Abstract
Nucleotide-binding proteins play pivotal roles in many cellular processes including cell signaling. However, targeted study of sub-proteome of nucleotide-binding proteins, especially protein kinases and GTP-binding proteins, remained challenging. Here, we reported a general strategy in using affinity-labeled chemical probes to enrich, identify, and quantify ATP- and GTP-binding proteins in the entire human proteome. Our results revealed that the ATP/GTP affinity probes facilitated the identification of 100 GTP-binding proteins and 206 kinases with the use of low mg quantities of lysate of HL-60 cells. In combination with the use of SILAC-based quantitative proteomics method, we assessed the ATP/GTP binding selectivities of nucleotide-binding proteins at the global proteome scale. Our results confirmed known and, more importantly, unveiled new ATP/GTP-binding preferences of hundreds of nucleotide-binding proteins. Additionally, our strategy led to the identification of three and one unique nucleotide-binding motifs for kinases and GTP-binding proteins, respectively, and the characterizations of the nucleotide binding selectivities of individual motifs. Our strategy for capturing and characterizing ATP/GTP-binding proteins should be generally applicable for those proteins that can interact with other nucleotides.
Introduction
Adenine and guanine nucleotides are abundant and they bind to numerous proteins involved in pivotal cellular processes, including cell signaling, proliferation, differentiation, and apoptosis1. Despite the importance of nucleotide-binding proteins in cellular functions, the current picture of nucleotide-protein interactions is far from complete. Therefore, comprehensive identification of ATP/GTP-binding proteins and dynamic analysis of nucleotide-protein interactions at the proteomic scale are important for understanding better the regulatory mechanisms of nucleotide-binding proteins.
The development of mass spectrometry (MS) instrumentation and bioinformatic tools provides the opportunity to identify and quantify up to several thousand proteins in complex samples2. However, proteomic studies of specific family of proteins, including nucleotide-binding proteins, by MS are still a big challenge owing to the extreme complexity of the proteome and the relatively low abundance of some proteins. This limitation can be partially overcome by combining MS with various separation techniques, such as polyacrylamide gel electrophoresis (PAGE)3 or multi-dimensional liquid chromatography4. However, none of these approaches permit selective enrichment of nucleotide-binding proteins from cell lysates.
Affinity chromatography is commonly used for fractionating complex protein mixture to yield functional sub-groups of proteins. Ito et al.5 used γ phosphate-linked ATP media to enrich ATP-binding proteins from the soluble fraction of Arabidopsis mitochondria. Additionally, Mann et al.6 employed kinase-selective affinity column with immobilized kinase inhibitors as capture ligands to facilitate the identification and quantification of approximately 200 protein kinases. On the other hand, chemical tagging methods involving specific labeling of proteins with functional similarities have emerged as an important technique in targeted proteomics7. For example, 5’-p-fluorosulfonylbenzoyladenosine, a reactive ATP analog, was employed as an activity-based probe to target nucleotide-binding proteins from whole cell lysates8. Additionally, a photo-reactive GTP analog possessing a diazirine moiety was developed for the detection of GTP-binding proteins9. Others and we also reported the application of biotin-conjugated acyl nucleotide probe for the enrichment and identification of ATP-binding proteins from complex protein mixtures10. In principle, this reactive affinity probe-based enrichment strategy, which involved labeling reaction, enzymatic digestion, affinity purification and LC-MS/MS analysis should be generally applicable for the identification and characterization of other nucleotide-binding proteins.
Here, we extended the use of the biotin-based nucleotide affinity probes as acylating agents to selectively label and enrich ATP- and GTP-binding proteins from the entire human proteome. Together with the use of extensive separation techniques, the method allowed for the identification of a significant number of nucleotide-binding proteins. In addition, the nucleotide-binding protein enrichment approach, along with quantitative proteomics using stable isotope labeling by amino acids in cell culture (SILAC)11, facilitated the characterizations of nucleotide-protein interactions at the entire proteome scale.
Experimental Details
Cell Lysate Preparation and Labeling with Nucleotide Affinity Probe
The biotinylated nucleotide affinity probes were prepared according to previously published procedures with minor modifications (see Supporting Information)10a. HL-60 cells (ATCC, Manassas, VA) were cultured in Iscove’s modified minimal essential medium (IMEM) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA) and penicillin (100 IU/mL). For SILAC experiments, the IMEM medium without L-lysine or L-arginine was custom-prepared according to ATCC formulation. The complete light and heavy IMEM media were prepared by the addition of light or heavy lysine and arginine, along with dialyzed FBS (Invitrogen), to the above lysine, arginine-depleted medium. The HL-60 cells were cultured in heavy IMEM medium for at least 5 cell doublings to achieve complete isotope incorporation. Approximately 2×107 cells were harvested and washed three times with cold PBS. The cells were then lysed in 1 mL lysis buffer, which contained 0.7% CHAPS, 50 mM HEPES (pH 7.4), 0.5 mM EDTA, 100 mM NaCl, and 10 µL (1:100) protease inhibitor cocktail on ice for 30 min. The cell lysates were centrifuged at 16000g at 4°C for 30 min, and the resulting supernatants were collected and subjected to gel filtration separation using NAP-25 columns (Amersham Biosciences) to remove free endogenous nucleotides. Cell lysates were eluted into a 2-mL buffer containing 50 mM HEPES (pH 7.4), 75 mM NaCl, and 5% glycerol. The resulting proteins in cell lysates were quantified using Quick Start Bradford Protein Assay (Bio-Rad, Hercules, CA) and stored at −80°C.
Immediately prior to the labeling reaction, MgCl2, MnCl2, and CaCl2 were added to the concentrated cell lysate until their final concentrations reached 50, 5, and 5 mM, respectively. Approximately 1 mg cell lysate was treated with biotin-nucleotide affinity probe at concentrations ranging from 15 to 100 µM. Labeling reactions were carried out at room temperature with gentle shaking for 25 min. After the reaction, the remaining probes in the cell lysates were removed by buffer exchange with 25 mM NH4HCO3 solution (pH 8.5) using Amicon Ultra-4 filter (10,000 NMWL, Millipore).
In-solution Enzymatic Digestion and Affinity Purification
After addition of 8 M urea for protein denaturation and dithiothreitol (DTT) and iodoacetamide (IAM) to reduce and block cysteines, the labeled proteins were digested with modified sequencing-grade trypsin (Roche Applied Science) at an enzyme/substrate ratio of 1:100 in 25 mM NH4HCO3 (pH 8.5) and at 37°C for overnight. The peptide mixture was subsequently dried in a Speed-vac and redissolved in 1 mL of 100 mM potassium phosphate and 0.15 M NaCl (pH 7.5, PBS buffer). Avidin-agarose resin (Sigma-Aldrich) was used to capture the biotin-labeled peptides. Prior to the binding, 200 µL resin was washed with 20 mM potassium phosphate and 0.15 M NaCl (pH 7.5) for 3 times. After adding to the digested peptide solution, the mixture was then incubated at 25°C for 1 hr with gentle shaking. To remove the unbound peptides, agarose resin was washed sequentially with 3 mL PBS buffer and 3 mL pure H2O. Following washing, the labeled peptides were eluted with 1% TFA in CH3CN/H2O (7/3, v/v) at 65°C. The eluates were dried in a Speed-vac and stored at −20°C prior to LC-MS/MS analysis. The details of LC-MS/MS conditions and database search parameters are shown in online Supporting Information.
