Abstract
As the molecular marker density grows, there is a strong need in both genome-wide association studies and genomic selection to fit models with a large number of parameters. Here we present a computationally efficient generalized ridge regression (RR) algorithm for situations in which the number of parameters largely exceeds the number of observations. The computationally demanding parts of the method depend mainly on the number of observations and not the number of parameters. The algorithm was implemented in the R package bigRR based on the previously developed package hglm. Using such an approach, a heteroscedastic effects model (HEM) was also developed, implemented, and tested. The efficiency for different data sizes were evaluated via simulation. The method was tested for a bacteria-hypersensitive trait in a publicly available Arabidopsis data set including 84 inbred lines and 216,130 SNPs. The computation of all the SNP effects required <10 sec using a single 2.7-GHz core. The advantage in run time makes permutation test feasible for such a whole-genome model, so that a genome-wide significance threshold can be obtained. HEM was found to be more robust than ordinary RR (a.k.a. SNP-best linear unbiased prediction) in terms of QTL mapping, because SNP-specific shrinkage was applied instead of a common shrinkage. The proposed algorithm was also assessed for genomic evaluation and was shown to give better predictions than ordinary RR.
Keywords: generalized ridge regression, genome-wide association study, heteroscedastic effects model, linear mixed model, GenPred, Shared data resources
HIGH-dimensional problems are increasing in importance in genetics, computational biology, and other fields of research where technological developments have greatly facilitated the collection of data (Hastie et al. 2009). In genome-wide association studies (GWAS) and genomic selection (GS), the number of observations n is generally in the order of hundreds/thousands whereas the number of marker effects to be fitted p is in the order of hundreds of thousands. This is a rather extreme p ≫ n problem, and the methods developed for analyses of the data need to be computationally feasible. At the same time the models fitted should be flexible enough to capture the important genetic effects that are often quite small (Hayes and Goddard 2001).
Methodologies regarding high-dimensional genomic data focus on both detection and prediction purposes. There is currently a trend that GWAS and GS could potentially apply the same framework of models. Such models fit the whole genome based on penalized likelihood or Bayesian shrinkage estimation (see the review by de los Campos et al. 2013). Ordinary GWAS usually avoids high-dimensional models and turns the problem into multiple testing instead (e.g., the review by Kingsmore et al. 2008). The tests of all the SNPs (single nucleotide polymorphisms) are dismembered. Such routine sacrifices both detective and predictive power. Using detected QTL (quantitative trait loci that are genome-wide significant), the prediction can be rather poor, which led to the insignificant application of marker-assisted selection (MAS) (Dekkers 2004). GS, however, has been practically useful by incorporating a large amount of small genetic effects unmappable from GWAS or QTL analysis. There are a number of whole-genome models where not only the individual predictors (breeding values) but also the SNP effects can be estimated, e.g., SNP–best linear unbiased prediction (BLUP) and different kinds of Bayesian models (e.g., Meuwissen et al. 2001; Xu 2003; Yi and Xu 2008; Gianola et al. 2009; Habier et al. 2011). The whole genome models are powerful; nevertheless, there are problems that limit its wide usage: (1) computation for these models including all the SNPs can be intensive, whereas efficiency is required in practice so that prediction can be obtained in early life of the individuals (de los Campos et al. 2013); (2) fitting large-p small-n models requires variable selection or shrinkage estimation, and the significant threshold for the shrinkage estimates of SNP effects is difficult to specify, which is an issue that limits the usage of such models in gene mapping; and (3) the fitting of Bayesian models is performed using randomization/simulations, where in application, mixing of the Markov chain Monte Carlo (MCMC) algorithm can become poor in case of high-dimensional models.
Linear mixed models (LMM) have been proposed for GS (SNP-BLUP; Meuwissen et al. 2001) and ridge regression (RR) for GWAS (Malo et al. 2008). LMMs and RR are fundamentally the same since they fit a penalized likelihood using a quadratic penalty function (see the Appendix for more details). It is well established (Hastie et al. 2009) that RR can be fitted for p ≫ n in a computationally efficient way using singular-value decomposition (SVD) of the design matrix, which, for instance, has been applied to expression arrays in genetics (Hastie and Tibshirani 2004). However, this approach assumes that the RR shrinkage parameter is constant for all p fitted parameters. In generalized RR the shrinkage parameter may vary between the parameters (Hoerl and Kennard 1970a,b). In both multilocus GWAS and GS, it is not reasonable to assume that shrinkage should be constant for all fitted SNP effects over the entire genome. This is because neither the gene effects are normally distributed nor are most markers linked to any functional gene (Meuwissen et al. 2001). To allow SNP-specific shrinkage, the previously mentioned Bayesian methods were developed.
There is a need for a method that is fast (efficient to perform), testable (can produce a genome-wide significance threshold for association study), deterministic (the same estimates are easy to replicate), and flexible (SNP-specific shrinkage can be easily applied). The aim of this article is to develop such a generalized RR method, which will be referred to as the heteroscedastic effects model (HEM), for p ≫ n high-dimensional problems, based on LMM theory. HEM approximates a previously proposed method (Rönnegård and Lee 2010; Shen et al. 2011) that was based on double hierarchical generalized linear models (DHGLM; Lee and Nelder 2006), but with a tremendous increase in computational speed for p ≫ n problems. An important contribution of the theory presented is a fast transformation of hat values (leverages) and prediction error variances of the random effects. The method has been implemented in the R (R Development Core Team 2010) package bigRR (available at https://r-forge.r-project.org/R/?group_id=1301).
Methods and Materials
Statistical models
Using Henderson’s mixed model equation:
We start by introducing the normal RR as a LMM. The theoretical basis of the connection between RR and LMM is given in the Appendix. The SNP effects are estimated as random effects, i.e., so-called SNP–BLUP. We use the terms RR and SNP–BLUP interchangeably in this article. Given a phenotype vector y for n individuals, fixed effects data X, and the data for p SNPs along the genome Z, the normal LMM for SNP–BLUP can be written as
| (1) |
where , , β is the vector of fixed effects, and b is the vector of random SNP effects. The matrix Z has p columns for the SNPs where each column is usually coded as 0, 1, and 2, for the homozygote aa, the heterozygote Aa, and the other homozygote AA, respectively. However, here, we standardize the coding for Z based on VanRaden (2008) using the allele frequencies. This is essential in RR problems since the sizes of the estimated effects need to be comparable. Although the models are introduced in the simple normal LMM notation, the method is developed for generalized distributions of phenotypes (see also Fitting algorithm and the Appendix).
It is well known that the fixed effects β and random effects b can be estimated jointly via Henderson’s mixed model equation (MME; Henderson 1953)
| (2) |
where , determined by the variance component estimators, is the shrinkage parameter for the random SNP effects. λ is analogous to the one in the penalized likelihood for RR. In terms of estimating SNP effects for QTL mapping, such an MME for RR is not appropriate because the same magnitude of shrinkage is applied to all the SNPs (Xu 2003). Hence, the markers are regarded a priori with no difference. Since most of the loci in the genome are supposed to contribute little to the observed phenotype, those SNPs should be penalized more in the analysis. This is one of the fundamental ideas from which the current Bayesian methods are developed (e.g., Meuwissen et al. 2001; Xu 2003).
