Abstract
The safety of HIV-1 based vectors was evaluated during the production of transgenic sheep. Vectors were introduced into the perivitelline space of in vivo derived one-cell sheep embryos by microinjection then transferred into the oviducts of recipient females. At 60–70 days of gestation, a portion of the recipients were euthanized and tissues collected from both surrogates and fetuses. Other ewes were allowed to carry lambs to term. Inadvertent transfer of vector from offspring to surrogates was evaluated in 330 blood and tissue samples collected from 57 ewes that served as embryo recipients. Excluding uterine contents, none of the samples tested positive for vector, indicating that that the vector did not cross the fetal maternal interface and infect surrogate ewes. Evaluating ewes, fetuses and lambs for replication competent lentivirus (RCL); 84 serum samples analyzed for HIV-1 capsid by ELISA and over 600 blood and tissue samples analyzed by quantitative PCR for the VSV-G envelopes revealed no evidence of RCL. Results of these experiments provide further evidence as to the safety of HIV-1 based vectors in animal and human applications.
Keywords: transgenic sheep, lentiviral vectors, replication competent lentivirus, embryo microinjection
Introduction
The ultimate goal of many in vitro assays for manipulating gene expression is the transition to in vivo applications such as somatic cell gene therapy or genetic engineering of plants and animals. In order to make this transition, stable and effective delivery and expression of the transgene is necessary. To accomplish this goal, lentiviral vectors have become a popular tool of choice since they permit entry of the transgene into the cell as RNA that is then reverse transcribed into DNA and integrated into the genome.(Manjunath et al. 2009) In the case of transgenic animal production, this integration allows for stable transgene expression throughout the life of the animal. In addition, expression of the transgene in the F1 generation from transgenic animals using lentiviral vectors suggests a lack of silencing of the transgene in the germline.(Lois et al. 2002) This is advantageous over other retroviral systems that have shown a lack of expression both in the F0 and F1 generations.(Jahner et al. 1982)
Although lentiviral vectors are designed using exogenous retroviral elements, the components that make the virus replication competent are removed to ensure the vectors are unable to replicate. (Dull et al. 1998) Moreover, lentiviral vectors are designed to contain a self-inactivating 3′ long terminal repeat (LTR) region that, when transferred to the 5′ LTR during reverse transcription and integration, results in transcriptional inactivation.(Miyoshi et al. 1998; Zufferey et al. 1998) These measures are in place to ensure safe, stable, and efficient delivery of transgenes both in vitro and in vivo.
In spite of these precautionary steps, there is still some concern that recombination of lentiviral vectors with wild-type viruses or endogenous retroviral elements may allow the integrated provirus genome to become a replication competent lentivirus (RCL), perhaps resulting in disease. In addition, when using lentiviral vectors to produce transgenic animals there is also the concern of inadvertent transfer of vector sequences to non-target tissues. Since embryos are implanted shortly after injection with high-titer lentivirus, exposure to surrogate mothers could occur after embryo transfer and/or fetal cells could potentially migrate across the placenta prior to or during delivery resulting in the introduction of vector into the surrogate. Given a major focus of the research conducted in our laboratory involves the production of transgenic animals by infecting preimplantation-stage embryos with a recombinant lentivirus followed by embryo transfer, we investigated the potential for our vector to give rise to RCL and/or infect our surrogate ewes after embryo transfer. To evaluate this safety concern, tissues were collected from pregnant recipients at 60–70 days of gestation or post-lambing. Serum was analyzed for RCL using a p24 ELISA and tissue samples were evaluated using real time quantitative PCR (qPCR). In select animals, a stringent biologic assay currently used to certify HIV-1 vectors for human use was also performed.(Cornetta et al. 2011). Tissue samples collected from recipient ewes was also extensively analyzed to assess the risk of inadvertent transfer of vector sequences from the embryo/fetus to their surrogate mothers.
This analysis found no evidence of RCL in transgenic animals generated by lentiviral gene transfer or the recipient ewes. Moreover, there was no evidence that transgene vector sequences were transferred to the surrogate mothers. These findings build confidence in the safety of lentiviral-mediated transgene delivery to produce genetically modified livestock and provide additional supportive data for human gene therapy applications.
