Abstract
The stress-induced heat shock protein 70 (HSP70) is an ATP-dependent molecular chaperone that plays a key role in refolding misfolded proteins and promoting cell survival following stress. HSP70 is marginally expressed in non-transformed cells, but is greatly overexpressed in tumor cells. Silencing HSP70 is uniformly cytotoxic to tumor but not normal cells; therefore, there has been great interest in the development of HSP70 inhibitors for cancer therapy. Here we report that the HSP70 inhibitor 2-phenylethynesulfonamide (PES) binds to the substrate-binding domain of HSP70, and requires the C-terminal helical ‘lid’ of this protein (amino acids 573-616) in order to bind. Using molecular modeling and in silico docking, we have identified a candidate binding site for PES in this region of HSP70, and we identify point mutants that fail to interact with PES. A preliminary structure-activity relationship analysis has revealed a derivative of PES, 2-(3-chlorophenyl) ethynesulfonamide (PES-Cl), which shows increased cytotoxicity and ability to inhibit autophagy, along with significantly improved ability to extend the life of mice with pre-B cell lymphoma, compared to the parent compound (p=0.015). Interestingly, we also show that these HSP70 inhibitors impair the activity of the Anaphase Promoting Complex/Cyclosome (APC/C) in cell-free extracts, and induce G2/M arrest and genomic instability in cancer cells. PES-Cl is thus a promising new anti-cancer compound with several notable mechanisms of action.
Keywords: Phenylethynesulfonamide, HSP70, HSP72, lymphoma, autophagy, HSP90
Introduction
The HSP70 family is comprised of a minimum of eight proteins that serve as molecular chaperones. In general each member is believed to possess distinct function and/or cellular localization. The major stress-induced cytosolic form is denoted here as HSP70, but is sometimes called HSP72, or HSP70A1A. HSP70 is marginally expressed in non-transformed cells but is frequently overexpressed in tumors and tumor cell lines, where it plays a key role in aiding survival under adverse conditions, including nutrient deprivation, proteotoxic stress and hypoxia (1). Consistent with its non-essential but stress-inducible role, knock-out mice for HSP70 are viable and fertile, but are more susceptible to certain stresses (2). The overexpression of HSP70 occurs in many different tumor types, and in general high levels of this protein are correlated with poor prognosis, increased tumor grade and drug resistance (for review see 3). Silencing of HSP70 is cytotoxic to a wide variety of cancer but not normal cells (4). Additionally, neutralization of HSP70 with a peptide containing a portion of apoptosis-inducing factor (AIF) has anti-tumor effects in xenograft models of cancer (5, 6). For these reasons, identifying small molecule inhibitors of HSP70 is an area of active interest in the cancer research community.
We previously identified 2-phenylethynesulfonamide (PES) as a potent and selective inhibitor of HSP70 (7). We found that PES is cytotoxic to tumor cell lines but markedly less toxic to non-transformed cells, including primary and immortalized human fibroblasts and immortalized breast epithelial cells (7). Consistent with a cancer-specific role for HSP70 in the control of lysosome integrity (8), we found that inhibition of HSP70 by PES leads to impaired autophagy (7). HSP70 is an important co-chaperone for HSP90, and we showed that treatment of cells with PES leads to sequestration of several HSP90 client proteins into an inactive, insoluble compartment; these include the HSP90 clients HER-2, AKT and CDK-4 (9). In a pre-clinical model of pre-B cell lymphoma, we showed that intra-peritoneal administration of PES markedly extends the lifespan of mice (7). More recently, others have shown that PES is cytotoxic to acute myeloid leukemia, acute lymphoid leukemia (10), and chronic lymphocytic leukemia (11), but is significantly less toxic to normal hematopoietic cells (10). The combined promising pre-clinical data on PES support the rationale for a more in-depth mechanistic analysis of this compound.
Materials and Methods
Cell culture, western blot analysis, PES-binding assays
H1299 and HeLa cells were obtained from the American Type Culture Collection and were used within six months of receipt; these were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS (Hyclone) and 100 units of penicillin/streptomycin. A375, 1205Lu, WM1366, 451Lu and 1617 cells were obtained from the Herlyn laboratory (Wistar Institute) and authenticated by genotype analysis; these were maintained in DMEM supplemented with 5% FBS (Hyclone) and 100 units of penicillin/streptomycin. The BRAF inhibitor resistant cell lines, 451Lu-R and 1617-R, were obtained from the Villanueva laboratory (Wistar Institute) and were used within six months of receipt; these were maintained in DMEM containing 5% FBS, 100 units of penicillin/streptomycin containing 1uM of the BRAF inhibitor SB-590885 (Tocris). Primary human neonatal epidermal melanocytes (1° melanocytes) were obtained from the Herlyn laboratory, and were cultured in M254CF media (Invitrogen) as described (12). Eμ-myc lymphoma cells were cultured as described (13). All cell lines were kept at 37°C in an atmosphere supplemented with 5% CO2. For treatment with PES or PES-Cl, stock solutions were made in DMSO and diluted in PBS; the final concentration of DMSO was less than 0.4%. Western analysis was performed using the following antibodies at supplier-recommended dilutions: Hsc70 (ADI-SPA-819D, Enzo Life Sciences), p62/SQSTM1 (sc-28359, Santa Cruz Biotechnology), LC3 (NB100-2331, Novus), Her-2 (791-100, Vertana), CDK4 (sc-601, Santa Cruz), cyclin B (554177, Pharmingen), actin (AC-15, Sigma), HA-tag (3742S, Cell Signaling), CHIP (2080S, Cell Signaling), cleaved lamin A (2035S, Cell Signaling), Hsp70 (4873S, Cell Signaling), and cleaved caspase 3 (9661S, Cell Signaling). For binding assays, PES and PES-Cl were biotinylated, and binding assays were performed using HSP70 deletion constructs, exactly as described (7).
Plasmid constructs
Deletion mutants of human HSP70 were made using the following primers:
a.a (386-543) 5′-gccaagaacgccctgtagtcctaggccttcaacatgaag-3′;
a.a (386-509) 5′-caacgacaagggctaatagcgcctgagcaagg-3′;
a.a (386-616) 5′-ccgctcgagctaaccaccggcaccctggtacagtccgctgat-3′;
a.a (386-602) 5′-cgctcgagctacacctgctccagctcctccctcctgtgctcaaa-3′;
a.a (386-573) 5′-ccgctcgagctacttgtccagcaccttcttcttgtcggcctcgct-3′.
