Abstract
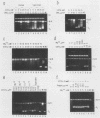
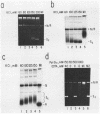
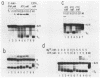
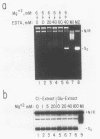
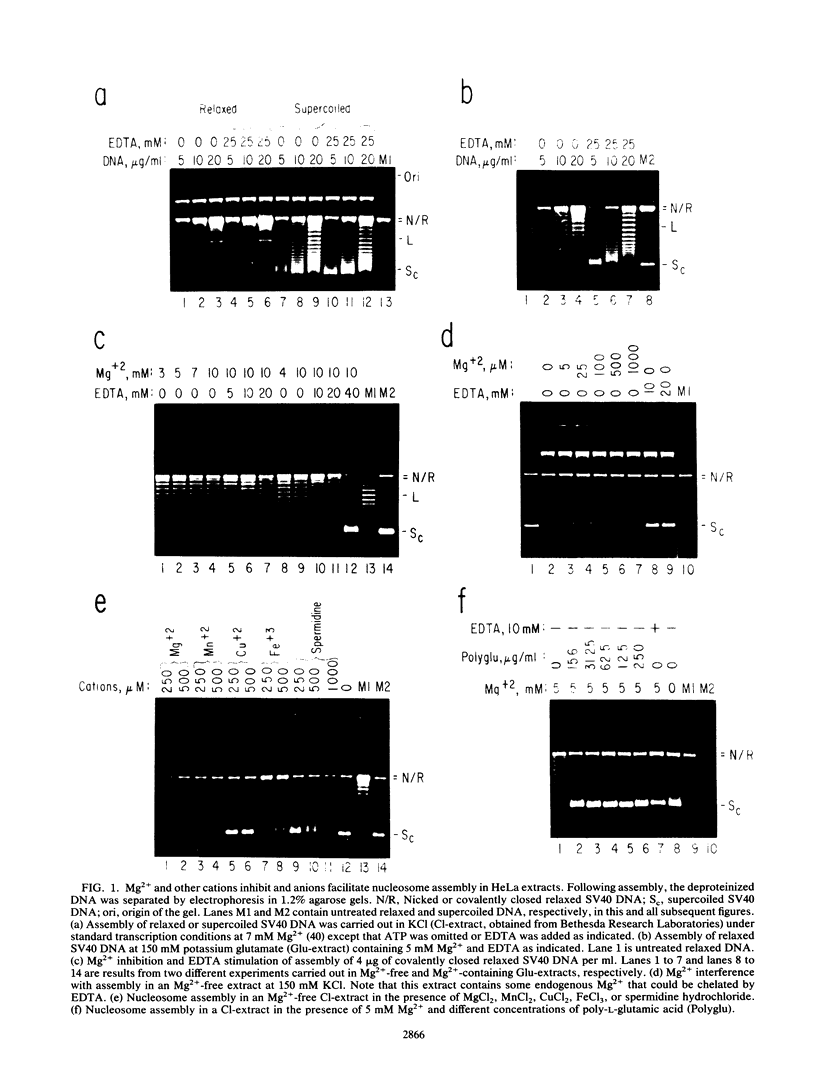
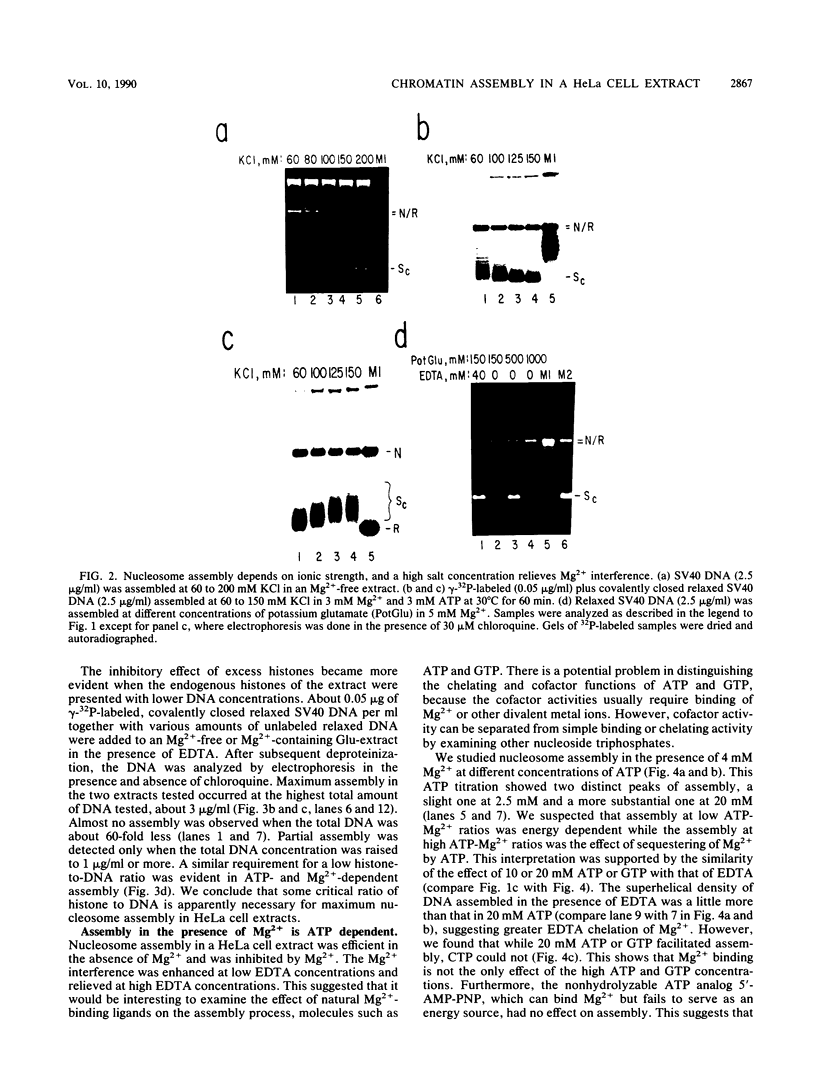
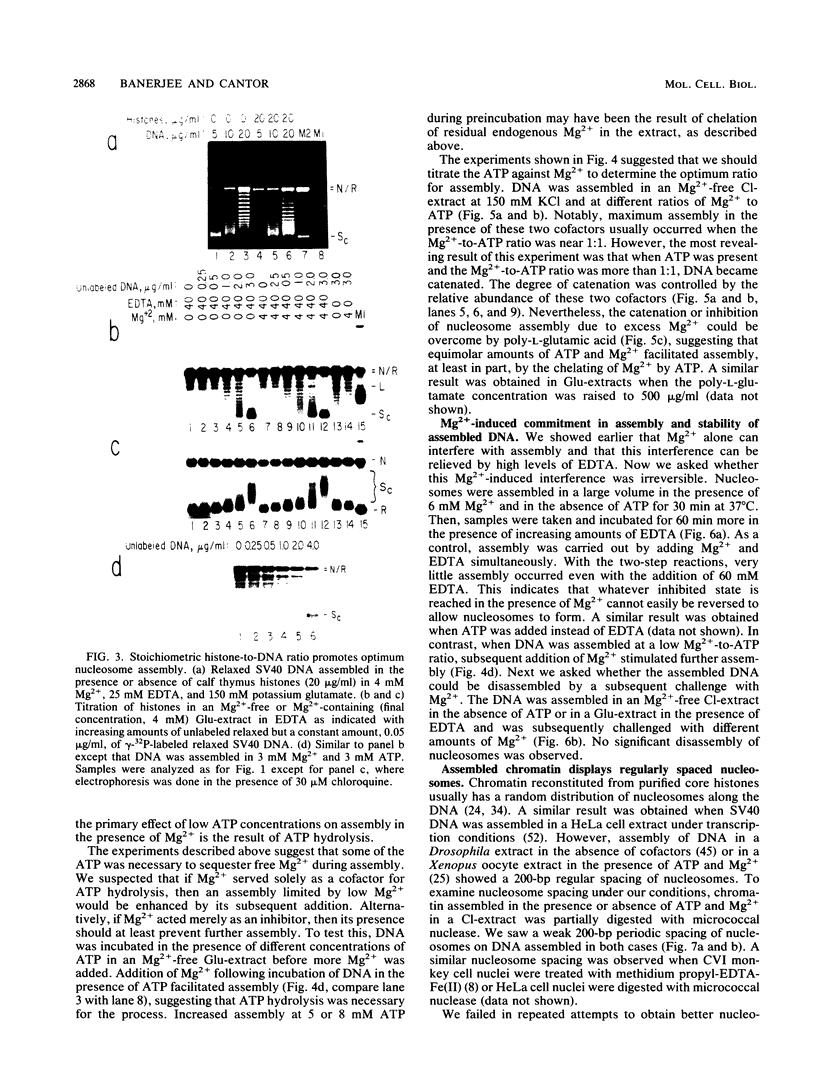
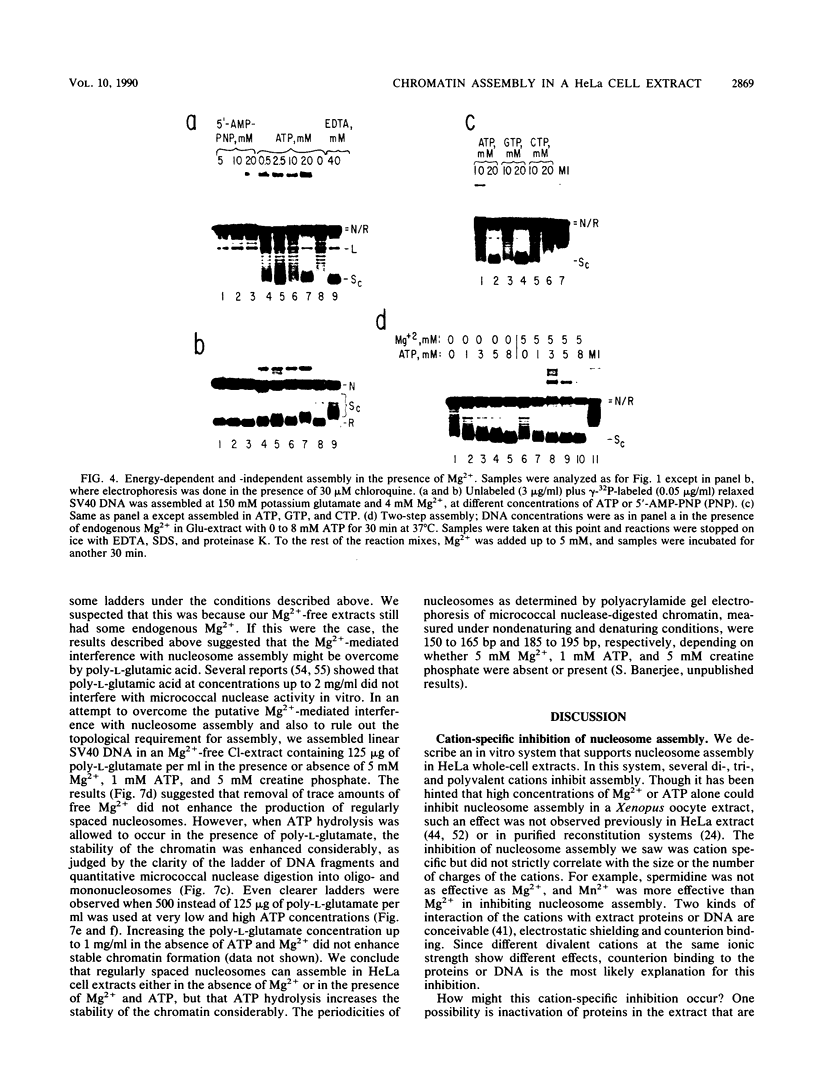
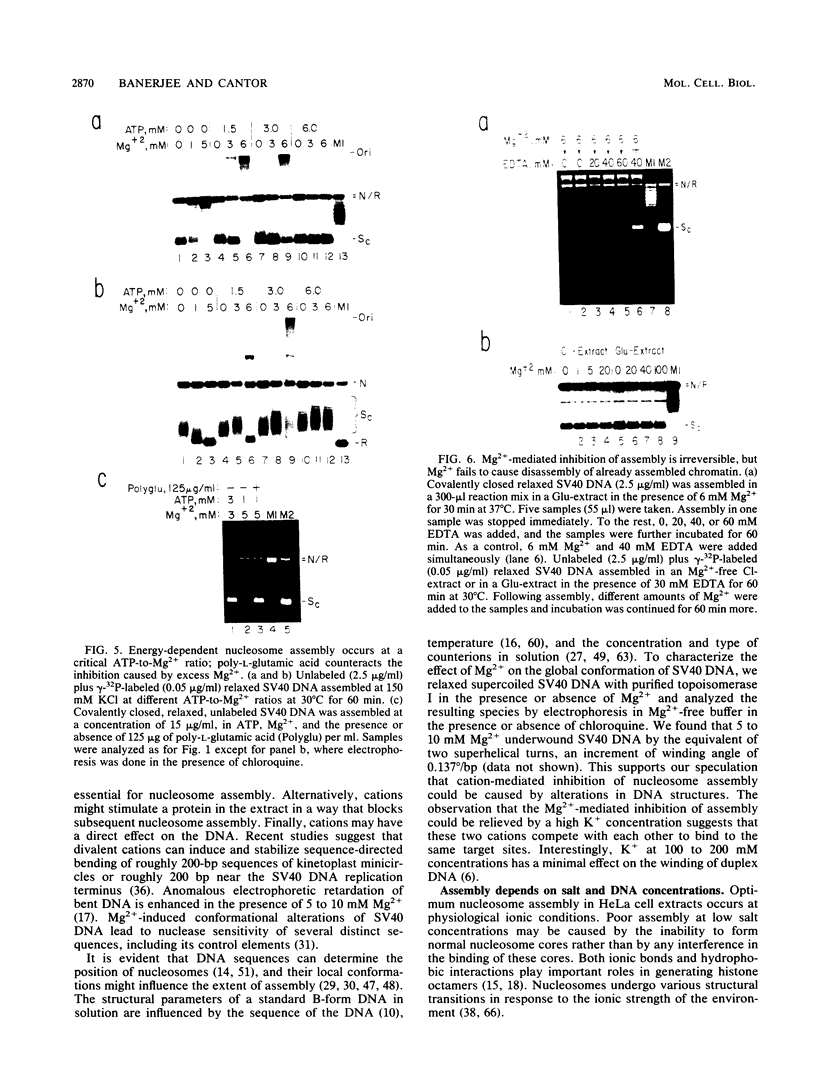
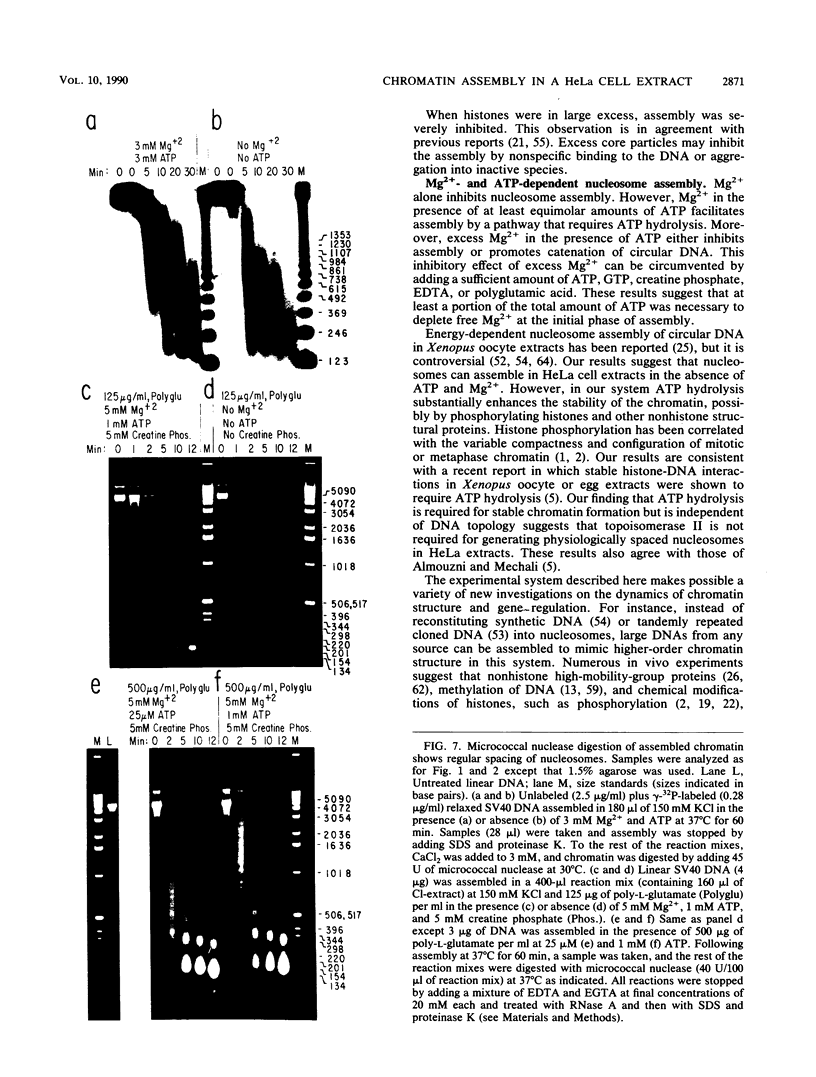
We report here a mammalian cell-free system that can support chromatin assembly. Effective nucleosome assembly in HeLa cell extracts occurred at 125 to 200 mM KCl or potassium glutamate. At this physiological K+ ion concentration, two types of chromatin assembly were observed. The first was interfered with by Mg2+. Other cations such as Mn2+, Ca2+, Fe3+, and spermidine also inhibited this type of nucleosome assembly. The second type of assembly occurred in the presence of Mg2+ and at least equimolar ATP. However, even in the presence of ATP, excess Mg2+ inhibited assembly and promoted catenation of DNA; these effects could be circumvented by excess ATP, GTP, EDTA, or polyglutamic acid. The critical DNA concentration for optimum assembly in both pathways suggested a stoichiometric association of histones with DNA. The spacing of nucleosomes formed by both types of assembly on linear and circular DNA was reasonably regular, but chromatin assembled in the presence of ATP and Mg2+ was more stable.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ajiro K., Borun T. W., Cohen L. H. Phosphorylation states of different histone 1 subtypes and their relationship to chromatin functions during the HeLa S-3 cell cycle. Biochemistry. 1981 Mar 17;20(6):1445–1454. doi: 10.1021/bi00509a007. [DOI] [PubMed] [Google Scholar]