MATERIALS AND METHODS
Production of recombinant lentivirus
Second generation lentiviral plasmids used in this experiment contained either the elongation factor 1-alpha or cytomegalovirus promoter driving expression of a short hairpin RNA (shRNA) designed to inhibit Foot and Mouth Disease virus replication, or silence the expression of myostatin; both vectors also contained the GFP gene. Recombinant lentivirus was produced as described by Miyoshi et al. (Miyoshi et al. 1998) using the appropriate lentiviral plasmid. Lentiviral plasmid (10μg) was co-transfected into HEK 293T cells along with 10μg pCMV R8.91 (packaging plasmid) and 1μg pMDG. The resulting lentivirus was ultracentrifuged at approximately 50,000 × g for 1.5 hours and resuspended in 30μL sterile PBS with 8μg/mL hexadimethrine bromide for microinjection. This resulted in a recombinant lentiviral titer of 1 × 109 IU/mL that is capable of infection and stable integration of GFP into the genome of primary cells or early stage embryos. This modified vector was selected based on the safety of the self-inactivating lentivirus it produces and previous reports that it has been transmitted through the germline and expressed in F1 progeny (Miyoshi et al. 1998; Carmell et al. 2003). Titer of infectious particles was determined by analysis of GFP expression using FACS. HEK293T cells were plated at a density of 25,000 cells/well in a 48-well plate and transduced with serial dilutions of concentrated lentivirus. Serial dilutions of 0.1, 0.01, 0.001, and 0.0001 were used and all transductions were done with 8μg/mL polybrene. Media was refreshed 18 hours later and cells were maintained in culture for 3 days before evaluation. Titer was calculated as follows:
Only concentrated virus with a titer at or above 1 × 109 IU/mL was used for microinjection of embryos. Concentrated virus was frozen at −80°C in 3–5μl aliquots until needed.
Production and transfer of transgenic embryos
Work was approved by the Texas A&M Animal Care and Use Committee. Ewes were synchronized to estrus using an Eazi-Breed Controlled Intravaginal Drug Releasing (CIDR) Device for 12 days. Superovulation of donor ewes was achieved via twice daily (0700 and 1900h) injections of follicle stimulating hormone (FSH) (Bioniche, Belleville, Ontario, Canada) over a four-day period from Days 9 to 12 after CIDR insertion. Dosage decreased daily (40, 30, 25 and 20 mg, respectively) with a total dosage of 115 mg. On day 11, the CIDR was removed, ewes were administered 15 mg prostaglandin F2 alpha (Lutalyse®, and mated to fertile Suffolk rams. Embryos were collected from donor females via midventral laparotomy, approximately 60 h after CIDR removal. Oviducts were cannulated through the fimbriated end with a sterile plastic catheter. Collection medium was then injected into the oviduct using a syringe fitted with a needle inserted at the utero-tubal junction. Approximately 20 ml of medium was flushed (retrograde wash) through each oviduct. The medium was collected through the plastic catheter which was directed into a sterile container.
One-cell fertilized sheep embryos were identified and isolated with the aid of a dissecting microscope. After microinjection with lentivirus, embryos (3–4 per recipient) were surgically transferred into the oviducts of embryo recipient ewes, ipsilateral to the corpus hemorragicum or dominant follicle.
Recipient ewes were synchronized to estrus using procedures similar to those employed for embryo donors except FSH was not administered to induce superovulation and estrus was determined by housing them with vasectomized rams fitted with marking chalk and observing for mating behavior.
Zygotes were surgically flushed from superovulated donor ewes 24 hours post mating. Micromanipulation was then used to inject approximately 20 picoliters of high titer (109 particles/ml) recombinant lentivirus under the zona pellucida (into the perivitelline space) of the embryos. Following injection, these were surgically transferred back into the oviducts of recipient ewes. Each ewe received 3–4 embryos. Approximately 35 days after embryo transfer ultrasonography was utilized to identify pregnant recipients. At approximately 70 days of gestation, a portion of the pregnant ewes were euthanized and complete hysterectomies performed on the ewes to collect tissues from the fetus(s), placenta and surrogate ewe for analysis. The remaining ewes were maintained throughout gestation and allowed to lamb.
Tissue collection
Tissues collected from recipient ewes for analysis included liver, lung, kidney, lymph node, mammary gland, ovary, skeletal muscle, placentome (when available) and uterus. Tissues were cut into 3–5 mm pieces and preserved in All Protect tissue reagent (QIAGEN, Hilden, Germany). All utensils were treated with 5% bleach and flamed between tissues, and different utensils were used for each animal. In addition, blood was collected from each recipient for analysis. For recipients allowed to lamb, only blood was collected. For recipients euthanized at 60–70 days gestation, fetuses were dissected, and the tissues were collected for analysis of transgene expression. Blood was also collected when possible. For lambs that were born live, blood and an ear biopsy were collected. Lambs that were delivered dead were dissected and tissues collected as described for fetuses.
