into GR5, and copy number and integration at pyroA were confirmed using qRT-PCR and PCR, respectively.
Construction of the matA frameshift mutation allele
Plasmid pWP3 carries the coding region of matA flanked by 1-kb upstream and 1.8-kb downstream genomic sequences plus pyrG as a prototrophy marker (Pyrzak et al. 2008). The frameshift mutation was created in the matA transgene carried on pWP3. A G base was added after the second in-frame ATG (codon 8) of the matA-coding region. The mutation was introduced with the Site-directed Mutagenesis Kit (New England BioLabs) using primers pWp3matAF1: P-ctgtatcgattgctatgGaaatcaccaacac and pWp3matAR1: P-cagccattttggcacttc. The capitalized “G” indicates the extra base that introduces the frameshift and loss of the native MatA protein. To confirm that the frameshift mutation results in the absence of a functional MatA protein, we transformed pWP3 (+ frameshift mutation) into A. nidulans strain UI465 matA(0). All recovered transformants were sterile, lacking fruiting bodies. The resulting plasmid pWP3 (+ frameshift mutation) was subsequently transformed into the matA wild-type GR5 strain to test the effect of the frameshift mutation upon gene silencing.
5′-FOA selection
The ectopically introduced matA transgene was flanked by homologous pyrG89/pyrG sequences; therefore, it could be efficiently evicted by homologous mitotic recombination between flanking sequences. A. nidulans strain UI432 carrying the endogenous matA and ectopic matA transgene was used to select for mitotic recombination events that resulted from excision of the ectopic matA allele and functional pyrG, leaving the pyrG89 allele. Selection of pyrimidine auxotrophic excisants was accomplished by a modification of the (5-FOA) counterselection scheme of Boeke et al. (1984). A total of 106 conidia/per plate were spread onto 5% agar plates containing appropriately supplemented minimal media and 5-FOA (0.1 mg/ml). Plates were incubated at 37° for 2 days.
Comparative RT-qPCR transcript analysis
The relative RT-qPCR method was used to assess the developmental expression of matA and gprA. Total RNA was extracted from undifferentiated hyphae and from reproductive tissue at 2, 4, and 6 days PI of sexual development. Total RNA was extracted and treated with DNase-I and reverse-transcribed from oligo(dT) primers using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). Primers framed a target sequence that crossed an intron, and the possible DNA contamination was diagnosed by agarose gel analysis. Necessary validation tests, analysis of the actin expression, and primer specificity have been previously performed, and the protocol for RT-qPCR has been established (Pyrzak et al. 2008).
Relative quantitation of transcript levels was determined by the threshold cycle (ΔΔ CT) method expressed as a difference in target gene expression with respect to an endogenous control (actin) in different samples. Wild-type hyphal RNA was used as the reference RNA. The expression of the matA gene was assessed using the primers AnmatAF33 (5′ ccgcacgcatcaccgagctcc 3′) and AnmatAR29 (5′ ggtgtgcgcagaacacgcaga 3′). The expression of gprA was analyzed with primers AngprAF3 (5′ cgggccattctcgaattcag 3′) and AngprAR2 (5′ gagggcaacgatggtcaaga 3′). Each complementary DNA sample was assayed in triplicate, and RNAs were obtained from three separate biological samples.
Light microscopy
A. nidulans strains were induced for sexual development on plates with solid complete medium. Plates with mature fruiting bodies were photographed using a Zeiss SV8 Stereomicroscope and Nikon Cool Pix 5400 camera. The internal content of fruiting body tissue was examined by differential interference contrast optics using a Zeiss Axioplan. Nuclei were visualized using a water solution (1 µg/ml) of 4′,6-diamino-2-phenylindole (DAPI) (Sigma) staining. Individual cleistothecia were cleaned, suspended in a water drop or in DAPI staining, and crushed under a coverslip on a glass slide. Photomicrographs were taken with either a Nikon Cool Pix 5400 camera or Photometrics CoolSnap ES camera and Metavue software (Universal Imaging).
Results
Duplication of the matA gene suppresses expression of matA, blocks entry into meiosis, and results in barren cleistothecia
The A. nidulans strains UI432 and UI471, containing duplications of the matA gene, were constructed in two different ways to exclude the possibility that genetic manipulation or randomly generated mutations could affect matA gene function. The UI432 is the progeny from a cross, whereas UI471 was created by transformation (described in Materials and Methods). An extra copy of the wild-type matA allele was integrated ectopically on chromosome I by homologous recombination at the pyrG locus using the pWP3 vector. In both strains, identical genomic sequences were introduced that included the matA transcriptional unit flanked by 1 kb of upstream and 1.8 kb of downstream genomic regions. The presence of the extra matA gene at the ectopic position was confirmed by Southern blot analysis (data not shown).
Duplication of the matA gene did not affect vegetative growth, mating, or formation of the fruiting body, but specifically interfered with the development of internal ascogenous tissue, asci, and ascospores. The UI432 strain, at 6 days post induction (PI) of sexual development, formed cleistothecia containing only undeveloped ascogenous tissue. Differentiation of asci and ascospores was blocked completely (Figure 1). Analysis of nuclear distribution revealed that sexual differentiation was arrested at the stage of dikaryotic cells and croziers, preceding karyogamy and meiosis. Cytological analyses suggest that matA silencing correlates with failure of nuclear movement from crozier into zygote and failure in karyogamy, resulting in the absence of zygotes (Figure 2).
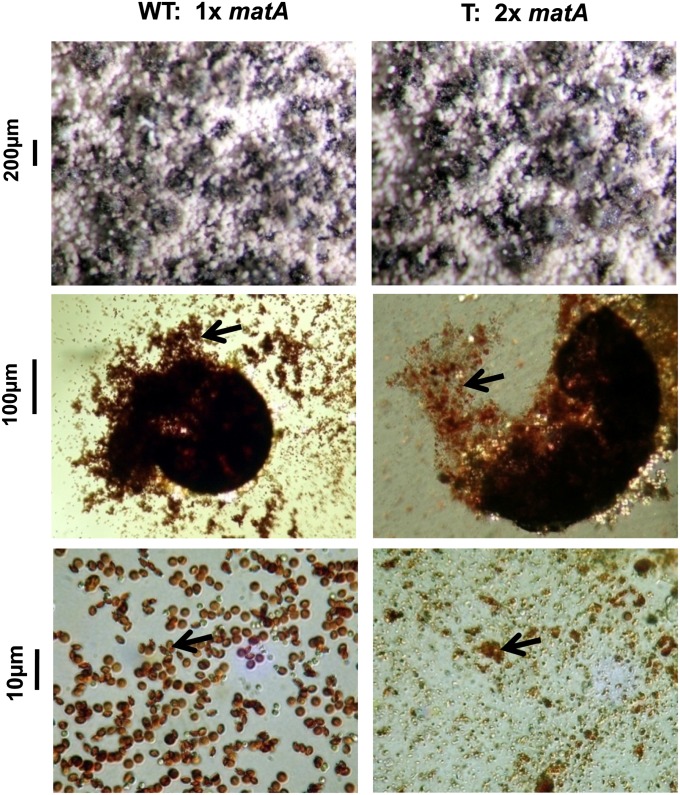
Figure 1.
Duplication of the matA gene in the haploid genome of A. nidulans prevents ascospore formation. Differentiation of cleistothecia and ascospores are compared in the wild type (WT) and a transformant with a matA duplication (T). (Top panels) Mature, dark-pigmented cleistothecia and mature white conidiophores. (Middle panels) (Left) Contents of the individual broken cleistothecium with mature ascospores (arrow). (Right) Undeveloped ascogenous tissue with orange debris (arrow). (Bottom panels) (Left) Mature ascospores (arrow) produced by wild-type strain. (Right) Debris (arrow) with absence of ascospores. Magnification bars are shown.