- Ajiro K., Nishimoto T. Specific site of histone H3 phosphorylation related to the maintenance of premature chromosome condensation. Evidence for catalytically induced interchange of the subunits. J Biol Chem. 1985 Dec 15;260(29):15379–15381. [PubMed] [Google Scholar]
- Almouzni G., Méchali M. Assembly of spaced chromatin involvement of ATP and DNA topoisomerase activity. EMBO J. 1988 Dec 20;7(13):4355–4365. doi: 10.1002/j.1460-2075.1988.tb03334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Bauer W. Supercoiling in closed circular DNA: dependence upon ion type and concentration. Biochemistry. 1978 Feb 21;17(4):594–601. doi: 10.1021/bi00597a006. [DOI] [PubMed] [Google Scholar]
- Been M. D., Burgess R. R., Champoux J. J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3097–3114. doi: 10.1093/nar/12.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R., Cantor C. R., Axel R. Nucleosomes are phased along the mouse beta-major globin gene in erythroid and nonerythroid cells. Cell. 1986 Mar 14;44(5):697–704. doi: 10.1016/0092-8674(86)90835-4. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bram S. Variation of type-B DNA x-ray fiber diagrams with base composition. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2167–2170. doi: 10.1073/pnas.70.7.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Chao M. V., Gralla J., Martinson H. G. DNA sequence directs placement of histone cores on restriction fragments during nucleosome formation. Biochemistry. 1979 Mar 20;18(6):1068–1074. doi: 10.1021/bi00573a021. [DOI] [PubMed] [Google Scholar]