Quantitative PCR (qPCR)
DNA was analyzed by qPCR based on modification of a previously published method.(Sastry et al. 2002) Primers and probe for the VSV-G envelope were forward 5′-tgcaaggaaagcattgaacaa-3″ (600 nM), reverse 5′-gaggagtcacctggacaatcact-3′ (600 nM) and probe 5′-6FAM-aggaacttggctgaatccaggcttcc-TAMRA-3′ (200 nM). Primers and probe for the vector sequence were forward 5′-acctgaaaggcaaagggaaac-3′ (600 nM), reverse 5′-cacccatctctctccttctagcc-3′ (600 nM), and probe 5′-6FAM-agctctctcgacgcaggactcggc-TAMRA-3′ (200 nM). Duplicate or triplicate reactions, each containing 0.2 μg of DNA, are initiated at 50 °C for 2 min, 95 °C for 10 min then 40 cycles of 95 °C for 15 sec and 60 °C for 1 min. Log dilutions of a VSV-G or transgene plasmid controls (between 101 and 105) are introduced into genomic DNA and used to establish the standard curve; the slope of the standard curve must be within −4.3823 to −3.7157 for the assay to be acceptable. The reactions are run as multiplex reaction with primers and probes for the GAPDH gene [forward 5′-ggcgccaagagggtcat-3′ (50 nM), reverse 5′-gtggttcacgcccatcaca-3′ (50 nM), and probe 5′-VIC-tctctgcaccttctg-MGB-3′ (100 nM)]. Before a VSV-G or vector results are considered negative, the amount of DNA must be confirmed; specifically the DNA content is considered adequate if 2 of the 3 replicates have GAPDH Ct values less than or equal to 2 standard deviations above the mean of control samples. Negative and positive controls must provide expected results for the assay to be acceptable. The level of sensitivity is 10 copies per 0.2 μg of DNA. qPCR analysis was performed under Good Manufacturing Practice (GMP) procedures on file with the US FDA.
RCL Assays
Serum was analyzed for the presence of HIV-1 virus using the Alliance HIV-1 p24 ELISA kit according to manufacturer’s specifications (Perkin Elmer, Waltham, MA). Triplicate samples were analyzed and duplicate samples above the lower limits of detection were required to consider the assay positive for RCL. The biologic RCL assay utilized the previously published method in which the test material is incubated with C8166 cells, a human T cells line shown to effectively amplify HIV-1 virus.(Cornetta et al. 2011) Briefly, C8166 cells were then cultured for three weeks to amplify virus, after which time cell-free culture media was used to infect naïve C8166 cells. After an additional week of culture, the C8166 cells are then analyzed for RCL using p24 ELISA analysis of the culture media and PCR of cellular DNA using primers for vector/viral sequences. Both the p24 evaluation and the biologic assay were performed under GMP procedures on file with the US FDA.
RESULTS
Transgenic sheep production
In an effort to produce genetically modified sheep with improved production characteristics, a total of 87 recipient ewes received 300 embryos microinjected with self-inactivating second generation lentivirus encoding short hairpin RNAs designed to inhibit Foot and Mouth Disease virus replication, or silence the expression of myostatin (to stimulate muscle development), in addition to Green Fluorescent Protein (GFP). Of these, 47 were pregnant as determined by ultrasound on day 35 of gestation. Euthanasia was performed on 26 ewes between day 60 and 70 of gestation, and 41 fetuses were recovered of which 14 were confirmed to be transgenic. The remaining 21 ewes were allowed to lamb resulting in 33 lambs of which 14 were transgenic. Euthanasia and analysis was also performed on 10 ewes that were deemed not pregnant on day 35 of gestation, the remaining non-pregnant ewes were not analyzed in this analysis.
Safety Analysis of Recipients of Transgenic Embryos
In this study we evaluated surrogates for two theoretical risks, inadvertent development of RCL and transfer of vector sequences to non-target tissues in recipient ewes. To monitor for RCL, ewes were analyzed for HIV-1 p24 capsid using a commercially available ELISA serum. Blood and organs were also assessed for RCL using qPCR for the VSV-G envelope that is used to pseudotype vector particles. As summarized in Table 1, 57 ewes received lentiviral transduced embryos, of which 10 had no offspring, 23 had offspring but were not transgenic, and 24 ewes had at least one transgenic offspring. Serum was available from 55 ewes, all were negative for p24 antigen. Similarly, blood cells were available from 57 ewes, all were negative by qPCR analysis for VSV-G. Ewes were sacrificed and samples were taken from liver, lung, ovary, spleen, muscle, heart, skin, kidney, and mammary gland; all 288 samples tested for VSV-G env were negative (ewe specific testing is documented in supplemental Table 1).