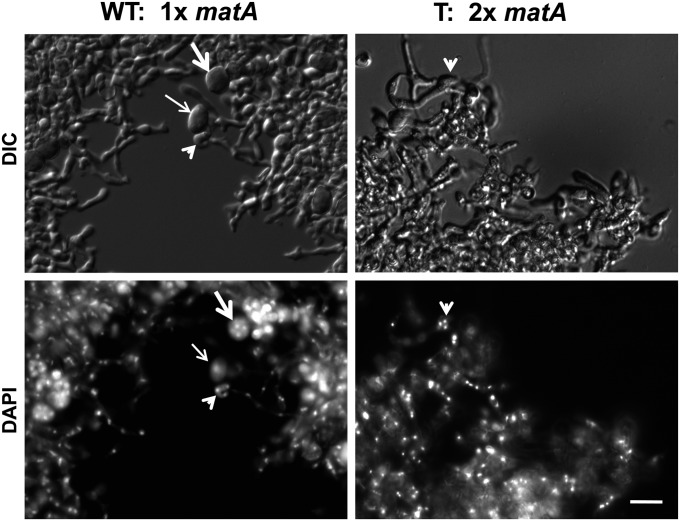
Figure 2.
Nuclei carrying duplications of matA do not undergo karyogamy and meiosis. Development of the ascogenous tissue and distribution of nuclei were analyzed at 4 days PI of sexual development. Nuclei are visualized with DAPI. Different stages of ascus development are shown in the wild-type (WT) strain. Prezygotic cell (arrowhead), zygote (thin arrow), and ascus with ascospores (thick arrow) are indicated. Strains carrying a matA gene duplication differentiate ascogenous tissue with normal nuclear distribution up to the prezygotic stage (arrowhead). Nuclei do not undergo karyogamy and meiosis; therefore, neither zygotes nor asci are recognized. Magnification bar: 10 µm.
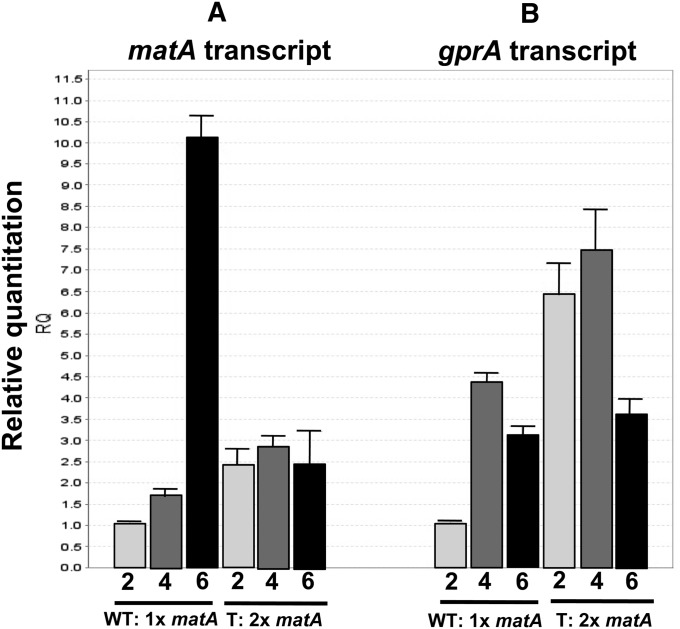
The level of the matA transcript was analyzed at three developmental time points: 2, 4, and 6 days PI of sexual development in UI432 and wild-type GR5. During the early stages of sexual differentiation (day 2 and 4 days PI), duplication of the matA gene did not alter developmental matA expression or sexual phenotype. The twofold increase in matA transcript abundance was apparently the result of two matA copies each being expressed at wild-type levels. This increased level of matA transcript did not affect early-to-mid sexual development, and normal abundance and morphology of fruiting bodies were observed. At the later developmental stages (6 days PI) when croziers and asci are differentiating in the wild-type strain, there was a characteristic 10-fold upregulation of matA expression (Figure 3) relative to the early developmental time point or 75-fold upregulation relative to undifferentiated hyphae (Figure 4B), which is consistent with previous observations. However, at later developmental stages (6 days PI), strains with 2× matA gene dosage showed a dramatic suppression of total matA expression, suggesting that transcription from both matA copies is affected. Alteration of the developmental expression pattern is observed during the time at which karyogamy and zygote formation would be occurring and correlates with aborted development of ascogenous hyphae and the complete absence of asci and ascospores. Therefore, from induction until day 4, matA transcription from both resident and ectopic loci was unaffected by silencing mechanisms and similar to the single copy in wild type. However, matA silencing was triggered by molecular events correlated with ascogenous hyphae at the dikaryotic stage, immediately preceding karyogamy and meiosis (Figure 3). The efficiency of silencing by the duplicated matA gene was ∼100%, meaning virtually every transformant carrying a duplication of matA had a complete absence of asci and ascospores.
Figure 3.
(A and B) Duplication of matA triggers gene silencing and has a downstream effect on the gprA target gene. Developmental expression of the matA transcript (A) and downstream target gprA (B) over the time course of 2, 4, and 6 days PI of sexual development.
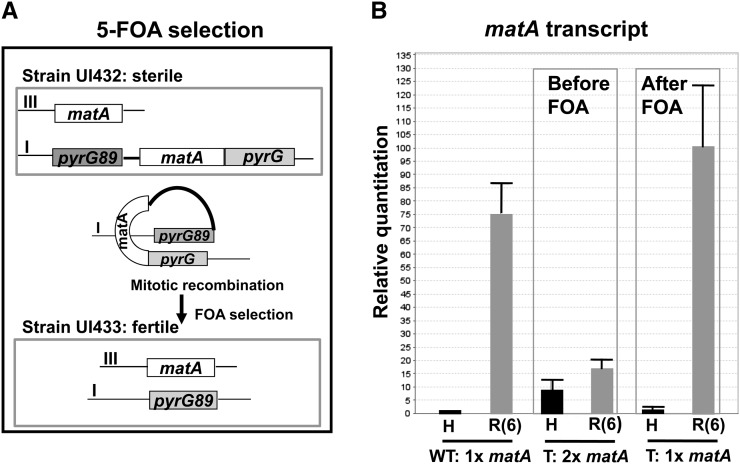
Figure 4.
(A and B) Ectopically introduced matA transgene induces gene silencing at both resident and ectopic matA. (A) Schematic representation of the removal of the ectopic matA transgene using 5′-FOA selection. (B) Expression analysis of the matA gene in a strain having duplication (before FOA) and a strain that was recovered upon excision of the ectopic matA (after 5′-FOA). Transcript levels were analyzed in undifferentiated hyphae (H) and in reproductive tissue 6 days PI of sexual development (R6).
Duplication of matA affects expression of the pheromone receptor gene gprA
Pheromone receptor signaling during sexual development in A. nidulans and other homothallic fungi is essential to control recognition mechanisms between sexually compatible cells (Poggeler 2000; Seo et al. 2004; Mayrhofer and Poggeler 2005; Mayrhofer et al. 2006; Poggeler et al. 2006a,b). Homologs of the budding yeast pheromone receptor genes gprA (α-factor receptor) and gprB (a-factor receptor) in homothallic A. nidulans have been previously reported (Seo et al. 2004). Our previous studies have determined that the MatA transcription factor modulates expression of the gprA gene during sexual development having activator and/or a combination of activator/inhibitor functions (Czaja et al. 2011).