Results and Discussion
1. Design of nucleotide-affinity probe
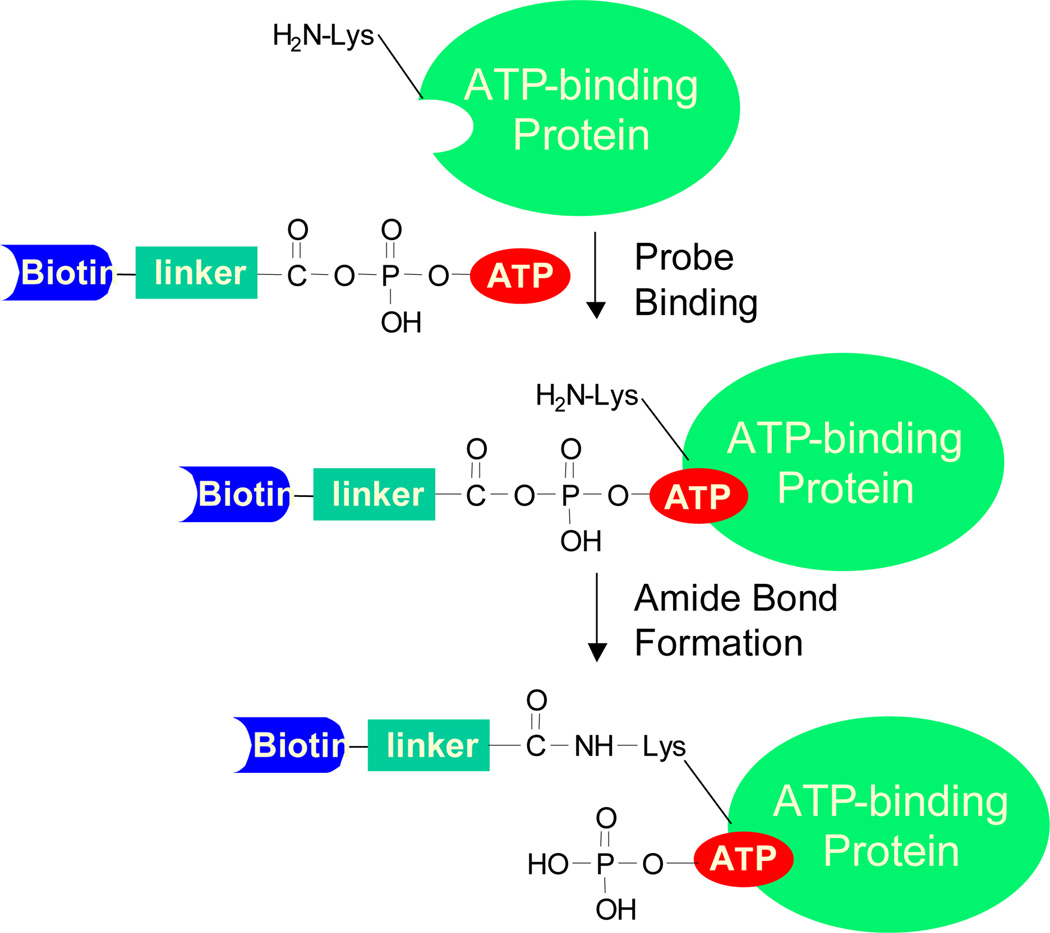
Bioinformatics studies showed that most ATP/GTP-binding proteins carry a consensus amino acid sequence motif termed phosphate-binding loop (P-loop), which constitutes the nucleotide-binding site and is responsible for ATPase/GTPase activity12. It has been frequently observed that there is at least one lysine residue at nucleotide binding sites. For example, a conserved motif of GxxxxGK, in which ‘x’ represents any amino acid, is often found in the P-loop region of ATP/GTP-binding proteins13. In light of these previous findings, a nucleotide analog bearing an acyl phosphate moiety is developed to target the lysine residue in the nucleotide-binding site (Figure S1). These nucleotide-binding probes contain three components, namely, a binding moiety consisted of nucleotide of interest (e.g., GTP or ATP), an enrichment moiety of biotin or its analog desthiobiotin which facilitates downstream purification, and an acyl phosphate group which targets side-chain amino group of the P-loop lysine residue to form a stable amide bond. For instance, when the ATP affinity probe interacts with ATP-binding proteins, the ATP moiety binds to the P-loop region, which facilitates the ε-amino group of the P-loop lysine residue to react with the acyl phosphate component to yield a stable amide bond (Figure 1). After the reaction, the protein mixture is digested with trypsin and the resulting biotin-labeled peptides can be enriched using an avidin agarose column. Finally, the affinity-purified peptides with the biotin tag are analyzed by LC-MS/MS (Figure S2). In this vein, it is worth noting that biotin conjugation with the side chain of cysteine or lysine was found not to result in significant alteration in peptide backbone fragmentation upon low-energy collision activation and peptide identification based on the resulting MS/MS14. We also did not observe any apparent difference in the percentage of spectra with successful peptide identification for the biotin-conjugated peptides vs. unmodified peptides.
Figure 1.
A schematic diagram showing the reaction between biotin-LC-ATP affinity probe with an ATP-binding protein.
Initial experiment with 50 and 100 µM biotin-LC-ATP/GTP affinity probe and 1-mg cell lysate led to the identification of similar numbers of ATP- or GTP-binding proteins, though more nucleotide-binding proteins apart from ATP/GTP-binding proteins were identified in 100 µM probe experiment (Figure S3). However, if the GTP probe concentration was decreased to 15 µM, the total number of identified GTP-binding proteins decreased by more than 4 fold (Data not shown). Thus, we employed a probe concentration of 100 µM for subsequent experiments.
Due to the very high affinity and specificity of biotin-avidin interaction, biotinylation is the most widely used chemical modification for the affinity purification and detection of proteins and peptides15. However, the extremely high stability of biotin-avidin complex renders it difficult to elute the biotinylated molecules from the avidin resin. In this vein, a similar ATP affinity probe carrying a desthiobiotin in lieu of biotin has been introduced by Pierce (Active X). The desthiobiotin moiety with lower affinity to avidin agarose may afford a better recovery of labeled peptides and higher detection sensitivity, thereby minimizing sample loss during the avidin enrichment step16. We compared the performance of biotin-LC- and desthiobiotin-based nucleotide affinity probes, and it turned out that there is no substantial difference for these two types of probes in the identification of nucleotide-binding proteins (Figure S3).