Point mutants of the denoted amino acids in HSP70 were made with the Quick change II site directed mutagenesis kit (Agilent) using the following oligonucleotides: A397R (for 5′-gctgctggacgtgcgtcccctgtcgctg-3′, rev 5′-cagcgacaggggacgcacgtccagcagc-3′); L401 (for 5′-ctcccctgtcgcgggggctggagac-3′, rev 5′-gtctccagcccccgcgacaggggag-3′); Y431 (for 5′-gcagatcttcaccaccgcctccgacaaccaaccc-3′, rev 5′-gggttggttgtcggaggcggtggtgaagatctgc-3′); G470L (for 5′-ctccggcccccaggctagtgccccagatcga-3′, rev 5′-tcgatctggggcactagcctgggggccggag-3′); N505A (for 5′-caagatcaccatcaccgccgacaagggccgcctg-3′, rev 5′-caggcggcccttgtcggcggtgatggtgatcttg-3′); D506A (for 5′-accatcaccaacgccaagggccgcctg-3′, rev 5′-caggcggcccttggcgttggtgatggt-3′); L510E (for 5′-cgacaagggccgcgagagcaaggaggag-3′, rev 5′-ctcctccttgctctcgcggcccttgtcg-3′); A538I (for 5′-cgcgagagggtgtcaatcaagaacgccctgga-3′, rev 5′-tccagggcgttcttgattgacaccctctcgcg-3′); N548A (for 5′-ggagtcctacgccttcgccatgaagagcgccgtg-3′, rev 5′-cacggcgctcttcatggcgaaggcgtaggactcc-3′); M549S (for 5′-tcctacgccttcaacagcaagagcgccgtggag-3′, rev 5′-ctccacggcgctcttgctgttgaaggcgtagga-3′); Y545D (for 5′-acgccctggagtccgacgccttcaacatg-3′, rev 5′-catgttgaaggcgtcggactccagggcgt-3′); A552K (for 5′-gccttcaacatgaagagcaaggtggaggatgaggggctc-3′, rev 5′-gagcccctcatcctccaccttgctcttcatgttgaaggc-3′); L558R (for 5′-ggaggatgaggggcgcaagggcaagatca-3′, rev 5′-tgatcttgcccttgcgcccctcatcctcc-3′); I607D (for 5′-ggtgtgtaaccccatcgacagcggactgtaccag-3′, rev 5′-ctggtacagtccgctgtcgatggggttacacacc-3′); Y611S (for 5′-tcatcagcggactgagccagggtgccggtg-3′, rev 5′-caccggcaccctggctcagtccgctgatga-3′)
Synthesis of PES-Cl
Chlorosulfonamide was synthesized according to (14); the ethynesulfonamide was synthesized in two steps from (3-chlorophenylethynyl) trimethylsilane. Chlorosulfonyl isocyanate (14,266-2; lot STBB0539AO), (3-chlorophenylethynyl) trimethylsilane (597708-1G; lot 16126BB) and titanium chloride (Aldrich 20,856-6; lot 9526) was purchased from Aldrich. Formic acid was purchased from Sigma (F0507-100ML; lot 039K0108). Potassium fluoride (Acros 20135-0250; lot B0126755B) was dried under vacuum at room temperature just prior to use. Dichloromethane, nitromethane and dioxane were dried by storage over calcium hydride. LC-MS was performed using a Waters 2545 binary gradient module and a 2487 dual wavelength detector set to 254 and 365, a 2424 ELS detector and a 3100 MS detector. The gradient was linear 5% MeOH 95% H2O to 95% MeOH 5% H2O over 15 minutes. The column was a Waters Delta Pak C-18 15u 100A 3.9 × 300mm (catalog number 11797) run at a flow rate of 0.8 ml per minute. Preparative chromatography was run on a Waters LC-MS system using a prep pak C-18 delta-pak column (47 × 300 mm). UV detection was set at 254l. The flow rate was 30 ml per minute. 1H NMR was performed on a Bruker WB Advance 300 MHz instrument.
Pre-clinical analysis of PES and PES-Cl
Eμ-myc mice of 8 weeks of age were injected intra-peritoneally with vehicle (n=19 mice), PES (20 mg/kg; n=20 mice) and PES-Cl (20 mg/kg; n=21 mice). Mice were treated once per week for a total of 20 weeks, and were observed daily for signs of discomfort or disease; at the earliest signs of lethargy or obvious lymph node mass totaling 5% of body weight, animals were euthanized. The study was concluded at 34 weeks (6 weeks after the final treatment). All animal experiments adhered to the guidelines set forth by the Institutional Animal Care and Use Committee at Fox Chase Cancer Center.
In vitro cyclin B1 degradation assay
Cells extracts were made as previously described (15, 16). Degradation assay reactions contained 10 uL HeLa cell extract, 1 uL of 10X degradation reaction mixture (100 mM Tris-HCl (pH7.6), 50 mM MgCl2, 10 mM DTT, 10 mg/ml ubiquitin, 100 mM phosphocreatine, 1 mg/ml creatine phosphokinase and 5 mM ATP) and the indicated concentration of drug. Reactions were incubated at 30° and samples were withdrawn every 30 minutes for 3 hours. The collected samples were immediately mixed with an equal volume 2X SDS sample buffer and resolved by SDS-PAGE.
Time-lapse video microscopy
Hela, Hela-GFP-H2B, H1299 and H1299-GFP-H2A cell were seeded on No. 1.5 coverslips (18×18 mm) in Hepes-buffered media. Cells were treated with 2 mM thymidine for twenty hours, allowed to recover for 4 hours and treated with the indicated drug for one hour prior to the start of imaging. Cells were observed with an Eclipse TE200S inverted microscope, images were captured overnight at 37°C, at 5 minutes intervals and processed with the MetaMorph software (Molecular Dynamics). All analysis was performed with the MetaMorph software and figures were compiled with Photoshop (Adobe).
Molecular Modeling, in silico docking
The human sequences of Hsp70 (Uniprot code P08017: residues 391 to 607) and Hsc71 (Uniprot code P11142: residues 361 to 613) were aligned to two different homologous proteins of known structure (E. coli DNA-K and Bovine Hsc71 substrate binding domains, PDB codes 1DKZ and 1YUW, respectively) using the program MolIDE (17, 18). Side chain conformations were predicted with the program SCWRL4 (19), which sampled rotamers for all side chains, and takes into account both potential hydrogen bonding and short range van Der Waals interactions, while avoiding clashes within the generated model. When needed, the SCWRL4-generated models were subjected to a simple minimization using Chimera (UCSF), prior to docking routines with the inhibitor PES using the program AutoDock.
Results
Previously we showed that a biotinylated version of the HSP70 inhibitor PES (Biotin-PES, Supp. Fig. 1A) can pull down HSP70 from cell extracts (7). We next wanted to test whether PES binds directly to HSP70, and whether un-biotinylated compound could compete this interaction. Toward this end we purified recombinant human HSP70 from bacteria, and used this protein in pull down assays with Biotin-PES, as described (7). Biotin-PES was consistently able to pull-down purified HSP70; moreover, we were able to efficiently compete this interaction using untagged compound (Fig. 1A). These data support the premise that PES binds directly to HSP70, and that this interaction is specific.
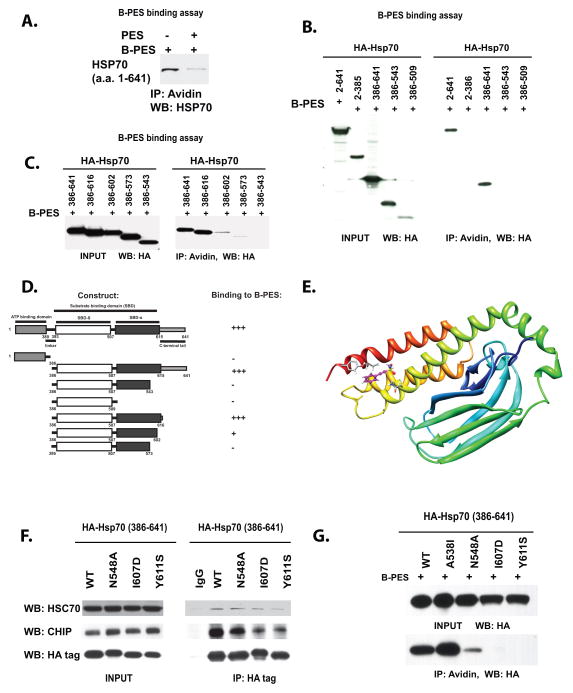
Figure 1. Binding to PES requires residues within a C-terminal helical bundle of HSP70.