Here we demonstrate that altered transcriptional expression of matA affects regulation of the downstream target gene gprA. The gprA transcript level was analyzed in the wild-type strain and a transgenic strain carrying both an endogenous and an ectopic matA gene. In wild type, gprA expression peaks at day 4 PI with a 4.5-fold upregulation relative to day 2. The gprA transcript levels decreases after 4 days PI. By contrast, the strain carrying the matA gene duplication showed a significant 6.5-fold upregulation of gprA abundance at day 2 PI and 2-fold at day 4 PI relative to wild-type expression levels. Even though the silencing seems to be not active early at 2 or 4 days, there is a significant upregulation of gprA. This early upregulation can be attributed to the fact that there is a double amount of MatA protein coming from two matA copies that potentially can boost expression of gprA. The decrease in gprA expression between day 4 and day 6 PI was much greater in the strain with two copies of matA and reflects the silencing of matA expression at this developmental time point (Figure 3). Therefore, duplication of the matA gene and the resulting silencing significantly altered developmental expression of gprA.
Deletion of the duplicated copy of matA restores full functionality of the endogenous matA gene
The ectopically integrated matA copy was flanked by the homologous pyrG and pyrG89 sequences on chromosome I. Mitotic recombination between pyrG flanking sequences during 5′-FOA counterselection resulted in excision of both the extra matA copy and the pyrG allele from chromosome I (Figure 4A). Southern blot analysis confirmed that pyrG89 auxotrophs had only the endogenous matA allele on chromosome III (data not shown). These strains were further analyzed for phenotype and matA expression.
All recombinant strains had restored wild-type phenotype with normal differentiation of fertile fruiting bodies containing ascospores. matA transcript levels in undifferentiated hyphae and reproductive tissue were compared for the wild-type strain (GR5), matA gene duplication strain (UI432), and a strain derived from UI432 after 5′-FOA counterselection (UI433). In undifferentiated hyphae, there was an eightfold increase in matA transcript levels in the UI432 strain relative to wild type. Elevated hyphal expression might be the result of expression coming from two matA genes and/or regulatory derepression associated with ectopic matA copy. This later possibility is more likely and is consistent with our previous data showing similar levels of elevated hyphal matA expression from a single ectopic copy of matA (Czaja et al. 2011). By contrast, expression from both endogenous and ectopic matA genes was dramatically suppressed during the latter stages of development (Figure 4B). Expression was only 20% that of the wild type at 6 days PI. The wild-type matA expression profile was restored after removal of the ectopic matA (UI433, Figure 4B). Therefore, the introduction of an extra matA copy alters functional expression of both the resident gene and the ectopic transgene.
Silencing by the matA transgene is position-independent
The matA transgene was integrated into additional chromosomal positions to determine if MatIS was a unique feature of matA transgene integration at the pyrG locus. A construct having the pyroA marker and the same matA sequences found in pWP3 was integrated at the pyroA4 locus of strain GR5 (Table 1). The presence of the matA transgene flanked by pyroA sequences caused MatIS silencing with identical efficiency compared to integration at the pyrG locus described above (Table 2). Integration of the matA transgene at the matA locus resulted in tandem duplication of matA sequences and MatIS silencing with the same efficiency (Table 2). Therefore, MatIS was identical whether the additional matA transgene was located between duplicated marker sequences at the pyrG (chromosome I) and pyroA (chromosome IV) loci or as tandem copies at the resident matA locus (chromosome III).
Table 2. Fertility in strains carrying an extra copy of matA.
| Additional copy |
Sexual cycle |
|||||
|---|---|---|---|---|---|---|
| Resident Gene | rsdAArgo | Chromosome I | Chromosome III | Chromosome IV | Cleistothecium | No. of ascospore/cleistothecium (%) |
| matA (chromosome III)wt | + | − | − | − | + | 100a |
| matA(0) (chromosome III) | + | − | − | − | − | − |
| matA(0) (chromosome III) | + | matA | − | − | + | 100 |
| matA (chromosome III) | + | matA | − | − | + | 0 |
| matA (chromosome III) | + | − | − | matA | + | 5 |
| matA (chromosome III) | + | − | matA | − | + | 7 |
| matA (chromosome III) | − | − | − | − | + | 100b |
| matA (chromosome III) | − | − | − | matA | + | 0.2c |
| matA (chromosome III) | − | − | − | matA | + | 0.1c |
rsdAArgo column: +, wt; −, null. Cleistothecium column: +/− indicate presence of absence of fruiting bodies.
117,000 ascospores/cleistothecium.
133,000 ascospores/cleistothecium.
Two independently isolated strains.
Neither duplicated matA-coding DNA nor promoter region are sufficient to induce gene silencing
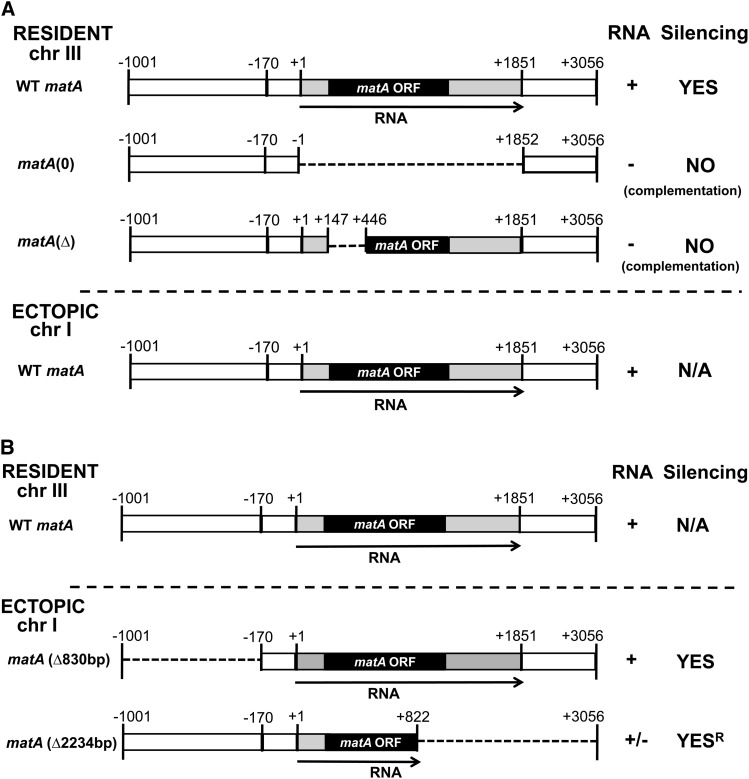
Gene-silencing phenomena (RIP, MIP, quelling) are specifically induced by duplicated DNA sequences. To determine the role of homologous DNA sequence in matA HDGS, we performed gene complementation studies using matA gene variants lacking different portions of the DNA sequence. First, we used the UI465 matA(0) strain that carries a deletion of the entire matA transcription unit (from +1 to +1851 nt) at the resident locus. UI465 is sterile and does not differentiate fruiting bodies or ascospores (Czaja et al. 2011). UI465 was complemented with an ectopically integrated matA transgene (from −1001 to +3056 nt) (Figure 5A). Therefore, the only DNA homology was outside the matA transcription unit. The matA(0) deletion was functionally complemented by the matA transgene, and no silencing effect was observed. Mating-type function and fruiting body formation and fertility were fully recovered. This observation demonstrates that DNA homology outside the matA transcription unit does not play a role in gene silencing (Figure 5A). A second approach used a strain carrying a 299-nt partial deletion of the matA-coding region (+148 to +446) at the resident locus. Strain UI464 (matAΔ) was also sterile, having a phenotype identical to that of the matA(0) deletion strain (Czaja et al. 2011). UI464 was complemented by a matA transgene (from −1001 to +3056 nt) integrated ectopically. In this case, there was a perfect DNA homology spanning the region between −1001 nt and +3056 nt except for 299 nt of the matA-coding region that was deleted at the resident locus. In all transformants analyzed, the transgene fully complemented the matAΔ deletion. Wild-type fertility was restored with development of normal numbers of cleistothecia and ascospores. No detectable gene silencing was observed (Figure 5A). Therefore, DNA homology corresponding to either the 5′ putative promoter coding or 3′ regions was not sufficient to trigger gene silencing, unless the silencing mechanism is specified by the 299 bp that are lacking in both matAΔ and matA(0) strains (Figure 5A). Further analyses of the 5′ and 3′ flanking regions revealed more details underlying gene silencing induced by duplication of matA. A transgene lacking 5′ regulatory flanking sequences, matA(Δ830 bp), was integrated ectopically into the background of wild-type matA. Gene silencing was still observed. The degree of silencing was unaffected by this deletion; mating-type function was impaired, and barren fruiting bodies were formed. Therefore, duplication of the 5′ regulatory region is dispensable for the silencing effect (Figure 5B). Interestingly, a transgene lacking approximately one-half of the C-terminal coding region plus 3′ flanking sequences was still able to induce silencing when integrated ectopically into the background of wild-type matA. However, mating-type function was not completely suppressed since some ascospores were observed although at very low levels (20–30% of wild type). Collectively, these data provide a strong argument that homology at the DNA level was not directly involved in triggering gene silencing (Figure 5B).