Table 1.
Evaluation of Replication Competent Virus and Vector Sequences in Ewes Receiving Lentiviral-Treated Embryos; All samples were negative by the respective assay.
| Number of Animals | p24 ELISA | Blood VSV- G qPCR | Organ VSV- G qPCR | Blood Vector qPCR | Organ Vector qPCR | |
|---|---|---|---|---|---|---|
| Surrogates without offspring | 10 | 10 | 10 | 63 | 10 | 62 |
| Surrogates with negative offspring | 23 | 23 | 23 | 126 | 23 | 123 |
| Surrogates with at least one positive offspring | 24 | 22 | 24 | 99 | 24 | 96 |
| Total | 57 | 55 | 57 | 288 | 57 | 281 |
Organs include from liver, lung, ovary, spleen, muscle, heart, skin, kidney, mammary gland and surrogate uterus. Details are provides in supplemental Tables 1 and 2.
The risk of inadvertent transfer of vector to the surrogate was assessed by qPCR in the 57 treated surrogates. As shown in Table 1, blood cells from 57 surrogates were negative for vector sequences. Analysis of 273 organ samples was also negative for vector sequences. Taken together, there was no evidence of RCL in surrogates receiving transgenic embryos injected with recombinant lentiviral vectors and no evidence of RCL.
RCL Evaluation of Transgenic Offspring
The risk that offspring of embryos exposed to lentiviral gene transfer may also harbor RCL was evaluated. For this evaluation, 41 fetuses were harvested from pregnant ewes and 280 blood and organ samples were evaluated for VSV-G env, all were negative (Table 2). Additional ewes were allowed to proceed to term and 108 blood and organ samples were analyzed from 33 lambs, again all were negative for VSV-G env (Table 2). Serum from 23 fetuses and 6 lambs were available for p24 ELISA analysis, all were negative. Finally, one non-transgenic and two transgenic lambs were analyzed by a biologic RCL assay used to certify lentiviral vectors for human use, (Cornetta et al. 2011) no evidence of RCL was detected.
Table 2.
Evaluation of Replication Competent Virus in Fetal Tissues and Lambs; All samples were negative by the respective assay.
| Number of Animals | Organ VSV- G qPCR | Uterine Contents VSV-G qPCR | p24 ELISA | Blood VSV- G qPCR | |
|---|---|---|---|---|---|
| Fetuses without evidence of vector sequences | 27 | 144 | 57 | 3 | 3 |
| Transgenic fetuses | 14 | 75 | 32 | 3 | 2 |
| Lambs without evidence of vector sequences | 19 | 31 | 12 | 12 | 13 |
| Transgenic Lambs | 14 | 14 | 4 | 11 | 13 |
| Total | 74 | 264 | 105 | 29 | 31 |
Organs include from liver, lung, ovary, spleen, muscle, heart, skin, kidney, uterus, and mammary gland. Uterine contents include placenta, placentome, cotelydon, and interplacentomal tissue.
Gene Transfer Assessment in Fetal and Lamb Tissues
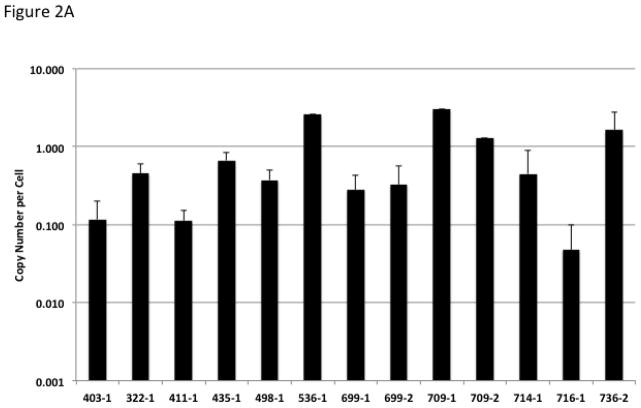
Multiplex qPCR using primers for the vector sequence along with primers for the GAPDH gene was used to assess the copy number in organ samples from fetal tissue. Figure 2A illustrates the mean copy number of vector in fetal organs. While there were differences in copy number between animals, for an individual animal the copy number was similar between organs. The majority of organs demonstrate copy number of approximately 1–3 copies per cell. Interestingly, a few animals demonstrate less that one copy number per cell suggesting partial mosaicism. Similarly, qPCR from lamb blood noted a subset of animals that had less than one integration per cell (Figure 2B).
