Abstract
Knowing the distribution of fitness effects (DFE) of new mutations is important for several topics in evolutionary genetics. Existing computational methods with which to infer the DFE based on DNA polymorphism data have frequently assumed that the DFE can be approximated by a unimodal distribution, such as a lognormal or a gamma distribution. However, if the true DFE departs substantially from the assumed distribution (e.g., if the DFE is multimodal), this could lead to misleading inferences about its properties. We conducted simulations to test the performance of parametric and nonparametric discretized distribution models to infer the properties of the DFE for cases in which the true DFE is unimodal, bimodal, or multimodal. We found that lognormal and gamma distribution models can perform poorly in recovering the properties of the distribution if the true DFE is bimodal or multimodal, whereas discretized distribution models perform better. If there is a sufficient amount of data, the discretized models can detect a multimodal DFE and can accurately infer the mean effect and the average fixation probability of a new deleterious mutation. We fitted several models for the DFE of amino acid-changing mutations using whole-genome polymorphism data from Drosophila melanogaster and the house mouse subspecies Mus musculus castaneus. A lognormal DFE best explains the data for D. melanogaster, whereas we find evidence for a bimodal DFE in M. m. castaneus.
Keywords: distribution of fitness effects, deleterious mutations, gamma distribution, discrete models, multimodality, fixation probability
NEW mutations generate genetic variation in the genome of every species. For example, it has been estimated that a newborn human has ∼70 new mutations that originated in its parents’ germlines (Keightley 2012). The fitness effects of new mutations can range from deleterious to neutral and to advantageous, and the relative frequencies of their effects is known as the distribution of fitness effects (DFE) of new mutations. Inferring the properties of the DFE is a long-standing goal of evolutionary genetics and is key to several important questions, including the evolution of sex and recombination, the prevalence of Muller’s ratchet, and the constancy of the molecular clock (Charlesworth 1996; Eyre-Walker and Keightley 2007).
A number of methodologies have been developed to infer the DFE based on DNA sequence data (Sawyer et al. 2003; Nielsen and Yang 2003; Piganeau and Eyre-Walker 2003; Loewe et al. 2006; Eyre-Walker et al. 2006; Keightley and Eyre-Walker 2007; Boyko et al. 2008; Schneider et al. 2011; Wilson et al. 2011). All of these assume that there is a neutrally evolving class of sites and contrast patterns of polymorphism and/or divergence from an outgroup with that of a tightly linked focal site class. Selection affecting the focal sites is expected to alter the pattern of polymorphism compared to that of the neutral class. A distribution of selection coefficients is then fitted to the data and its properties inferred. The three most widely used methods are those developed by Eyre-Walker et al. (2006), Keightley and Eyre-Walker (2007), and Boyko et al. (2008). Keightley and Eyre-Walker (2007) use a Wright–Fisher transition-matrix approach (Ewens 1979), whereas Eyre-Walker et al. (2006) and Boyko et al. (2008) use a diffusion approximation (Sawyer and Hartl 1992; Williamson et al. 2005). All three methods have been reported to give similar results, but make slightly different assumptions. For example, they differ in the way in which they model demographic changes (e.g., population size changes). Eyre-Walker et al. (2006) use a heuristic approach, whereas the other two explicitly model some simple demographic scenarios. It is necessary to model demographic change, because this is known to alter patterns of polymorphism in ways that can resemble selection. Because these methods use allele-frequency information (summarized as the site-frequency spectrum or SFS), they are expected to be sensitive to demographic change.
Several studies have employed the above methods to infer properties of the DFE of amino acid-changing mutations. In these analyses, a gamma distribution of fitness effects has often been assumed, since it is a flexible distribution with two parameters, the shape (b) and the scale (a). For example, for amino acid-changing mutations in Drosophila melanogaster, the shape parameter has been estimated to be ∼0.4 (implying a leptokurtic distribution), and most (>90%) new mutations are inferred to be moderately to strongly deleterious, with effective strength of selection Nes > 10 (Keightley and Eyre-Walker 2007; Eyre-Walker and Keightley 2009). In humans, the DFE appears to be more even more leptokurtic than in Drosophila (i.e., the estimated shape parameter is ∼0.2), and only ∼60% of mutations appear to be moderately to strongly deleterious (Eyre-Walker et al. 2006; Keightley and Eyre-Walker 2007; Boyko et al. 2008; Eyre-Walker and Keightley 2009). Differences between Drosophila and humans in the properties of the DFE have been attributed to a difference in their effective population size (Ne), the former being at least 2 orders of magnitude larger (Eyre-Walker et al. 2002). An effect attributable to Ne has also been observed in several other species. For example, Ne in wild house mice is substantially larger than humans but smaller than Drosophila, and ∼70–80% of amino acid mutations are estimated to be moderately to strongly deleterious (Halligan et al. 2010; Kousathanas et al. 2011). Capsella grandiflora and Aribidopsis thaliana are two plant species with large and small Ne, respectively, and ∼86% and ∼66% of amino acid mutations are estimated to be moderately to strongly deleterious, respectively (Foxe et al. 2008; Slotte et al. 2010).
Most of the above methods assume that the DFE can be approximated by a certain type of mathematical distribution, such as the gamma distribution. One would like, however, to have a more general approach to obtain information about the DFE without needing to assume an explicit distribution. Steps in this direction were taken by Keightley and Eyre-Walker (2010), who examined a model of multiple discrete selection coefficients rather than assuming a continuous distribution. However, Keightley and Eyre-Walker (2010) did not examine the performance of their models when the true distribution deviated from a gamma distribution. Boyko et al. (2008) also fitted several types of distributions and combinations of continuous distributions and discrete fixed effects when inferring the DFE for amino acid-changing mutations in humans. Wilson et al. (2011) recently developed a new method that assumes a series of discrete fixed selection coefficients, the density associated with each selection coefficient estimated as a parameter. However, due to the complexity of the model, Wilson et al. (2011) needed to assume constant population size.
Although several different types of parametric and nonparametric DFE models have been fitted to DNA polymorphism data, to our knowledge their performance in cases where the true DFE is bimodal or multimodal has not previously been investigated. In this study, we use simulations to examine cases in which the true DFE is unimodal, bimodal, or multimodal. We analyze simulated data assuming six models for the DFE. The first two are parametric unimodal distributions: the lognormal and the gamma distribution. The third model is a parametric distribution that can be bimodal: the beta distribution. The fourth model is a discrete point mass distribution of selection coefficients where the locations and the probability densities of each point mass (or “spikes”) are estimated parameters. We refer to this model as the spikes model, which is similar to the discretized model used by Keightley and Eyre-Walker (2010). The fifth model (“steps” model’) consists of multiple continuous, uniform distributions (or steps), the boundaries and probability densities of which are estimated parameters. The sixth model is a variant of the model used by Wilson at al. (2011) and assumes six fixed selection coefficients where only their probability densities are estimated parameters. We refer to this model as the “fixed six-spikes” model. We use simulations to test the performance of the six models assuming various scenarios for the complexity of the true DFE. We go on to fit the six models to protein polymorphism data sets from D. melanogaster and Mus musculus castaneus, each containing sequences of several thousand protein-coding genes.
Materials and Methods
Population genetic model and assumptions
In this study, we extend the methods developed by Keightley and Eyre-Walker (2007) to infer the DFE of new mutations based on the allele frequency distribution of polymorphic nucleotide sites among individuals sampled from a population. This approach is based on Wright–Fisher population genetics theory and makes a number of assumptions. We assume that sites are unlinked and have the same mutation rate and that polymorphic sites are biallelic. We assume that there are two classes of sites in the genome, one “neutral” and one “selected.” The fates of new mutations in the neutral class are affected only by genetic drift. New mutations at selected sites are assumed to be unconditionally deleterious and to have additive effects on fitness. We define the selection coefficient s as the fitness reduction experienced by the homozygote for the mutant allele compared to the homozygote for the wild-type allele. Therefore, the fitnesses of the wild-type, heterozygote, and mutant homozygote are 1, 1 − s/2 and 1 − s, respectively.