Figure 5.
(A and B) Transgene-derived matA transcript is involved in gene silencing. Schematic summary of the complementation studies is presented. (A) Resident matA wild-type allele or deletion mutant alleles matA(0) or matAΔ, respectively, were complemented in separate experiments by a complete matA transgene that was introduced ectopically. (B) matA deletions introduced ectopically into a wild-type matA [chromosome (chr) III] background. Genetic distance is marked (−1001 to +3056 bp). The solid bar indicates the matA-coding region. The shaded flanking regions represent 5′ and 3′ UTRs. Chromosomal position in the genome is indicated (chr III, chr I). Deleted regions of matA sequence are indicated by dashed lines. RNA status and silencing effect associated with each complementation experiment are shown: present (+), absent (−), silencing present (YES), no silencing (NO), does not apply (N/A), and 70–80% reduced fertility (R).
Silencing is not a function of MatA protein but may be mediated by matA RNA
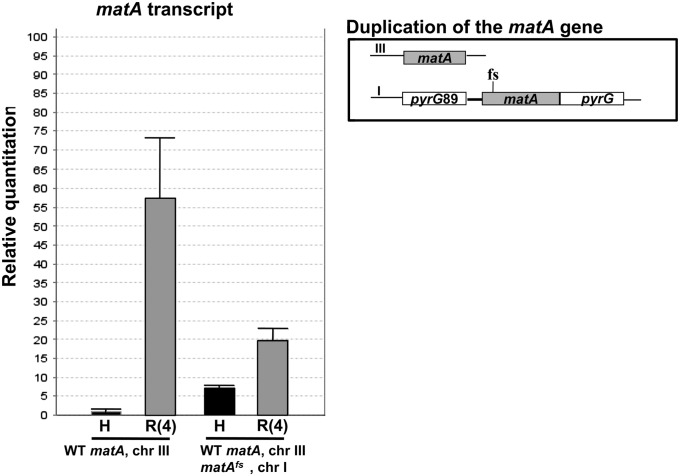
To determine the molecular component (RNA or protein) involved in gene silencing, we created the strain UI470 having a matA gene duplication, where only the resident matA allele can be translated into a functional MatA protein. The ectopic matA transgene (matAfs) carries a frameshift mutation at the eighth codon and therefore expresses an RNA that cannot be translated into a functional MatA protein (Figure 6). We confirmed this by showing that the matAfs allele is unable to complement the matA(0) deletion strain. UI470 expressed elevated hyphal levels of matA RNA similar to that observed for other strains with two copies of matA due to derepression of the ectopic allele (compare Figure 6 to Figure 4B). However, only wild-type levels of MatA protein would be translated from matA RNA expressed from the resident gene. If the interference phenomenon is mediated by the dosage of MatA protein, then the strain with the frameshift mutation should have a wild-type phenotype. However, if RNA triggers interference, this strain should form barren cleistothecia. UI470 was induced to undergo sexual development to test these two alternatives. Differentiation of sexual reproductive tissues in this strain showed all of the hallmarks of interference with abundant barren cleistothecia that lacked meiotic progeny or ascospores. Notably, it is not simply elevated matA RNA levels that trigger silencing. We have previously shown that the matA (Δ830 bp) construct is capable of complementing the matA(0) strain when integrated ectopically. Deletion of 830 bp of the upstream regulatory region of matA resulted in a 140-fold increase of matA RNA abundance in both hyphal and sexually differentiated tissue relative to the wild type. However, silencing is not triggered in this strain, and fertility is like the wild type (Czaja et al. 2011).Therefore, neither MatA protein nor the level of matA RNA per se is involved in gene silencing. It appears that the transgene-derived RNA itself triggers the gene-silencing phenomenon.
Figure 6.
MatA protein is not involved in HDGS phenomenon. Graphic representation of the wild-type matA gene at the resident locus and the ectopically integrated matAfs transgene carrying a frameshift mutation and therefore deficient in native MatA protein. Analyses of matA transcript level in the undifferentiated hyphae (H) and in the reproductive tissue 4 days PI of sexual development (R4).
matA transcript levels in UI470 were analyzed in undifferentiated hyphae and reproductive tissue. Similar to our observations above, interference was restricted to the later stages of the reproductive cycle. Total transcript levels expressed from both resident and ectopic copy in the reproductive tissue was only 36% of wild type, which is consistent with our previous data (Figure 6). Therefore, the matA RNA triggers interference and alters functional matA expression at the transcriptional or post-transcriptional level.
MatIS is not dependent upon Argonaute and RNAi
Other reported cases of HDGS silencing systems in fungi are RNA-mediated and require functional RNAi. RNA-mediated transcriptional and post-transcriptional gene silencing are dependent upon the presence of an Argonaute protein as a components of a RITS or RISC complex, respectively. A. nidulans has a single functional Argonaute protein encoded by rsdA, which is required for RNAi (Hammond and Keller 2005; Hammond et al. 2008). Deletion of rsdA does not affect vegetative growth, conidiation, or sexual reproduction (Hammond et al. 2008). We also observed that loss of rsdA function had no effect upon sexual fertility (Table 2). We integrated a matA transgene at the pyroA4 locus of the rsdAΔ strain RTMH200.10, which has a functional resident matA. We found that the transformed strain was infertile and had ascospore yields similar to that of an rsdA strain with the transgene at the pyroA locus (UI480 vs. UI482; Table 2). Therefore, MatIS was fully effective in a strain lacking an Argonaute protein required for RNA-mediated RISC or RITS silencing.
MatIS is not a general phenomenon induced by duplication of sex-related genes
MatIS is not a general phenomenon that is induced in response to duplication of sex-related genes. matB is the α-box mating-type gene that is required for sexual induction and ascosporogenesis in A. nidulans. tubB encodes a meiosis-specific α-tubulin. Induction of the sexual cycle and development of the fruiting body is normal in a tubBΔ strain, but development of ascogenous tissue is blocked premeiotically at a stage prior to karyogamy (Kirk and Morris 1991). This phenotype appears microscopically identical to that of our strains expressing a MatIS response. Sex-induced silencing of tubB expression due to tubB gene duplication should therefore also cause sterility. However, we observed that additional copies of either tubB or matB did not cause induction of MatIS and infertility.