Figure 2.
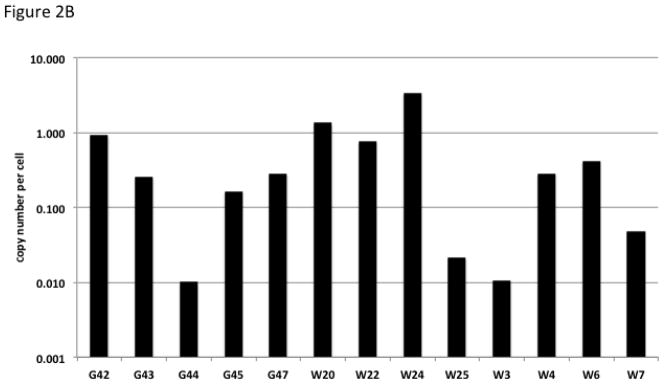
Copy Number Estimate for Transgenic Animals. (A) Values represent the vector copy number per cell for transgenic fetuses. Fetuses had between one and eight organs available for analysis, the standard deviation is displayed for fetuses in which multiple organs were analyzed. Error bars represent standard deviation of the mean for animals with multiple organs available for analysis. (B) Values represent the vector copy number per cell from blood of transgenic lambs.
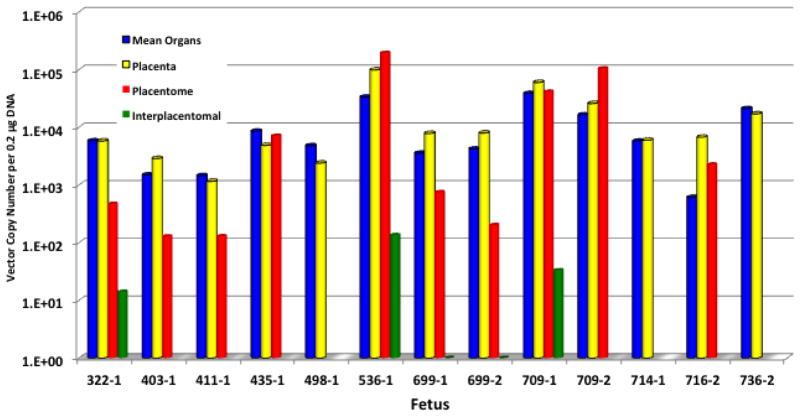
While a limited number of material uteruses were evaluated for vector sequences, all samples were negative for vector. More extensive analysis was performed on uterine contents and as shown in Figure 3, the vector copy number detected in the placenta corresponds to the level detected in fetal organ tissue from the animal. Copy number in the placentomal and interplacentomal tissues was generally lower than that of the placenta, consistent with the combined maternal and fetal origins of these tissues. Interestingly, evidence of mosaicism was suggested by one lamb with transgenic placental tissue (5.67 × 103 copies/0.2 μg of DNA) but no evidence of vector within fetal tissues (data not shown).
Figure 3.
Copy Number Estimate in Uterine Contents of Transgenic Animals. Values represent the vector copy number per 0.2 μg of DNA for placenta, placentomal and interplacentomal tissues. The mean copy number of internal organs is also provided for comparison. In general the placenta and internal organs are similar in copy number. The placentomal and interplacentomal tissues are generally lower in copy number consistent with the fetal and maternal origins of these structures.