2. Extensive prefractionation for Large-scale profiling of nucleotide-binding proteins from the whole human proteome
Owing to the extreme complexity of human proteome, MS-based identification of specific subfamilies of proteins, including nucleotide-binding proteins, from complex samples necessitates the use of powerful separation techniques, such as SDS-PAGE or multi-dimensional LC for sample prefractionation3–4. Thus, we employed three different pre-fractionation methods, i.e. SDS-PAGE, offline SCX chromatography and online 2D-LC with SCX-C18 separation, for experiments with desthiobiotin-based ATP affinity probe. For SDS-PAGE fractionation, we separated ATP affinity probe-treated whole cell lysate using 12% SDS-PAGE, cut the gel into 8 slices, digested the proteins in-gel with trypsin, enriched the biotin-labeled peptides from each fraction using avidin agarose, and analyzed the enriched peptides with LC-MS/MS. This method allowed for the identification of 1072 proteins with biotin-labeled peptides (Figure S4). Offline SCX separation, during which the digested peptide mixture from labeled whole cell lysate was separated into 6 fractions and then subjected individually to avidin enrichment, led to the identification of 953 proteins with biotin-labeled peptides from 1-mg cell lysates (Figure S4). On the other hand, online 2D-LC system with SCX and C18 separation results in the identification of 946 proteins with the probe-labeled peptides (Figure S4). Relative to the results obtained from the unfractionated lysates, the total number of probe-labeled proteins increased by approximately two-fold with each single prefractionation method and almost a 3-fold increase was achieved by combining all three prefractionation strategies.
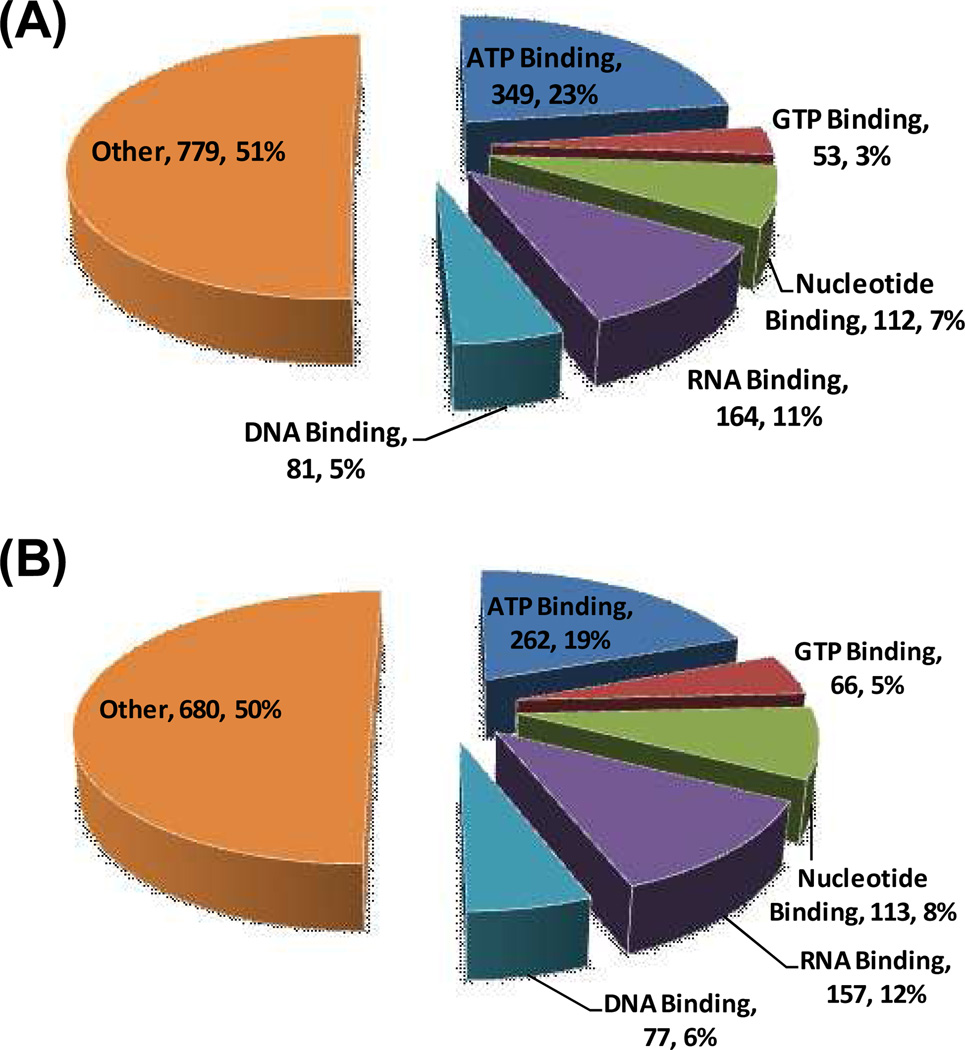
Altogether we identified a total of 1538 unique proteins with desthiobiotin modification from these three separation strategies (Figure 2 and Table S3). Among these proteins, 349 (23%), 168 (11%) and 53 (3%) are known ATP-binding proteins, kinases, and GTP-binding proteins, respectively. Additionally, more than 510 known nucleotide-binding proteins were unambiguously detected, highlighting the great potential of this method in nucleotide-binding protein studies at the global proteome scale. We also subjected biotin-based GTP-probe-labeled peptides to online 2D-LC-MS/MS analysis alone and we were able to identify a total of 1355 proteins, among which 66 (5%) are known GTP-binding proteins (Figure 2 and Table S3) and 441 possess nucleotide-binding capability. The relatively low percentage of known GTP-binding proteins among all the identified proteins could be attributed, in part, to the presence of many as-yet characterized or annotated GTP-binding proteins.
Figure 2.
A summary of proteins identified with biotin-ATP probe (A) and biotin-GTP probe (B) from HL-60 whole cell lysate with extensive separations.
3. A strategy to compare the relative ATP/GTP-binding affinities of proteins from whole cell lysates
Nucleotide-binding affinity and phosphohydrolase activity are key features of various enzymes such as kinases, helicases and G proteins. However, the selectivity in binding toward different nucleotides, is an often overlooked but non-trivial property of nucleotide-binding proteins. Both ATP and GTP are purine nucleoside triphosphates and share similar structures as well as biological functions. Some kinases, e.g. casein kinase 217, may employ GTP as phosphate donor to phosphorylate substrates, whereas ATP may also bind to GTP-binding proteins to regulate their activities. Therefore, direct comparison of the binding affinity of signal transduction proteins such as kinases and G-proteins toward ATP/GTP is essential for elucidating better the underlying mechanism of signal activation and transduction in cells. Although various assays can be used to assess the binding selectivities of nucleotides for individual proteins, systematic comparison of ATP- and GTP-binding affinities of proteins, especially at the entire proteome scale, has not yet been performed.