Description of the modeled distributions of selection coefficients
New mutations affecting the selected class of sites are sampled from a probability distribution. We investigated six models for this probability distribution: the first is a lognormal distribution, which has two parameters: the mean or location (μ) and the standard deviation or scale (σ). The second is a gamma distribution, which has two parameters: the shape (b) and the scale (a). The third model is the beta distribution, which has two shape parameters (k1, k2). The fourth model (spikes model) assumes m mutational effects classes (spikes), which are modeled as point masses. For each mutational effect class i (i = 1…m), the location si and the probability density (pi) are estimated parameters, for a total of 2m − 1 parameters. The fifth model (steps model) assumes m mutational effects classes, and each class i (i = 1...m) is modeled as a uniform distribution where the minimum and maximum values (Nesi−1 and Nesi, respectively) and the probability density (pi) are estimated parameters. The minimum value of the first step is fixed to zero. We assume that the start of each step is the end of the previous, that is, for step i, Nesi = Nesi−1, ensuring that there are no overlapping steps. The total number of parameters to be estimated is m for the minimum and maximum values values of the steps plus m − 1 for the probability density of each step, giving a total of 2m − 1 parameters. The sixth model (fixed six-spikes) assumes six mutational effects classes (spikes), modeled as point masses arbitrarily fixed at Nes1 = 0, Nes2 = 1, Nes3 = 5, Nes4 = 10, Nes5 = 50, Nes6 = Ne. The probability densities of the fixed point masses are estimated parameters, for a total of five parameters.
Demographic model
Following Keightley and Eyre-Walker (2007), we also incorporate a simple demographic model of a step change from population size N1 to population size N2 at some time t in the past. N1 is fixed at 100, the parameter t is estimated relative to N2, and the parameter N2 is estimated relative to N1 (i.e., the magnitude of the size change is estimated). There may be little information with which to estimate the relative values of N1 and N2 so we also compute a weighted recent effective population size Nw,
| (1) |
where and (Eyre-Walker and Keightley (2009)). We also incorporate a parameter f0, which is the proportion of unmutated sites. Under selective neutrality and stationary equilibrium, 1 − f0 is proportional to the product of the mutation rate and the persistence time of a new mutation.
Generation of the expected allele-frequency vector and computation of likelihood
We assume that at some point in the past, a population of size N1 was at mutation–selection–drift equilibrium. This population then experienced a size change (either expansion or contraction) to size N2 t generations from the present. Throughout this period, new mutations arise, which are neutral for the neutral class of sites and deleterious with selection coefficients s sampled from a probability distribution f(s) for the selected class. Following Keightley and Eyre-Walker (2007), we employ Wright–Fisher transition matrix methods to generate the expected allele frequency distribution at the present time for a set of parameter values f0, t, N2, and a given s value, and we store it in vector v(s). The lognormal, gamma, spike, and step distributions can potentially have substantial parts of their density at s > 1. We modeled the contribution of mutations for s > 1 assuming that their frequency in the population goes down in proportion to the expectation at mutation–selection balance, following Keightley and Eyre-Walker (2007). The expected mean allele-frequency distribution z is obtained by integrating over the distribution of selection coefficients for all elements of v(s),
| (2) |
where Θ represents the parameters of the distribution of selection coefficients (e.g., a and b for the gamma distribution).
The numbers of derived alleles in a sample of nT alleles constitute the SFSs and are stored in vectors q(N) and q(S) for the selected and neutral sites, respectively. Numbers of alleles are binomial draws from a diploid population of size N2. Since we do not distinguish between the derived and ancestral states, we use only folded SFSs. We fold the SFS and the allele-frequency vector z as follows:
| (3) |
| (4) |
Under the assumption that numbers of derived alleles are binomially distributed, we compute the log likelihood of the observed allele frequency distributions (i.e., SFSs) for neutral and selected sites as
| (5) |
(Keightley and Eyre-Walker 2007), where b〈i|n, p〉 is the binomial probability for i derived alleles in a sample of n alleles with probability of occurrence p. We find the set of the parameter values that best fits the observed SFSs by maximizing the sum of the log likelihoods calculated for the neutral and selected classes of sites.
Likelihood maximization
The parameters to be estimated are f0, N2, t, plus additional parameters, depending on the selection model implemented (Table 1). Maximization of the likelihood was done using a custom likelihood search algorithm for N2, and the SIMPLEX algorithm (Nelder and Mead 1965) for the remaining parameters. To increase the speed of the maximization procedure, we first estimated the demographic parameters N2 and t and the parameter f0 from the neutral SFS. We assumed the maximum likelihood (ML) estimates of N2 and t when estimating the parameters from the selected SFS.
Table 1. The selection models investigated in this study.
| DFE Model | No. Parameters | Parameters |
|---|---|---|
| Lognormal | 2 | μ, σ (location, scale) |
| Gamma | 2 | a, b (scale, shape) |
| Beta | 2 | k1, k2 (shape 1, shape 2) |
| Spike | 2m − 1 | For i (i = 1…m), Nesi |
| For i (i = 1…m − 1), pi | ||
| Step | 2m − 1 | For i (i = 1…m) Nesi |
| For i (i = 1…m − 1), pi | ||
| Six-fixed spikes | 5 | For i (i = 1…5), pi |
We generated starting values for the location parameters of the spikes and the steps by using a power series,
| (6) |
where Ne = Nw as calculated by Equation 1 and r is a pseudorandom deviate from a normal distribution with a mean 0 and standard deviation 0.1. This power series was devised empirically and has several desirable properties: the term Nei/m places the spikes or steps at a reasonable distance from each other; the last spike or step is placed at Ne, therefore avoiding generating extremely large Nes values; the pseudorandom normal deviate r adds noise in the placement of the spikes/steps.
The starting values for the relative probability densities of the steps were set to 1/m. As the number of parameters increases, the possibility of multiple local maxima also increases. To ensure that the global maximum had been found, we performed 10 starts of the maximization algorithm for each run, each time using a different seed for the pseudorandom number generator. We recorded the ML estimates that gave the highest likelihood in these runs.
Implementation of the model
Our simulations used a forward Wright–Fisher simulator to generate SFSs and we then used ML to fit demographic and selection models and estimate the parameters. This was implemented in a recoded version of the C program DFE-alpha (Eyre-Walker and Keightley 2009). This version implements all of the models we describe, can be used to analyze SFS data sets in a similar way to DFE-alpha, and will be made available via the authors’ website.
Simulations assuming a constant population size
We simulated SFS data sets assuming a diverse set of distributions of selection coefficients, including unimodal, bimodal, and multimodal distributions. We performed simulations in which we assumed a constant population size (N1 = N2 = 100). We used 106 neutral and 106 selected sites and sampled 64 alleles. Parameter f0 was set to 0.9. We also compared simulations in which we assumed different numbers of sequenced alleles (8, 16, 32, 64, 128, and 256), while assuming a set number of sites (106). For each simulated data set, we performed 100 replicate simulations.
Simulations assuming variable population size
We modeled population size changes as step changes from an initial population of size N1 = 100 at stationary equilibrium. Time is expressed in units of N1. We simulated two demographic histories: a population expansion and a bottleneck. The simulated expansion was a step change to size N2 (N2/N1 = 3.1), at time t2/N1 = 1. The simulated bottleneck was a reduction in population size N2/N1 = 0.72 at time t2/N1 = 1.1 and a subsequent expansion with a step change in size N3/N1 = 3.8 at time t3/N1 = 0.11. The parameters for the two simulated demographic scenarios were chosen to match the inferred histories of real populations. The simulated expansion matches that inferred for a population of wild mice (Halligan et al. 2010) and for the American population of humans with African ancestry (Boyko et al. 2008). The bottleneck scenario matches that inferred for the American population of humans with European ancestry (Boyko et al. 2008). For these simulations we assumed a gamma DFE with a = 0.05 and b = 0.5. For each simulated data set we used 106 neutral and 106 selected sites, sampled 64 alleles, and performed 20 replicate simulations.