A) Pull-down assay of purified, recombinant HSP70 with biotinylated PES (B-PES). 238 nM of purified HSP70 protein was incubated with 0.25 mM biotinylated PES (B-PES), in the absence or the presence of 0.125 mM untagged PES. Immunoprecipitation-western blot (IP-WB) analysis reveals direct interaction between B-PES and HSP70.
B) Pull-down assays of H1299 cells transfected with HA-tagged full-length and deletion mutants of HSP70, followed by treatment with 20 uM biotinylated Biotin-PES (B-PES). The HA-tagged mutants were precipitated with avidin beads, eluted and detected with an anti-HA antibody following imunoblotting. Input is shown on the left panel; immunoprecipitation (IP) with avidin is depicted on the right.
C) Pull-down assays of additional C-terminal deletion mutants of HA-tagged HSP70, performed as in B. Input is shown in the left panel; immunoprecipitation with avidin beads is depicted in the right panel.
D) Summary of the results of multiple independent B-PES binding assays performed as in B.
E) Molecular model of the substrate-binding domain of human HSP70; depiction of one pose for PES binding from an in silico analysis of potential PES docking sites. The depicted pose highlights three potential interaction residues; clockwise from top these are Y611, I607 and N548.
F) Immunoprecipitation-western analysis depicting the interaction of HSP70 point mutants with the co-chaperones CHIP and Hsc70. Input is shown on the left, and the co-precipitated CHIP and Hsc70 are depicted on the right. Mouse immunoglobulin (IgG) is the negative control.
G) PES-binding analysis of C-terminal point mutants of HA-tagged HSP70, performed as in (B). Input is shown on the top panel, immunoprecipitation with avidin beads (IP) is on the bottom.
We next sought to narrow down the region of HSP70 responsible for interaction with PES. We previously identified the substrate binding domain (SBD) of HSP70 (amino acids 386-641) as required for PES binding (7). The substrate binding domain of HSP70 can be divided into two subdomains: a beta-sandwich region that maps to amino acids 393-507 (SBD-beta), and a C-terminal alpha-helical “lid” that maps to amino acids 507-616 (SBD-alpha) (20). We generated deletion mutants of these sub-domains and used these in pull-down assays with biotinylated PES. To ensure that these deletion mutants were properly folded, we first checked them for their ability to interact with APAF1 in transfected cells. Several deletion mutants of SBD-alpha (encoding amino acids 386-616, 386-602 and 386-573) retained the ability to interact with APAF1, indicating that they were not grossly misfolded (data not shown). In contrast, deletion mutants in SBD-beta were unable to bind to APAF1 or CHIP, suggesting they were misfolded (data not shown); these were therefore not analyzed further. In biotinylated PES pull-down assays, the HSP70 construct encoding amino acids 386-616 showed strong binding to PES, while that encoding amino acids 386-602 demonstrated markedly impaired binding, and those encoding 386-573 and 386-543 consistently failed to bind (Fig. 1B, C). The combined mapping data indicate that HSP70 requires SBD-alpha (the C-terminal helical lid, including amino acids 573-616) in order to interact with PES (Fig. 1D).
We next chose to use in silico docking in order to identify potential PES binding sites in the SBD of HSP70. Toward this end, the human sequences of HSP70 (Uniprot code P08017: residues 391 to 607) and Hsc70 (Uniprot code P11142: residues 361 to 613) were aligned to two different homologous proteins of known structure (the E. coli DNA-K and Bovine Hsc70 substrate binding domains, PDB codes 1DKZ and 1YUW, respectively) using the program MolIDE (17, 18). Side chain conformations were predicted with the program SCWRL4 (19), and the SCWRL4-generated models were subjected to a simple minimization using Chimera (UCSF). Following generation of this model, in silico docking routines with PES were generated using the program AutoDock. This- analysis revealed three potential PES interaction sites within the SBD of HSP70. We identified predicted contact residues for all three potential docking sites, and generated HSP70 point mutants for each residue; these point mutants were then used in Biotin-PES pull down assays. We found that three different point mutants comprising one of the docking sites all showed markedly impaired PES binding; notably, this docking site was located in the C-terminal helical domain, so this site was consistent with our mapping data. In this docking model, residues N548, I607 and Y611 all make significant contacts with PES (Fig. 1E). Mutagenesis of each of these residues produced HSP70 proteins that showed either normal, or somewhat decreased, interaction with Hsc70 and CHIP (Fig. 1F), but which showed markedly impaired binding to biotinylated PES (Fig. 1G). In contrast, the majority of the other point mutants showed normal ability to bind to PES (data not shown). Our combined data support this binding pocket in the C terminal helical domain of HSP70 as the potential binding site for PES.
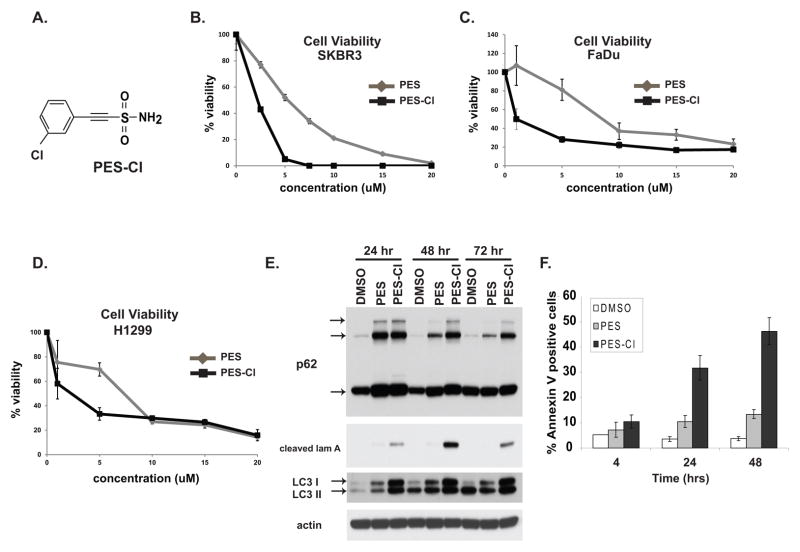
A logical next step for these studies was to perform a preliminary structure-activity relationship for PES. Toward this goal we identified and obtained three analogues of PES from the National Cancer Institute chemical repository (Supp. Fig. 1B). These compounds were compared to PES for cytotoxicity and the ability to inhibit autophagy. Two of these compounds contained a reduced acetylene group and had greatly decreased cytotoxicity, along with decreased ability to inhibit autophagy (Supp. Figs. 1C, D). We next synthesized several derivatives of PES; three of these derivatives replaced the amide group of PES with a pyrrolidine ring (PES-P). One of these derivatives introduced a chloride at the meta position of the phenyl ring (PES-PCl), and another introduced a carboxymethyl group at the para position (PES-PCM) (Supp. Fig. 2A). We found that the compounds with the pyrrolidine ring generally showed reduced cytotoxicity compared to PES; however, the compound containing a pyrrolidine ring plus a chloride on the phenyl ring (PES-PCl) had comparable cytotoxicity (Supp. Figs. 2B, C). Additionally, PES-PCl was consistently able to induce programmed cell death, an attribute unique to this derivative (Supp. Fig. 2C). These data prompted us to synthesize and test a PES derivative containing only a chloride on the phenyl ring, or 2-(3-chlorophenyl) ethynesulfonamide, hereafter referred to as PES-Cl.