- Daban J. R., Cantor C. R. Role of histone pairs H2A,H2B and H3,H4 in the self-assembly of nucleosome core particles. J Mol Biol. 1982 Apr 25;156(4):771–789. doi: 10.1016/0022-2836(82)90141-3. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S., Wang J. C. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J Mol Biol. 1985 Nov 5;186(1):1–11. doi: 10.1016/0022-2836(85)90251-7. [DOI] [PubMed] [Google Scholar]
- Dieterich A. E., Axel R., Cantor C. R. Salt-induced structural changes of nucleosome core particles. J Mol Biol. 1979 Apr 25;129(4):587–602. doi: 10.1016/0022-2836(79)90470-4. [DOI] [PubMed] [Google Scholar]
- Draetta G., Beach D. Activation of cdc2 protein kinase during mitosis in human cells: cell cycle-dependent phosphorylation and subunit rearrangement. Cell. 1988 Jul 1;54(1):17–26. doi: 10.1016/0092-8674(88)90175-4. [DOI] [PubMed] [Google Scholar]
- Farzaneh F., Zalin R., Brill D., Shall S. DNA strand breaks and ADP-ribosyl transferase activation during cell differentiation. Nature. 1982 Nov 25;300(5890):362–366. doi: 10.1038/300362a0. [DOI] [PubMed] [Google Scholar]
- Gargiulo G., Worcel A. Analysis of the chromatin assembled in germinal vesicles of Xenopus oocytes. J Mol Biol. 1983 Nov 5;170(3):699–722. doi: 10.1016/s0022-2836(83)80128-4. [DOI] [PubMed] [Google Scholar]
- Gautier J., Matsukawa T., Nurse P., Maller J. Dephosphorylation and activation of Xenopus p34cdc2 protein kinase during the cell cycle. Nature. 1989 Jun 22;339(6226):626–629. doi: 10.1038/339626a0. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Rouvière-Yaniv J., Yaniv M., Brutlag D. Nicking-closing enzyme assembles nucleosome-like structures in vitro. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3779–3783. doi: 10.1073/pnas.76.8.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Goodwin G. H., Johns E. W. Are the high mobility group non-histone chromosomal proteins associated with 'active' chromatin? Biochim Biophys Acta. 1978 Jun 22;519(1):279–284. doi: 10.1016/0005-2787(78)90081-3. [DOI] [PubMed] [Google Scholar]
- Hanlon S., Chan A., Berman S. Specific cation effects on conformational transitions of DNA in aqueous solutions. Biochim Biophys Acta. 1978 Jul 24;519(2):526–536. doi: 10.1016/0005-2787(78)90105-3. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hovatter K. R., Martinson H. G. Ribonucleotide-induced helical alteration in DNA prevents nucleosome formation. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1162–1166. doi: 10.1073/pnas.84.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. The terminus of SV40 DNA replication and transcription contains a sharp sequence-directed curve. Cell. 1988 Feb 26;52(4):535–544. doi: 10.1016/0092-8674(88)90466-7. [DOI] [PubMed] [Google Scholar]
- Iacono-Connors L., Kowalski D. Altered DNA conformations in the gene regulatory region of torsionally-stressed SV40 DNA. Nucleic Acids Res. 1986 Nov 25;14(22):8949–8962. doi: 10.1093/nar/14.22.8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezetic J. A., Luse D. S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986 Apr 11;45(1):95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Finch J. T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978 Oct 5;275(5679):416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Laundon C. H., Griffith J. D. Cationic metals promote sequence-directed DNA bending. Biochemistry. 1987 Jun 30;26(13):3759–3762. doi: 10.1021/bi00387a003. [DOI] [PubMed] [Google Scholar]