Apart from nucleotide selectivity, the nucleotide-binding site, or ‘nucleotide-interacting residues', is another aspect of significant interest in nucleotide-protein interaction studies. The ATP/GTP binding sites are particularly important because many anti-cancer drugs also target nucleotide-binding sites in ATPases and GTPases18. The experimental determination of residues that interact with ATP/GTP often relies on X-ray crystal structure or site-directed mutagenesis, which is costly and time-consuming. We demonstrated previously that lysine residues labeled by the biotin-ATP probe may represent the specific nucleotide-binding site of proteins, which directly interacts with γ-phosphate group of the bound ATP10a. We reason that this site-directed labeling strategy using nucleotide-affinity probe, in combination with quantitative proteomics, may allow for the assessment of the selectivities of ATP and GTP in binding toward specific sequence segments of nucleotide-binding proteins.
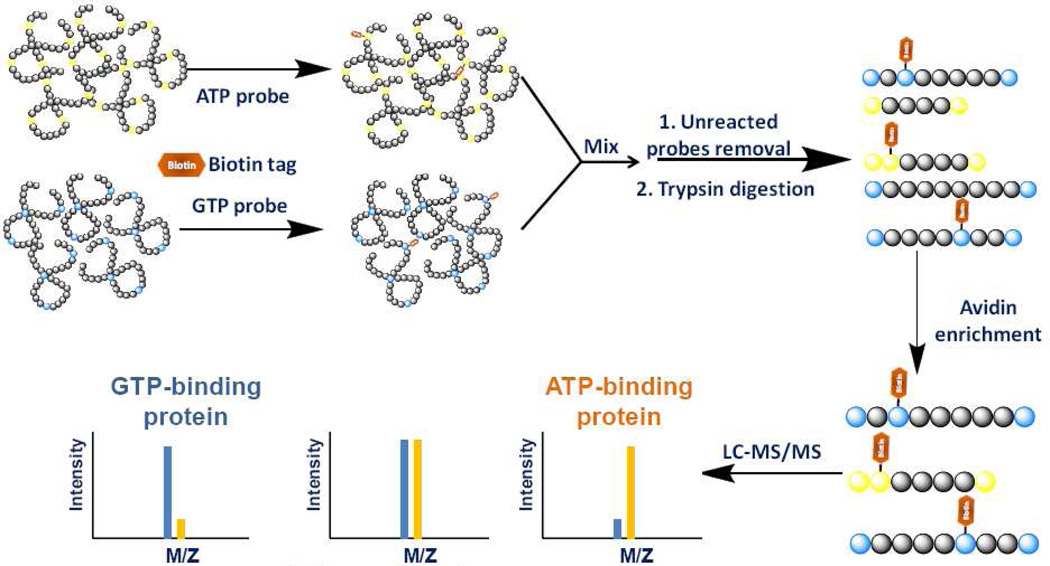
To explore the nucleotide-binding affinity of ATP/GTP-binding proteins at specific binding sites and to uncover novel nucleotide-binding targets, we employed SILAC together with our nucleotide affinity probe to investigate the nucleotide-binding property of proteins at the whole proteome scale. In a pilot experiment, light- and heavy-labeled cell lysates were treated with the same concentrations of biotin-based ATP and GTP probes (100 µM each), respectively. After the reaction, light- and heavy-labeled cell lysates were mixed prior to any further steps of sample manipulation as described in regular affinity experiments. To minimize the bias introduced by SILAC, we also performed reverse SILAC experiment, where heavy and light cell lysates were treated with ATP and GTP probes, respectively (Figure 3). All the biotin-labeled peptides were directly analyzed by 1D-LC/MS/MS. Peak intensity ratios of light and heavy biotin-labeled tryptic peptides were subsequently used to derive ATP/GTP binding affinity ratio, RATP/GTP, which reflects the relative binding affinities of ATP and GTP towards specific lysine residues in proteins of interest.
Figure 3.
A general strategy for comparing ATP/GTP-binding properties of proteins from SILAC cell lysates.
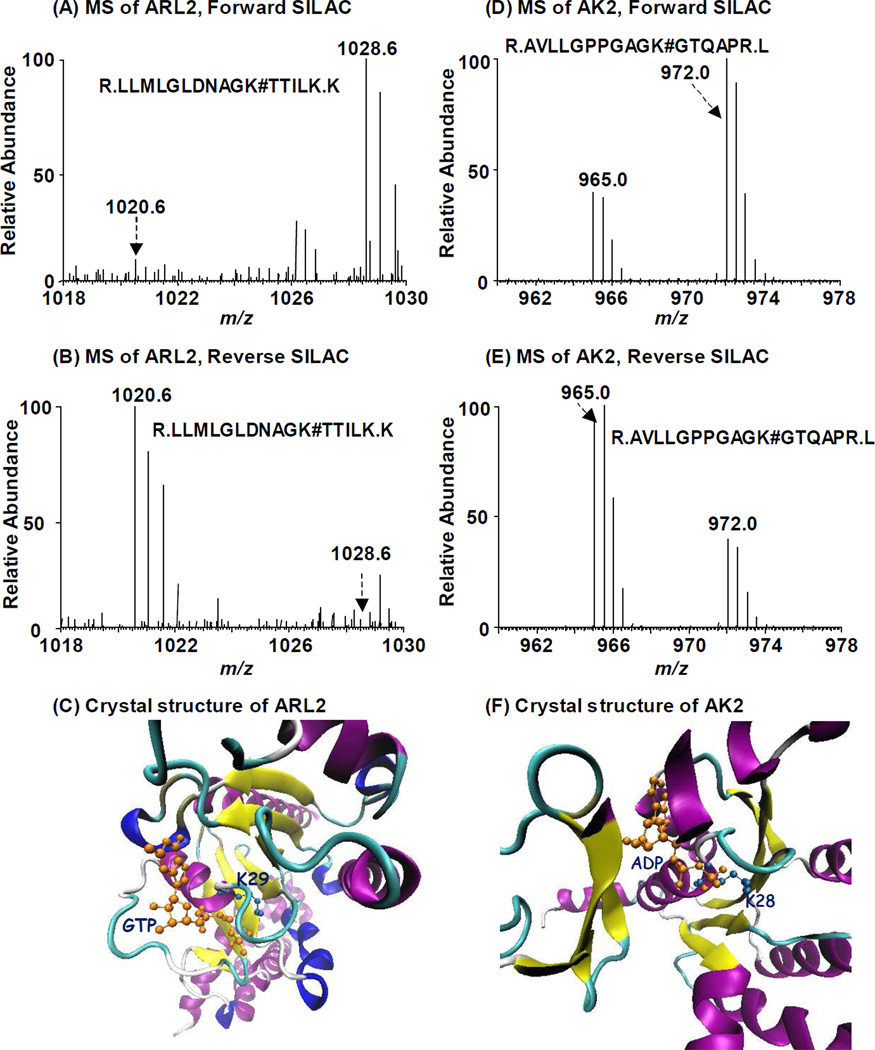
The median RATP/GTP ratio for all the quantified peptides from a single protein was employed to represent its relative affinity towards ATP and GTP. The medium was selected to minimize the effect of outlier and non-specific binders19. For example, ADP-ribosylation factor-like protein 2 (ARL2), a member of the Ras small GTPase superfamily, exhibits strong binding towards GTP in both forward and reverse SILAC labeling experiments as the median of RATP/GTP for all the detected biotin-labeled peptides from ARL2 is only 0.11. In this regard, one biotin-labeled lysine with K29 being conjugated, i.e., R.LLMLGLDNAGK#TTILK.K, was found for ARL2 (Figure 4, Figure S5). The crystal structure of the ARL2-GTP complex (PDB entry: 3DOE) revealed that K29 contacts directly the γ-phosphate group of GTP20, demonstrating that biotin-labeled lysine with RATP/GTP ratio deviating substantially from 1 corresponds to real nucleotide-binding site, or more precisely, the lysine residue that is in close proximity to the γ-phosphate group of the nucleotide.