HDGS in A. nidulans is recessive and does not spread in the heterokaryon
PTGS has been frequently correlated with the ability of gene silencing to be transmitted across nuclei and spread both within a common cytoplasm and systemically between cells. We have tested the ability of the matA-induced gene silencing to diffuse between silenced nuclei and wild-type nuclei sharing common cytoplasm in the reproductive heterokaryotic hypha. Stable heterokaryons were established between the parental strains UI432 and RTMH 202.11. The UI432 contains a matA duplication, is a white conidiating strain, and makes fruiting bodies but no ascospores. The RTMH 202.11 contains a single matA gene, is a yellow conidiating strain, and makes fruiting bodies and ascospores. If silencing can spread between the nuclei in the heterokaryon, there will be only barren cleistothecia. Instead, we observed abundant fertile and crossed cleistothecia produced by heterokaryons. Ascospores of crossed cleistothecium were analyzed for recombinants between conidia color markers. Ascospores were plated on selective media, which selects against parental genotypes. Colonies produced by ascospores were of white, yellow, and green color, which indicates a successful cross and recombination between genetic markers and between two parental strains. The presence of crossed cleistothecia with recombinant progeny indicates that the silencing effect does not spread between nuclei. This observation demonstrates that the silencing effect is not diffusible and not propagated between nuclei but is apparently a nucleus-restricted phenomenon.
Discussion
Our findings identify a novel gene-silencing phenomenon associated with mating-type function that is induced by duplication of the matA gene and operates exclusively during the premeiotic sexual stage in A. nidulans. We refer to this phenomenon as MatIS (defined above).
Duplication of the matA gene impairs late sexual development in A. nidulans
We have demonstrated that duplication of matA (one resident + one ectopic copy) interrupts normal patterns of matA expression, resulting in a dramatic decrease in matA mRNA abundance during late sexual development. Low levels of the matA expression during the prezygotic stage of cleistothecium development contribute to failure in karyogamy and meiosis and aborted development of asci and ascospores. Fruiting body differentiation in A. nidulans is coordinated by a mat-regulated pheromone signal transduction pathway that includes GprA, a homolog of the budding yeast α-factor receptor Ste2. GprA is specifically required for mating and self-fertilization (Seo et al. 2004; Yu 2006; Harris et al. 2009). We have previously observed a direct correlation between matA transcript level and cleistothecium differentiation in A. nidulans, suggesting that fruiting body development is regulated by specific threshold levels of matA expression (Pyrzak et al. 2008). It has been proposed that higher levels of mat HMG expression are required to regulate correct distribution and segregation of nuclei at the prezygotic stage (Debuchy 1999; Coppin and Debuchy 2000). We observed that reduced levels of matA transcript due to gene duplication caused significant upregulation of gprA transcription over the course of development. Our data suggest that significant alteration in matA expression at this critical stage has an adverse impact on karyogamy and meiosis, resulting in barren fruiting bodies.
Duplication of matA gene induces homology-dependent silencing of mating-type gene function
The silencing phenomenon observed in A. nidulans displays unique characteristics but also shares some common features with HDGS phenomena that have been reported in several species of fungi, plants, and animals (Bingham 1997; Cogoni and Macino 1999a). Generally, HDGS as a result of the introduction of a transgene(s) involves silencing of a target locus by an unlinked silencing locus. Both the transgene and unlinked homologous endogenous copy of a gene are silenced by DNA methylation and TGS and/or mRNA degradation and PTGS. Silencing in most cases is reversible upon removal of the transgene (Furner et al. 1998; Mourrain et al. 2007).
Similarly, in A. nidulans, silencing was induced by the introduction of a matA transgene. However, unlike other reports of HDGS, MatIS is confined to a specific stage of development. Furthermore, we have demonstrated that the sum of transcription from both transgene and resident matA genes was greatly reduced, suggesting mutual silencing of both matA copies. Therefore, the presence of a duplicated matA was essential to maintain the silenced status of both matA alleles. Removal of the ectopic matA transgene upon 5′-FOA treatment eliminated silencing and restored a normal expression pattern of the endogenous matA and a wild-type sexual phenotype.
Because matA encodes a master regulator of sexual development, we analyzed the role of MatA protein as a potential trigger of the MatIS silencing effect. The failure of zygote and ascospore formation could be easily explained by gene duplication causing an excessively elevated level of MatA protein and the disruption of finely tuned spatiotemporal patterns of target sex-specific gene expression. Phenotypic and transcriptional analyses of the A. nidulans strain with both an intact resident matA gene and a frameshift mutation in the ectopic matA copy excluded the possibility that protein encoded by a duplicated matA gene was involved in the silencing phenomenon. This observation is consistent with HDGS phenomena reported in other eukaryotic organisms where proteins encoded by duplicated genes were not involved in the gene-silencing effect.
Silencing is mediated by transgene-derived matA RNA
The homology between repeated DNA segments of genes appears to be the molecular trigger in many reported cases of HDGS. It has been determined that coding-region homology acts as the trigger for gene silencing in both quelling and meiotic silencing in N. crassa (Cogoni 2001; Shiu and Metzenberg 2002). DNA fragments with a minimal size of 132 bp of coding-region homology were necessary and sufficient to trigger quelling in the vegetative stage. A more complex scenario has been described for meiotic silencing during the sexual stage. DNA unpairing, or lack of the DNA homology, between alleles during chromosome alignment was the signal for silencing of all homologous gene sequences prior to meiosis in the zygotic cell. We observed that neither DNA homology within 5′ and 3′ noncoding regulatory regions nor homology within the coding region play roles in premeiotic silencing in A. nidulans. A comparison of the matA deletion variants [matA(0) and matAΔ] that lack a functional matA transcript— but that retain partial DNA homology to matA regulatory and/or coding regions—demonstrates that, in contrast to RIP, MIP, and quelling, the DNA component was not sufficient to induce MatIS silencing. Furthermore, we have also shown that DNA unpairing does not trigger silencing in A. nidulans, as sexual outcrosses between one parental genome containing a duplication of matA and another parental genome having only a wild-type resident matA resulted in a wild-type sexual phenotype. Therefore, in contrast to RIP, MIP, quelling and meiotic silencing caused by matA duplication in A. nidulans is driven by transcribed RNA and represents a novel mechanism for HGDS.
High-efficiency silencing depends upon qualitative features of the matA transcript, not RNA abundance
Silencing efficiency varies greatly from gene to gene, and the limiting factors are not fully understood. The strength of the transgene promoter, transgene copy number, or formation of antisense RNA can contribute to silencing efficiency (Que et al. 1997; Vaucheret and Fagard 2001; Fulci and Macino 2007). The introduction of exogenous transgenes in N. crassa is necessary, but not sufficient, to trigger gene silencing, and only a portion of transformants (typically 30%) containing duplicated sequences show silencing by quelling (Cogoni et al. 1996). The unexpected formation of transgenic sense and/or antisense RNA from promoterless constructs was implicated in low silencing efficiency (Cogoni and Macino 1997b). Both high copy number and a tandem sequence arrangement were important for triggering quelling. By contrast, the introduction of a single copy of the matA transgene is necessary and sufficient to trigger silencing in A. nidulans with an efficiency of nearly 100%. The integration site of matA transgene into the genome was carefully designed, and only sense, transgene-derived mRNA was expected to be transcribed. Therefore, it is highly unlikely that an unexpected antisense RNA plays a role in gene silencing in A. nidulans. MatIS was also independent of the transgene integration site and therefore not a function of chromosomal position.