- Libertini L. J., Small E. W. Effects of pH on low-salt transition of chromatin core particles. Biochemistry. 1982 Jul 6;21(14):3327–3334. doi: 10.1021/bi00257a013. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Samuels M., Sharp P. A. In vitro transcription: whole-cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- Manning G. S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978 May;11(2):179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- Matsui T. Transcription of adenovirus 2 major late and peptide IX genes under conditions of in vitro nucleosome assembly. Mol Cell Biol. 1987 Apr;7(4):1401–1408. doi: 10.1128/mcb.7.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mertz J. E. Linear DNA does not form chromatin containing regularly spaced nucleosomes. Mol Cell Biol. 1982 Dec;2(12):1608–1618. doi: 10.1128/mcb.2.12.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T., Hsieh T. S., Brutlag D. Extracts of Drosophila embryos mediate chromatin assembly in vitro. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5510–5514. doi: 10.1073/pnas.76.11.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T., Wiegand R., Brutlag D. Ribonucleic acid and other polyanions facilitate chromatin assembly in vitro. Biochemistry. 1981 Apr 28;20(9):2594–2601. doi: 10.1021/bi00512a035. [DOI] [PubMed] [Google Scholar]
- Nickol J., Martin R. G. DNA stem-loop structures bind poorly to histone octamer cores. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4669–4673. doi: 10.1073/pnas.80.15.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poljak L. G., Gralla J. D. The SV40 termination region exhibits an altered helical DNA conformation. Nucleic Acids Res. 1987 Jul 10;15(13):5433–5442. doi: 10.1093/nar/15.13.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Satchwell S. C., Travers A. A. Asymmetry and polarity of nucleosomes in chicken erythrocyte chromatin. EMBO J. 1989 Jan;8(1):229–238. doi: 10.1002/j.1460-2075.1989.tb03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant A., Bohmann D., Zentgraf H., Weiher H., Keller W. A transcription enhancer acts in vitro over distances of hundreds of base-pairs on both circular and linear templates but not on chromatin-reconstituted DNA. J Mol Biol. 1984 Dec 15;180(3):577–600. doi: 10.1016/0022-2836(84)90028-7. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Stein A., Bina M. A model chromatin assembly system. Factors affecting nucleosome spacing. J Mol Biol. 1984 Sep 15;178(2):341–363. doi: 10.1016/0022-2836(84)90148-7. [DOI] [PubMed] [Google Scholar]
- Stein A., Whitlock J. P., Jr, Bina M. Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5000–5004. doi: 10.1073/pnas.76.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986 May 23;45(4):555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- Sugden B., Keller W. Mammalian deoxyribonucleic acid-dependent ribonucleic acid polymerases. I. Purification and properties of an -amanitin-sensitive ribonucleic acid polymerase and stimulatory factors from HeLa and KB cells. J Biol Chem. 1973 Jun 10;248(11):3777–3788. [PubMed] [Google Scholar]