Figure 4.
Forward- and reverse-SILAC combined with LC-MS/MS for the quantitative comparison of ATP/GTP binding affinity towards ADP-ribosylation factor-like protein 2 (A, B) and adenylate kinase 2 (C, D). # indicates the biotin-labeling site. Crystal structures of ARL2 bound with GTP (E) and AK2 bound with ATP (F) demonstrate the direct contact of nucleotide with identified lysine residues.
It is worth noting that the nucleotide affinity probes with an acyl phosphate linkage have relatively high reactivity; therefore, apart from the lysine residue(s) located in the nucleotide binding site, other lysine residues may also be labeled with biotin because of their non-specific electrostatic interaction with phosphate group of the probe10a. Nevertheless, because of the structural similarity of ATP and GTP probes, peptides housing these non-specifically labeled lysine residues are expected to exhibit a RATP/GTP ratio being close to unity. For instance, we found two biotin-labeled lysine sites in adenylate kinase 2 (AK2, MS/MS shown in Figure S5), where K28 displays a clear ATP binding preference with an RATP/GTP of 2.3. This is consistent with the observation that K28 is responsible for ATP binding and phosphohydrolase activity (PDB entry: 3TLX, Figure 4). The other labeling site (K85), however, exhibits similar ATP/GTP binding affinity (RATP/GTP=1.09). This finding, together with the absence of prior report supporting K85’s involvement in ATP binding, suggests that modification of K85 is likely derived from non-specific interaction. Meanwhile, the median RATP/GTP ratio for all peptides identified for AK2 is 2.26 (Table S2), supporting that the median RATP/GTP ratio of all the quantified peptides from a single protein allows for the prediction of the protein’s overall nucleotide-binding property.
4. Global profiling of ATP/GTP binding selectivity of nucleotide-binding proteins from the whole human proteome
Previous studies revealed that the cellular concentration of ATP is approximately 6–7 times higher than GTP21. Thus, those proteins with similar or even slightly better affinity towards GTP at the same concentrations of ATP and GTP may be associated with more ATP than GTP in cells. To better mimic physiological conditions and to identify true novel GTP-binding proteins, we carried out similar ATP/GTP competition experiment using ~ 7:1 (100:15 µM) desthiobiotin-based ATP/GTP probe concentration ratio. We employed online 2D-LC-MS/MS with SCX-C18 biphasic column in 7:1 ATP/GTP competition experiment to obtain better coverage of nucleotide-binding proteins. As a result, a larger number (a total of 1695) of proteins were successfully quantified. For example, we were able to determine the RATP/GTP ratios of 206 kinases from four trials of 7:1 ATP/GTP competition experiments (Figure S6 and Table S5). Notably, these 206 kinases, including 153 protein kinases, were identified from a single cell line (HL-60 cells) and is estimated to cover approximately half of the expressed kinome in a given mammalian cell, which consists of up to 300 distinct protein kinases22. Moreover, only approximately 1 mg light and heavy cell lysates were used in our approach, whereas 20–40 mg cell lysates were used for kinase enrichment previously22–23. This demonstrates the sensitivity of our approach in detecting kinases from complex whole cell lysates.
As expected, a vast majority of RATP/GTP ratios for the quantified nucleotide-binding proteins are well above 1. For example, the average RATP/GTP ratio for all the proteins with ATP-binding GO annotation (GO: 0005524) reaches 6.08 in 7:1 ratio experiment, which is accompanied with an increase in average RATP/GTP ratio for all the known kinases to 12.2. These results suggest that most purine nucleotide-binding proteins bind preferentially to ATP in vivo. Because of the pivotal role of nucleotide binding in regulating kinases activity in signal transduction cascades, we next sought to systematically assess the ATP/GTP binding affinity of known kinases from LC-MS/MS results based on 7:1 ATP/GTP competition experiment. A heatmap was generated to better visualize the relative ATP/GTP binding affinity of the 206 identified kinases in 7:1 ATP/GTP competition experiment according to the median RATP/GTP ratios for all the quantified peptides for each kinase (Figure S6). White box was set for a minimum RATP/GTP ratio of 0. Because the instrument and software limitations in detecting extremely large ratios in SILAC experiment, a RATP/GTP ratio of 7 or greater was represented with dark blue box. Some insights into ATP/GTP selectivity of kinases can be gleaned from the heatmap. As shown in Figure S6, even though most kinases display blue or dark blue color (i.e., with preferential binding to ATP over GTP), kinases from different subgroups with various structure and function assume distinct ATP/GTP binding selectivity. For example, all four identified NimA-related kinases (NEK1, NEK3, NEK7, NEK9) show dark blue color in our heatmap, indicating their significant preference toward ATP-binding. This is consistent with the fact that these four kinases are known to adopt ATP as phosphate donor24. However, N-acetyl-D-glucosamine (GlcNAc) kinase (NAGK) and nucleoside diphosphate kinases (NME1, NME2) display light blue color in the heatmap, translating to moderate ATP/GTP binding selectivity. Indeed, previous studies indicate that NAGK and NME kinases may have the capacity to bind GTP, and GTP may serve as the phosphate donor for NAGK to convert endogenous GlcNAc to GlcNAc-6-phosphate25.