We next compared the properties of PES-Cl (Fig. 2A) with the parent compound PES using assays for cell viability, apoptosis, autophagy inhibition, and the ability to reduce the levels of HSP90 client proteins. Cell viability assays indicated that PES-Cl has an IC50 in tumor cells that is up to 8-fold reduced relative to the parent compound PES (Fig. 2B–D). We also found that PES-Cl has superior ability to cause increased p62SQSTM1 and LC3 accumulation, indicators of autophagy inhibition (Fig. 2E). Notably, unlike PES, treatment of cells with PES-Cl was accompanied by robust programmed cell death, as determined by the appearance of cleaved lamin A (Fig. 2E) and Annexin V positive cells (Fig. 2F). Like PES, we found that PES-Cl showed comparable ability to bind to HSP70 (Supp. Fig. 3A), limited cytotoxicity to non-transformed human fibroblasts (Supp. Fig. 3B), and consistent ability to down-regulate the soluble levels of HSP90 client proteins (Supp. Fig. 3C). In silico docking revealed that PES-Cl has the potential to dock in the identical site on the C-terminal helical bundle in HSP70 (Supp. Fig. 3D). These data indicated that PES-Cl might prove to be a superior anti-cancer compound.
Figure 2. PES-Cl shows increased cytotoxicity and superior ability to inhibit autophagy.
A) Chemical structure of PES-Cl.
B, C, D) Cell viability analysis (MTT) in the breast carcinoma line SKBR3 (B), the head and neck squamous cell carcinoma line FaDu (C), and the lung adenocarcinoma line H1299 (D), treated with PES or PES-Cl. Cells were treated with the indicated dose for forty eight hours. Data are the averaged results from three independent experiments, and error bars mark standard deviations.
E) Immunoblot analysis for the autophagy markers p62SQSTM1 and LC3 I and II, as well as caspase-cleaved lamin A (cleaved lam A) in H1299 cells treated with 10 uM PES or PES-Cl for the indicated timepoints. The arrows mark monomeric and oligomeric forms of p62, as well as cleaved and lipidated forms of LC3 I and II.
F) Quantification of Annexin V positive H1299 cells following treatment with 10 uM PES or PES-Cl for the indicated timepoints. Data are the averaged results from three independent experiments, and error bars mark standard deviations.
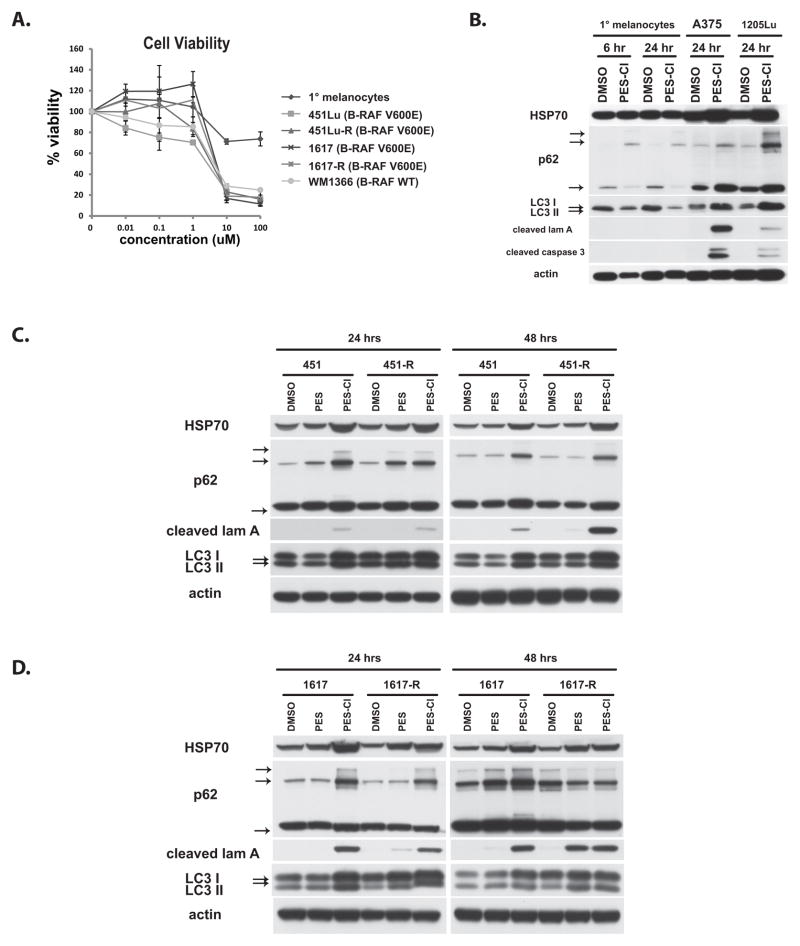
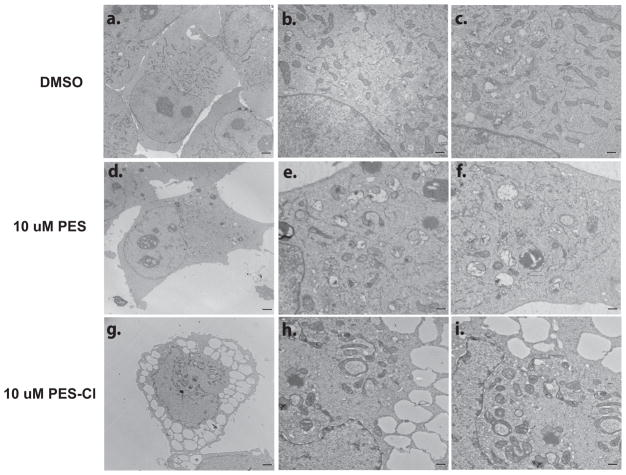
In order to further test the potential of PES-Cl as a promising anti-cancer compound, we chose to compare the cytotoxicity of PES-Cl in primary melanocytes to that in five different melanoma cell lines. Two of these melanoma lines have acquired resistance to BRAF inhibitors (451Lu-R and 1617-R), and one is intrinsically resistant (WM1366), therefore giving us the opportunity to analyze the efficacy of PES-Cl in what are otherwise chemo-resistant cancer lines (20). We found that the IC50 for PES-Cl in all five melanoma cell lines is between 2–5 uM; in contrast the IC50 for primary melanocytes is over 100 uM (Fig. 3A). The increased cytotoxicity in melanomas compared to primary melanocytes is associated with increased inhibition of autophagy and increased apoptosis, as assessed by p62SQSTM1 accumulation and the appearance of caspase-cleaved lamin A and caspase-3 in melanomas (Fig. 3B). Interestingly, the BRAF-inhibitor-resistant melanomas showed either indistinguishable or increased apoptosis in response to PES-Cl, compared to parental cells (Fig. 3C, D). Electron microscopy analysis supported the premise that PES-Cl has superior ability to inhibit autophagy compared to PES, as evidenced by a significantly greater accumulation of autophagosomes in tumor cells (Fig. 4). The combined data indicate that PES-Cl retains all of the activities of PES (including the preferential death of tumor over normal cells), but in addition shows superior cytotoxicity and the unique ability to induce apoptosis.
Figure 3. Efficacy of PES-Cl in melanoma.
A) Cell viability analysis (MTT) of primary neonatal epidermal melanocytes (black diamonds), and melanoma lines 451Lu (BRAF V600E mutant), 1617 (BRAF V600E mutant) and WM1366 (wt BRAF, mutant NRas) as well as two melanoma lines that are resistant to the BRAF inhibitor SB-590885 (451Lu-R and 1617-R). Cells were treated with the indicated doses for forty eight hours. Data are the averaged results from three independent experiments, and error bars mark standard deviations.
B) Immunoblot analysis of autophagy (p62SQSTM1, LC3 I and II) and apoptosis (cleaved lamin A, cleaved caspase 3) markers in primary (1°) melanocytes and melanomas A375 and 1205Lu treated with 10 uM PES-Cl for the indicated times. The arrows mark monomeric and oligomeric forms of p62, as well as LC3 I and II.