- Tegtmeyer P., Dohan C., Jr, Reznikoff C. Inactivating and mutagenic effects of nitrosoguanidine on simian virus 40. Proc Natl Acad Sci U S A. 1970 Jul;66(3):745–752. doi: 10.1073/pnas.66.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Variation of the average rotation angle of the DNA helix and the superhelical turns of covalently closed cyclic lambda DNA. J Mol Biol. 1969 Jul 14;43(1):25–39. doi: 10.1016/0022-2836(69)90076-x. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Weintraub H. Isolation of actively transcribed nucleosomes using immobilized HMG 14 and 17 and an analysis of alpha-globin chromatin. Cell. 1981 Feb;23(2):391–400. doi: 10.1016/0092-8674(81)90134-3. [DOI] [PubMed] [Google Scholar]
- Wilson R. W., Bloomfield V. A. Counterion-induced condesation of deoxyribonucleic acid. a light-scattering study. Biochemistry. 1979 May 29;18(11):2192–2196. doi: 10.1021/bi00578a009. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Andrews M. T., Crawford E., Losa R., Brown D. D. Negative supercoiling is not required for 5S RNA transcription in vitro. Cell. 1987 May 8;49(3):301–303. doi: 10.1016/0092-8674(87)90279-0. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Roeder R. G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987 Nov 20;51(4):613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Dattagupta N., Hogan M., Crothers D. M. Structural changes of nucleosomes in low-salt concentrations. Biochemistry. 1979 Sep 4;18(18):3960–3965. doi: 10.1021/bi00585a018. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]