We also found a significant number of kinases in the same subfamily exhibiting differential selectivity toward binding to ATP and GTP. For instance, among 7 casein kinases detected (shown as CSNK in heatmap), four of them belong to casein kinases 1 and all show dark blue colors with strong preference for ATP binding. However, the other three identified casein kinases display significantly lighter blue color and all of them belong to casein kinase 2. This is consistent with the previous findings that casein kinase 2 can effectively utilize both ATP and GTP in the phosphotransferase reaction, whereas casein kinase 1 is only known to bind to ATP17, 26. Among the cyclin-dependent kinases, CDK5, CDK9 and CDK12 display strong preference for GTP binding; in contrast, CDK6, CDK7, CDK8 and CDK10 exhibit more favorable binding toward ATP than GTP. Taken together, our SILAC-based competition experiment enables global profiling of binding selectivity of kinases toward ATP and GTP.
On the other hand, even in 7:1 ATP/GTP competition experiment, some proteins still display preferential binding toward GTP with RATP/GTP ratios smaller than one. Therefore, we considered those proteins with medium RATP/GTP ratio smaller than 1 in 7:1 ATP/GTP competition experiment as true GTP-binding proteins, which include 119 unique proteins (Table S4). GO analysis using DAVID27 showed that 42 out of those 119 proteins were with GTP-binding GO (GO: 0005525), suggesting a 14-fold increase in enrichment factor relative to entire human proteome with a p-value of 6.2E-34. Other than the known GTP-binding proteins, 14 known ATP-binding proteins including 6 kinases still show median ratio < 1. Six of these 14 proteins with known ATP-binding GO were also detected in 1:1 ATP/GTP competition experiment and all of them show clear preference for GTP binding, demonstrating the reproducibility of our ATP/GTP competition approach (Table S4). Furthermore, aside from AK3, AK4 and SUCLG2, the remaining 11 proteins with ATP-binding GO annotation were not annotated as GTP-binding proteins. However, there are still a few reports supporting the GTP binding affinity of some of these proteins. For example, it was found that GTP serves as a slightly better phosphate donor for deoxycytidine kinase (DCK) than ATP, though most studies only considered ATP as its phosphate donor28. In our 7:1 ATP/GTP competition experiments, we found that DCK exhibits preferential binding toward GTP over ATP with the biotin-labeled peptide (K.ISIEGNIAAGK#STFVNILK.Q). In another example, methylmalonic aciduria type A (MMAA) is annotated as an ATP-binding protein in Uniprot (Accession #: Q495G5). However, 7:1 ATP/GTP competition experiments led to the identification of only one peptide from MMAA (R.VGLSGPPGAGK#STFIEYFGK.M) with a very small RATP/GTP ratio of 0.19. This is in keeping with a recent study revealing that MMAA shares similar sequence motif as GTPase chaperons and possesses GTPase activity29. Therefore, ATP/GTP competition experiment not only confirms the nucleotide-binding affinity of known ATP/GTP-binding proteins, but also serves as a discovery tool to explore novel nucleotide-binding proteins without prior knowledge of GO annotation.
5. Discovery of nucleotide binding motif in proteins
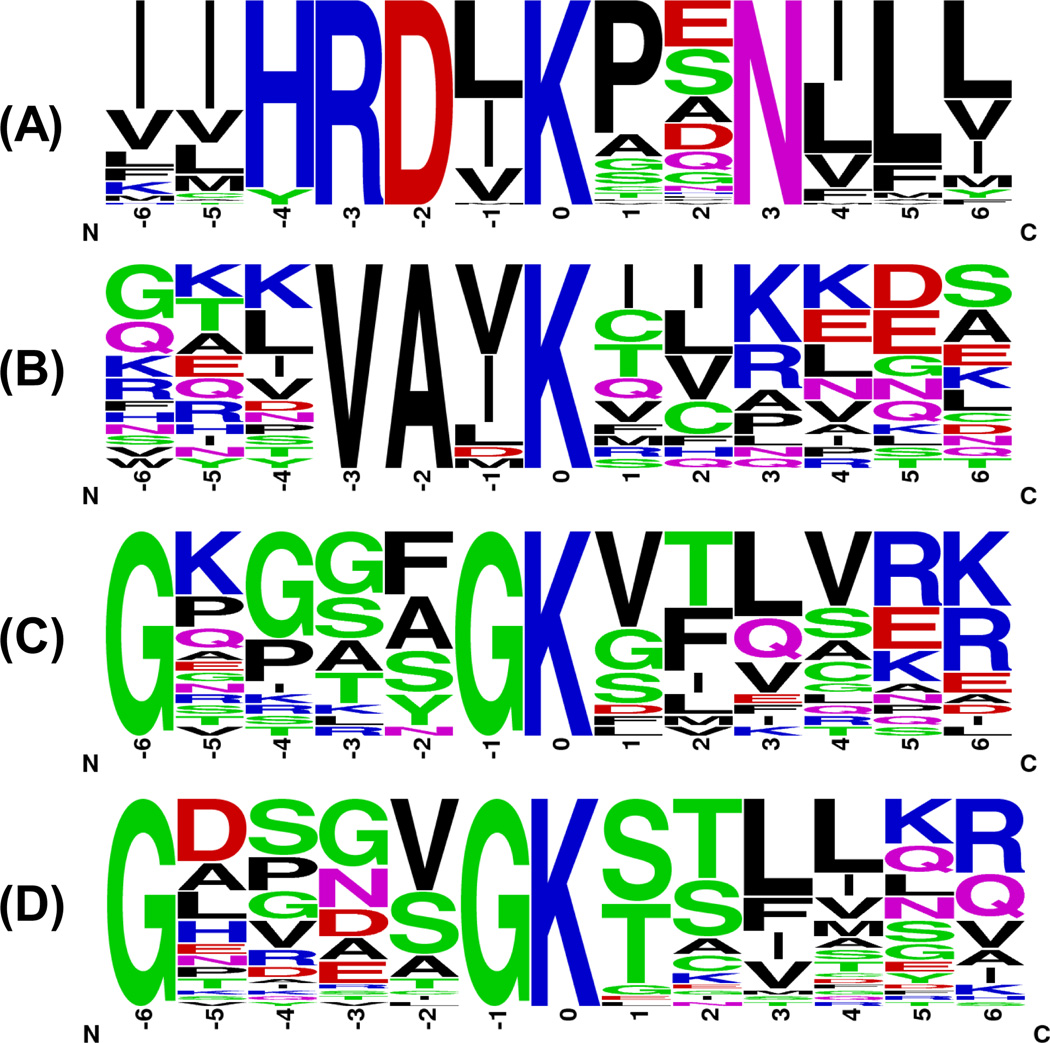
As mentioned above, our nucleotide competition strategy distinguishes from other nucleotide-binding assay by virtue of site-specific differentiation of ATP and GTP binding. In this regard, we sought to analyze local sequence context surrounding the biotin-modified lysine, which is considered as the nucleotide-binding site in proteins (Figure 5). First, we examined all the biotin-labeled peptides from known ATP-binding proteins. The well-known P-loop sequence motif of GxxxxGK was successfully identified by motif-X30 search with a 9.9-fold enrichment with respect to the occurrence frequency in our input sequence compared to the entire proteome. In addition, another two unique motifs of HRDxKxxN and VAxK, which were identified with enrichment factors (352 and 12.3, respectively) greater than that for the well-known GxxxxGK motif, are derived exclusively from kinases. For peptides from GTP-binding protein group, only GxxxxGK motif was identified, which agrees with the previous observation that GxxxxGK motif is the conserved nucleotide binding site in GTP-binding proteins12.