Abnormally high levels of mRNA generated from either a single-copy gene with a strong promoter or from multicopy transgenes have been implicated in the gene silencing of homologous sequences in plants and fungi (Napoli et al. 1990; Que et al. 1997). In our previous studies, we have demonstrated that overexpression of the ectopically introduced matA transgene in the undifferentiated hyphae (∼135-fold) and in the reproductive tissue (∼112-fold), as compared to the wild-type hyphal level, did not induce gene silencing (Czaja et al. 2011). Therefore, unlike cosuppression and quelling, silencing of the mating-type gene appears to be independent of the overall levels of matA transcript.
The formation of a sense aberrant RNA (aRNA) produced from, or induced by, the transgene has been proposed to trigger homology-dependent gene silencing (Cogoni 2002; Nakayashiki et al. 2005). The origin and nature of aRNA are not well understood. Chromosomal location of the transgene could potentially have some effect on the expression or processing of RNA transcript. It has been suggested that methylation of transgene DNA could contribute aRNA (Baulcombe 1996). It has been proposed that aRNA might be recognized by RdRP, leading to synthesis of complementary RNA strand and double-stranded RNA formation (Lindbo et al. 1993). Double-stranded RNA would trigger homologous mRNA degradation affecting both endogenous and transgenic RNA simultaneously. The A. nidulans genome encodes two RdRPs that hypothetically could be implicated in the aRNA-induced PTGS. However, Hammond and Keller (2005) have shown that RdRPs are not required for RNAi in A. nidulans.
One interesting feature of the matA transcript is its unusually long 3′ UTR. 3′ UTRs have been demonstrated to be important in regulation of gene expression where they play a role in translational efficiency and/or mRNA localization (Guo and Sherman 1996; Antic and Keene 1997; Long et al. 1997, 2001). It has been demonstrated in the fungus Cochliobolus heterostrophus that truncation of the mat HMG 3′ UTR results in formation of barren fruiting bodies (Wirsel et al. 1998). Our data suggest another potential role for the 3′ UTR in gene silencing. Remarkably, MatIS silencing efficiency by an ectopic transgene was reduced upon deletion of DNA sequences of the 3′ flanking region that included part of the C-terminal coding region and 3′ UTR. Cleistothecia were not completely barren, and a low, but significant, level of ascospores (20–30% of wild type) was observed. By contrast, a full-length, but nonfunctional, matAfs transcript is capable of triggering high-efficiency silencing and suppression of mating-type function. These observations, when taken together with results from ectopic transgenes in matA(0) and matAΔ strains, indicate that a full-length matA transgene-derived transcript is required for triggering efficient gene silencing. They further suggest that the matA 3′ UTR plays at least a partial role in driving MatIS.
matA silencing appears to be a specialized feature of mating-type regulation
Gene-silencing phenomena such as RIP and MIP seem to be general silencing mechanisms in Neurospora and Ascobolus, with the ability to affect any type of repeated gene. Exceptionally, N. crassa sexual-cycle-specific genes, in particular the mating-type genes mat-a (the matA homolog) and mat-A are protected from meiotic silencing. In A. nidulans, repeated or duplicated genes typically do not trigger silencing. Documented cases of HGDS in A. nidulans have not been reported, and matA mating-type repeat-induced silencing reported here is unique. Furthermore, duplication of two other sex-induced genes critical to a fertile sexual cycle, matB and tubB, did not induce silencing at any developmental stage. Thus, silencing associated with matA gene duplication appears to be a specialized feature of mating-type regulation and represents an example of a silencing phenomenon distinct from other reported examples of HDGS.
In heterothallic species, the correct genetic organization of mating-type alleles is essential for efficient fertilization and development of fertile fruiting bodies (Coppin et al. 1997). Illegitimate fusions between the same mating types in both a HMG × a HMG and Aα × Aα combinations resulted in the failure of ascogenous hyphae development and the formation of enlarged, barren fruiting bodies in Aspergillus stercorarius (Coppin et al. 1997). Similarly, the artificial association of both mating types in the same nucleus in heterothallic N. crassa resulted in self-mating and differentiation of nonfertile, barren perithecia. Therefore, gene silencing associated with altered mating-type gene status could also function in other fungi where genetic manipulation of mat loci resulted in impaired fertility.
The silencing of the sex-specific matA gene might be functionally linked to sexual-cycle-specific, genome quality control mechanisms that protect genome integrity and prevent genetic aberration to be passed to progeny. Recent findings demonstrated a link between transgene-induced RNAi gene silencing and genome defense during sexual reproduction in Cryptococcus neoformans (Wang et al. 2010). A. nidulans is a haploid homothallic (self-fertile) fungus, and its sexual identity is determined by a single copy of matA (HMG-box) and matB (α-box) mating-type genes. Mating-type genes are involved in the fine-tuning and balanced expression of the sex-specific target genes. The precise dosage of mating-type genes and their products might be critical in regulating balanced expression of target genes. Changes in the gene dosage, such as matA gene duplication, could trigger genome quality control mechanisms that would consequently both silence homologous genes and block meiosis to prevent transmission of an abnormal status of mating-type genes to the next generation of progeny. However, MatIS appears to be confined to the matA gene, is RNAi independent, and therefore differs from SIS in C. neoformans.
A model for nucleus-restricted RNA-mediated premeiotic silencing in A. nidulans
Gene-silencing during the sexual stage has been reported in N. crassa (RIP), and meiotic silencing has been reported in A. immersus (MIP) and C. neoformans (SIS). Both RIP and MIP operate before karyogamy, and homologous repeated genes are methylated and silenced transcriptionally (TGS). By contrast, meiotic silencing occurs after karyogamy, and unpairing between homologous genes triggers PTGS. Furthermore, it has been demonstrated that unpaired mating-type genes in N. crassa are immune to meiotic silencing (Shiu et al. 2001).
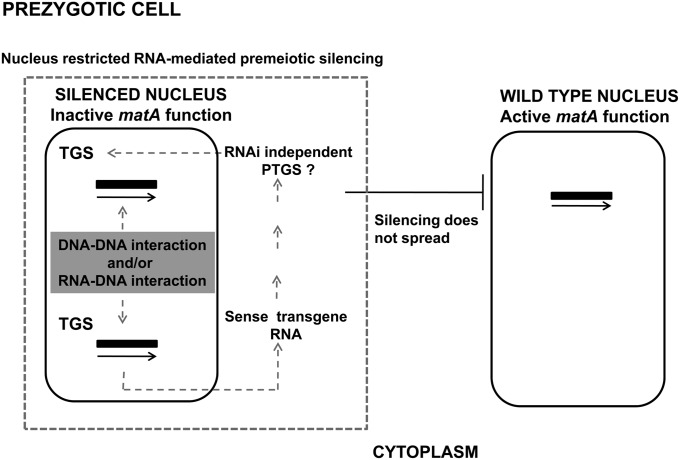
Silencing of the matA gene occurs before karyogamy and appears mechanistically similar to RIP or MIP, suggesting transcriptional pairwise mat silencing. This could potentially involve DNA–DNA and/or RNA–DNA interaction between matA homologous sequences on different chromosomes (trans-inactivation) and sequence-specific de novo methylation of a homologous transgene and an endogenous counterpart (Kooter et al. 1999; Vaucheret and Fagard 2001) (Figure 7). This notion might be further supported by the fact that the cytosine methyltransferase homolog dmtA in A. nidulans is essential during sexual development (Lee et al. 2008). However, neither widespread DNA methylation nor active MIP or RIP has been reported in any Aspergillus species. This suggests the possibility that de novo DNA methylation by DmtA might occur transiently during the sexual phase, and, when needed, it could be involved in HDGS as a part of the genome defense system (Lee et al. 2003). Moreover, silencing of mating-type function in A. nidulans does not spread between nuclei but has a recessive and nucleus-restricted character, which might further support TGS as the mechanism involved in MatIS.