From (2), it is clear that to obtain different shrinkage for different SNPs, the λI part should be replaced so that the p numbers on the diagonal are not identical. An essential question here is how much shrinkage should be given to each SNP. We propose a generalized RR solution to this problem, which is presented as the following HEM. We use the MME and fit a generalized RR after the ordinary RR in (2).
| (3) |
where λ is a vector of p shrinkage parameters with its jth element . The SNP-specific variance component is calculated as
| (4) |
where for the jth SNP, is the BLUP from (2), and hjj, known as the hat value, is the (n + j)th diagonal element of the hat matrix H = T(T′T)−1T′, where
| (5) |
Such a quantity (4) is useful because it (1) directly tells how much shrinkage should be given to each SNP and (2) makes the entire procedure deterministic and repeatable. This simple way of setting the shrinkage parameters is an approximation of previously established theory for double hierarchical generalized linear models (Lee and Nelder 2006) applied in Rönnegård and Lee (2010) and Shen et al. (2011), where bj depended on a second-layer model including random effects and was estimated iteratively until convergence. Using (4) the shrinkage for each SNP in HEM is computed directly without iteration.
Transformation via the animal model:
Stranden and Garrick (2009) showed that the computations for an animal model including a genomic relationship matrix is equivalent to fitting an LMM including random SNP effects. Below we exploit this fact to derive the algorithm for HEM. A major contribution of the theory below is the derivation of a simple equation to compute the hat values for the SNP effects from the hat values for the animal effects (in step 5 of Fitting algorithm below). It should also be noted that the generalized RR part of the HEM algorithm does not use singular-value decomposition as proposed by Hastie and Tibshirani (2004) and Hastie et al. (2009) and described in the Appendix, but rather uses transformation between equivalent models as described below.
From (3) and (4), the generalized RR method, HEM, is quite easy to describe mathematically. However, to obtain estimated effects for all the SNPs, the matrix Z′Z + diag(λ) (size p × p) is too large to invert. So we need to make the equations computationally simple. This can be done by connecting (3) to an animal model. Let us define G = ZZ′, which is a matrix representing genomic kinship that indicates the relatedness between individuals. The animal model
| (6) |
contains as random effects for n individuals. The size of the MME for such an animal model is much smaller than (2), i.e.,
| (7) |
Because of the mathematical equivalence, the λ in (7) is identical to that in (2). In fact, there is no need to directly fit the MME (2) or (3) because all the information that links the estimated individual effects to the SNP effects are contained in the single genotype matrix Z. After fitting the animal model via (7), one can show that the following transformation holds (e.g., Pawitan 2001, p. 446),
| (8) |
where only the matrices Z′ (size p × n) and G−1 (size n × n) are required, given that G is full ranked. Since n ≪ p, the MME (2) and HEM (3) can be solved a lot faster by the “animal model (7) + transformation (8)” procedure. For HEM, the hat value for each SNP is required in the MME (3), and we show that fortunately, a similar transformation can be applied for transforming the hat values of the animal effects to the SNP effects as well (see Fitting algorithm and the Appendix). Besides the efficiency, by avoiding huge matrices, a significant amount of memory can be saved so that almost any large number of SNPs can be loaded simultaneously.
To evaluate the run time for fitting a linear mixed model or RR using our algorithm, we simulated a standard-normally distributed phenotype with sample sizes varied from 100 to 1000. Marker genotypes were also simulated and the number of markers varied from 10K to 1M.
Fitting algorithm
Below we present the fitting algorithm for HEM. Steps 1–4 fit SNP–BLUP and steps 5–8 fit a generalized RR. The algorithm also includes a Cholesky decomposition of the genomic relationship matrix G to simplify the computations and the transformation of hat values (in step 5).
Given a phenotype vector y (size n × 1) that belongs to any GLM (generalized linear model) family, e.g., binary, poisson, gamma, etc., fixed effects design matrix X (size n × k), the SNP genotype matrix Z (size n × p), the SNP–BLUP (RR), and HEM (generalized RR) can be computed as follows:
Calculate G = ZZ′, its Cholesky decomposition L s.t. LL′ = G, and its inverse G−1.
Fit a GLMM with response y, fixed effects X and random effects design matrix L. Because of mathematical equivalence, this fits the animal model (6) as a GLMM with correlated random effects.
From step 2, store the estimated variance components , , and the animal effects . Calculate .
Transform back to the SNP effects .
- Define
and divide the inverse of Cv into blocks(9)
Define a transformation matrix M = Z′G−1L. Calculate the hat value for each random SNP effect as(10)
where Mj is the jth row of the transformation matrix M.(11) - Define a diagonal matrix W with each diagonal element
and update G to be G* = ZWZ′. Calculate G*−1, and L* s.t. .(12) Fit a GLMM with response y, fixed effects X, and random effects design matrix L*.
From step 7, transform the updated individual effects back to the SNP effects .
In this algorithm, GLMMs are estimated based on penalized quasi-likelihood (PQL) for MME (see R package hglm and its algorithm in Rönnegård et al. 2010). To be comparative to MME for normal LMM, the notation is used in the algorithm even for GLMM to denote the residual dispersion parameter. Theoretical details about the transformations are given in the Appendix.
Randomization test
Specifying a significance threshold for any whole genome model has been a challenging problem. The predictors in LMM or GLMM for the random effects (i.e., BLUP) have “prediction errors” (e.g., Pawitan 2001), which could be used to construct “t-like” statistics. But this is properly applicable only when the number of random effects predictors is small, namely, when shrinkage does not affect much of the test statistic distribution since the random effects estimates are not so different from those if all the effects are estimated as fixed. But this is not proper anymore when the number of explanatory variables or genetic markers is much more than the number of individuals, because the estimated effects are too much biased from their real genetic effects (Zeng 1993; Rodolphe and Lefort 1993). So when a whole genome of markers is fitted together, the model ends up with too much shrinkage to make the t-distribution hold. Hence, current Bayesian methods (e.g., Xu 2003) just set up an empirical LOD score threshold (e.g., Che and Xu 2012) using the suggestions by Kidd and Ott (1984) and Risch (1991). Nevertheless, the genome-wide significance test can actually be practically important. Here, randomization/permutation is a solution if the computation is not too intensive for fitting all the markers. Since the HEM algorithm proposed in this article is computationally efficient, a genome-wide significance threshold can be determined by randomization test.
In the analysis of the Arabidopsis thaliana GWAS data using HEM, the permutation test was performed to determine a 5% genome-wide significance threshold for QTL detection, where the phenotype was permuted 1000 times, and the 95% quantile of the maximum effects was calculated as the threshold.
Data
We applied HEM on three data sets. Using HEM, we searched for significant SNPs in a publicly available A. thaliana GWAS data set. In the other two data sets the predictive power of HEM in GS was assessed.
A. thaliana GWAS data:
Atwell et al. (2010) performed GWA studies for 107 phenotypes of A. thaliana and successfully detected a set of candidate genes. Using the heteroscedastic effects model, we analyzed one defense-related binary trait out of their 107 published phenotypes: hypersensitive response to the bacterial elicitor AvrRpm1. The reason for choosing this trait is because it is under regulation of a known candidate gene RPM1 and so we can validate our HEM method in terms of QTL detection. A total of 84 ecotypes were phenotyped (28 controls and 56 cases). The genotype data are from a 250K SNP chip including 216 130 available SNPs (http://arabidopsis.usc.edu).