Figure 5.
Unique binding motifs found for known kinases (A, B, C) and known GTP-binding proteins (D) in 7:1 ATP/GTP competition experiment.
The proximity of nucleotide γ-phosphate to the catalytic domain is a prerequisite for the enzymatic activity of kinases. Previous studies revealed that there are three major sequence components in kinases that are essential for nucleotide binding31. First, the glycine-rich loop of subdomain I at the N-terminus of the kinase domain contains the consensus motif of GxxxxGKV. The second part of the kinase domain that has been implicated in ATP binding localizes in a region of subdomain II with an invariant lysine residue. In addition, most kinases show another conserved motif of HRDxKxxN located in subdomain VIB, which participates in ATP binding32. It is of note that the nucleotide-binding site of kinases or other nucleotide-binding proteins are conformationally dynamic, and ATP may bind to different sites in nucleotide-binding proteins under different physiological states18. Therefore, it is very difficult to predict the most relevant amino acid residues involved in ATP binding solely based on motif analysis, especially a significant number of kinases possess at least two of the three possible binding sequences. Interestingly, our motif analysis of peptides from ATP-binding protein groups in 7:1 ATP/GTP competition experiments uncovers the aforementioned three possible ATP binding sites in kinases successfully (K in VAxK motif corresponds to the invariant lysine in subdomain II). The motif with the most significant ATP-binding preference may represent the most relevant region involved in kinase’s binding toward γ-phosphate of ATP, thereby providing important knowledge basis for designing mutants to modulate kinase activity.
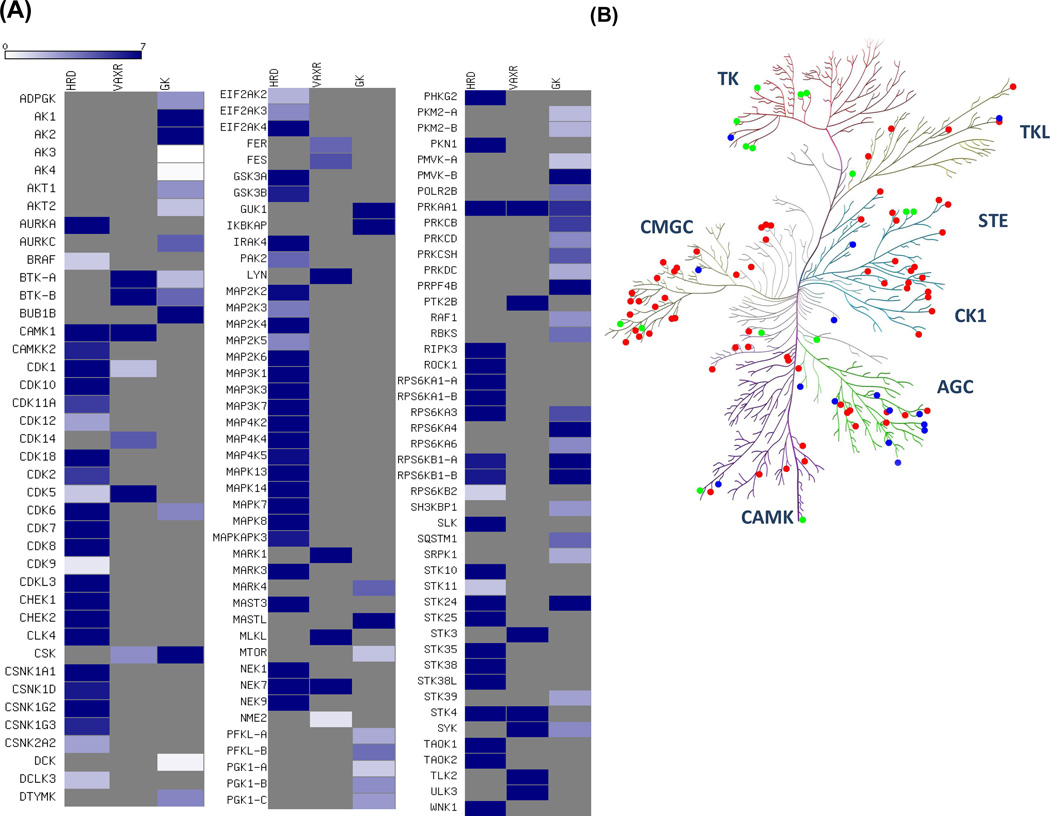
Among all of 206 kinases identified, 123 contained the biotin-labeled peptides covering at least one of these three motifs in 7:1 ATP/GTP competition experiment. A heatmap was created to better visualize the ATP/GTP binding selectivity of these three motifs for each kinase (Figure 6A). Meanwhile, we map the kinases to the human kinome dendrogram with color annotation to specify the motif with the most profound ATP/GTP selectivity (i.e., with the largest RATP/GTP ratio). As shown in Figure 6B, the reactive lysine most frequently resides in the consensus motif of HRDxKxxN in kinases, except for tyrosine kinases. For example, both GxxxxGK and HRDxKxxN motifs are present in ribosomal protein S6 kinase alpha-1 (RPS6KA1), whereas K94 in RPS6KA1 is the equivalent lysine in the VAxK motif except that V is replaced with a Y. However, two biotin-labeled peptides derived from the HRDxKxxN motif, namely, R.DLK#PENILLDEEGHIK.L including K189 (RATP/GTP ratio= 61.3) and R.DLK#PSNILYVDESGNPECLR.I including K537 (RATP/GTP ratio= 51.2), were detected in our ATP/GTP competition experiment with significant selectivity of ATP over GTP. This finding is consistent with the X-ray crystal structure of RPS6KA1 (PDB entry: 2Z7Q, residue 33–353), which reveals the close proximity of K189 to the reactive γ-phosphate group of ATP; however, K94 and K75 in the VAxK and GxxxxGK motifs are more proximal to non-transferable β- and α-phosphates, respectively (Figure S7).
Figure 6.
(A) A heatmap displaying the relative selectivity of ATP/GTP towards each possible binding motif of kinases. Dark blue and white indicate significant ATP-binding preference with high RATP/GTP ratio and significant GTP-binding preference with small RATP/GTP ratio, respectively (See scale bar above the heatmap). (B) Protein kinases were mapped to the human kinome dendrogram with color annotation to specify the motif with the highest RATP/GTP ratio. Red, green and blue represent kinases with HRDxKxxN, VAxK, and GxxxxGK motifs, respectively.