C, D) Immunoblot analysis of autophagy (p62SQSTM1, LC3 I and II) and apoptosis (cleaved lamin A) markers in parental 451 and BRAF-inhibitor resistant 451-R melanoma lines (C) as well as 1617 parental and BRAF-inhibitor resistant 1617-R melanoma lines (D) treated with 10 uM PES or PES-Cl for the indicated times. The arrows mark monomeric and oligomeric forms of p62, as well as LC3 I and II.
Figure 4. Enhanced autophagosome accumulation in cells treated with PES-Cl.
Electron micrographs of H1299 cells treated with 10 uM PES or PES-Cl for twenty four hours. Scale bar in a, d and g is 2 microns. Scale bar in b, c, e, f, h and i is 500 nm.
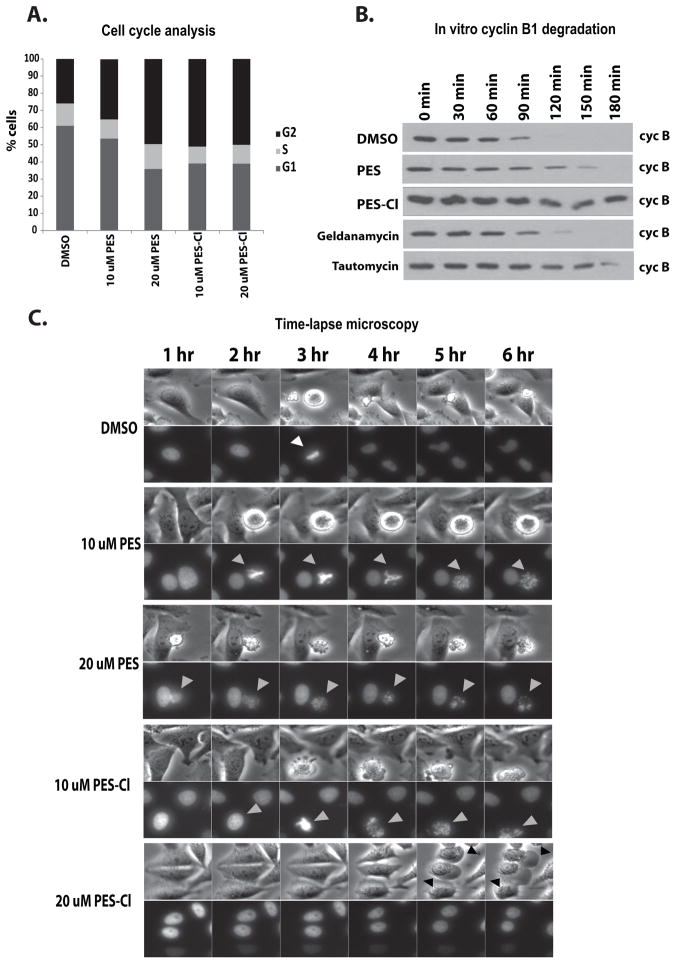
In the course of these analyses we noted apparent mitotic defects in cells exposed to PES-Cl. Therefore, we performed cell cycle analysis of tumor cells treated with both HSP70 inhibitors. This analysis revealed that treatment of tumor cells with these compounds led to a significant accumulation of cells in the G2/M phase of the cell cycle (Fig. 5A). These data are consistent with the finding that silencing HSP70 leads to G2/M arrest (4). In order to probe the possible mechanism underlying G2/M arrest by HSP70 inhibitors, we noted a previous study suggesting that an ATP-dependent chaperone controls the Anaphase Promoting Complex/Cyclosome (APC/C) (16). To test the hypothesis that this ATP-dependent chaperone was HSP70, we monitored cyclin B1 degradation in cell-free extracts made from synchronized HeLa cells. In this assay, extracts were made from HeLa cells synchronized in G1/S using thymidine block; following addition of ATP, the activity of the APC/C is monitored by the degradation of cyclin B1. Notably, the ability of the extracts to activate the APC/C and degrade cyclin B was markedly inhibited by PES-Cl, to levels comparable to the positive control tautomycin (Fig. 5B). These data suggested that these compounds were affecting a biochemical process required for mitotic exit and thus sustaining the mitotic checkpoint. To probe this finding further, HeLa cells stably transfected with GFP-tagged histone H2B were synchronized in G1/S following thymidine block, released, and then treated with PES or PES-Cl, followed by time-lapse video microscopy. This analysis revealed that the majority of PES and PES-Cl-treated cells arrested at G2/M with condensed chromosomes, but that in a percentage of cases these arrested cells appeared to transit through mitosis, resulting in defects in chromosome segregation (Fig. 5C, grey arrows) and eventual cell death.
Figure 5. Treatment with PES or PES-Cl causes G2/M cell cycle arrest and inhibition of APC/C function.
A) Asynchronous HeLa cells were treated with DMSO, PES or PES-Cl at the doses indicated and analyzed for cell cycle stage by propidium iodide staining. Data are the average results from three independent experiments.
B) The degradation of cyclin B in synchronized HeLa cell extracts was monitored by immunoblot analysis. Equal amounts of extract were analyzed at the timepoints indicated following treatment of extracts with DMSO or 10 uM PES or PES-Cl. Treatment with 10 uM Geldanamycin or 150 nM Tautomycin served as positive controls.
C) Time-lapse video microscopy of HeLa cells that were released from synchronization in S phase following thymidine block and treated with the indicated concentrations of DMSO, PES or PES-Cl for 3 hours. Top panels: phase contrast images. Bottom panels: GFP-H2B fluorescence. Gray arrowheads: treated cells with chromosome abnormalities. Representative images at the indicated time points are shown.
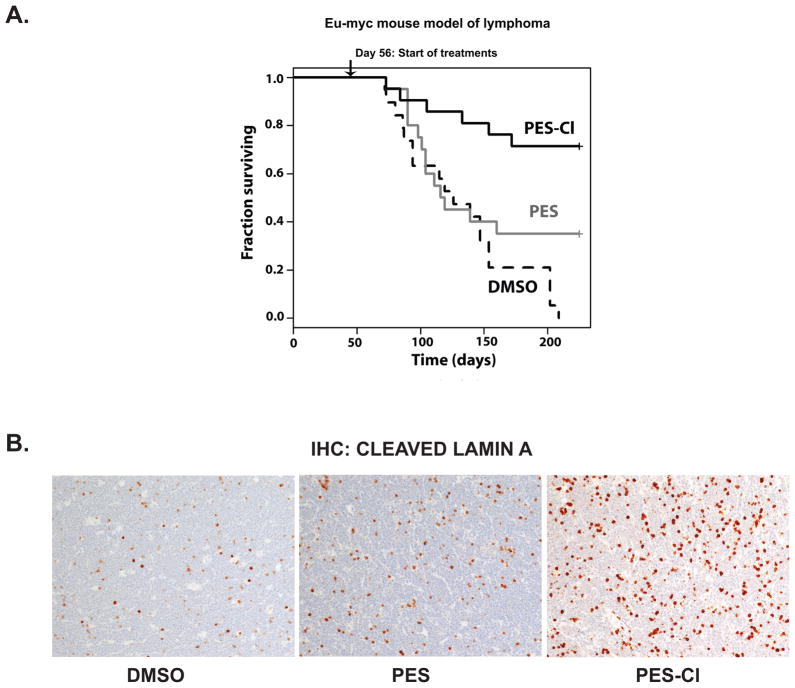
Previously we showed that PES was efficacious as an anti-cancer agent in Eμ-myc mice, which develop pre-B cell lymphoma (7); this transgenic tumor model appears to be particularly sensitive to the effects of autophagy inhibition (22). To compare the efficacy of PES and PES-Cl in lymphoma we treated Eμ-myc mice of 8 weeks of age and no evidence for cancer with dilution vehicle (n=19 mice), PES (20 mg/kg; n=20 mice) and PES-Cl (20 mg/kg; n=21 mice), administered by intra-peritoneal injection once per week for 20 weeks. Whereas all of the vehicle-treated mice succumbed to lymphoma by day 210, 35% of the PES-treated mice survived to this time point, and 71.4% of mice treated with PES-Cl survived (Fig. 6A). The difference between PES and PES-Cl was significant (p=0.015) and the difference between vehicle and PES-Cl was highly significant (p<0.000006). To confirm that PES and PES-Cl functioned as predicted in these B cell lymphomas, we isolated tumor-containing lymph nodes of approximately equal-size from control, PES- and PES-Cl treated mice, and assessed the level of apoptosis in these tumors 24 hours after treatment. Notably, whereas there were low levels of cells positive for cleaved lamin A in the control and PES-treated mice, there was a marked increase in apoptotic cells in the PES-Cl treated mice (Fig. 6B). These data indicate that PES-Cl functions as expected to induce programmed cell death in the tumors of treated mice.