Simulations with linkage
We used C++ program SLiM, developed by Philip Messer and available at http://www.stanford.edu/∼messer/software.html to perform simulations with linkage (Messer 2013). We simulated 1-Mbp-long chromosomes. Each chromosome had 20 loci. Each locus consisted of 10 exons of length 100 bp each alternating with 1-kbp introns. The loci were at a distance of 40 kbp from each other. We used exonic sites and the first 100 bp of introns as selected and neutral sites respectively. We simulated a population of size N = 100 for 10N generations to reach stationary equilibrium and sampled 64 chromosomes every 2N generations for 100N generations to obtain polymorphism data for a total of 106 selected and 106 neutral sites. We assumed a mutation rate 4Neμ = 1% and simulated various levels of linkage between sites by assuming recombination rates (4Ner) varying between 10−5 and 1. We performed three types of simulations, varying the properties of the DFE for selected sites: First, we assumed a gamma DFE (a = 0.05, b = 0.5), second we assumed that 97% of sites were under negative selection (gamma DFE; a = 0.05, b = 0.5) and 3% were under positive selection (single spike DFE; Nes1 = 10), and third we assumed a bimodal DFE consisting of two spikes of selection coefficients (Nes1 = 0, Nes2 = 10, p1 = 0.2). We performed 20 replicate runs for each simulation type.
Evaluation of model performance
We are interested in knowing how well the mean effect (), the mean fixation probability of a new deleterious mutation relative to a neutral mutation (), and the proportion of mutations falling into five Nes categories (0.0–0.1, 0.1–1.0, 1.0–10.0, 10.0–100.0, >100.0) are estimated. and are important quantities for several questions, including inferring the proportion of mutations fixed by positive selection and the rate of adaptive relative to neutral evolution (i.e., α and ωα, respectively; Eyre-Walker and Keightley 2009; Gossmann et al. 2010). was calculated by taking the arithmetic average of the selection coefficients over the range of s between 0 and 100 (i.e., the Nes range was between 0 and 104, for Ne = 100). was calculated by integrating over the DFE, as in Eyre-Walker and Keightley (2009),
| (7) |
where u (Ne, s), is the fixation probability of a new deleterious mutation (Fisher 1930; Kimura 1957, 1962).
To assess the accuracy in recovering the properties (X) of the simulated distributions, we compared estimates (Xi) vs. true values (Xtrue). For and , we calculated the relative error as
| (8) |
We compared the goodness of fit between models by comparing their likelihoods and by comparing Akaike information criterion (AIC) scores. The AIC score penalizes parameter-rich models as
| (9) |
where k is the number of parameters in the model, and L is the maximum likelihood for the estimated model. We considered an AIC difference >2 as significant when comparing models. For the spike/step models we increased the number of fitted spike/steps until an improvement of <2 AIC units was obtained.
Drosophila and house mouse data sets
We analyzed polymorphism data for protein-coding genes of D. melanogaster and M. m. castaneus using the six approaches described above. We also fitted a simple demographic model of a step change in population size. For D. melanogaster, we analyzed a data set of 17 genomes from individuals originating in East Africa (haploid Rwanda lines from the Drosophila Population Genomics Project (DPGP; release v. 2.0, http://www.dpgp.org/dpgp2/DPGP2.html; Pool et al. 2012). The data set was compiled as in Campos et al. (2012), but we did not use a minimum quality cut-off. It included polymorphism data for 8367 autosomal genes orthologous between D. melanogaster and D. yakuba. For M. m. castaneus, we used a data set of 20 genomes from individuals sampled in northwest India (Halligan et al. 2010; D.L. Halligan, A. Kousathanas, R.W. Ness, H. Li, B. Harr, L. Eory, T. M. Keane, D. J. Adams, P. D. Keightley, unpublished data). The data set included polymorphism data for 18,671 autosomal genes orthologous between M. m. castaneus and rat. CpG dinucleotides have substantially higher mutation rates in mammals (Arndt et al. 2003) and their frequencies differ between coding and noncoding DNA. Therefore for M. m. castaneus, we restricted the analysis to non-CpG-prone sites (sites not preceded by C or followed by G). To calculate α and ωa we used the divergences at nonsynonymous and synonymous sites between D. melanogaster and D. yakuba and between M. m. castaneus and rat, as follows,
