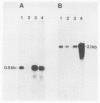
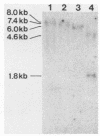
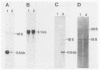
Abstract
Replication-dependent histone mRNAs are prime examples of nonpolyadenylated mRNAs. We isolated and characterized cDNAs and a genomic clone for a replication-dependent histone H2A.1 mRNA which segregated into the poly(A)+ fraction during mRNA isolation through an oligo(dT)-cellulose column. However, the results of sequencing of the genomic clone suggested that the mRNA did not contain a poly(A) tail. Instead, the genomic sequence revealed a nonterminal oligo(A) tract directly upstream from the typical 3'-terminal hairpin loop of replication-dependent histone mRNAs. The nonterminal oligo(A) tract consisted of 14 adenylate residues interrupted by one guanylate residue (A4GA10). We concluded that this short oligo(A) stretch mediated binding of the mRNA to oligo(dT) even after stringent washes with 0.1 M NaCl, indicating that rather short oligo(A) sequences can ensure binding to oligo(dT)-cellulose. The cDNA and genomic clones contained an AAATAAG sequence at the end of the coding region. It has been suggested that this sequence contains a polyadenylation signal in some yeast and mouse transcripts, but it does not function as a polyadenylation signal in the histone transcript described in this paper.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Artishevsky A., Grafsky A., Lee A. S. Isolation of a mammalian sequence capable of conferring cell cycle regulation to a heterologous gene. Science. 1985 Nov 29;230(4729):1061–1063. doi: 10.1126/science.4059922. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantine J. E., Woodland H. R. Polyadenylation of histone mRNA in Xenopus oocytes and embryos. FEBS Lett. 1985 Jan 28;180(2):224–228. doi: 10.1016/0014-5793(85)81075-9. [DOI] [PubMed] [Google Scholar]
- Bannon G. A., Calzone F. J., Bowen J. K., Allis C. D., Gorovsky M. A. Multiple, independently regulated, polyadenylated messages for histone H3 and H4 in Tetrahymena. Nucleic Acids Res. 1983 Jun 25;11(12):3903–3917. doi: 10.1093/nar/11.12.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Berger S. L. Lymphocytes as resting cells. Methods Enzymol. 1979;58:486–494. doi: 10.1016/s0076-6879(79)58163-4. [DOI] [PubMed] [Google Scholar]
- Bird R. C., Jacobs F. A., Stein G., Stein J., Sells B. H. A unique subspecies of histone H4 mRNA from rat myoblasts contains poly(A). Proc Natl Acad Sci U S A. 1985 Oct;82(20):6760–6764. doi: 10.1073/pnas.82.20.6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Brush D., Dodgson J. B., Choi O. R., Stevens P. W., Engel J. D. Replacement variant histone genes contain intervening sequences. Mol Cell Biol. 1985 Jun;5(6):1307–1317. doi: 10.1128/mcb.5.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Sharp P. A., Guarente L. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature. 1988 Jul 7;334(6177):37–42. doi: 10.1038/334037a0. [DOI] [PubMed] [Google Scholar]
- Cabada M. O., Darnbrough C., Ford P. J., Turner P. C. Differential accumulation of two size classes of poly(A) associated with messenger RNA during oogenesis in Xenopus laevis. Dev Biol. 1977 Jun;57(2):427–439. doi: 10.1016/0012-1606(77)90227-5. [DOI] [PubMed] [Google Scholar]
- Challoner P. B., Moss S. B., Groudine M. Expression of replication-dependent histone genes in avian spermatids involves an alternate pathway of mRNA 3'-end formation. Mol Cell Biol. 1989 Mar;9(3):902–913. doi: 10.1128/mcb.9.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubet N., Chaboute M. E., Clément B., Ehling M., Philipps G., Gigot C. The histone H3 and H4 mRNAs are polyadenylated in maize. Nucleic Acids Res. 1988 Feb 25;16(4):1295–1304. doi: 10.1093/nar/16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chrysogelos S., Riley D. E., Stein G., Stein J. A human histone H4 gene exhibits cell cycle-dependent changes in chromatin structure that correlate with its expression. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7535–7539. doi: 10.1073/pnas.82.22.7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S., Wells J. R. A gene-specific promoter element is required for optimal expression of the histone H1 gene in S-phase. EMBO J. 1988 Jan;7(1):49–56. doi: 10.1002/j.1460-2075.1988.tb02782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson J. B., Yamamoto M., Engel J. D. Chicken histone H3.3B cDNA sequence confirms unusual 3' UTR structure. Nucleic Acids Res. 1987 Aug 11;15(15):6294–6294. doi: 10.1093/nar/15.15.6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. 1979 Sep 11;7(1):179–192. doi: 10.1093/nar/7.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin M. E., Chakravarti S., Bartos B. B., Liu S. H., Friedman R. L., Chung A. E. Amino acid sequence and domain structure of entactin. Homology with epidermal growth factor precursor and low density lipoprotein receptor. J Cell Biol. 1988 Dec;107(6 Pt 2):2749–2756. doi: 10.1083/jcb.107.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst S. G., Miller H., Brenner C. A., Nocente-McGrath C., Francis S., McIsaac R. Characterization of a cDNA clone coding for a sea urchin histone H2A variant related to the H2A.F/Z histone protein in vertebrates. Nucleic Acids Res. 1987 Jun 11;15(11):4629–4644. doi: 10.1093/nar/15.11.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrner K., Yarger J., Hereford L. Yeast histone mRNA is polyadenylated. Nucleic Acids Res. 1980 Dec 11;8(23):5725–5737. doi: 10.1093/nar/8.23.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROBSTEIN C. Trans-filter induction of tubules in mouse metanephrogenic mesenchyme. Exp Cell Res. 1956 Apr;10(2):424–440. doi: 10.1016/0014-4827(56)90016-7. [DOI] [PubMed] [Google Scholar]