Discussion
The development of successful and efficient methods for genetic engineering of livestock will undoubtedly afford many benefits to agriculture, animal and human health. A new and extremely promising method for producing genetically engineered livestock involves the utilization of lentiviral vectors. This approach will undoubtedly play a major role in both the production of genetically modified animals in addition to their broad utilization for gene therapy in veterinary medicine to treat animal diseases. Numerous studies have now been completed which demonstrate the utility and effectiveness of using lentiviral vectors for the delivery of transgenes into different mammalian cell lines and embryos, sometimes with increases in efficiency up to 100-fold. (Hasuwa et al. 2002; Rubinson et al. 2003; Scherr et al. 2003)
In this study, 54% of the recipient ewes became pregnant after implantation with embryos that had been injected with our recombinant lentivirus. This is comparable to pregnancy rates that would be expected following the transfer of non-injected embryos. In addition, 34% of the fetuses and 42% of the lambs were transgenic. Although this represents a lower rate of transgenisis than previous reports involving cattle, mice and pigs which document rates up to 80%, the efficiency of producing transgenic sheep reported here is still much higher than when compared to methods involving pronuclear injection (Pursel and Rexroad 1993; Golovan et al. 2001; Behboodi et al. 2005; Maga et al. 2006) and more efficient and less expensive than genetically engineering cell lines followed by somatic cell nuclear transfer. (Denning et al. 2001; Lois et al. 2002; Kuroiwa et al. 2004) Further development of this technology requires careful analysis of the safety concerns inherent in using viral vectors. A major concern addressed in this study is the potential of RCL developing during vector production or through recombination with endogenous sheep DNA. After extensive analysis of 57 surrogates, 41 fetuses and 33 lambs no evidence of RCL could be documented. This analysis includes three different methods for RCL detection. Samples were analyzed by the p24 ELISA used in screening human for evidence of the HIV-1 viral capsid. Second, a highly sensitive qPCR method that detects the VSV-G envelope was used. Most investigators believe that should a RCL develop, it is most likely to occur during vector production and would contain the VSV-G envelope used to pseudotype vector particles. Presently the in vivo fate of a VSV-G pseudotyped RCL is unknown; it is possible that the fusogenic nature of the VSV-G envelope would limit virus propagation but this envelope would also allow a RCL to infect a much broader range of cell types than the native HIV-1 envelope. Currently the FDA requires post-trial monitoring of patients for RCL after treatment with lentiviral vectors and an acceptable method is PCR for the VSV-G envelope. Therefore, this method of detection was utilized in our study. Thirdly, the most sensitive assay currently available that is required to certify vector products for human use was utilized in select fetuses. The lack of RCL is consistent with other studies evaluating vectors generated by the transient transfection method.(Escarpe et al. 2003; Sastry et al. 2003; Manilla et al. 2005; Cornetta et al. 2011)
Another safety concern is that lentiviral vector sequences could be mobilized to surrogate animals. Such transfer could occur by three mechanisms; (1) mobilization of vector by RCL, (2) vector particles introduced into the maternal circulation during the time of embryo implantation, or (3) fetal cells migrating across the placenta prior to or during delivery. To evaluate this risk, we utilized qPCR directed at vector sequences and performed an extensive evaluation of surrogate animals. Analyzing of blood and organ samples from 57 ewes found no evidence of vector sequences in the surrogate. These findings indicate that if there is any transfer of vector sequences into the surrogates, it is a rare event. The latter finding has significance when considering quarantine requirements for surrogates after they have delivered transgenic animals.
When evaluating offspring, 14 fetuses and 14 lambs were transgenic. Samples from the surrogate uterus were negative for vector sequences (Table 1 and supplemental Table 2). As expected, placenta and placentome were found to contain vector sequences given the close fetal-maternal connections during pregnancy. The placentome is a unique placental structure present in ruminant species in which the fetal side of the placenta (cotyledon) is fused with the maternal uterine tissue (caruncle) at discrete locations within the placenta. Small ruminants, including sheep, develop a synepitheliochorial placenta. As development ensues, there is a transient loss of the maternal uterine luminal epithelium (LE) resulting from fusion of binucleated cells of the conceptus trophoblast with the maternal LE. The placenta type is referred to as cotyledonary and characterized by the presence of placentomes which are vascularized structures formed from the interdigation of aglandular maternal caruncules with fetal cotyledons. They are fully functional by gestational day 60 and represented as distinct contacts between uterus and placenta. It is estimated that placentomes serve as the source for 85% of the blood flow to the placenta during late gestation. The anatomical structure of these tissues is consistent with the copy number analysis demonstrating similar copy number in fetal organs and placenta with lower copy number in structures that are of fetal and maternal origin.
While the focus of this study was an evaluation of safety, a subset of animals was selected to quantitate gene transfer. Interestingly, the level of gene transfer for an individual animal was similar for each organ tested. The finding that animals had one or more copies per cell indicates that gene transfer can occur very early in embryo development.
In summary, an extensive evaluation of transgenic animals and their surrogates found no evidence of RCL or transfer of inadvertent vector sequences. These studies support the continued development of this technology for transgenic livestock. The data also provides additional safety information relevant to human gene therapy applications.