| (10) |
| (11) |
where dN and dS are the nucleotide divergences between the focal species and the outgroup at nonsynonymous and synonymous sites, respectively.
Results
We simulated SFS data sets, choosing the parameters of the simulated distributions to create three different scenarios for their complexity (i.e., unimodality, bimodality, and multimodality). We also aimed at generating distributions that were biologically plausible. We then examined the performance of several models incorporating parametric or nonparametric distributions. We considered four main criteria for evaluating the performance of the tested models: the log-likelihood score, the accuracy in estimating the mean effect of a new mutation (), the accuracy in estimating the average fixation probability of a new mutation (), and the accuracy in estimating the proportion of mutations in five Nes ranges. Estimates for the parameters of each of the six tested models for each simulation set (SIM1, SIM2, SIM3) are shown in Supporting Information, Table S1.
A gamma distribution simulated (SIM1)
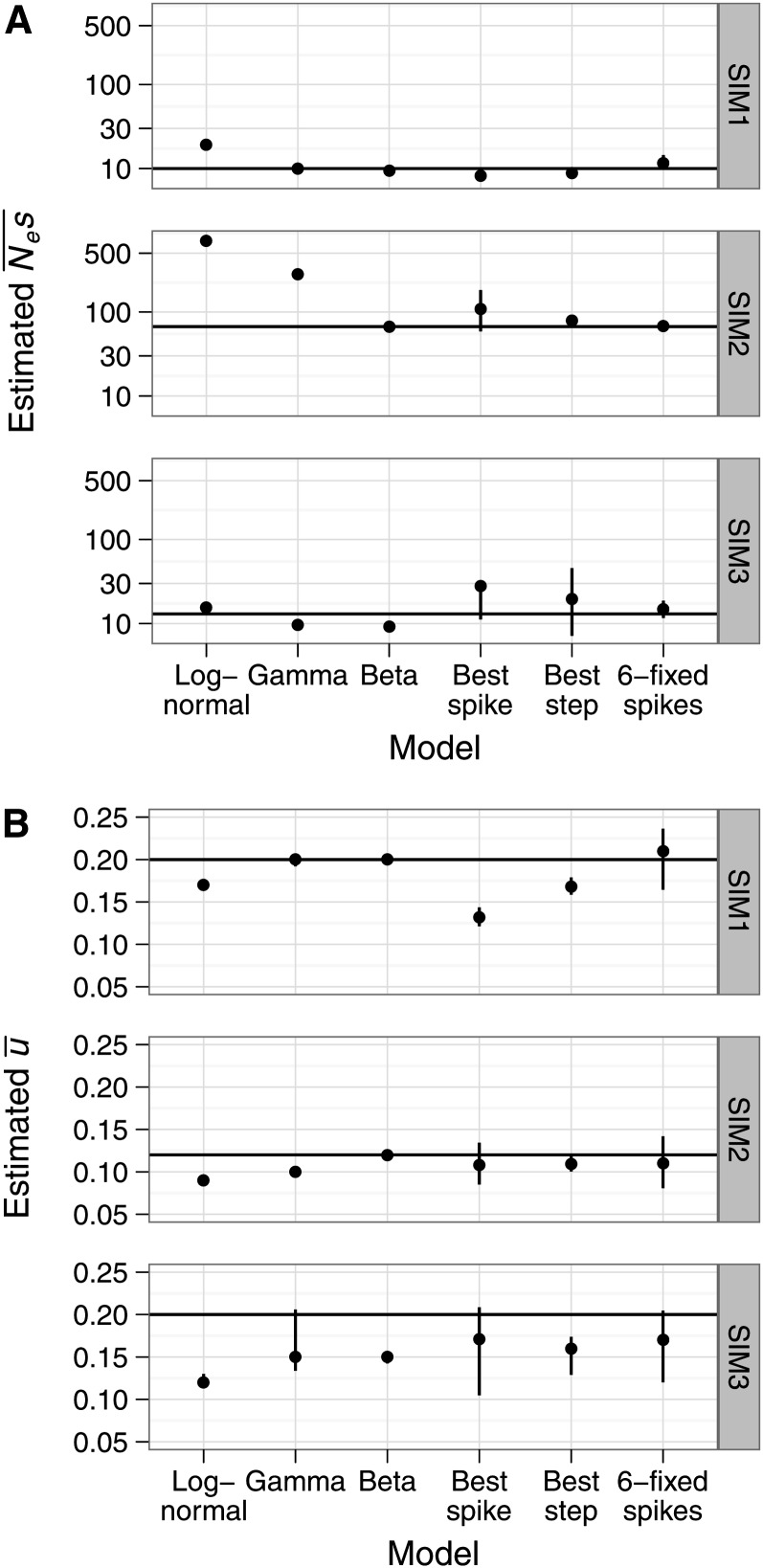
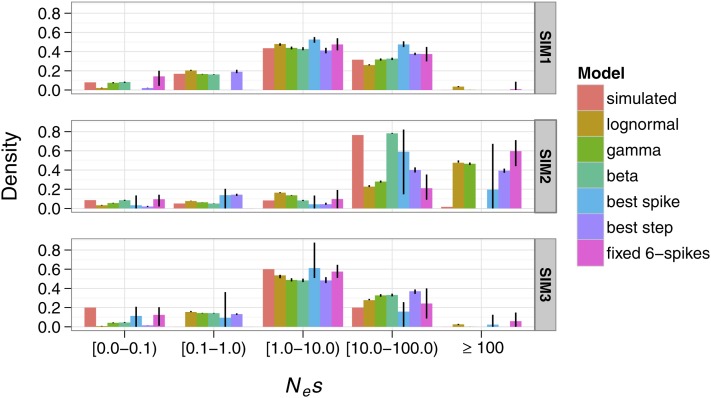
To approximate a realistic scenario for protein-coding loci, where current information suggests a leptokurtic DFE and most sites under strong negative selection, we simulated a gamma DFE with scale a = 0.05 and shape b = 0.5 (SIM1; Figure 1). As expected, the gamma model gave the best fit to the data, accurately estimating and (SIM1; Table 2). The lognormal model performed poorly, overestimating and underestimating , while the beta model gave a good fit (ΔAIC from the best-fitting model was −0.5) and accurately estimated and (SIM1; Figure 2, A and B, respectively). Based on their AIC scores, the best-fitting variable spike and variable steps models were the two-spike and two-step models, respectively (SIM1; Table 2), and these models fitted only slightly worse than the gamma model. However they did not recover and as accurately as the gamma (SIM1; Figure 2, A and B, respectively). All models tested performed well in accurately recovering the proportions of mutations in the Nes ranges we examined (Figure 3). However, the lognormal and all the nonparametric models did not succeed in accurately assigning the proportions of mutations in the Nes ranges 0.0–0.1 and 0.1–1.0, presumably because there is little information to discriminate between these categories. In contrast, the gamma and beta models performed almost perfectly in assigning the proportions of mutations to these categories.
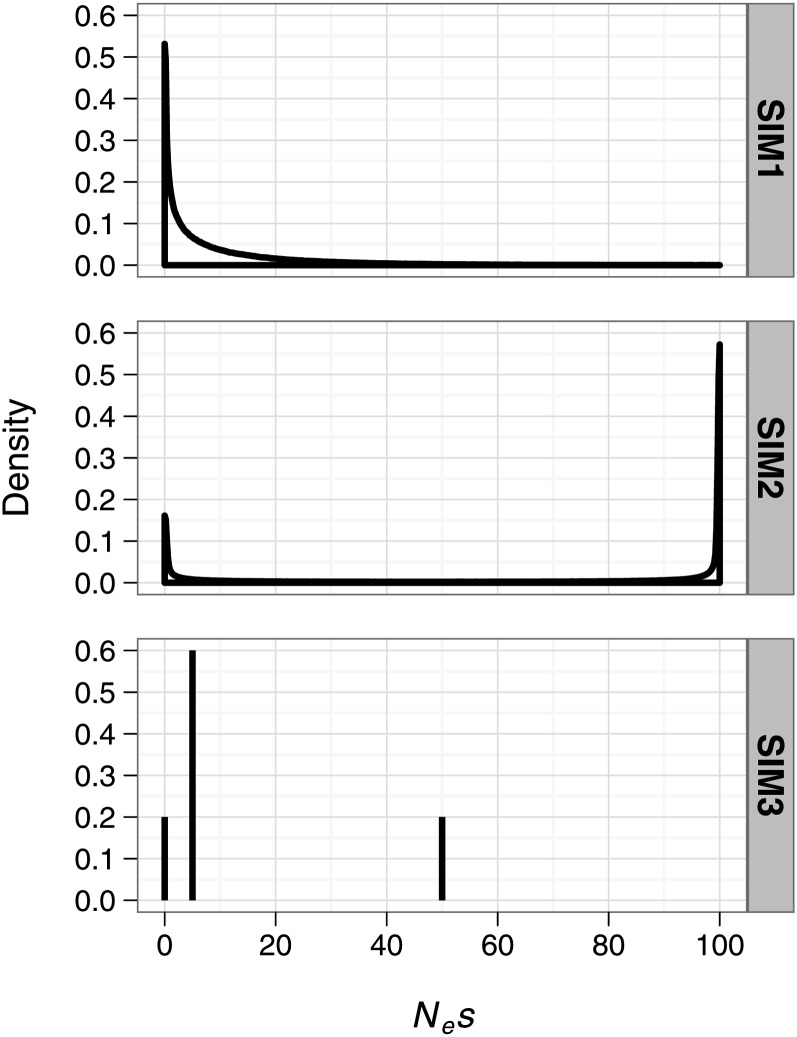
Figure 1.
The simulated DFEs. For SIM1, we simulated a gamma DFE with scale a = 0.05 and shape b = 0.5. For SIM2, we simulated a beta DFE with shape parameters k1 = 0.2 and k2 = 0.1 scaled to the Nes interval [0, 100]. For SIM3, the DFE was composed of three selection coefficients, Nes1 = 0, Nes2 = 5, Nes3 = 50, with probability densities p1 = 0.2, p2 = 0.6, p3 = 0.2.
Table 2. Goodness-of-fit statistics for the models tested for each simulation set.
| Simulation | Model | ΔlogL | ΔAIC |
|---|---|---|---|
| SIM1 (gamma) | Lognormal | −13.9 | −27.8 |
| Gamma | −0.02 | 0.0 | |
| Beta | −0.3 | −0.5 | |
| Best spike (2) | −1.5 | −4.9 | |
| Best step (2) | 0.0 | −2.0 | |
| Six-fixed spikes | −0.6 | −7.1 | |
| SIM2 (bimodal beta) | Lognormal | −300.0 | −597.2 |
| Gamma | −46.4 | −89.9 | |
| Beta | −1.4 | 0.0 | |
| Best spike (3) | 0.0 | −3.1 | |
| Best step (2) | −1.3 | −1.8 | |
| Six-fixed spikes | −3.5 | −10.2 | |
| SIM3 (three-spike multimodal) | Lognormal | −29.5 | −53.0 |
| Gamma | −6.9 | −7.8 | |
| Beta | −8.2 | −10.4 | |
| Best spike (3) | 0.0 | 0.0 | |
| Best step (3) | −0.7 | −1.3 | |
| Six-fixed spikes | −0.6 | −1.3 |
The statistics reported are the mean log-likelihood and the mean AIC score difference from the highest scoring model over 100 simulation replicates. A sequencing effort of 64 alleles and 106 neutral and selected sites were assumed. Only results for the best-fitting spike and step model, based on the AIC criterion, are shown.