Figure 6. PES-Cl significantly promotes survival in the Eμ-myc mouse model of lymphoma.
A) Eight week old Eμ-myc mice were treated once per week with intra-peritoneal injections of DMSO (n=19), 20 mg/kg PES (n=20) or 20 mg/kg PES-Cl (n=21), for a total of 20 treatments. All treatments were terminated at day 209. Log rank analysis of data: DMSO vs. PES (p = 0.0839); PES vs. PES-Cl (p = 0.0151) and DMSO vs. PES-Cl (p = 4.7 × 10−6).
B) Immunohistochemistry for cleaved lamin A in lymph nodes from tumor-bearing Eμ-myc mice twenty-four hours after treatment with DMSO, PES or PES-Cl (10% DMSO in PBS, or 20 mg/kg PES or PES-Cl in 10% DMSO). Samples shown are from equal-sized tumors from 2 mice each and are representative of multiple views from each tumor.
Discussion
The heat shock proteins are attractive targets for anti-cancer therapeutics. Considerable efforts have been made toward the generation and testing of inhibitors of HSP90. In the clinic these compounds have not been entirely promising (23, 24); in part, this is believed to be because inhibition of HSP90 causes activation of the transcription factor HSF-1, which transactivates HSP70. In fact, for most HSP90 inhibitors, the ability to activate HSF1 and upregulate HSP70 is a biomarker for activity (24). Because of this phenomenon, some researchers have proposed that combining HSP90 and HSP70 inhibitors together should increase efficacy. Notably however, most HSP70 inhibitors (including PES (9)) inhibit both HSP70 and HSC70, the constitutively expressed homologue. Powers and colleagues have shown that inhibition of both HSP70 and HSC70 leads to concomitant inhibition of HSP90; this is because HSP70 and HSC70 are critical co-chaperones for HSP90 (26). Indeed, we have previously shown that PES can sequester and inactivate HSP90 client proteins (9). Therefore, we propose that HSP70 inhibitors alone will show marked anti-cancer efficacy, and that this class of compounds may one day overtake the use of HSP90 inhibitors in the clinic.
Our data indicate that PES-Cl may be a more efficacious anticancer compound, compared to the parental compound PES. While PES-Cl showed only a modest decrease in IC50 relative to the parent compound, it conferred a marked and statistically significant increase in survival in our B cell lymphoma model (35% survival at the end of the study for PES, compared to 71% for PES-Cl; p=0.015). The discrepancy between in vitro and in vivo results may reflect differences in short term versus long term assays, or may reflect an impact of HSP70 on the tumor micro-environment, as has been demonstrated (27). In our pre-clinical model, there are several reasons why PES-Cl might show improved efficacy over the parent compound. The presence of the extra halogen in PES-Cl could increase the stability of the interaction with HSP70. Alternatively this chloride could increase membrane permeability or absorption, or may alter cytochrome P450 metabolism (28). Interestingly, our data also indicate that PES-Cl, unlike PES, can induce programmed cell death. We find that PES-Cl is a superior inhibitor of autophagy, and autophagy inhibition is known to induce programmed cell death (22, 29). However, at present we cannot rule out the possibility that the ability of PES-Cl to induce apoptosis is due to a unique action of this compound, and is the predominant mode of cytotoxicity. This possibility remains to be explored.
The Anaphase Promoting Complex/Cyclosome (APC/C) is the most complex ubiquitin ligase known, consisting of more than 11 subunits. Several years ago Hershko and colleagues showed that APC/C was inactive in mitotic extracts from nocodazole-arrested HeLa cells (30), but that addition of ATP converted APC/C to its active form, and allowed it to mediate the degradation of cyclin B1 in cell-free extracts (31). Notably, this group discovered that ATP hydrolysis was required for APC/C activation, suggesting the involvement of a required ATPase (16). Because we find that PES-Cl efficiently inhibits this process, our data indicate that this ATPase may be HSP70; alternatively, the process may be regulated in some way by HSP70. To date, there are few other examples of pharmacological inhibitors of the APC/C; one of these, TAME (tosyl-L-arginine methyl ester) was identified by virtue of its ability to inhibit cyclin B1 degradation in Xenopus extracts (32). Mechanistically, TAME prevents Cdc20 association with the APC/C (33), but at present, TAME has not been tested in pre-clinical models of cancer. Therefore, PES and PES-Cl appear to be unique inhibitors of APC/C function with promising pre-clinical activity. We are currently exploring the mechanism whereby these compounds inhibit APC/C.
In addition to PES and PES-Cl, there are at least two other HSP70 inhibitors that have been identified and studied for anti-cancer activity. One of these is the adenosine analog VER-155008, and the other is YM-1, a derivative of the cationic rhodacyanine dye MKT-077. Like PES and PES-Cl, these compounds are cytotoxic to a broad range of tumor cells, and show limited cytotoxicity to normal, non-transformed cells (34–38). Unlike PES and PES-Cl, however, there are limited pre-clinical data on their efficacy. Additionally, to date only PES and PES-Cl have been shown to inhibit autophagy, induce apoptosis (PES-Cl), inhibit HSP90 client protein function, induce G2/M arrest, and inhibit the activity of the Anaphase Promoting Complex/Cyclosome (APC/C). It is of note that HSP70 is also implicated in immune destruction of tumor cells (26); therefore, of the ten ‘hallmarks’ of cancer outlined by Hanahan and Weinberg (39), HSP70 inhibitors have the potential to target at least four (cell death, cellular energetics, proliferative signaling and immune destruction). These compounds therefore represent an exciting new class of anticancer drugs.
Supplementary Material
Supplemental Figure 1. Analysis of PES derivatives for effects on cell viability and inhibition of autophagy.
A) Chemical structure of biotinylated PES
B) Chemical structure of PES and derivatives NSC85560, NSC166772 and NSC303578 (nomenclature is from the National Cancer Institute Drug Repository).
C) Cell viability of H1299 and PANC1 cells after 48 hour treatment with 10 uM PES, NSC85560 (855600), NSC166777 (16677) and NSC303578 (303578). Data are the averaged results from three independent experiments, and error bars mark standard deviations.