GSA common simulated data:
To compare different newly developed genomic evaluation methods, the Genetics Society of America (GSA) provides several common data sets for authors to analyze and report their results (Hickey and Gorjanc 2012). We chose the simulated livestock data structure to assess our method.
The total number of segregating sites across the genome was approximately 1.67 million. A random sample of 60,000 segregating sites was selected from the sequence to be used as SNPs on a 60K SNP array. In addition, a set of 9000 segregating sites were randomly selected from the sequence to be used as candidate QTL in two different ways: (1) a randomly sampled set, and (2) a randomly sampled set with the restriction that their minor allele frequencies (MAFs) should not exceed 0.30. Four different traits were simulated, assuming an additive genetic model. The first pair of traits was generated using the 9000 unrestricted QTL. For the first trait (PolyUnres), the allele substitution effect at each QTL was sampled from a standard normal distribution. For the second trait (GammaUnres) a random subset of 900 of the candidate QTL was selected with allele substitution effects sampled from a gamma distribution with a shape parameter of 0.4, scale parameter of 1.66 (Meuwissen et al. 2001), and a 50% chance of being positive or negative. The second pair of traits (PolyRes and GammaRes) was generated in the same way as the first pair except that the candidate QTL have the restriction that their MAF not be >0.30. Phenotypes with a heritability of 0.25 were generated for each trait.
Training and validation subsets of the data were extracted for training and validation. The training set comprised the 2000 individuals in generations 4 and 5. The validation set comprised 1500 individuals sampled at random from generation 6, 8, and 10 (500 individuals from each generation). We fit a whole-genome model using HEM and compare the prediction performance in the validation data set.
QTLMAS data:
The third data set used in this article was simulated for the 14th QTLMAS workshop (http://jay.up.poznan.pl/qtlmas2010/; Szydlowski and Paczyńska 2011). A pedigree consisting of 3226 individuals in five generations (F0–F4) was simulated, where F0 contained 5 males and 15 females. Each female mated once and gave birth to around 30 progeny. Two traits were simulated, where one was quantitative (QT), and the other was binary (BT). Young individuals (F4 generation, individuals 2327–3226) had no phenotypic records. The genome was about 500-Mb long, consisting of five chromosomes, each of which contained about 100 Mb. Each individual was genotyped for 10,031 biallelic SNPs that densely distributed along the genome. Regarding recombination rate, 1 cM was assumed to be 1 Mb; therefore, the size of the genome is about 500 cM.
Thirty-seven QTL were simulated along the genome, and no QTL existed on chromosome 5 (Table 1). All the QTL controlled QT, including 30 additive loci, 2 epistatic pairs, and 3 imprinted loci. Of the 30 additive QTL, 22 also controlled BT, namely that pleiotropic effects existed for the two traits. QT was mainly controlled by additive QTL 14 and 17, as well as the two epistatic pairs. BT was mainly controlled by additive QTL 14. Due to epistasis and imprinting, QT had a more complicated genetic architecture than BT. We have published (Shen et al. 2011) our previous analysis of this data set using a DHGLM (Lee and Nelder 2006). Considering HEM as a simple and efficient substitution of DHGLM, we reanalyzed the data by fitting all the markers and compared the results with the previous results.
Table 1. Genetic models of simulated quantitative trait for the QTLMAS 2010 data (Szydlowski and Paczyska 2011).
| QTL | Chra | SNPb | Distc (bp) | Freqd | Adde | QTL Variance | Type |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 152 | 788 | 0.45 | 1.93 | 1.84 | Additive |
| 2 | 1 | 960 | 27540 | 0.64 | −1.56 | 1.13 | Additive |
| 3 | 1 | 1106 | 11564 | 0.67 | −1.56 | 1.09 | Additive |
| 4 | 1 | 1226 | 5083 | 0.30 | −1.68 | 1.19 | Additive |
| 5 | 2 | 2036 | 1852 | 0.84 | −1.92 | 0.97 | Additive |
| 6 | 2 | 2675 | 17414 | 0.38 | 1.02 | 0.48 | Additive |
| 7 | 2 | 3114 | 128504 | 0.14 | 1.69 | 0.69 | Additive |
| 8 | 2 | 3414 | 111127 | 0.47 | −1.32 | 0.87 | Additive |
| 9 | 2 | 3534 | 0 | 0.66 | 0.25 | 0.03 | Additive |
| 10 | 2 | 3553 | 15051 | 0.39 | 0.85 | 0.34 | Additive |
| 11 | 2 | 3946 | 96549 | 0.26 | 1.01 | 0.40 | Additive |
| 12 | 2 | 3959 | 1516 | 0.27 | 1.69 | 1.13 | Additive |
| 13 | 3 | 4318 | 8609 | 0.17 | −1.98 | 1.10 | Additive |
| 14 | 3 | 4483 | 17356 | 0.51 | 3.00 | 4.5 | Additive (major controled) |
| 15 | 3 | 4615 | 44344 | 0.23 | −1.11 | 0.43 | Additive |
| 16 | 3 | 4980 | 5654 | 0.60 | −0.73 | 0.26 | Additive |
| 17 | 3 | 5488 | 0 | 0.53 | 3.00 | 4.49 | Additive (major controled) |
| 18 | 3 | 5616 | 2462 | 0.49 | −0.76 | 0.29 | Additive |
| 19 | 3 | 5722 | 184175 | 0.83 | −0.95 | 0.26 | Additive |
| 20 | 3 | 5858 | 28506 | 0.88 | 1.66 | 0.58 | Additive |
| 21 | 3 | 6022 | 0 | 0.57 | 0.65 | 0.21 | Additive |
| 22 | 4 | 6224 | 45783 | 0.24 | 1.25 | 0.57 | Additive |
| 23 | 4 | 6423 | 4137 | 0.47 | 0.96 | 0.46 | Additive |
| 24 | 4 | 6684 | 40188 | 0.37 | 0.56 | 0.14 | Additive |
| 25 | 4 | 6833 | 650 | 0.53 | −0.31 | 0.05 | Additive |
| 26 | 4 | 6870 | 43162 | 0.28 | 1.55 | 0.96 | Additive |
| 27 | 4 | 6982 | 20468 | 0.6 | 0.93 | 0.42 | Additive |
| 28 | 4 | 7013 | 36740 | 0.57 | −0.22 | 0.02 | Additive |
| 29 | 4 | 7446 | 119 | 0.55 | 0.75 | 0.28 | Additive |
| 30 | 4 | 8024 | 27501 | 0.89 | 1.92 | 0.72 | Additive |
| 31 | 1 | 939 | 0 | 0.54 | 0 | 7.01 | Epistatic (pair 1)f |
| 32 | 1 | 959 | 0 | 0.56 | 0 | Epistatic (pair 1) | |
| 33 | 2 | 2715 | 0 | 0.5 | 0 | 4.18 | Epistatic (pair 2)g |
| 34 | 2 | 2727 | 0 | 0.51 | 0 | Epistatic (pair 2) | |
| 35 | 2 | 3102 | 0 | 0.55 | 0 | 2.16 | Imprintedh |
| 36 | 2 | 3623 | 0 | 0.54 | 0 | 2.20 | Imprintedh |
| 37 | 2 | 3776 | 0 | 0.56 | 0 | 2.17 | Imprintedh |
Chromosome number. No QTL on chromosome 5.