Supplementary Material
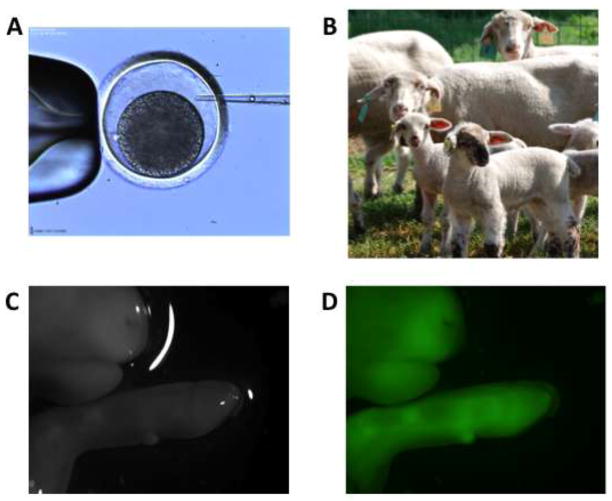
Figure 1.
Embryo Injection of Lentiviral Vectors Containing the gene for Green Fluorescent Protein. (A) Microinjection of vector into the perivitelline space of one-cell sheep embryo; (B) Transgenic sheep and their surrogates; (C) fetal sheet under normal light conditions, and D) confirmation of transgenic status of a fetal sheep using fluorescent lighting.
Acknowledgments
We wish to thank Troy Hawkins, Lisa Duffy and Sue Koop for assistance with p24 and RCL testing and Tom Spencer, Alison Wilkerson, Kimberly Green, Gayle Williamson, Jane Pryor, Michael Peoples, Katherine Dunlap, Sarah Black, Gregory Burns, Lisbeth Ramirez for assisting with embryo transfers and sample collection and processing. This project was supported by National Research Initiative Competitive Grant no. 2007-35205-17921, Biotechnology Risk Assessment Competitive Grant no. 2009-33120-20238 from the USDA National Institute of Food and Agriculture, with additional funding provided by the NHLBI National Gene Vector Biorepository (P40 HL024928).
Footnotes
CONFLICT OF INTEREST
KC is founder of Rimedion Inc. but is not employed by the company. There is no overlap or conflict with the work reported here.
References
- Behboodi E, Ayers SL, Memili E, O’Coin M, Chen LH, Reggio BC, Landry AM, Gavin WG, Meade HM, Godke RA, Echelard Y. Health and reproductive profiles of malaria antigen-producing transgenic goats derived by somatic cell nuclear transfer. Cloning Stem Cells. 2005;7:107–118. doi: 10.1089/clo.2005.7.107. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Zhang L, Conklin DS, Hannon GJ, Rosenquist TA. Germline transmission of RNAi in mice. Nat Struct Biol. 2003;10(2):91–92. doi: 10.1038/nsb896. [DOI] [PubMed] [Google Scholar]
- Cornetta K, Yao J, Jasti A, Koop S, Douglas M, Hsu D, Couture LA, Hawkins T, Duffy L. Replication competent lentivirus analysis of clinical grade vector products. Molecular Therapy. 2011;19:557–566. doi: 10.1038/mt.2010.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning C, Burl S, Ainslie A, Bracken J, Dinnyes A, Fletcher J, King T, Ritchie M, Ritchie WA, Rollo M, de Sousa P, Travers A, Wimut I, Clark AJ. Deletion of alpha(1,3)galactosyl transferase (GGTA1) gene and the prion protein (PrP) gene in sheep. Nat Biotechnol. 2001;19:559–562. doi: 10.1038/89313. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72(11):8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escarpe P, Zayek N, Chin P, Borellini F, Zufferey R, Veres G, Kiermer V. Development of a sensitive assay for detection of replication-competent recombinant lentivirus in large-scale HIV-based vector preparations. Molecular Therapy. 2003;8:332–341. doi: 10.1016/s1525-0016(03)00167-9. [DOI] [PubMed] [Google Scholar]
- Golovan SP, Meidinger RG, Ajakaiye A, Cottrill M, Wiederkehr MZ, Barney DJ, Plante C, Pollard JW, Fan MZ, Hayes MA, Laursen J, Hjorth JP, Hacker RR, Phillips JP, Fosberg CW. Pigs expressing salivary phytase produce low-phosphorus manure. Nat Biotechnol. 2001;19:741–745. doi: 10.1038/90788. [DOI] [PubMed] [Google Scholar]
- Hasuwa H, Kaseda K, Einarsdottie T, Okabe M. Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Lett. 2002;532:227–230. doi: 10.1016/s0014-5793(02)03680-3. [DOI] [PubMed] [Google Scholar]