Figure 2.
Summary statistics for the models tested for each simulation set. (A) Mean estimates of the mean effect of a new mutation () and (B) the probability of fixation of a new mutation (). Error bars are the 5th and 95th percentiles of estimates over 100 simulation replicates. The horizontal lines represent the simulated values. Only results for the best-fitting spike and step model, according to the AIC criterion, are shown. The y-axis is log scaled for panel A.
Figure 3.
The mean estimated proportions of mutations in five Nes ranges for SIM1, SIM2, and SIM3. We assumed a sequencing effort of 64 alleles and 106 neutral and selected sites. Error bars are the 5th and 95th percentiles of estimates over 100 simulation replicates.
A bimodal beta distribution simulated (SIM2)
We then investigated a beta distribution with shape parameters k1 = 0.2 and k2 = 0.1 and scaled to the Nes interval [0, 100] (SIM2; Figure 1). For this distribution, ∼10% of selected sites are under weak negative selection (Nes < 1), another 10% are under moderately strong negative selection (Nes = 1–10), and the remaining 80% are under very strong negative selection (Nes > 10). Such a bimodal distribution is intended to model protein-coding loci where amino-acid changing mutations are either neutral or strongly deleterious, with relatively few mutations of intermediate effect. As expected, the beta model had the best AIC score (SIM2; Table 2), recovering and accurately (SIM2; Figure 2, A and B, respectively). The unimodal lognormal and gamma models fitted the data very poorly (ΔAIC from beta = −597.2 for the lognormal and −89.9 for the gamma, SIM2; Table 2). was grossly overestimated by the lognormal and gamma models (SIM2; Figure 2A). However, was estimated relatively accurately by these models (SIM2; Figure 2B). The estimate for can be heavily influenced by a long tail in the fitted distribution whereas is mostly affected by effects in the Nes range 0–1. Therefore, the low accuracy of estimates from the lognormal and gamma models presumably reflects a bad fit to the “strong effects” part of the distribution (i.e., Nes > 10), but there is a reasonably good fit to the “nearly neutral effects” part of the distribution (i.e., 0 < Nes < 1). The best-fitting three-spike and two-step models and the fixed six-spike model fitted almost as well as the beta distribution (SIM2; Table 2). These nonparametric models accurately estimated and (SIM2; Figure 2, A and B, respectively). We observed that the lognormal, gamma, and nonparametric models assigned substantial proportions of mutations into the Nes >100 range (Figure 3), although the simulated distribution had a near-zero density in this range. Presumably, there is little information with which to precisely estimate the upper limit of the simulated distribution.
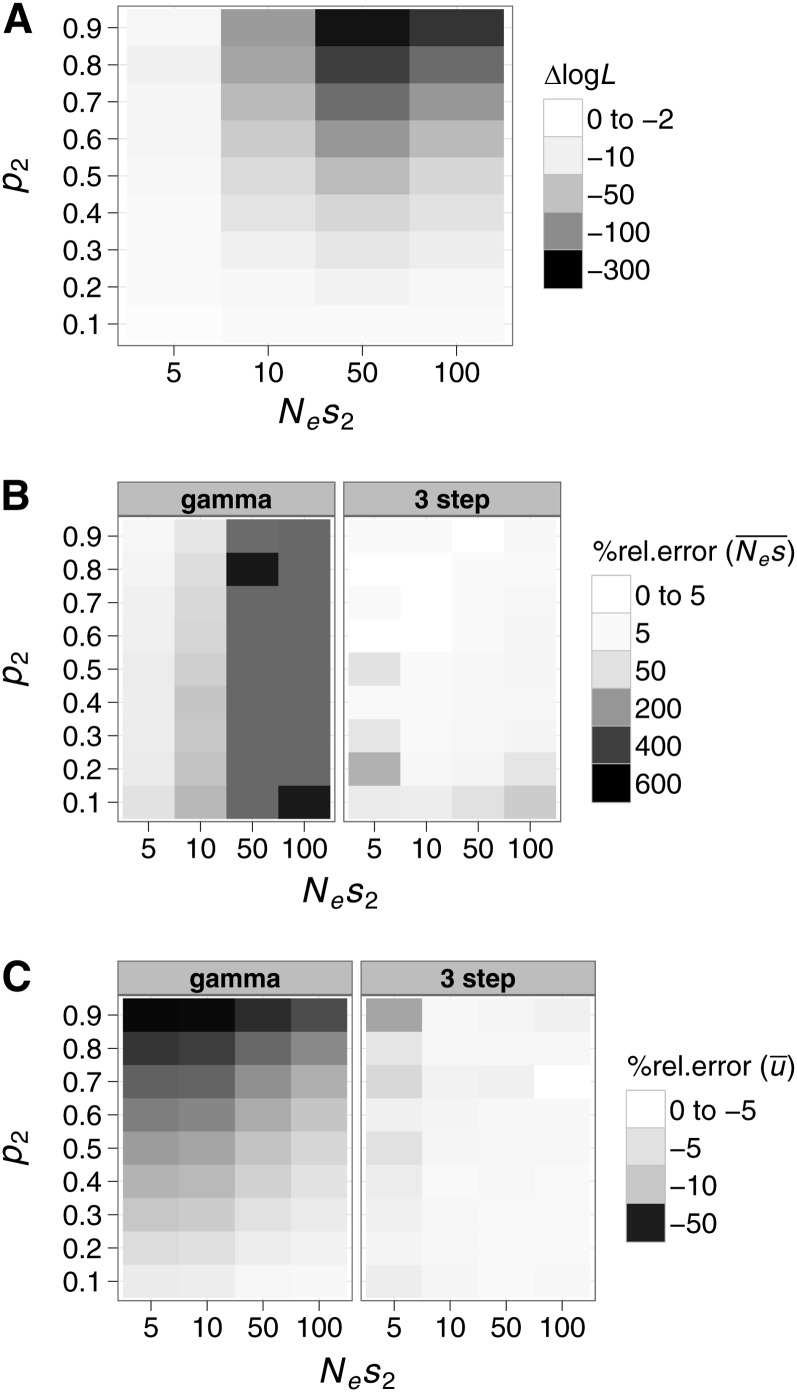
We also examined the performance of the models when varying the locations of the modes of a bimodal DFE. We investigated distributions with two classes of effects (two spike): The first class of mutations was assumed to be neutral with Nes1 = 0, and we varied the selection strength and probability density associated with the second class (Nes2 and p2, respectively). We then fitted the gamma and the three-step models to these distributions and compared their performance. In Figure 4A we show the ΔlogL between the three-step and gamma models for different combinations of values for Nes2 and p2. We found that for two-spike distributions, where Nes2 ≥ 10 and p2 ≥ 0.4, the three-step model significantly outperformed the gamma model (Figure 4A). Additionally, we examined the performance of the models in estimating and . We found that the gamma model overestimated when Nes2 ≥ 10 and underestimated for almost all parameter combinations of Nes2 and p2 (Figure 4, B and C, respectively), while the three-step model overestimated and underestimated when Nes2 < 10 (Figure 4, B and C, respectively).
Figure 4.
The performance of the gamma and three-step models when fitted to bimodal DFEs. We simulated two-spike DFEs with one spike fixed at Nes1 = 0 and we varied the selection strength (Nes2) and probability density (p2) of the second spike. (A) ΔlogL between the three-step and gamma models fitted to the simulated DFEs as a function of Nes2 and p2. We also compared the % rel. error in estimating (B) and (C) . Positive and negative values of % rel. error signify overestimation and underestimation of these parameters, respectively.