Figure 7.
Model of the nucleus-restricted, RNA-mediated premeiotic silencing in A. nidulans. Schematic representation of the prezygotic cell containing two haploid nuclei from a cross between two parental strains is shown. One nucleus carries duplication of the matA gene (solid bars); the other nucleus carries a single copy of matA. Gene silencing is mediated by a transgene-derived matA transcript and appears to be restricted to the nucleus of origin. The silencing effect does not spread between nuclei in the common cytoplasm of the syncytium or dikaryon. Therefore, a parental nucleus with a single wild-type matA gene retains active mat function that is fully able to complement the silenced matA function of the other parental nucleus, resulting in a wild-type sexual phenotype. Framed box with dashed line indicates a potential cytoplasmic compartment that might contribute to nucleus-restricted MatIS in the prezygotic cell. Refer to Discussion for details.
The observation that transgene-derived RNA appears necessary and sufficient for silencing suggests that a PTGS pathway could be potentially involved. PTGS-inducible RNAi has been reported in A. nidulans (Barton and Prade 2008). Components of post-transcriptional gene silencing such as Dicer, Argonaute proteins, and RdRPs have been identified in A. nidulans. However, they are not required for normal growth and sexual development under standard culture conditions (Hammond and Keller 2005; Hammond et al. 2008). This is in contrast to Neurospora, where components of the meiotic RNA silencing pathway are required for the completion of sexual development (Kelly and Aramayo 2007). We have demonstrated that the RNAi pathway is apparently not involved in MatIS. The fact that MatIS is recessive and does not spread and appears to be independent of RNAi argues against PTGS as a mechanism involved in silencing. Alternatively, MatIS may represent a new type of PTGS, where matA RNA does not diffuse and is degraded in a specific subcellular compartment close to the nucleus of origin in RNAi-independent manner.
Discovery of a novel, mating-type-specific HDGS system in homothallic A. nidulans opens a new line of investigation that may provide insights into molecular mechanisms underlying the regulation of mating-type function, sexual-cycle-specific gene-silencing phenomena, and genome surveillance in eukaryotes.
Acknowledgments
We thank Jose G. Alcocer for his technical assistance. This work was supported by grant #0318801 from the National Science Foundation and grant #P20 RR015587 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCRR or NIH.
Footnotes
Communicating editor: F. Winston
Literature Cited
- Antic D., Keene J. D., 1997. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am. J. Hum. Genet. 61: 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C., Faugeron G., Rossignol J. L., 1993. Methylation induced premeiotically in Ascobolus: coextension with DNA repeat lengths and effect on transcript elongation. Proc. Natl. Acad. Sci. USA 90: 4557–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton L. M., Prade R. A., 2008. Inducible RNA interference of brlAbeta in Aspergillus nidulans. Eukaryot. Cell 7: 2004–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. C., 1996. RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol. Biol. 32: 79–88. [DOI] [PubMed] [Google Scholar]
- Bingham P. M., 1997. Cosuppression comes to the animals. Cell 90: 385–387. [DOI] [PubMed] [Google Scholar]
- Boeke J., Lacroute F., Fink G., 1984. A positive selection for mutants lacking orotidine-5′ phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197: 345–346. [DOI] [PubMed] [Google Scholar]
- Catalanotto C., Pallotta M., ReFalo P., Sachs M. S, Vayssie L., et al. , 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24: 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H., Casas-Mollano J. A., 2006. On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 50: 81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champe S. P., Simon L. D., 2009, Cellular differentiation and tissue formation in the fungus Aspergillus nidulans, pp. 63–91 in Morphogenesis: An Analysis of the Development of Biological Form, edited by Rossomando E. F., Alexander S. Marcel Dekker, New York. [Google Scholar]
- Champe S. P., Nagle D. L., Yager L. N., 1994. Sexual sporulation. Prog. Ind. Microbiol. 29: 429–454. [PubMed] [Google Scholar]
- Chicas A., Cogoni C., Macino G., 2004. RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa. Nucleic Acids Res. 32: 4237–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C., 2001. Homology-dependent gene silencing mechanisms in fungi. Annu. Rev. Microbiol. 55: 381–406. [DOI] [PubMed] [Google Scholar]
- Cogoni C., 2002. Unifying homology effects. Nat. Genet. 30: 245–246. [DOI] [PubMed] [Google Scholar]
- Cogoni C., Macino G., 1997a Conservation of transgene-induced post-transcriptional gene silencing in plants and fungi. Trends Plant Sci. 2: 438–443. [Google Scholar]
- Cogoni C., Macino G., 1997b Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 94: 10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C., Macino G., 1999a Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni C., Macino G., 1999b Homology-dependent gene silencing in plants and fungi: a number of variations on the same theme. Curr. Opin. Microbiol. 2: 657–662. [DOI] [PubMed] [Google Scholar]
- Cogoni C., Irelan J. T., Schumacher M., Schmidhauser T. J., Selker E. U., et al. , 1996. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 15: 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Coppin E., Debuchy R., 2000. Co-expression of the mating-type genes involved in internuclear recognition is lethal in Podospora anserina. Genetics 155: 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppin E., Debuchy R., Arnaise S., Picard M., 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61: 411–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja W., Miller K. Y., Miller B. L., 2011. Complex mechanisms regulate developmental expression of the matA (HMG) mating type gene in homothallic Aspergillus nidulans. Genetics 189: 795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R., 1999. Internuclear recognition: a possible connection between euascomycetes and homobasidiomycetes. Fungal Genet. Biol. 27: 218–223. [DOI] [PubMed] [Google Scholar]
- Debuchy R., Turgeon B. G., 2006. Mating-type structure, evolution, and function in Euascomycetes, pp. 293–323 in The Mycota I, edited by U. Kües and R. Fischer. Springer-Verlag, Berlin; Heidelberg, Germany. [Google Scholar]
- Forrest E. C., Cogoni C., Macino G., 2004. The RNA-dependent RNA polymerase, QDE-1, is a rate-limiting factor in post-transcriptional gene silencing in Neurospora crassa. Nucleic Acids Res. 32: 2123–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman T., Pukkila P. J., 1993. De novo methylation of repeated sequences in Coprinus cinereus. Genetics 135: 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulci V., Macino G., 2007. Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr. Opin. Microbiol. 10: 199–203. [DOI] [PubMed] [Google Scholar]
- Furner I. J., Sheikh M. A., Collett C. E., 1998. Gene silencing and homology-dependent gene silencing in Arabidopsis: genetic modifiers and DNA methylation. Genetics 149: 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Sherman F., 1996. 3′-end-forming signals of yeast mRNA. Trends Biochem. Sci. 21: 477–481. [DOI] [PubMed] [Google Scholar]
- Hammond T. M., Keller N. P., 2005. RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases. Genetics 169: 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T. M., Bok J. W., Andrewski M. D., Reyes-Dominguez Y., Scazzocchio C., et al. , 2008. RNA silencing gene truncation in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell 7: 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. D., Turner G., Meyer V., Espeso E. A., Specht T., et al. , 2009. Morphology and development in Aspergillus nidulans: a complex puzzle. Fungal Genet. Biol. 46(Suppl. 1): S82–S92. [DOI] [PubMed] [Google Scholar]
