Abstract
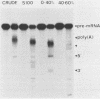
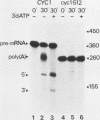
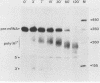
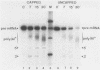
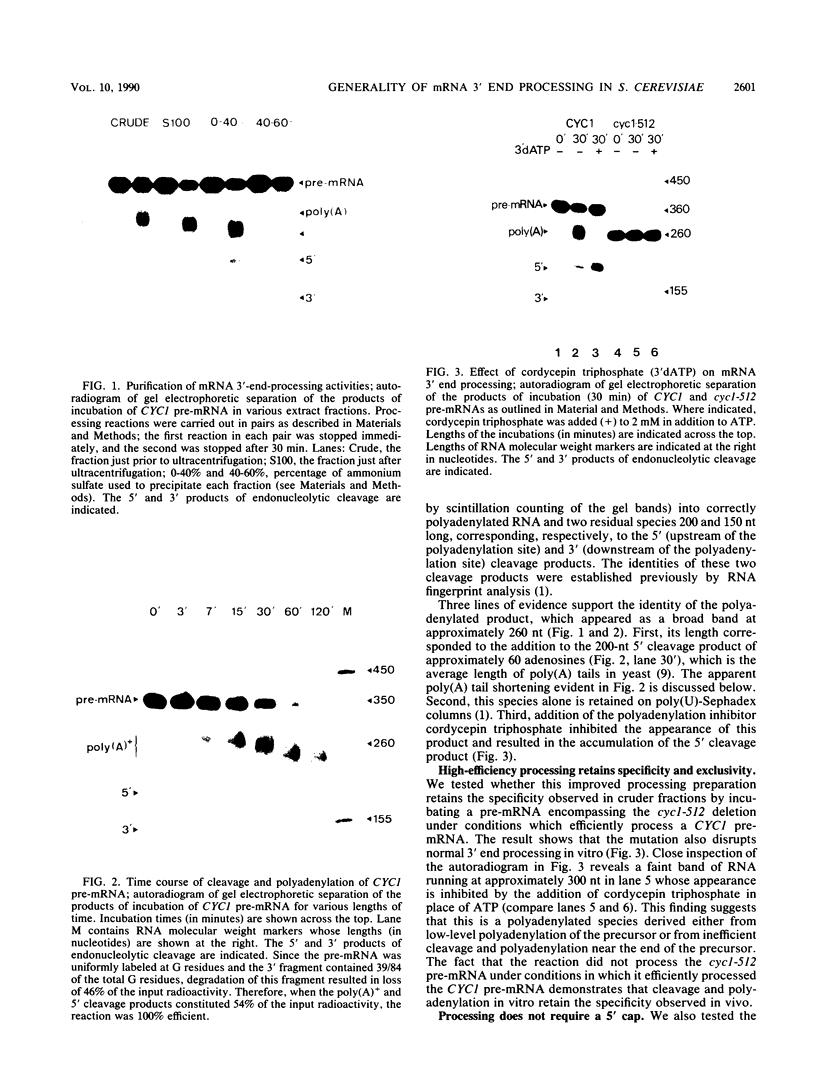
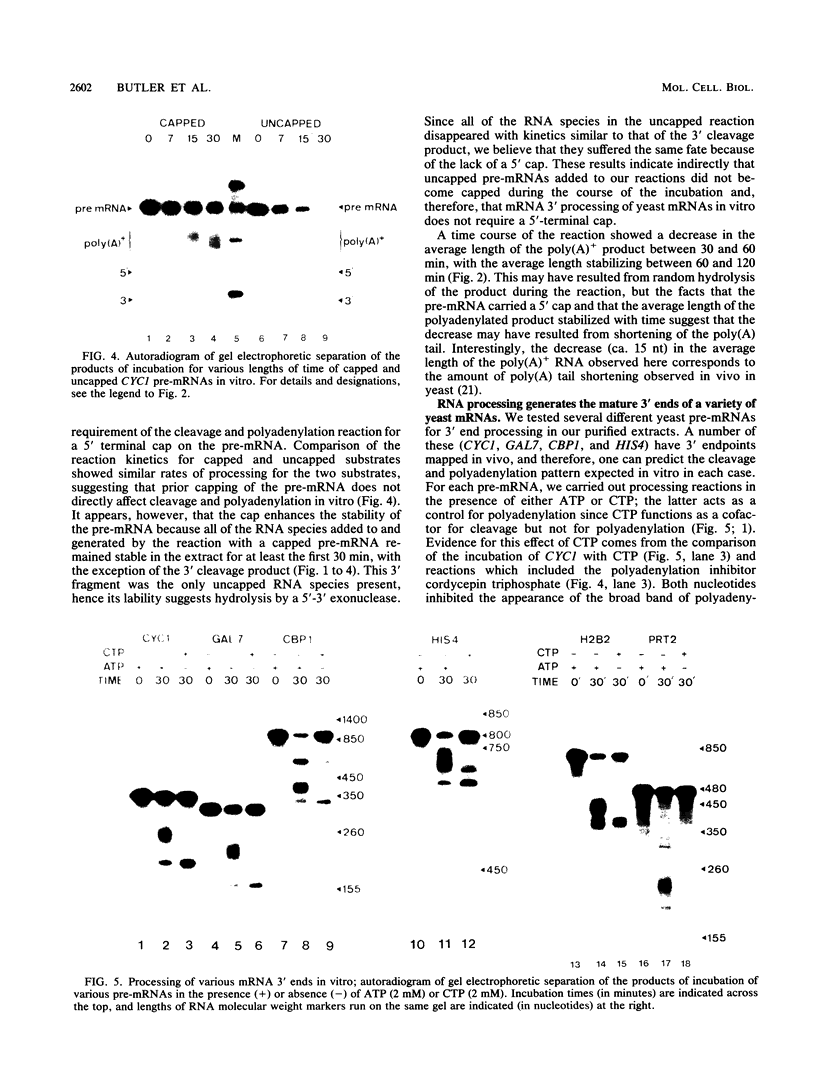
Ammonium sulfate fractionation of a Saccharomyces cerevisiae whole-cell extract yielded a preparation which carried out correct and efficient endonucleolytic cleavage and polyadenylation of yeast precursor mRNA substrates corresponding to a variety of yeast genes. These included CYC1 (iso-1-cytochrome c), HIS4 (histidine biosynthesis), GAL7 (galactose-1-phosphate uridyltransferase), H2B2 (histone H2B2), PRT2 (a protein of unknown function), and CBP1 (cytochrome b mRNA processing). The reaction processed these pre-mRNAs with varying efficiencies, with cleavage and polyadenylation exceeding 70% in some cases. In each case, the poly(A) tail corresponded to the addition of approximately 60 adenosine residues, which agrees with the usual length of poly(A) tails formed in vivo. Addition of cordycepin triphosphate or substitution of CTP for ATP in these reactions inhibited polyadenylation but not endonucleolytic cleavage and resulted in accumulation of the cleaved RNA product. Although this system readily generated yeast mRNA 3' ends, no processing occurred on a human alpha-globin pre-mRNA containing the highly conserved AAUAAA polyadenylation signal of higher eucaryotes. This sequence and adjacent signals used in mammalian systems are thus not sufficient to direct mRNA 3' end formation in yeast. Despite the lack of a highly conserved nucleotide sequence signal, the same purified fraction processed the 3' ends of a variety of unrelated yeast pre-mRNAs, suggesting that endonuclease cleavage and polyadenylation may produce the mature 3' ends of all mRNAs in S. cerevisiae.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Butler J. S., Platt T. RNA processing generates the mature 3' end of yeast CYC1 messenger RNA in vitro. Science. 1988 Dec 2;242(4883):1270–1274. doi: 10.1126/science.2848317. [DOI] [PubMed] [Google Scholar]
- Carswell S., Alwine J. C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989 Oct;9(10):4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofori G., Keller W. 3' cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988 Sep 9;54(6):875–889. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- Conway L., Wickens M. A sequence downstream of A-A-U-A-A-A is required for formation of simian virus 40 late mRNA 3' termini in frog oocytes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo J. D., Imperiale M. J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989 Nov;9(11):4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Green T. L., Hart R. P. Mutations in poly(A) site downstream elements affect in vitro cleavage activity. Mol Cell Biol. 1988 Apr;8(4):1839–1841. doi: 10.1128/mcb.8.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner B., Phillips S. L. Polyadenylate metabolism in the nuclei and cytoplasm of Saccharomyces cerevisiae. J Biol Chem. 1975 Jul 25;250(14):5640–5646. [PubMed] [Google Scholar]
- Henikoff S., Cohen E. H. Sequences responsible for transcription termination on a gene segment in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1515–1520. doi: 10.1128/mcb.4.8.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford L. M., Osley M. A., Ludwig T. R., 2nd, McLaughlin C. S. Cell-cycle regulation of yeast histone mRNA. Cell. 1981 May;24(2):367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- Humphrey T., Christofori G., Lucijanic V., Keller W. Cleavage and polyadenylation of messenger RNA precursors in vitro occurs within large and specific 3' processing complexes. EMBO J. 1987 Dec 20;6(13):4159–4168. doi: 10.1002/j.1460-2075.1987.tb02762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. J., Newman A. J., Cheng S. C., Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985 Nov 25;260(27):14780–14792. [PubMed] [Google Scholar]
- Manley J. L. Polyadenylation of mRNA precursors. Biochim Biophys Acta. 1988 May 6;950(1):1–12. doi: 10.1016/0167-4781(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Mayer S. A., Dieckmann C. L. The yeast CBP1 gene produces two differentially regulated transcripts by alternative 3'-end formation. Mol Cell Biol. 1989 Oct;9(10):4161–4169. doi: 10.1128/mcb.9.10.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985 Jul;41(3):845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Skolnik-David H., Sharp P. A. Sedimentation analysis of polyadenylation-specific complexes. Mol Cell Biol. 1988 Jan;8(1):226–233. doi: 10.1128/mcb.8.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne B. I., Guarente L. Mutational analysis of a yeast transcriptional terminator. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4097–4101. doi: 10.1073/pnas.86.11.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Guthrie C. A point mutation in the conserved hexanucleotide at a yeast 5' splice junction uncouples recognition, cleavage, and ligation. Cell. 1985 May;41(1):107–118. doi: 10.1016/0092-8674(85)90065-0. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sadofsky M., Alwine J. C. Sequences on the 3' side of hexanucleotide AAUAAA affect efficiency of cleavage at the polyadenylation site. Mol Cell Biol. 1984 Aug;4(8):1460–1468. doi: 10.1128/mcb.4.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano J. E., Adams D. E. Assembly of a polyadenylation-specific 25S ribonucleoprotein complex in vitro. Mol Cell Biol. 1988 May;8(5):2052–2062. doi: 10.1128/mcb.8.5.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A., Broach J. R. Signals for transcription initiation and termination in the Saccharomyces cerevisiae plasmid 2 micron circle. Mol Cell Biol. 1985 Oct;5(10):2770–2780. doi: 10.1128/mcb.5.10.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima M., Nogi Y., Fukasawa T. Primary structure of the Saccharomyces cerevisiae GAL7 gene. Yeast. 1985 Sep;1(1):67–77. doi: 10.1002/yea.320010108. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Ryner L. C., Manley J. L. Four factors are required for 3'-end cleavage of pre-mRNAs. Genes Dev. 1989 Nov;3(11):1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Ryner L. C., Manley J. L. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988 Mar 11;52(5):731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Yarger J. G., Armilei G., Gorman M. C. Transcription terminator-like element within a Saccharomyces cerevisiae promoter region. Mol Cell Biol. 1986 Apr;6(4):1095–1101. doi: 10.1128/mcb.6.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Elder R. T. Some of the signals for 3'-end formation in transcription of the Saccharomyces cerevisiae Ty-D15 element are immediately downstream of the initiation site. Mol Cell Biol. 1989 Jun;9(6):2431–2444. doi: 10.1128/mcb.9.6.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. Mutationally altered 3' ends of yeast CYC1 mRNA affect transcript stability and translational efficiency. J Mol Biol. 1984 Jul 25;177(1):107–135. doi: 10.1016/0022-2836(84)90060-3. [DOI] [PubMed] [Google Scholar]
- Zarkower D., Wickens M. A functionally redundant downstream sequence in SV40 late pre-mRNA is required for mRNA 3'-end formation and for assembly of a precleavage complex in vitro. J Biol Chem. 1988 Apr 25;263(12):5780–5788. [PubMed] [Google Scholar]
- Zarkower D., Wickens M. Specific pre-cleavage and post-cleavage complexes involved in the formation of SV40 late mRNA 3' termini in vitro. EMBO J. 1987 Dec 20;6(13):4185–4192. doi: 10.1002/j.1460-2075.1987.tb02765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]