- Graves R. A., Pandey N. B., Chodchoy N., Marzluff W. F. Translation is required for regulation of histone mRNA degradation. Cell. 1987 Feb 27;48(4):615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Green L. L., Dove W. F. Correlation between tubulin mRNA stability and poly(A) length over the cell cycle of Physarum polycephalum. J Mol Biol. 1988 Mar 20;200(2):321–328. doi: 10.1016/0022-2836(88)90244-6. [DOI] [PubMed] [Google Scholar]
- Hatch C. L., Bonner W. M. Sequence of cDNAs for mammalian H2A.Z, an evolutionarily diverged but highly conserved basal histone H2A isoprotein species. Nucleic Acids Res. 1988 Feb 11;16(3):1113–1124. doi: 10.1093/nar/16.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs D. R., Goodbourn S. E., Lamb J., Clegg J. B., Weatherall D. J., Proudfoot N. J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983 Nov 24;306(5941):398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Hraba-Renevey S., Kress M. Expression of a mouse replacement histone H3.3 gene with a highly conserved 3' noncoding region during SV40- and polyoma-induced Go to S-phase transition. Nucleic Acids Res. 1989 Apr 11;17(7):2449–2461. doi: 10.1093/nar/17.7.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., Colman A., Wells J. R. Chicken histone H5 mRNA: the polyadenylated RNA lacks the conserved histone 3' terminator sequence. Nucleic Acids Res. 1982 Nov 11;10(21):6777–6785. doi: 10.1093/nar/10.21.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. J., Chodchoy N., Marzluff W. F., Skoultchi A. I. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem-loop sequence at the 3' end of the mRNA. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6189–6193. doi: 10.1073/pnas.84.17.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber T., Angerer L. M., Angerer R. C., Childs G. A histone H1 protein in sea urchins is encoded by a poly(A)+ mRNA. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4123–4127. doi: 10.1073/pnas.85.12.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. J., Liu L., Marzluff W. F. Mouse histone H2A and H2B genes: four functional genes and a pseudogene undergoing gene conversion with a closely linked functional gene. Nucleic Acids Res. 1987 Apr 10;15(7):3023–3039. doi: 10.1093/nar/15.7.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Stauber C., Schindler R., Schümperli D. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3'-terminal part of the gene. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4389–4393. doi: 10.1073/pnas.82.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K., Deutzmann R., Aumailley M., Timpl R., Raimondi L., Yamada Y., Pan T. C., Conway D., Chu M. L. Amino acid sequence of mouse nidogen, a multidomain basement membrane protein with binding activity for laminin, collagen IV and cells. EMBO J. 1989 Jan;8(1):65–72. doi: 10.1002/j.1460-2075.1989.tb03349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss S. B., Challoner P. B., Groudine M. Expression of a novel histone 2B during mouse spermiogenesis. Dev Biol. 1989 May;133(1):83–92. doi: 10.1016/0012-1606(89)90299-6. [DOI] [PubMed] [Google Scholar]
- Mowry K. L., Oh R., Steitz J. A. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3'-end processing reaction. Mol Cell Biol. 1989 Jul;9(7):3105–3108. doi: 10.1128/mcb.9.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N., Sanders M., Newton A. Poly(adenylic acid) sequences in the RNA of Caulobacter crescenus. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2343–2346. doi: 10.1073/pnas.72.6.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvari R., Avivi A., Lentner F., Ziv E., Tel-Or S., Burstein Y., Schechter I. Chicken immunoglobulin gamma-heavy chains: limited VH gene repertoire, combinatorial diversification by D gene segments and evolution of the heavy chain locus. EMBO J. 1988 Mar;7(3):739–744. doi: 10.1002/j.1460-2075.1988.tb02870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro C., Mermod N., Andrews P. C., Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988 Jul 21;334(6179):218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- Scherer G., Telford J., Baldari C., Pirrotta V. Isolation of cloned genes differentially expressed at early and late stages of Drosophila embryonic development. Dev Biol. 1981 Sep;86(2):438–447. doi: 10.1016/0012-1606(81)90202-5. [DOI] [PubMed] [Google Scholar]
- Seiler-Tuyns A., Birnstiel M. L. Structure and expression in L-cells of a cloned H4 histone gene of the mouse. J Mol Biol. 1981 Oct 5;151(4):607–625. doi: 10.1016/0022-2836(81)90426-5. [DOI] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vournakis J. N., Gelinas R. E., Kafatos F. C. Short polyadenylic acid sequences in insect chorion messenger RNA. Cell. 1974 Nov;3(3):265–273. doi: 10.1016/0092-8674(74)90141-x. [DOI] [PubMed] [Google Scholar]
- Wells D., Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc Natl Acad Sci U S A. 1985 May;82(9):2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D., McBride C. A comprehensive compilation and alignment of histones and histone genes. Nucleic Acids Res. 1989;17 (Suppl):r311–r346. doi: 10.1093/nar/17.suppl.r311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. M., Shapiro D. L., Allis C. D., Gorovsky M. A. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 1988 Jan 11;16(1):179–198. doi: 10.1093/nar/16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]