D) Immunoblot analysis of H1299 cells following treatment with 10 uM of the compounds indicated for the timepoints indicated, using antisera to the autophagy (p62SQSTM1 and LC3) and apoptosis (cleaved lamin A) markers indicated, as well as HSP70 and actin (control). The arrows mark monomeric and oligomeric forms of p62SQSTM1, as well as cleaved and lipidated forms of LC3 I and II.
Supplemental Figure 2. PES-PCl induces apoptosis.
A) Chemical structures of PES and synthesized derivatives. P=pyrrolidine; CM= carboxymethyl; Cl=chloride.
B) MTT cell viability analyses of H1299 and PANC1 cells after 48 hour treatment with 10 uM PES, PES-P, PES-PCM and PES-PCl. The values shown are the averaged results from three independent experiments, and error bars mark standard deviations.
C) Immunoblot analysis of H1299 cells following treatment with 10 uM of the compounds indicated for the timepoints indicated, using antisera to the autophagy (p62SQSTM1 and LC3) and apoptosis (cleaved lamin A) markers indicated, as well as HSP70 and actin (control). The arrows mark monomeric and oligomeric forms of p62SQSTM1, as well as cleaved and lipidated forms of LC3 I and II.
Supplemental Figure 3. PES-Cl shows increased cytotoxicity compared to PES; PES-Cl retains the ability to bind to HSP70
A) Binding assays of H1299 cells transfected with HA-tagged full-length and truncated HSP70 encoding the amino acids (aa) shown, treated with 20 uM biotinylated PES (B-PES) and 20 uM PES-Cl (PES-Cl). Biotinylated PES complexes were precipitated with avidin beads, eluted and detected with an anti-HA antibody following imunoblotting. Input is shown on the top panel, immunoprecipitation (IP) with avidin is depicted on the bottom panel.
B) Cell viability of WI38 non-transformed fibroblasts after 48 hour treatment with 10 uM PES and PES-Cl. The values shown are the averaged results from three independent experiments, and error bars mark standard deviations.
C) Western blot analysis of HSP90 client proteins in soluble cell extract following 48 hours of treatment with 10 uM PES-Cl. Actin is shown as a loading control.
D) In silico docking of PES-Cl to the substrate binding domain of HSP70.
Acknowledgments
This research was supported by the National Cancer Institute through R01 CA139319 (M. Murphy), R01 CA118761 (D.L. George), by a pilot project from the Head and Neck Cancer Keystone at Fox Chase Cancer Center (G.M. Balaburski), and by R01 GM84453 (R.L. Dunbrack). S.H. was supported by T32CA009035-36 (Fox Chase Cancer Center). We thank Christine Eischen (Vanderbilt University Medical Center) for Eμ-myc lymphoma lines and Erica Golemis (Fox Chase Cancer Center) for support. This work utilized the Organic Synthesis Facility, the Molecular Modeling Facility and the Laboratory Animal Facility at Fox Chase Cancer Center; this work used the Electron Microscopy Core Facility at the Raymond and Ruth Perelman School of Medicine at the University of Pennsylvania. PES-pyrrolidine compounds were designed and synthesized at the Fox Chase Chemical Diversity Center, Doylestown PA.
Footnotes
Authors’ contributions:
Conception and Design: GMB, JIJL, NB, RLD, DLG and MEM
Development of Methodology: NB, SH, MDA, RLD
Acquisition of data: GMB, JIJL, NB, SH, MDA, GZ
Analysis and interpretation of data: all authors
Writing, review and/or revision of manuscript: all authors
Administrative, technical or material support: RLD, TY, DLG, MEM
Study supervision: RLD, TY, DLG, MEM
The authors declare there are no conflicts of interest
References
- 1.Daugaard M, Rohde M, Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–10. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 2.Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, Pandita TK. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol Cell Biol. 2004;24:899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powers MV, Jones K, Barillari C, Westwood I, van Montfort RL, Workman P. Targeting HSP70: the second potentially druggable heat shock protein and molecular chaperone? Cell Cycle. 2010;9:1542–50. doi: 10.4161/cc.9.8.11204. [DOI] [PubMed] [Google Scholar]
- 4.Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, Jäättelä M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005;19:570–82. doi: 10.1101/gad.305405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt E, Maingret L, Puig PE, Rerole AL, Ghiringhelli F, Hammann A, Solary E, Kroemer G, Garrido C. Heat shock protein 70 neutralization exerts potent antitumor effects in animal models of colon cancer and melanoma. Cancer Res. 2006;66:4191–7. doi: 10.1158/0008-5472.CAN-05-3778. [DOI] [PubMed] [Google Scholar]
- 6.Rérole AL, Gobbo J, De Thonel A, Schmitt E, Pais de Barros JP, Hammann A, Lanneau D, Fourmaux E, Deminov O, Micheau O, Lagrost L, Colas P, Kroemer G, Garrido C. Peptides and aptamers targeting HSP70: a novel approach for anticancer chemotherapy. Cancer Res. 2011;71:484–95. doi: 10.1158/0008-5472.CAN-10-1443. [DOI] [PubMed] [Google Scholar]
- 7.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nylandsted J, Gyrd-Hansen M, Danielewicz A, Fehrenbacher N, Lademann U, Høyer-Hansen M, Weber E, Multhoff G, Rohde M, Jäättelä M. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J Exp Med. 2004;200:425–35. doi: 10.1084/jem.20040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leu JI, Pimkina J, Pandey P, Murphy ME, George DL. HSP70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Mol Cancer Res. 2011;9:936–47. doi: 10.1158/1541-7786.MCR-11-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaiser M, Kuhnl A, Reins J, Fischer S, Ortiz-Tanchez J, Schlee C, Mochmann LH, Heesch S, Benlasfer O, Hofmann W-K, Thiel E, Baldus CD. Antileukemic activity of the HSP70 inhibitor pifithrin-μ in acute leukemia. Blood Cancer J. 2011;1:e28. doi: 10.1038/bcj.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steele AJ, Prentice AG, Hoffbrand AV, Yogashangary BC, Hart SM, Lowdell MW, Samuel ER, North JM, Nacheva EP, Chanalaris A, Kottaridis P, Cwynarski K, Wickremasinghe RG. 2-Phenylacetylenesulfonamide (PAS) induces p53-independent apoptotic killing of B-chronic lymphocytic leukemia (CLL) cells. Blood. 2009;114:1217–25. doi: 10.1182/blood-2008-11-190587. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Fukunaga-Kalabis M, Herlyn M. Isolation and cultivation of dermal stem cells that differentiate into functional epidermal melanocytes. Methods Mol Biol. 2012;806:15–29. doi: 10.1007/978-1-61779-367-7_2. [DOI] [PubMed] [Google Scholar]
- 13.Humbey O, Pimkina J, Zilfou JT, Jarnik M, Dominguez-Brauer C, Burgess DJ, Eischen CM, Murphy ME. The ARF tumor suppressor can promote the progression of some tumors. Cancer Res. 2008;68:9608–13. doi: 10.1158/0008-5472.CAN-08-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patrone JD, Yao J, Scott NE, Dotson GD. Selective inhibitors of bacterial phosphopantothenoylcysteine synthetase. J Am Chem Soc. 2009;131:16340–1. doi: 10.1021/ja906537f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–36. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miniowitz-Shemtov S, Teichner A, Sitry-Shevah D, Hershko A. ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc Natl Acad Sci U S A. 2010;107:5351–6. doi: 10.1073/pnas.1001875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Canutescu AA, Dunbrack RL., Jr SCWRL and MolIDE: computer programs for side-chain conformation prediction and homology modeling. Nat Protoc. 2008;3:1832–47. doi: 10.1038/nprot.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 19.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 20.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, Santiago-Walker AE, Letrero R, D’Andrea K, Pushparajan A, Hayden JE, Brown KD, Laquerre S, McArthur GA, Sosman JA, Nathanson KL, Herlyn M. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daugaard M, Rohde M, Jäättelä M. The heat shock protein 70 family: Highly homologousproteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–10. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 22.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pacey S, Gore M, Chao D, Banerji U, Larkin J, Sarker S, Owen K, Asad Y, Raynaud F, Walton M, Judson I, Workman P, Eisen T. A Phase II trial of 17-allylamino, 17-demethoxygeldanamycin (17-AAG, tanespimycin) in patients with metastatic melanoma. Invest New Drugs. 2012;30:341–9. doi: 10.1007/s10637-010-9493-4. [DOI] [PubMed] [Google Scholar]