The closest SNP marker index.
Distance from the QTL to the closest SNP marker.
Frequency of allele 1.
Additive effect: half the difference between homozygote means.
Extra effect of each haplotype 1–1: 4.00. Frequency of haplotype 1–1: 0.35.
Extra effect of each haplotype 1–1: 4.00. Frequency of haplotype 1–1: 0.17.
Extra effect of paternal allele 1: 3.0.
Results
Computational efficiency
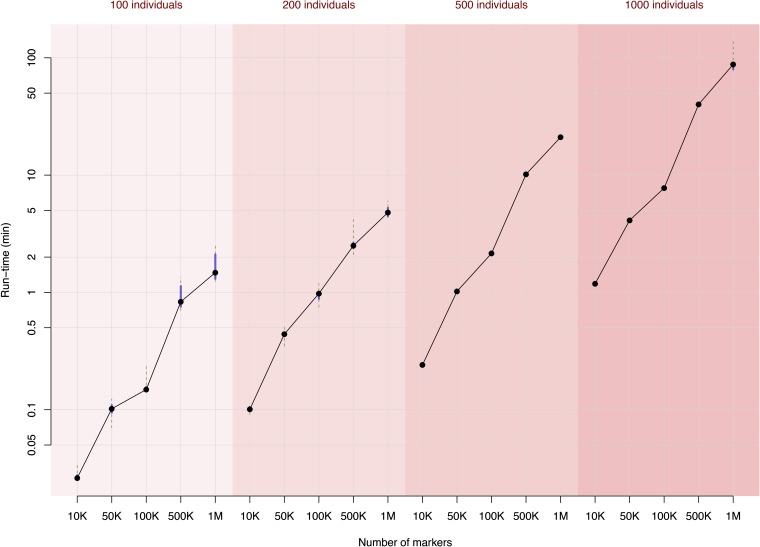
On a single Intel Xeon E5520 2.27-GHz CPU, the computation was fast, especially when the number of individuals was small (Figure 1), since the computation-demanding parts in the algorithm depend mainly on the sample size. For a population with 100 individuals, even when there are 1 million markers, estimation of all the effects along the genome takes <2 min.
Figure 1.
Run-time efficiency of the R package bigRR for different sizes of data. Each column in the figure was evaluated by 12 replicates, where the dot shows the median, the solid line shows the 25–75% quantile interval, and the dashed lines indicate the range from minimum to maximum.
Analysis of the Arabidopsis data
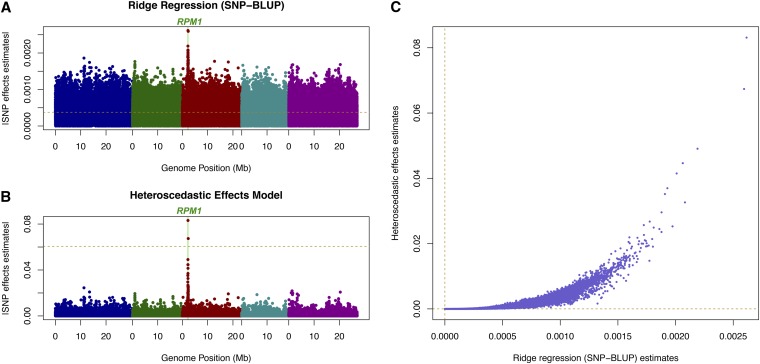
For the 84 individuals and 216,130 informative markers on the Arabidopsis trait AvrRpm1, the shrinkage effect was much stronger for HEM than SNP–BLUP (Figure 2). According to Atwell et al. (2010), this defense-related trait is essentially monogenically controlled by the gene RPM1. The analysis via a whole-genome model should validate such a strong monogenic effect in terms of QTL detection. In Figure 2, 5% genome-wide significance thresholds via permutation tests are provided for both SNP–BLUP and HEM. Here, SNP–BLUP is not appropriate for QTL mapping due to constant shrinkage along the genome (Xu 2003). By allowing different weights on different SNPs, HEM has the property that it shrinks the small effects down toward zero, highlights the QTL effects, and produces reasonable genome-wide significance threshold obtained by permutation testing.
Figure 2.
Estimated SNP effects for the Arabidopsis bacteria-hypersensitive trait AvrRpm1 (Atwell et al. 2010) using (A) ridge regression (SNP–BLUP) and (B) heteroscedastic effects model, which are plotted against each other in C. The horizontal dashed lines in A and B indicate the 5% genome-wide significance threshold from a randomization test using 1000 permutations.
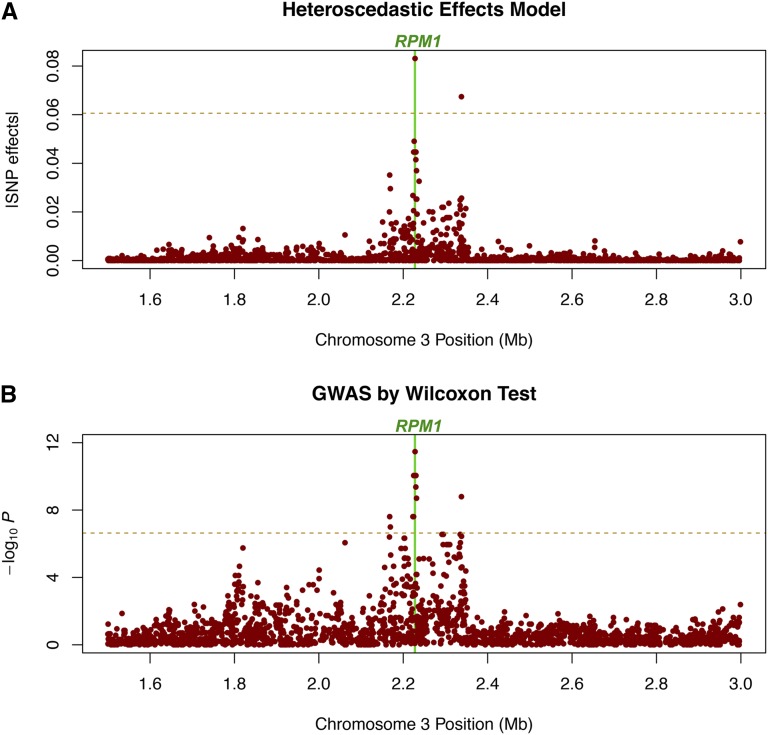
HEM also produces better resolution in mapping the candidate gene. A close-up of the region surrounding the RPM1 gene on chromosome 3 shows that the SNP with the largest −log10 P-value from the Wilcoxon GWAS (Atwell et al. 2010) also has the largest estimate from HEM, whereas the second largest estimate is found ∼0.1 Mb away from RPM1 (Figure 3). Hence, a ranking of the top estimates results in a similar ranking as for the −log10P-values from Atwell et al.’s GWAS, where HEM is better at separating the ranking of SNPs close to each other on the chromosome.