- Idnurm A., Walton F. J., Floyd A., Heitman J., 2008. Identification of the sex genes in an early diverged fungus. Nature 451: 193–196. [DOI] [PubMed] [Google Scholar]
- Kafer E., 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19: 33–131. [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Aramayo R., 2007. Meiotic silencing and the epigenetics of sex. Chromosome Res. 15: 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk K. E., Morris N. R., 1991. The tubB alpha-tubulin gene is essential for sexual development in Aspergillus nidulans. Genes Dev. 5: 2014–2023. [DOI] [PubMed] [Google Scholar]
- Kooter J. M., Matzke M. A., Meyer P., 1999. Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 4: 340–347. [DOI] [PubMed] [Google Scholar]
- Laudet V., Stehelin D., Clevers H., 1993. Ancestry and diversity of the HMG box superfamily. Nucleic Acids Res. 21: 2493–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Seong K. Y., Pratt R. J., Baker K., Aramayo R., 2004. Properties of unpaired DNA required for efficient silencing in Neurospora crassa. Genetics 167: 131–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Freitag M., Selker E. U., Aramayo R., 2008. A cytosine methyltransferase homologue is essential for sexual development in Aspergillus nidulans. PLoS ONE 3: e2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Lee T., Lee Y. W., Yun S. H., Turgeon B. G., 2003. Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol. Microbiol. 50: 145–152. [DOI] [PubMed] [Google Scholar]
- Lindbo J. A., Silva-Rosales L., Proebsting W. M., Dougherty W. G., 1993. Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. M., Singer R. H., Meng X., Gonzalez I., Nasmyth K., et al. , 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277: 383–387. [DOI] [PubMed] [Google Scholar]
- Long R. M., Gu W., Meng X., Gonsalvez G., Singer R. H., et al. , 2001. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol. 153: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac F., Wendel B., Goyon C., Faugeron G., Zickler D., et al. , 1997. A gene essential for de novo methylation and development in Ascobolus reveals a novel type of eukaryotic DNA methyltransferase structure. Cell 91: 281–290. [DOI] [PubMed] [Google Scholar]
- Mayrhofer S., Poggeler S., 2005. Functional characterization of an alpha-factor-like Sordaria macrospora peptide pheromone and analysis of its interaction with its cognate receptor in Saccharomyces cerevisiae. Eukaryot. Cell 4: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrhofer S., Weber J. M., Poggeler S., 2006. Pheromones and pheromone receptors are required for proper sexual development in the homothallic ascomycete Sordaria macrospora. Genetics 172: 1521–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B. L., Miller K. Y., Timberlake W. E., 1985. Direct and indirect gene replacements in Aspergillus nidulans. Mol. Cell. Biol. 5: 1714–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B. L., Miller K. Y., Roberti K. A., Timberlake W. E., 1987. Position-dependent and -independent mechanisms regulate cell-specific expression of the SpoC1 gene cluster of Aspergillus nidulans. Mol. Cell. Biol. 7: 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P., van Blockland R., Kooter J. M., Vaucheret H., 2007. A single transgene locus triggers both transcriptional and post-transcriptional silencing through double-stranded RNA production. Planta 225: 365–379. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H., Hanada S., Nguyen B. Q., Kadotani N., Tosa Y., et al. , 2005. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 42: 275–283. [DOI] [PubMed] [Google Scholar]
- Napoli C., Lemieux C., Jorgensen R., 1990. Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggeler S., 2000. Two pheromone precursor genes are transcriptionally expressed in the homothallic ascomycete Sordaria macrospora. Curr. Genet. 37: 403–411. [DOI] [PubMed] [Google Scholar]
- Poggeler, S., M. Nowrousian, and U. Kuck, 2006a Fruiting-body development in Ascomycetes, pp. 325–356 in The Mycota, Ed. 2, edited by U. Kües and R. Fischer. Springer: Berlin; Heidelberg, Germany; New York. [Google Scholar]
- Poggeler S., Nowrousian M., Ringelberg C., Loros J. J., Dunlap J. C., et al. , 2006b Microarray and real-time PCR analyses reveal mating type-dependent gene expression in a homothallic fungus. Mol. Genet. Genomics 275: 492–503. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J. A., Hemmons L. M., Mackdonald K. D., Bufton A. W., 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5: 141–238. [DOI] [PubMed] [Google Scholar]
- Pyrzak W., Miller K. Y., Miller B. L., 2008. Mating type protein Mat1–2 from asexual Aspergillus fumigatus drives sexual reproduction in fertile Aspergillus nidulans. Eukaryot. Cell 7: 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q., Wang H. Y., English J. J., Jorgensen R. A., 1997. The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell 9: 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurs T. A., Schaeffer E. A., Wessels J. G., 1997. Homology-dependent silencing of the SC3 gene in Schizophyllum commune. Genetics 147: 589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., 1990. Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 24: 579–613. [DOI] [PubMed] [Google Scholar]
- Selker E. U., 1997. Epigenetic phenomena in filamentous fungi: Useful paradigms or repeat-induced confusion? Trends Genet. 13: 296–301. [DOI] [PubMed] [Google Scholar]
- Seo J. A., Han K. H., Yu J. H., 2004. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 53: 1611–1623. [DOI] [PubMed] [Google Scholar]
- Shiu P. K., Metzenberg R. L., 2002. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics 161: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu P. K., Raju N. B., Zickler D., Metzenberg R. L., 2001. Meiotic silencing by unpaired DNA. Cell 107: 905–916. [DOI] [PubMed] [Google Scholar]
- Sohn K. T., Yoon K. S., 2002. Ultrastructural study on the cleistothecium development in Aspergillus nidulans. Mycobiology 30: 117–127. [Google Scholar]
- Vallim M. A., Miller K. Y., Miller B. L., 2000. Aspergillus SteA (sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 36: 290–301. [DOI] [PubMed] [Google Scholar]
- van West P., Kamoun S., van ’t Klooster J. W., Govers F., 1999. Internuclear gene silencing in Phytophthora infestans. Mol. Cell 3: 339–348. [DOI] [PubMed] [Google Scholar]
- van West P., Shepherd S. J., Walker C. A., Li S., Appiah A. A., et al. , 2008. Internuclear gene silencing in Phytophthora infestans is established through chromatin remodelling. Microbiology 154: 1482–1490. [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Fagard M., 2001. Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 17: 29–35. [DOI] [PubMed] [Google Scholar]
- Wang X., Hsueh Y. P., Li W., Floyd A., Skalsky R., et al. , 2010. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 24: 2566–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M., 2005. The role of the RNAi machinery in heterochromatin formation. Cell 122: 13–16. [DOI] [PubMed] [Google Scholar]
- Wirsel S., Horwitz B., Yamaguchi K., Yoder O. C., Turgeon B. G., 1998. Single mating type-specific genes and their 3′ UTRs control mating and fertility in Cochliobolus heterostrophus. Mol. Gen. Genet. 259: 272–281. [DOI] [PubMed] [Google Scholar]
- Wu J., Miller B. L., 1997. Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol. Cell. Biol. 17: 6191–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelton M. M., Hamer J. E., de Souza E. R., Mullaney E. J., Timberlake W. E., 1983. Developmental regulation of the Aspergillus nidulans trpC gene. Proc. Natl. Acad. Sci. USA 80: 7576–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H., 2006. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 44: 145–154. [PubMed] [Google Scholar]
- Yun S. H., Berbee M. L., Yoder O. C., Turgeon B. G., 1999. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. USA 96: 5592–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]