- 24.Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, Teitcher J, Wolchok JD, Germino FJ, Krown SE, Coit D, Rosen N, Chapman PB. Phase II trial of 17-allylamino-17 demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res. 2008;14:8302–7. doi: 10.1158/1078-0432.CCR-08-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powers MV, Clarke PA, Workman P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell. 2008;14:250–62. doi: 10.1016/j.ccr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, Boireau W, Rouleau A, Simon B, Lanneau D, De Thonel A, Multhoff G, Hamman A, Martin F, Chauffert B, Solary E, Zitvogel L, Garrido C, Ryffel B, Borg C, Apetoh L, Rébé C, Ghiringhelli F. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120:457–71. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandes MZ, Cavalcanti SM, Moreira DR, de Azevedo WF, Junior, Leite AC. Halogen atoms in the modern medicinal chemistry: hints for the drug design. Curr Drug Targets. 2010;11:303–14. doi: 10.2174/138945010790711996. [DOI] [PubMed] [Google Scholar]
- 29.Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D, Souquere S, Yoshimori T, Pierron G, Codogno P, Kroemer G. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–40. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braunstein I, Miniowitz S, Moshe Y, Hershko A. Inhibitory factors associated with anaphase- promoting complex/cylosome in mitotic checkpoint. Proc Natl Acad Sci U S A. 2007;104:4870–5. doi: 10.1073/pnas.0700523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eytan E, Braunstein I, Ganoth D, Teichner A, Hittle JC, Yen TJ, Hershko A. Two different mitotic checkpoint inhibitors of the anaphase-promoting complex/cyclosome antagonize the action of the activator Cdc20. Proc Natl Acad Sci U S A. 2008;105:9181–5. doi: 10.1073/pnas.0804069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verma R, Peters NR, D’Onofrio M, Tochtrop GP, Sakamoto KM, Varadan R, Zhang M, Coffino P, Fushman D, Deshaies RJ, King RW. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science. 2004;306:117–20. doi: 10.1126/science.1100946. [DOI] [PubMed] [Google Scholar]
- 33.Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, Hathaway N, Dimova N, Cuny GD, King RW. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–95. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wadhwa R, Sugihara T, Yoshida A, Nomura H, Reddel RR, Simpson R, Maruta H, Kaul SC. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 2000;60:6818–21. [PubMed] [Google Scholar]
- 35.Massey AJ, Williamson DS, Browne H, Murray JB, Dokurno P, Shaw T, Macias AT, Daniels Z, Geoffroy S, Dopson M, Lavan P, Matassova N, Francis GL, Graham CJ, Parsons R, Wang Y, Padfield A, Comer M, Drysdale MJ, Wood M. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother Pharmacol. 2010;66:535–45. doi: 10.1007/s00280-009-1194-3. [DOI] [PubMed] [Google Scholar]
- 36.Macias AT, Williamson DS, Allen N, Borgognoni J, Clay A, Daniels Z, Dokurno P, Drysdale MJ, Francis GL, Graham CJ, Howes R, Matassova N, Murray JB, Parsons R, Shaw T, Surgenor AE, Terry L, Wang Y, Wood M, Massey AJ. Adenosine-derived inhibitors of 78 kDa glucose regulated protein (Grp78) ATPase: insights into isoform selectivity. J Med Chem. 2011;54:4034–41. doi: 10.1021/jm101625x. [DOI] [PubMed] [Google Scholar]
- 37.Rousaki A, Miyata Y, Jinwal UK, Dickey CA, Gestwicki JE, Zuiderweg ER. Allosteric drugs: the interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J Mol Biol. 2011;411:614–32. doi: 10.1016/j.jmb.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koren J, 3rd, Miyata Y, Kiray J, O’Leary JC, 3rd, Nguyen L, Guo J, Blair LJ, Li X, Jinwal UK, Cheng JQ, Gestwicki JE, Dickey CA. Rhodacyanine derivative selectively targets cancer cells and overcomes tamoxifen resistance. PLoS One. 2012;7:e35566. doi: 10.1371/journal.pone.0035566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Analysis of PES derivatives for effects on cell viability and inhibition of autophagy.
A) Chemical structure of biotinylated PES
B) Chemical structure of PES and derivatives NSC85560, NSC166772 and NSC303578 (nomenclature is from the National Cancer Institute Drug Repository).
C) Cell viability of H1299 and PANC1 cells after 48 hour treatment with 10 uM PES, NSC85560 (855600), NSC166777 (16677) and NSC303578 (303578). Data are the averaged results from three independent experiments, and error bars mark standard deviations.
D) Immunoblot analysis of H1299 cells following treatment with 10 uM of the compounds indicated for the timepoints indicated, using antisera to the autophagy (p62SQSTM1 and LC3) and apoptosis (cleaved lamin A) markers indicated, as well as HSP70 and actin (control). The arrows mark monomeric and oligomeric forms of p62SQSTM1, as well as cleaved and lipidated forms of LC3 I and II.
Supplemental Figure 2. PES-PCl induces apoptosis.
A) Chemical structures of PES and synthesized derivatives. P=pyrrolidine; CM= carboxymethyl; Cl=chloride.
B) MTT cell viability analyses of H1299 and PANC1 cells after 48 hour treatment with 10 uM PES, PES-P, PES-PCM and PES-PCl. The values shown are the averaged results from three independent experiments, and error bars mark standard deviations.
C) Immunoblot analysis of H1299 cells following treatment with 10 uM of the compounds indicated for the timepoints indicated, using antisera to the autophagy (p62SQSTM1 and LC3) and apoptosis (cleaved lamin A) markers indicated, as well as HSP70 and actin (control). The arrows mark monomeric and oligomeric forms of p62SQSTM1, as well as cleaved and lipidated forms of LC3 I and II.
Supplemental Figure 3. PES-Cl shows increased cytotoxicity compared to PES; PES-Cl retains the ability to bind to HSP70
A) Binding assays of H1299 cells transfected with HA-tagged full-length and truncated HSP70 encoding the amino acids (aa) shown, treated with 20 uM biotinylated PES (B-PES) and 20 uM PES-Cl (PES-Cl). Biotinylated PES complexes were precipitated with avidin beads, eluted and detected with an anti-HA antibody following imunoblotting. Input is shown on the top panel, immunoprecipitation (IP) with avidin is depicted on the bottom panel.
B) Cell viability of WI38 non-transformed fibroblasts after 48 hour treatment with 10 uM PES and PES-Cl. The values shown are the averaged results from three independent experiments, and error bars mark standard deviations.
C) Western blot analysis of HSP90 client proteins in soluble cell extract following 48 hours of treatment with 10 uM PES-Cl. Actin is shown as a loading control.
D) In silico docking of PES-Cl to the substrate binding domain of HSP70.