Figure 3.
The significant association peak for the Arabidopsis bacteria-hypersensitive trait AvrRpm1 (Atwell et al. 2010) from (A) heteroscedastic effects model and (B) genome-wide association using Wilcoxon rank-sum test. The window of the candidate gene RPM1 is indicated as a vertical line.
Analysis of the GSA simulated data
HEM is able to fit the entire 60K SNP chip on the GSA simulated data. We analyzed all the 10 replicates of the four simulated traits and performed the prediction using both SNP–BLUP and HEM (Table 2). It is not surprising that HEM is generally better in prediction than SNP–BLUP because of more flexible shrinkage. It is noteworthy that such an advantage in prediction is clearer when the QTL effects are skewed (Gamma) than symmetrically (Normal) distributed. This is because the SNPs that have major genetic effects are highlighted more by the HEM shrinkage compared to SNP–BLUP. This is a good property for HEM since most of the time, one would expect the genes to carry skewed genetic effects (Hayes and Goddard 2001).
Table 2. Summary of breeding value estimation for the 10 replicates of validation samples of the GSA common simulated data set (Hickey and Gorjanc 2012).
| QTL |
Ridge Regression (SNP-BLUP) |
Heteroscedastic Effects Model |
Prediction | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | Restriction | Distribution | COR (SE) | MSE (SE) | COR (SE) | MSE (SE) | Enhancement (%) | ORCOR (P-value) | ORMSE (P-value) |
| PolyUnres | — | Normal | 0.2921 (0.0113) | 2.0967 (0.5133) | 0.2943 (0.0115) | 2.0245 (0.5082) | 0.74 | 8/10 (0.055) | 10/10 (0.001) |
| GammaUnres | — | Gamma | 0.2801 (0.0114) | 3.2790 (0.5930) | 0.3029 (0.0101) | 3.0752 (0.6036) | 8.14 | 9/10 (0.011) | 10/10 (0.001) |
| PolyRes | MAF ≤ 0.30 | Normal | 0.2546 (0.0089) | 1.1089 (0.1476) | 0.2587 (0.0089) | 1.0949 (0.1466) | 1.61 | 6/10 (0.172) | 9/10 (0.011) |
| GammaRes | MAF ≤ 0.30 | Gamma | 0.2679 (0.0122) | 2.1452 (0.4047) | 0.2850 (0.0117) | 2.0280 (0.3969) | 6.40 | 10/10 (0.001) | 10/10 (0.001) |
A 60K microarray was used to genotype the 2000 individuals in the training set and 1500 in the validation set. Each simulated trait has heritability of 25% under regulation of 9000 simulated QTL. COR, average correlation coefficients between TBV & GEBV; MSE, average mean squared errors between TBV and GEBV; SE, standard error. Prediction enhancement was calculated based on the improvement in COR. OR, outdoing rate: the frequency that the heteroscedastic effects model dominates ridge regression.
Analysis of the QTLMAS data
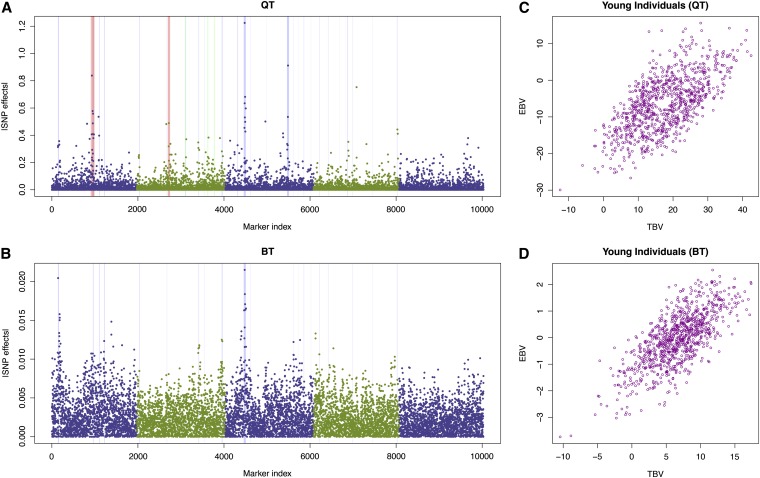
Here we focus on breeding value estimation for the QTLMAS 2010 data set that was previously analyzed by Shen et al. (2011). Due to the data size, the previous report could not fit all the markers into a DHGLM, but this is possible using HEM. Although HEM is theoretically an approximation of DHGLM, it is not worse than our previous DHGLM method in terms of breeding value estimation where the strongest effects match the simulated true QTL very well (Figure 4). By taking into account all the markers in the genome, HEM was able to improve the prediction of the young individuals compared to DHGLM. It did as good as the previous DHGLM for the binary trait and gave a correlation between the true breeding values (TBV) and estimated breeding values (EBV) of 0.72, whereas for the quantitative trait, HEM successfully raised the correlation between TBV and EBV from 0.60 to 0.64 compared to our previous report (Shen et al. 2011).
Figure 4.
Analysis of the 14th QTLMAS workshop (Szydlowski and Paczyńska 2011) common data set using HEM. The results of the quantitative trait (QT) and the binary trait (BT) are shown in the top and bottom, respectively. (A and B) The shrinkage estimates of the SNP effects across the genome. The red, green, and blue vertical bars indicate the simulated epistatic, imprinting, and addtive QTL, where the width of each bar is proportional to the corresponding QTL effect. Chromosomes are separated by the dual-colored dots. (C and D) Compare the estimated breeding values (EBV) with the true breeding values (TBV).
Discussion
The presented generalized RR algorithm, HEM, fits models in which the number of parameters p is much greater than the number of observations n. The focus of the article has been on applications in both GS and QTL detection, but the algorithm is expected to be of general use for applications of RR in other fields of research as well. The computational limitations of the bigRR package come mainly from the number of observations, and not the number of parameters. In our implementation of the algorithm, we used the hglm package (Rönnegård et al. 2010) in R for the variance component estimation, which is computationally feasible for data sets having up to 10,000 observations on a uni-core laptop computer. On any computer that has a CUDA-supported graphic card, an advanced version of the package can be required from the authors, which utilizes GPU for matrix calculation, accelerating the computation even more.
Compared to LMM and ordinary RR, the estimates from HEM are less sensitive to the assumption that the effects come from a common normal distribution and are therefore more robust. The method is computationally efficient due to its compression–decompression properties. In the Appendix, we show that the SNP-effects model (19) with p effects to be estimated is compressed into an animal model (20) whose size depends on n. In the decompression part, the estimated SNP effects can quickly be estimated through a simple transformation from individual effects to SNP effects. These estimates are used to update the matrix G, which is subsequently used in a compressed animal model. The final estimates are then computed by decompressing the animal model once more.