- Hawkins TB, Dantzer J, Peters B, Dinauer MC, Mockaitis K, Mooney S, Cornetta K. Identifying viral integration sites using SeqMap 2.0. Bioinformatics. 2011;27:720–722. doi: 10.1093/bioinformatics/btq722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahner D, Stuhlmann H, Stewart CL, Harbers K, Lohler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298(5875):623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Kasinathan P, Matsushita H, Sathiyaselan J, Sullivan EJ, Kakitani M, Tomizuka K, Ishida I, Robl JM. Sequential targeting of the gene encoding immunoglobulin-mu and prion protein in cattle. Nat Genet. 2004;36:775–780. doi: 10.1038/ng1373. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Dullmann J, Kamino K, von Neuhoff N, Schlegelberger B, Li Z, Baum C. Clonal Dominance of Hematopoietic Stem Cells Triggered by Retroviral Gene Marking. Science. 2005;308(5725):1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Geiger H, Li Z, Brugman MH, Chambers SM, Shaw CA, Pike-Overzet K, Ridder Dd, Staal FJT, Keudell Gv, Cornils K, Nattamai KJ, Modlich U, Wagemaker G, Goodell MA, Fehse B, Baum C. Retroviral vector insertion sites associated with dominant hematopoietic clones mark “stemness” pathways. Blood. 2007;109(5):1897–1907. doi: 10.1182/blood-2006-08-044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295(5556):868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Maga EA, Cullor JS, Smith W, Anderson GB, Murray JD. Human lysozyme expressed in the mammary gland of transgenic dairy goats can inhibit the growth of bacteria that cause mastitis and the cold-spoilage of milk. Foodborne Pathog Dis. 2006;3:384–392. doi: 10.1089/fpd.2006.3.384. [DOI] [PubMed] [Google Scholar]
- Manilla P, Rebello T, Afable C, Lu X, Slepushkin V, Humeau LM, Schonely K, Ni Y, Binder GK, Levine BL, Macgregor R, June CH, Dropulic B. Regulatory considerations for novel gene therapy products: A review of the process leading to the first clinical lentiviral vector. Human Gene Therapy. 2005;16:17–25. doi: 10.1089/hum.2005.16.17. [DOI] [PubMed] [Google Scholar]
- Manjunath N, Wu H, Subramanya S, Shankar P. Lentiviral delivery of short hairpin RNAs. Adv Drug Deliv Rev. 2009;61(9):732–745. doi: 10.1016/j.addr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72(10):8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivurs vector. Journal of Virology. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursel VG, Rexroad CEJ. Recent progress in the transgenic modication of swine and sheep. Mol Reprod Dev. 1993;36:251–254. doi: 10.1002/mrd.1080360223. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nature Genetics. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Sadat M, Dirscherl S, Sastry L, Dantzer J, Pech N, Griffin S, Hawkins T, Zhao Y, Barese C, Cross S, Orazi A, An C, Goebel WS, Yoder MC, Li X, Grez M, Cornetta K, Mooney S, Dinauer MC. Comparison of gamma-retroviral vector integration in post-transplant hematopoiesis in mice conditioned with either submyeloablative or ablative irradiation. Gene Therapy. 2009;16:1452–1464. doi: 10.1038/gt.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry L, Johnson T, Hobson MJ, Smucker B, Cornetta K. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Therapy. 2002;9:1155–1162. doi: 10.1038/sj.gt.3301731. [DOI] [PubMed] [Google Scholar]
- Sastry L, Xu J, Johnson T, Desai K, Rissing D, Marsh J, Cornetta K. Certification assays for HIV-1-based vectors: Frequent passage of gag sequences without evidence of replication competent viruses. Molecular Therapy. 2003;8:830–839. doi: 10.1016/j.ymthe.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Scherr M, Battmer K, Gasner A, Eder M. Modulation of gene expression by lentiviral-medisated delivery of small interfering RNA. Cell Cycle. 2003;2:251–257. [PubMed] [Google Scholar]
- Schmidt M, Hoffmann G, Wissler M, Müßig A, Lemke N, Glimm H, Hesemann CU, von Kalle C. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Human Gene Therapy. 2001;12:743–749. doi: 10.1089/104303401750148649. [DOI] [PubMed] [Google Scholar]
- Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig JCB, Lagresle-Peyrou C, Benjelloun F, Hacein-Bey-Abina S, Fischer A, Cavazzana-Calvo M, Bushman FD. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acid Research. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72(12):9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.