A three-spike multimodal distribution simulated (SIM3)
To examine a case in which the true DFE is more complex, we simulated a DFE comprising three selection coefficients, Nes1 = 0, Nes2 = 5, Nes3 = 50, with probability densities p1 = 0.2, p2 = 0.6, p3 = 0.2, respectively (SIM3; Figure 1). The choice of parameters was mainly based on generating three sufficiently distinct modes. As expected, a three-spike model gave the best fit according to the AIC criterion (SIM3; Table 2). The other nonparametric models fitted almost equally well (ΔAIC was −1.3 for both the three-step model and the fixed six-spike model, SIM3; Table 2). However, the lognormal, gamma and beta models gave a poorer fit than the nonparametric models (ΔAIC was −53, −7.8, and −10.4 for the lognormal, gamma, and beta models, respectively, SIM3; Table 2). However, we did not observe large differences in the accuracy of estimating and between the models tested (SIM3; Figure 2, A and B, respectively). The lognormal, best spike, best step, and fixed six-spike models slightly overestimated , whereas the gamma and beta models slightly underestimated (SIM3; Figure 2A). All models tested slightly underestimated (SIM3; Figure 2B).
The effect of increasing the allele sequencing effort
The primary goal of this section was to examine whether the general trends in the performance of the six models tested hold for different allele sequencing efforts. We compared the performance of the models for 8, 16, 32, 64, 128, and 256 alleles sequenced. For the gamma distribution (SIM1), increasing the sequencing effort led to more accurate estimates of for all models (SIM1; Figure S1A). Accuracy of estimating improved only marginally (SIM1; Figure S1B). For the beta distribution (SIM2), increasing the allele sequencing effort increased the accuracy of estimating (SIM2; Figure S1A), but the accuracy of estimating did not increase for the spike, step, and fixed six-spike models and surprisingly decreased for the lognormal and gamma models (SIM2; Figure S1B). This decrease can be explained if we consider that the overall fit of the gamma and lognormal models improves as the number of alleles sequenced is increased, but the fit of the models to the Nes range 0–1 worsens (the good fit of the models to the Nes range 0–1 is crucial for an accurate estimate of ). For the three-spike multimodal distribution (SIM3), we observed that the parametric lognormal, gamma, and beta models showed no improvement in accuracy for estimating and when increasing the number of alleles sequenced (SIM3; Figure S1A and Figure S1B, respectively). The spike, step, and fixed six-spikes models at low sequencing efforts (8–32 alleles) had an inferior performance compared to the parametric models (SIM3; Figure S1A and Figure S1B). However, as the number of alleles sequenced was increased to 64 or greater, the performance of these models became superior to the parametric models (SIM3; Figure S1A and Figure S1B).
The effect of incorporating a population size change
We then examined whether population size changes can affect the performance of the nonparametric relative to the parametric models by simulating two population histories: an expansion and a bottleneck. The expansion was a threefold step change in population size. The bottleneck was a long-lasting 30% reduction in population size, followed by a short-lived fourfold step expansion. For the selected sites, we assumed a gamma DFE with scale a = 0.05 and shape b = 0.5 (as for SIM1). Since our method can incorporate a model of a step change in population size, we fitted this model to the neutral data for both simulated histories. For the expansion scenario, the demographic parameters of the step change were accurately estimated and the performance of the different selection models was similar to SIM1 (Table S2). For the bottleneck scenario, the two-epoch demographic model appeared to mostly capture the second change in population size (Table S2). However, the nonparametric two-spike and two-step selection models fitted the data better than the parametric models (Table S2). Therefore, a long-lasting bottleneck followed by rapid expansion can produce a signal in the data that is not fully accounted for by the fitted two-step demographic scenario and can cause the spike and step models to overfit the data and produce spurious evidence for multimodality. Other population histories such as a bottleneck followed by long-lasting recovery or expansion gave similar results to the two-step expansion scenario (result not shown).
The effect of linkage and selection
In our simulations we have assumed that sites are unlinked, but genomes of real organisms can exhibit various amounts of linkage. We performed simulations assuming a range of recombination rates between sites to examine how linkage can affect the performance of the three-step model in detecting a bimodal DFE. This performance is assessed by a significantly better fit of the three-step model than the gamma model.
First, we investigated whether background selection alone could produce a spurious signature of a bimodal DFE by simulating a gamma DFE with a = 0.05 and b = 0.5. We observed a better fit of the three-step model than the gamma model for high levels of linkage (Figure S1C, top). However, when we fitted a demographic model of a step change to the neutral sites, a procedure that has been suggested to control for the effects of linkage (Messer and Petrov 2012), the three-step and gamma models fitted the data equally well at all levels of linkage (Figure S1C, bottom).
Second, we examined whether positive selection could produce a signature of a bimodal DFE. We simulated a gamma DFE with a = 0.05 and b = 0.5 for negatively selected mutations and a single spike for positively selected mutations with selection strength Nesa=10 and probability density pa = 0.03, which is similar to what has been observed for protein-coding genes in D. melanogaster (Schneider et al. 2011). We observed very similar results to those we obtained by assuming only negative selection (Figure S1D). Therefore fitting a demographic model to the neutral sites is essential for controlling the effects of linkage in producing spurious evidence of a bimodal DFE.
Third, we investigated whether linkage could affect our power to detect a multimodal DFE with the nonparametric steps model. We simulated a bimodal two-spike DFE with Nes1 = 0, Nes2 = 10 with probability densities p1 = 0.2, p2 = 0.8, respectively. We found that strong linkage can reduce the ΔlogL between three-step and gamma models (Figure S1E, top). The results were similar when we also fitted a demographic model of a step change to the neutral sites (Figure S1E, bottom). Therefore, a true bimodal DFE would be harder to detect in genomic regions that exhibit strong linkage.
Analysis of protein polymorphism data sets from D. melanogaster and M. m. castaneus
To account for demographic effects on our inferences of selection we fitted a step change in population size to synonymous sites. The step-change model inferred a population expansion for both D. melanogaster and M. m. castaneus (Table S3) and fitted very well to the data (Figure S2). We then fitted the lognormal, gamma, beta, variable spike, variable step, and fixed six-spike models to nonsynonymous sites. For each data set, we computed ΔlogL, ΔAIC scores, the proportions of mutations falling into four Nes ranges (0–1, 1–10, 10–100, >100), , and (Table 3).
Table 3. Results from the analysis of protein-coding loci in D. melanogaster and M. m. castaneus.
| Species | Model | Δ log L | ΔAIC |
Nes |
α | ωa | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| [0-1) | [1-10) | [10-100) | ≥100 | ||||||||
| D. melanogaster | Lognormal | −0.8 | 0.0 | 0.044 | 0.064 | 0.11 | 0.78 | 1359.2 | 0.050 | 0.62 | 0.082 |
| Gamma | −3.3 | −5.1 | 0.049 | 0.055 | 0.12 | 0.78 | 1624.1 | 0.054 | 0.59 | 0.079 | |
| Beta | −94.2 | −187.0 | 0.064 | 0.025 | 0.043 | 0.87 | 94.6 | 0.066 | 0.50 | 0.067 | |
| Best spike (3) | 0.0 | −4.5 | 0.063 | 0.00 | 0.10 | 0.84 | 275.2 | 0.063 | 0.52 | 0.069 | |
| Best step (2) | −3.2 | −7.0 | 0.023 | 0.097 | 0.058 | 0.82 | 289.4 | 0.039 | 0.70 | 0.10 | |
| six-fixed spikes | −72.3 | −144.6 | 0.070 | 0.00 | 0.048 | 0.88 | 96.8 | 0.070 | 0.47 | 0.063 | |
| M. m. castaneus | Lognormal | −23.9 | −41.8 | 0.17 | 0.052 | 0.061 | 0.72 | 1298.9 | 0.16 | 0.30 | 0.070 |
| Gamma | −21.2 | −36.4 | 0.17 | 0.050 | 0.065 | 0.71 | 1840.1 | 0.16 | 0.29 | 0.069 | |
| Beta | −4.4 | −2.9 | 0.18 | 0.016 | 0.022 | 0.78 | 141.2 | 0.18 | 0.22 | 0.052 | |
| Best spike (3) | 0.0 | 0.0 | 0.19 | 0.00 | 0.12 | 0.69 | 755.4 | 0.19 | 0.20 | 0.047 | |
| Best step (2) | −2.8 | −1.6 | 0.18 | 0.0098 | 0.10 | 0.71 | 237.4 | 0.19 | 0.20 | 0.047 | |
| Six-fixed spikes | −2.9 | −5.8 | 0.19 | 0.0053 | 0.02 | 0.78 | 142.6 | 0.19 | 0.20 | 0.046 | |
Log-likelihood and AIC score differences from the highest scoring model, estimated proportion of mutations falling into four Nes ranges, estimated mean effects of a new mutation (), estimated mean probability of fixation of a new mutation (), and estimates of α and ωa are shown. Only results for the best-fitting spike and step models, based on the AIC criterion, are shown.