RR is known to be able to address collinearity (Hoerl and Kennard 1970b). It avoids computational trouble for an ill-conditioned data matrix and also solves the problem due to ill-conditioned Fisher’s information matrix (e.g., in Poisson and binomial GLM) (Hastie et al. 2009). In our analysis of the Arabidopsis data, we found that HEM seems to have a good performance in terms of correctly fine-mapping functional loci. This suggests that when linkage disequilibrium (LD) exists around a QTL, a clearer signal could be identified, which is a good property of the method, although further investigations are required to verify this property of HEM. The improvement of a stronger feature selection method compared to RR depends on the underlying genetic architecture. For a trait with only a few QTL, an even stronger feature selection method, e.g., least absolute shrinkage and selection operator (LASSO) (Tibshirani 1996), may perform much better. However, many complex traits, such as human height, have been shown to be very polygenic (e.g., Yang et al. 2011). At present, it is even more challenging in terms of both QTL mapping and genomic prediction when there are so many small and even undetectable QTL.
The method combines two ideas from our earlier articles. First, in Rönnegård and Lee (2010) and Shen et al. (2011), a DHGLM (Lee and Nelder 2006) was proposed. This model fits SNP-specific variance components using a random effects model also for the second level in Equation 30 (see the Appendix). By introducing HEM as a simplification of the DHGLM, one achieves a dramatic gain in speed. Second, Christensen and Lund (2010) suggest in their discussion that G could be weighted by a diagonal matrix D calculated from SNP effects and note, “However, incorporating uncertainty on such estimated SNP effects into the method seems less straight-forward.” Here, we have incorporated this uncertainty by using prediction error variances in the estimation of D through computations of hat values (Equation 12). Calculation of the prediction error variances is an important part of HEM. It should also be noted that HEM is based on fitting MME for an animal model and that GS problems involving both genotyped and nongenotyped individuals can be solved following the method by Christensen and Lund (2010).
Zhang et al. (2010) proposed a two-stage method, similar to ours, where the SNP-specific shrinkage parameters were calculated from the squared estimated SNP effects from a preliminary RR analysis (the method was referred to as TAP–BLUP by the authors). There are two important differences though. First, they did not include the uncertainty in the estimated SNP effects, which produces biased results. The calculations of prediction error variances in our proposed method are therefore an important contribution. Furthermore, in their preliminary RR analysis, a user-defined shrinkage parameter had to be given, and in their simulation study they used the true simulated values to calculate this shrinkage. In HEM, the shrinkage is estimated from the data.
The well-known Sherman–Morrison–Woodbury formula (Sherman and Morrison 1950; Golub and Van Loan 1996) can be used to invert a big p × p matrix of low rank (e.g., Rönnegård et al. 2007) and could therefore be a possible alternative to our implementation. However, to obtain all the SNP effects using this formula, the big p × p matrix still needs to be stored, which is avoided in HEM. Furthermore, an important part of HEM is a transformation algorithm to obtain the hat values for all the SNPs, while this is not straightforward using the Sherman–Morrison–Woodbury formula.
The proposed HEM is intended to be capable of addressing both feature selection and prediction. Certainly, such a universal capacity is fulfilled with price-biased estimates due to shrinkage. Many statistical methods are intended to simultaneously perform feature selection and prediction, such as ridge regression or SNP–BLUP, LASSO, their combination elastic net (Zou and Hastie 2005), our proposed generalized ridge method HEM, and all the series of Bayesian methods in the genomic prediction area (e.g., Meuwissen et al. 2001). Taking the SNP–BLUP for instance, especially for p ≫ n cases, so much shrinkage is given to each effect estimate, sacrificing the unbiasedness (BLUE) of the effects, to save degrees of freedom so that the model is estimable. Fortunately, combining all the “overshrunk” estimates, we are able to obtain a good prediction even when there are a lot of small effects undetectable (e.g., the results in human height by Yang et al. 2010, 2011).
HEM can be used to fit all SNP effects in a single model and the estimated effects can be used to rank interesting SNPs for further investigation in GWAS. Furthermore, using the computational advantage of HEM, we are able to calculate genome-wide significance thresholds using permutation testing.
A possible extension of the method would be to apply a more general autoregressive smoothing along each chromosome for the shrinkage values using DHGLM (applied in Rönnegård and Lee 2010; Shen et al. 2011). An important development would be to implement a computationally fast full DHGLM algorithm.
Acknowledgments
X.S. is funded by a Future Research Leaders grant from Swedish Foundation for Strategic Research (SSF) to Örjan Carlborg. L.R. is funded by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS).
Appendix
An Example in R
Here we include the R code for fitting SNP–BLUP and the HEM using the Arabidopsis data as an example. The code generates subfigures of Figure 2 in black and white. The code can also be found as an embedded example in the bigRR package.
install.packages(’bigRR’, repos = ’http://r-forge.r-project.org’)
require(bigRR)
data(Arabidopsis)
X <- matrix(1, length(y), 1)
SNP.BLUP.result <- bigRR(y = y, X = X, Z = scale(Z), family = binomial(link = ’logit’))
HEM.result <- bigRR.update(SNP.BLUP.result, scale(Z), family = binomial(link = ‘logit’))
dev.new(); plot(SNP.BLUP.result$u)
dev.new(); plot(HEM.result$u)
dev.new(); plot(SNP.BLUP.result$u, HEM.result$u)
Definitions of the Statistical Terminologies Used
Ridge regression (RR): A shrinkage estimation method often used for fitting more explanatory variables than the number of observations. A common shrinkage is applied to all the effects, and the magnitude of shrinkage is usually determined via cross validation (see also Månsson and Shukur 2011, for a recent review on RR methods for binary data).
Linear mixed model (LMM): A linear (regression) model including fixed and random effects. Treating effects as random, the model can handle more parameters than the number of observations. It provides shrinkage estimates for the random effects with a common magnitude of shrinkage, whereas there is no shrinkage applied on the estimated fixed effects. Furthermore, the covariates for the fixed effects and the random effects are assumed to be independent. The likelihood for LMM is equivalent to the ridge regression penalized likelihood but the magnitude of shrinkage is determined by the variance component estimates in LMM.
Generalized RR: A ridge regression method allowing different magnitudes of shrinkage for different explanatory variables.
Heteroscedastic effects model (HEM): A generalized RR method based on LMM theory, where the magnitudes of shrinkage for different effects are determined by the LMM random effects estimates and model hat values.
Double hierarchical generalized linear model (DHGLM): A double-layer random effects model, which allows fitting any variance component in an LMM using another random effects model, so that the second layer of the model determines the weights in the first layer. The full model is established using the hierarchical likelihood (h-likelihood), and statistical inference can be performed on the basis of the extended likelihood theory (Bjørnstad 1996; Lee et al. 2007). Fitting DHGLM in GWAS was proposed by Rönnegård and Lee (2010) and Shen et al. (2011).
Reducing the Dimension in Ordinary Ridge Regression Using Singular-Value Decomposition
Computationally fast methods for fitting RR in p ≫ n problems have been proposed based on SVD. These methods were developed for RR but not generalized RR. To clarify the difference between our approach and algorithms using SVD for RR, we also describe the latter below. A RR model is given by
| (A1) |
where β are fixed effects estimated without shrinkage and b are effects estimated with shrinkage. When β is simply an intercept term, the model can be reformulated by centering Z and the estimates of b are given by
| (A2) |
where Ip is the identity matrix with the subscript p denoting the size, and λ is the shrinkage parameter. Let Z = UDV′ be the SVD of Z and define R = UD; then (Hastie and Tibshirani 2004; Hastie et al. 2009)
| (A3) |
which reduces the size of the matrix to be inverted from p × p to n × n. Hence the parameter space is rotated to reduce the dimension and assumes that λ is a constant.