For D. melanogaster, we found that the best-fitting model according to the AIC citerion was the lognormal model, the gamma model having a slightly worse fit (ΔAIC from the lognormal was −5.1 units; Table 3). However, the estimated proportion of mutations in the examined Nes ranges, and , were very similar between these two models (Table 3). All models estimate that ∼2–7% of new mutations are nearly neutral (Nes 0–1), a further ∼4–20% are moderately to strongly deleterious (Nes 1–100), and ∼80–90% are very strongly deleterious (Nes >100). The beta and six-fixed spike models gave a substantially poorer fit than the lognormal model (ΔAIC to lognormal was −187 units; Table 3). The main discernible difference was a ∼10 times lower estimated for the beta and fixed six-spikes models than the lognormal model. The beta and fixed six-spike models do not allow selection strength Nes> Ne and their poor fit may be a consequence of a substantial proportion of mutational effects lying in that range.
For M. m. castaneus, the best-fitting model according to the AIC criterion was the three-spike model (Table 3). The estimated parameter values were Nes1 = 2.3 × 10−12, Nes2 = 16.4, Nes3 = 1056, with probability densities p1 = 0.19, p2 = 0.12, p3 = 0.69, respectively (Table S3). The fixed six-spike, two-step, and beta models fitted only slightly worse than the three-spike model, while the lognormal and gamma models had substantially worse fits (Table 3). The parameter estimates of the three-spike model together with the good fit of the beta model support a bimodal DFE in M. m. castaneus. The DFE is inferred to have a peak at near neutrality (Nes 0–1) of density ∼20%, and another peak at very strongly deleterious to lethal effects (Nes > 100) with density ∼70% (Table 3). Intermediate effects (Nes 1–100) are inferred to have a density of ∼10% (Table 3).
The average fixation probability of a new deleterious mutation () is an important quantity, since it can be used to estimate the fraction of adaptive substitutions between two species (Eyre-Walker and Keightley 2009). We calculated α and ωa (Equations 10 and 11) by using the estimated for each model (Table 3). For D. melanogaster, we obtained values of α in the range 0.47–0.7 and ωa 0.063–0.1 from the different models (Table 3). For M. m. castaneus, the lognormal and the gamma models gave slightly lower estimates for and therefore higher estimates for α and ωa (0.30 and 0.070, respectively; Table 3) than the best-fitting three-spike model (0.20 and 0.047, respectively; Table 3).
Discussion
In this study, we have examined the performance of several models incorporating parametric and nonparametric distributions for inferring the properties of the DFE. Since the true DFE is of unknown complexity and can have multiple modes, our purpose was to examine the performance of the different models when the true DFE was unimodal, bimodal, or multimodal. We investigated parametric distributions, including the unimodal lognormal and gamma distributions, which are widely used to model the DFE, and the beta distribution, which can also take a bimodal shape. We also examined the performance of custom nonparametric models, including discretized distributions, where the selection coefficients are modeled as point masses, or uniform distributions, that are either variable or fixed. Spike or step models with two or more classes of effects performed almost as well as the gamma model for cases in which the true DFE was a gamma distribution. When the true DFE was a bimodal beta distribution, we found that the lognormal and gamma models fitted poorly and produced inaccurate estimates of , , and the density in several Nes ranges, most notably mutations with Nes > 100. When we simulated a more complex DFE, the biases affecting estimates of and from the lognormal and gamma models were not as pronounced. Accuracy in estimating and seems to depend mostly on the density of the extreme tails of the DFE, irrespectively of its complexity. In our simulations, we frequently observed that a particular model could have a good overall fit, but perform relatively poorly for parts of the DFE that are crucial for estimating or . For example, we consistently observed that was not estimated with high accuracy if the models fitted were different from that simulated. Presumably, the SFS contains limited information about mutations with very small selective effects in the Nes range 0–1 implying that estimation of strongly depends on the properties of the distribution assumed. Since can be used for calculating the proportion of adaptive substitutions (α) and the rate of adaptive evolution (ωa), underestimation of would lead to overestimation of α and ωa (and vice versa). When we examined a series of bimodal DFEs in which we varied the locations and densities of the two modes of the DFE, we observed substantial underestimation of by the gamma model for cases where one mode of the DFE was at Nes = 0 with density <30% and the other mode was at a weakly to moderately deleterious effect with density >70%. Therefore, if the true DFE is bimodal, underestimation of by the gamma model would be expected for genomic regions in which most of the sites are under selection, such as protein-coding genes or conserved noncoding elements, but not for genomic regions in which most of the sites are evolving neutrally such as UTRs and introns.
We also applied the parametric and nonparametric models to infer the DFE for amino acid-changing mutations in D. melanogaster and the house mouse M. m. castaneus, based on data from several thousand autosomal protein-coding genes. In D. melanogaster, we found that the lognormal model gave the best fit to the data, a result that is consistent with a previous study (Loewe and Charlesworth 2006). The estimate for was 1360 by the best-fitting lognormal model. This estimate is similar to estimates obtained from a smaller data set of Shapiro et al. (2007) analyzed by Keightley and Eyre-Walker (2007). If we assume that the DFE for amino acid-changing mutations in Drosophila is lognormal and that Ne is of the order 0.7 × 106 (Halligan et al. 2010), then the mean selection coefficient of new deleterious amino-acid changing mutations for D. melanogaster is of the order 2 × 10−3. We also estimate that α and ωa are 0.62 and 0.082, respectively. Reassuringly, the choice of the distribution to model the DFE does not strongly affect and consequently α and ωa. Regardless of the model assumed, α > 0.47 and ωa > 0.063, supporting the presence of highly effective positive selection in D. melanogaster, as several other researchers have inferred (Sella et al. 2009).
In M. m. castaneus, we found that a three-spike model gave the best fit to the SFS. The beta distribution also fitted almost as well as the three-step model, while the lognormal and gamma models gave substantially poorer fits. These observations suggest that the DFE for new deleterious amino-acid changing mutations in M. m. castaneus is bimodal, with 20% of the distribution’s density attributable to weakly deleterious mutations (Nes 0–1) and 70% to very strongly deleterious mutations (Nes > 100). We also obtained estimates for α and ωa, of 0.20 and 0.046, respectively. We observed differences among the estimates of α and ωa between different models, the lognormal and gamma models producing higher estimates than the best-fitting three-spike and beta models. Underestimation of by the gamma and lognormal models was observed in simulations in which the true DFE was a bimodal beta of similar properties to the inferred DFE for M. m. castaneus. It seems likely that fitting a lognormal or a gamma distribution to the DFE leads to overestimation of α and ωa. Halligan et al. (2010), who fitted a gamma distribution to a small gene sample from M. m. castaneus, obtained estimates for α larger (α = 0.37 for non-CpG-prone sites and using rat as outgroup) than those obtained in the present study.