Note that the equivalence between LMM and RR has conditions, especially in terms of the assumptions. In an LMM, covariates are separated into fixed and random effects, where the inference of the fixed effects, based on the marginal likelihood, gives unbiased estimates, while shrinkage estimates are obtained for the random effect. In RR, all the covariates are penalized, without separation of fixed and random effects. Philosophically, an LMM considers the random effects as a sample drawn from an underlying distribution with a dispersion parameter to be estimated, whereas ridge regression is simply a computational method that provides estimates when the model is oversaturated. Only when the selected penalty parameter in RR equals the ratio of the variance components in the corresponding LMM, they become mathematically the same.
Generalized Ridge Regression and Linear Mixed Models
In the following subsections, methods from LMM theory will be used to develop a fast generalized RR algorithm for p ≫ n, where λ is allowed to be a vector of length ≤p (Hoerl and Kennard 1970a,b). Consider the linear mixed model
| (A4) |
where , , β is a vector of fixed effects, and b is a random effect. This is equivalent to the above RR model and give the same estimates for a known λ.
The differences between LMM and RR are found in the estimation techniques used. For LMM, λ is given by the variance component estimated using restricted maximum likelihood (REML) with , whereas for RR λ is computed using the generalized cross-validation (GCV) function GCV(λ) = e′e/(n − d.f.e), where d.f.e is the effective degrees of freedom (Hastie et al. 2009). These two methods tend to give similar estimates of λ (see Pawitan 2001, p. 488).
In LMMs it is possible to include several variance components, which is equivalent to defining λ as a vector. This is possible in generalized RR (Hoerl and Kennard 1970b) but the dimension reduction based on SVD assumes a constant λ. In generalized RR we have
| (A5) |
where K = diag(λ) and λ is the vector of shrinkage values. Below, we present how LMM theory can be used to reduce dimension from p to n also for the case of λ being a vector of length p, and thereafter propose a method to give suitable values for λ.
The Linear Mixed Model Approach
Here, we consider the estimation of an LMM with linear predictor η and a diagonal weight matrix D (size p × p) for the random effects
| (A6) |
where Σ is a diagonal matrix of weights and φ is the dispersion parameter equal to , and is equivalent to the weight matrix W in the Fitting algorithm. This notation allows for a later extension to a generalized linear mixed model (GLMM). The diagonal matrices K and D are related as .
To derive a computationally efficient implementation of the algorithm for p ≫ n, we present equivalent models to model (18) and show how the estimates of the effects, and their associated prediction error variances, can be transformed between these. Prediction error variances are important to compute since they are the basis for calculations of standard errors and d.f.e.
Three different, but equivalent, specifications of the random effects are used and are referred to as the SNP model, the animal model, and the Cholesky model. For all three models the linear predictor η is the same:
- SNP model
(A7) - Animal model
(A8) - Cholesky model
(A9)
The use of equivalent LMMs in the research field of animal breeding and quantitative genetics is well established (Lynch and Walsh 1998; Rönnegård and Carlborg 2007). The contribution of this article is to present how LMM theory can be used for generalized RR, to show how the prediction error variances can be transformed between models, to implement the theory in a computationally efficient R package bigRR (including GLMM), and to apply it to a novel heteroscedastic effects model presented further below.
Different Mixed Model Equations for the Equivalent Models
For LMM Henderson’s MME are used to estimate both the fixed and random effects for given variance components. They can also be used iteratively to estimate variance components as implemented in the R package hglm (Rönnegård et al. 2010). Although the models above are equivalent, the MME are different.
SNP model
For the SNP Model we have the MME
| (A10) |
These MMEs are of size (k + p) × (k + p), where k is the number of columns in X. Hence, the size of the equations are very large for high-dimensional data.
Animal model
Let the random effects a be individual effects for each observation and G = ZZ′ the correlation matrix between these. Then G is relatively small (n × n) and the MME are
| (A11) |
of size (k + n) × (k + n). Hence, the size of these MME is much smaller than Equation A10 for P ≫ n.
Cholesky model
In a third equivalent model we define LL′ = G (where L has size n × n) and the random effects v are individual independent random effects. The MME are
| (A12) |
of size (k + n) × (k + n).
Transformation of Effects Between Equivalent Models
For P ≫ n, the size of the MME in models (A11) and (A12) are much smaller than in model (A10). The random effects can be transformed between these equivalent models (Lynch and Walsh 1998; Nagamine 2005) so that the estimated SNP effects can easily be calculated from the individual effects in model (A11)
| (A13) |
Furthermore, we have so that
| (A14) |
The matrix Z is moderately large (n × p) but the transformation is a simple cross-product. Hence, the calculations can be made in parts without reading all of Z into memory. They can also easily be parallelized if necessary.
Transformation of Prediction Error Variances Between Equivalent Models
Not only the estimates, but also the prediction error variances (i.e., the diagonal elements of ), are important to compute to allow for model checking and inference. In the Cholesky model (Equation A12), let Cv be
| (A15) |
Decompose the inverse of Cv as
| (A16) |
Then the prediction covariance matrix is (Henderson 1984). Define the jth diagonal element,, as . Then these elements can be calculated separately as
| (A17) |
where Mj is the jth row of the transformation matrix M = Z′G−1L.
Extension to Penalized GLM Estimation
For penalized generalized linear models (i.e., GLMM), the expectation of y is connected to the linear predictor η through a link function g(.) such that E(y) = g(η). PQL estimation uses the same MME as above with a working weight matrix Σ and y being replaced by an adjusted response z, where both Σ and z are updated iteratively until convergence (Breslow and Clayton 1993). Such a penalized likelihood is similar to the one of ridge regression, where the sum of squared effects are used as a penalty term (Hastie et al. 2009). The penalty parameter is estimated as the ratio of the two dispersion parameters in the mixed model setting. For GLMM, the left-hand side of the MME can be described by the above formulae, e.g., Equations A10–A12, and the same transformations can be applied. The algorithm was implemented in the R (R Development Core Team 2010) package bigRR and uses the hglm (Rönnegård et al. 2010) package to estimate the variance components and individual effects a.
Using Generalized Ridge Regression to Calculate Heteroscedastic SNP Effects
Here, we consider the estimation of the hierarchical model
| (A18) |
having a second-level model
| (A19) |
where uj are fixed effects in the linear predictor for the SNP variances, and j is an index for the p different SNPs. The model is saturated and , so are updated as
| (A20) |
where hjj are the hat values for the random effects (Lee et al. 2006). The hat values are related to the prediction error variance as .
In the current article, we consider estimation where the SNP-specific variance components are updated twice and refer to it as the heteroscedastic effects model, which gives an increased shrinkage for small SNP effects compared to ordinary RR.
Here the transformation of prediction error variances between the Cholesky model (Equation A12) and the SNP models (Equation A10) are derived. Let Cv be the left-hand side of the MME from the Cholesky model (Equation A12)
| (A21) |
Decompose the inverse of Cv as
| (A22) |
Then the prediction covariance matrix is (Henderson 1984). Furthermore, let Cb be the left-hand side of the MME from the SNP model (Equation A10)
| (A23) |
Decompose the inverse of Cb as
| (A24) |
Then the prediction covariance matrix is (Henderson 1984)
| (A25) |
Define M to be the matrix transforming effects v to b in Equation A14 so that M = Z′G−1L; then
| (A26) |
Combining these two equations, we get
| (A27) |
i.e.,
| (A28) |
so
| (A29) |
Footnotes
Communicating editor: F. Zou
Literature Cited
- Atwell S., Huang Y. S., Vilhjalmsson B. J., Willems G., Horton M., et al. , 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørnstad J. F., 1996. On the generalization of the likelihood function and the likelihood principle. J. Am. Stat. Assoc. 91: 791–806. [Google Scholar]
- Breslow N. E., Clayton D. G., 1993. Approximate inference in generalized linear mixed models. J. Am. Stat. Assoc. 88: 9–25. [Google Scholar]
- Che X., Xu S., 2012. Generalized linear mixed models for mapping multiple quantitative trait loci. Heredity 109: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen O. F., Lund M. S., 2010. Genomic prediction when some animals are not genotyped. Genet. Sel. Evol. 42: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Hickey J. M., Pong-Wong R., Daetwyler H. D., Calus M. P. L., 2013. Whole genome regression and prediction methods applied to plant and animal breeding. Genetics 193: 327–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers J. C. M., 2004. Commercial application of marker- and gene-assisted selection in livestock: strategies and lessons. J. Anim. Sci. 82: E313–E328. [DOI] [PubMed] [Google Scholar]
- Gianola D., de los Campos G., Hill W., Manfredi E., Fernando R., 2009. Additive genetic variability and the bayesian alphabet. Genetics 183: 347–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub G., Van Loan C., 1996. Matrix Computations, Ed. 3 Johns Hopkins University Press, Baltimore. [Google Scholar]
- Habier D., Fernando R., Kizilkaya K., Garrick D., 2011. Extension of the bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., 2004. Efficient quadratic regularization for expression arrays. Biostatistics 5: 329–340. [DOI] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., Friedman J., 2009. The Elements of Statistical Learning. Springer-Verlag, Berlin. [Google Scholar]
- Hayes B., Goddard M., 2001. The distribution of the effects of genes affecting quantitative traits in livestock. Genet. Sel. Evol. 33: 209–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson C. R., 1953. Estimation of variance and covariance components. Biometrics 9: 226–252. [Google Scholar]
- Henderson, C. R., 1984 Applications of Linear Models in Animal Breeding. University of Guelph, Guelph, Ontario, Canada.
- Hickey J. M., Gorjanc G., 2012. Simulated data for genomic selection and genome-wide association studies using a combination of coalescent and gene drop methods. G3: Genes, Genomes, Genetics 2: 425–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerl A., Kennard R., 1970a Ridge regression: applications to nonorthogonal problems. Technometrics 12: 69–82. [Google Scholar]
- Hoerl A., Kennard R., 1970b Ridge regression: biased estimation for nonorthogonal problems. Technometrics 12: 55–67. [Google Scholar]
- Kidd K., Ott J., 1984. Power and sample size in linkage studies: Human Gene Mapping 7 (1984): Seventh International Workshop on Human Gene Mapping. Cytogenet. Cell Genet. 37: 510–511. [Google Scholar]
- Kingsmore S. F., Lindquist I. E., Mudge J., Gessler D. D., Beavis W. D., 2008. Genome-wide association studies: progress and potential for drug discovery and development. Nat. Rev. Drug Discov. 7: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Nelder J. A., 2006. Double hierarchical generalized linear models (with discussion). Appl. Stat. 55: 139–185. [Google Scholar]
- Lee Y., Nelder J. A., Noh M., 2007. H-likelihood: problems and solutions. Stat. Comput. 17: 49–55. [Google Scholar]
- Lee Y., Nelder J. A., Pawitan Y., 2006. Generalized Linear Models with Random Effects - Unified Analysis via h-Likelihood, Chapman & Hall, London. [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Malo N., Libiger O., Schork N. J., 2008. Accommodating linkage disequilibrium in genetic-association analyses via ridge regression. Am. J. Hum. Genet. 82: 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Månsson K., Shukur G., 2011. On ridge parameters in logistic regression. Commun. Stat. 40: 3366–3381. [Google Scholar]
- Meuwissen T., Hayes B., Goddard M., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine Y., 2005. Transformation of QTL genotypic effects to allelic effects. Genet. Sel. Evol. 37: 579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawitan Y., 2001. In All Likelihood: Statistical Modelling and Inference Using Likelihood. Oxford University Press, Oxford. [Google Scholar]
- R Development Core Team , 2010. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Risch N., 1991. A note on multiple testing procedures in linkage analysis. Am. J. Hum. Genet. 48: 1058–1064. [PMC free article] [PubMed] [Google Scholar]
- Rodolphe F., Lefort M., 1993. A multi-marker model for detecting chromosomal segments displaying QTL activity. Genetics 134: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnegård L., Carlborg Ö., 2007. Separation of base allele and sampling term effects gives new insights in variance component QTL analysis. BMC Genet. 8 . 10.1186/1471-2156-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnegård, L., and Y. Lee, 2010 Hierarchical generalized linear models have a great potential in genetics and animal breeding. Proceedings of the World Congress on Genetics Applied to Livestock Production, Leipzig, Germany. [Google Scholar]
- Rönnegård L., Mischenko K., Holmgren S., Carlborg Ö., 2007. Increasing the efficiency of variance component quantitative trait loci analysis by using reduced-rank identity-by-descent matrices. Genetics 176: 1935–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnegård L., Shen X., Alam M., 2010. hglm: a package for fitting hierarchical generalized linear models. R J. 2: 20–28. [Google Scholar]
- Shen X., Rönnegård L., Carlborg Ö., 2011. Hierarchical likelihood opens a new way of estimating genetic values using genome-wide dense marker maps. BMC Proceedings 5(Suppl. 3): S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman J., Morrison W. J., 1950. Adjustment of an inverse matrix corresponding to a change in one element of a given matrix. Ann. Math. Stat. 21: 124–127. [Google Scholar]
- Stranden I., Garrick D., 2009. Technical note: derivation of equivalent computing algorithms for genomic predictions and reliabilities of animal merit. J. Dairy Sci. 92: 2971–2975. [DOI] [PubMed] [Google Scholar]
- Szydlowski M., Paczyńska P., 2011. QTLMAS 2010: simulated dataset. BMC Proc. 5(Suppl. 3): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R., 1996. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58: 267–288. [Google Scholar]
- vanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Xu S., 2003. Estimating polygenic effects using markers of the entire genome. Genetics 163: 789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Manolio T. A., Pasquale L. R., Boerwinkle E., Caporaso N., et al. , 2011. Genome partitioning of genetic variation for complex traits using common snps. Nat. Genet. 43: 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi N., Xu S., 2008. Bayesian LASSO for quantitative trait loci mapping. Genetics 179: 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z.-B., 1993. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90: 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Liu J., Ding X., Bijma P., de Koning D.-J., et al. , 2010. Best linear unbiased prediction of genomic breeding values using a trait-specific marker-derived relationship matrix. PLoS ONE 5: e12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Hastie T., 2005. Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 67: 301–320. [Google Scholar]