There are some potential caveats to our study. First, our models do not incorporate genetic linkage in the inference method. We investigated whether linkage and background or/and positive selection can affect inferences from the models tested and found that under moderate linkage, spurious evidence for multimodality can be produced (assessed by a better fit of spike/step models to data than unimodal distributions). We can account for the effects of linkage, however, by fitting a simple demographic model to the neutral class of sites (as is also suggested by Messer and Petrov 2012). Second, our two-epoch demographic model is not sufficient for more complex demographic histories, such as bottlenecks. Assuming a more realistic population history of a long-lasting bottleneck followed by a rapid expansion, we found that the spike/step models can overfit the data, producing spurious evidence for multimodality of the DFE. Therefore, when inferring the DFE using spike/step models it is necessary to fit a three-epoch model to data from populations that have experienced bottlenecks. A three-epoch model can be incorporated into the inference procedure of our method, but due to computational limitations it was not feasible to investigate its performance in simulations. However, a three-epoch model fitted only slightly better to the folded synonymous SFS for D. melanogaster and M. m. castaneus than a two-epoch model (ΔlogL between the two-epoch and three-epoch model was 3 and 7, respectively; result not shown). Therefore, we do not expect a substantial effect of the demographic history on our inferences of selection in these populations. Third, the fact that we infer a bimodal DFE for M. m. castaneus does not necessarily rule out a more complex DFE. It appears that there is limited information in the SFS, and our simulations indicate that at best three modes can be inferred, even for very large data sets. It is likely that the precise shape of the DFE cannot accurately be determined based on SFS data alone, as has been shown for the demographic history of a population (Myers et al. 2008).
In conclusion, we have shown that nonparametric discretized models, such as the spike and step models, can perform as well or better than parametric distributions, such as the gamma. They produce accurate estimates of the important parameters, notably and , and increasing the numbers of alleles sequenced will increase their performance. These models can also help in determining whether the DFE has multiple modes. We note that we have examined only one particular case of each type of distribution (unimodal, bimodal, multimodal) and we do not consider the particular simulated examples as representatives of all possible unimodal, bimodal, and multimodal distributions. However, our results are relevant in showing the limitations of fitting relatively inflexible distributions, such as the gamma distribution to the DFE, and illustrate the advantages of using a more general model such as the spike or step model to infer the DFE. Fitting the spike or the step model with different numbers of classes of mutational effects can be informative about the complexity of the DFE and identifying which Nes ranges we have little information on.
Acknowledgments
We thank Dan Halligan, Adam Eyre-Walker, Brian Charlesworth, Laurence Loewe, and two anonymous reviewers for helpful comments on earlier versions of the manuscript and for helpful discussions. We thank Jose Campos for compiling the DPGP2 protein-coding data. We acknowledge funding from grants from the Biotechnology and Biological Sciences Research Council (BBSRC) and the Wellcome Trust. A.K. is funded by a BBSRC postgraduate studentship.
Footnotes
Communicating editor: S. I. Wright
Literature Cited
- Arndt P. F., Petrov D. A., Hwa T., 2003. Distinct changes of genomic biases in nucleotide substitution at the time of mammalian radiation. Mol. Biol. Evol. 20: 1887–1896. [DOI] [PubMed] [Google Scholar]
- Boyko A. R., Williamson S. H., Indap A. R., Degenhardt J. D., Hernandez R. D., et al. , 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, J. L., K. Zeng, D. J. Parker, B. Charlesworth, and P. R. Haddrill, 2012 Codon usage bias and effective population sizes on the X chromosome vs. the autosomes in Drosophila melanogaster Mol. Biol. Evol., http://mbe.oxfordjournals.org/content/early/2013/01/20/molbev.mss222. [DOI] [PMC free article] [PubMed]
- Charlesworth B., 1996. The good fairy godmother of evolutionary genetics. Curr. Biol. 6: 220. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 1979. Mathematical Population Genetics. Springer-Verlag, Berlin. [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2009. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol. 26: 2097–2108. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., Smith N. G. C., Gaffney D., 2002. Quantifying the slightly deleterious mutation model of molecular evolution. Mol. Biol. Evol. 19: 2142–2149. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Woolfit M., Phelps T., 2006. The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics 173: 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford. [Google Scholar]
- Foxe J. P., Dar V.-N., Zheng H., Nordborg M., Gaut B. S., et al. , 2008. Selection on amino acid substitutions in Arabidopsis. Mol. Biol. Evol. 25: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann T. I., Song B.-H., Windsor A. J., Mitchell-Olds T., Dixon C. J., et al. , 2010. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 27: 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan D. L., Oliver F., Eyre-Walker A., Harr B., Keightley P. D., 2010. Evidence for pervasive adaptive protein evolution in wild mice. PLoS Genet. 6: e1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., 2012. Rates and fitness consequences of new mutations in humans. Genetics 190: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177: 2251–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2010. What can we learn about the distribution of fitness effects of new mutations from DNA sequence data? Philos. Trans. R. Soc. B. Biol. Sci. 365: 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1957. Some problems of stochastic processes in genetics. Ann. Math. Stat., 882–901. [Google Scholar]
- Kimura M., 1962. On the probability of fixation of mutant genes in a population. Genetics 47: 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A., Oliver F., Halligan D. L., Keightley P. D., 2011. Positive and negative selection on noncoding DNA close toprotein-coding genes in wild house mice. Mol. Biol. Evol. 28: 1183–1191. [DOI] [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., 2006. Inferring the distribution of mutational effects on fitness in Drosophila. Biol. Lett. 2: 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., Bartolomé C., Nöel V., 2006. Estimating selection on nonsynonymous mutations. Genetics 172: 1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer, P. W., 2013 SLiM: simulating evolution with selection and linkage. arXiv:1301.3109 http://arxiv.org/abs/1301.3109. [DOI] [PMC free article] [PubMed]
- Messer, P. W., and D. A. Petrov, 2012 The McDonald–Kreitman test and its extensions under frequent adaptation: problems and solutions. arXiv:1211.0060 http://arxiv.org/abs/1211.0060.
- Myers S., Fefferman C., Patterson N., 2008. Can one learn history from the allelic spectrum? Theor. Popul. Biol. 73: 342–348. [DOI] [PubMed] [Google Scholar]
- Nelder J. A., Mead R., 1965. A Simplex method for function minimization. Comput. J. 7: 308–313. [Google Scholar]
- Nielsen R., Yang Z., 2003. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol. Biol. Evol. 20: 1231–1239. [DOI] [PubMed] [Google Scholar]
- Piganeau G., Eyre-Walker A., 2003. Estimating the distribution of fitness effects from DNA sequence data: implications for the molecular clock. Proc. Natl. Acad. Sci. USA 100: 10335–10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., et al. , 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8: e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S., Hartl D. L., 1992. Population genetics of polymorphism and divergence. Genetics 132: 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S., Kulathinal R., Bustamante C., Hartl D., 2003. Bayesian analysis suggests that most amino acid replacements in Drosophila are driven by positive selection. J. Mol. Evol. 57: S154–S164. [DOI] [PubMed] [Google Scholar]
- Schneider A., Charlesworth B., Eyre-Walker A., Keightley P. D., 2011. A method for inferring the rate of occurrence and fitness effects of advantageous mutations. Genetics 189: 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G., Petrov D. A., Przeworski M., Andolfatto P., 2009. Pervasive natural selection in the Drosophila genome? PLoS Genet. 5: e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Huang W., Zhang C., Hubisz M. J., Lu J., et al. , 2007. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. U.S.A 104: 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Foxe J. P., Hazzouri K. M., Wright S. I., 2010. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol. Biol. Evol. 27: 1813–1821. [DOI] [PubMed] [Google Scholar]
- Williamson S. H., Hernandez R., Fledel-Alon A., Zhu L., Nielsen R., et al. , 2005. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl. Acad. Sci. USA 102: 7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. J., Hernandez R. D., Andolfatto P., Przeworski M., 2011. A population genetics–phylogenetics approach to inferring natural selection in coding sequences. PLoS Genet. 7: e1002395. [DOI] [PMC free article] [PubMed] [Google Scholar]