The identification of HRDxKxxN motif with a moderate RATP/GTP ratio could raise the possibility that other sequence may contribute to ATP binding, even though HRDxKxxN is the only motif identified. For instance, one peptide with K416 residing in the HRDxKxxN motif of double-stranded RNA-activated protein kinase (PKR or EIF2AK2) was quantified with a moderate RATP/GTP ratio of 2.08, whereas another peptide containing K296 (K.TYVIK#R.V) without any clear motif feature shows a much larger RATP/GTP ratio of 61.4. Site-directed mutagenesis studies showed that substitution of K296, but not K416, with an arginine gave rise to a catalytically dead mutant that completely abolishes the kinase activity of PKR33. These examples demonstrate the capability of our approach in differentiating the selectivity of nucleotide binding affinity to both known binding motifs and previously unrecognized nucleotide-binding site of kinases. The latter facilitates the identification of the lysine residue that is most relevant for ATP binding and hydrolysis even in the absence of structural information of the protein.
Most identified tyrosine kinases carry the VAxK, instead of HRDxKxxN, as the motif exhibiting the greatest selectivity toward ATP binding. For instance, protein tyrosine kinase Lyn has one biotin-modified peptide K.VAVK#TLKPGTMSVQAFLEEANLMK.T with the VAxK motif, in which the biotin-labeled K275 is proximal to the transferable phosphate (PDB entry: 2ZV8). Careful examination of all the tyrosine kinases identified from our competition experiment revealed that the lysine residue in the HRDxKxxN motif is substituted with an arginine in tyrosine kinases. This arginine is in closer proximity to γ-phosphate of ATP than the lysine residue in the VAxK motif (Figure S7). However, arginine does not react with our amine-reactive probe and K275 is the only available reactive lysine in Lyn sequence. Interestingly, the VAxK motif peptide is rarely detected for serine-threonine kinases, despite the fact that they are generally present in the primary sequence of these kinases, which distinguishes serine-threonine protein kinases from tyrosine kinases.
GxxxxGK motif generally serves as the most selective binding motif for metabolic kinases, which lack the HRDxKxxN and VAxK motifs. For example, we detected four kinases in the adenylate kinase (AK) subfamily and biotin-labeled peptides from all of them contain a lysine in the GxxxxGK motif, which was found to be the closest lysine to γ-phosphate of ATP as revealed by crystal structure analysis12. This finding allowed us to precisely characterize the ATP/GTP selectivity of proteins in the same subfamily with similar structures by targeting unique nucleotide binding motif. AK1 and AK2, which use ATP as the phosphate donor, are colored in dark blue in heatmap. By contrast, AK3 and AK4 with very similar binding sequences of R.AVIMGAPGSGK#GTVSSR.I (RATP/GTP =0.05) and R.AVILGPPGSGK#GTVCQR.I (RATP/GTP =0.11) exhibit robust GTP binding preference. Similar to AK2, AK3 is a phosphotransferase enzyme located in mitochondria and it induces AMP phosphorylation; however, AK3 can only use GTP in lieu of ATP as the phosphate donor34. Although AK4 is annotated as an ATP-AMP transphosphorylase in Uniprot (Accession #: P27144), recombinant human AK4 exhibited greater efficiency in phosphorylating AMP in the presence of GTP than in the presence of ATP35. Moreover, Blast search revealed that AK4 has approximately 60% sequence similarity to AK3 but only around 40% similarity to AK2. Besides, we also note that most protein kinases exhibiting the greatest probe-labeling selectivity toward GxxxxGK motif belong to the AGC kinase group. For example, protein kinase C beta (PKCb) and protein kinase C delta (PKCd), belonging to AGC kinase group, only have one peptide within GxxxxGK motifs quantified with RATP/GTP ratio of 5.26 and 3.27, respectively, despite the fact that they both possess the HRDxKxxN motif. This finding suggests that the lysine in GxxxxGK motif may play an important role in deciding the specificity of nucleotide binding of protein kinase C in aqueous solution. However, further experiments are needed to validate this notion.
The capabilities of our approach in assessing quantitatively the nucleotide binding selectivities towards unique binding motifs can also be extended to GTP-binding proteins. In this vein, GxxxxGK is the only conserved nucleotide-binding motif for GTP-binding proteins, and a heatmap was generated to better visualize the ATP/GTP binding selectivity of this motif for each identified known GTP-binding protein (Figure S8 and Table S5). As depicted on the map, the GxxxxGK motif in most known GTP-binding proteins, including four heterotrimeric G proteins as well as four translation initiation and elongation factors, shows significant binding preference toward GTP. In addition, 21 small GTPases, including 14 Rab proteins, have been quantified and the GxxxxGK motif in all of them displays significant binding preference to GTP. In contrast, GxxxxGK motif in GTP-binding protein 9 (GTPBP9) exhibits significant preference toward ATP-binding. GTPBP9 is the human homologue of bacterial YchF, which is considered a GTPase and involved in protein translation. However, Roland et al.36 found that GTPBP9 binds and hydrolyzes ATP more efficiently than GTP and hence renamed human GTPBP9 to Obg-like ATPase 1. Our result further demonstrated the ATP binding preference for GTPBP9 protein, which furnishes another line of evidence to support that the GxxxxGK motif is the actual ATP-binding site in GTPBP9. The identification of a GxxxxGK motif with preferential binding toward ATP in a previously documented GTP-binding protein further illustrates the ability of our approach in exploiting novel nucleotide-protein interactions and in uncovering the actual nucleotide binding sites.
Conclusions
Here, we report a general strategy using affinity chemical probe to enrich and identify ATP/GTP-binding proteins in human cell proteome. Our ATP/GTP affinity probe mainly has two applications. First, it can be used as an enrichment method to facilitate the subsequent LC-MS/MS identification and quantification of ATP/GTP-binding proteins at the global proteome scale; more than 100 GTP-binding proteins and 206 kinases from cell lysate of a single cell line could be identified and quantified with low mg quantity of lysate. Therefore, the ATP and GTP affinity probes constitute powerful reagents for the enrichment and subsequent detection of nucleotide-binding proteins from human proteome, which will enable sensitive and accurate profiling of cell signaling pathway perturbed by extracellular stimuli.
Second, this approach, in conjunction with quantitative proteomics, can be employed to characterize, at the entire proteome level, nucleotide-protein interactions and to identify specific nucleotide-binding sites in proteins. ATP/GTP binding selectivity of nucleotide-binding proteins in the entire proteome was systematically investigated by quantifying peak intensity ratios of light and heavy biotin-labeled tryptic peptides from ATP or GTP probe reaction, demonstrating the great potential of our ATP/GTP affinity probes in quantitative proteomic analysis. A key advantage in charactering nucleotide-protein interaction with our affinity probe over traditional binding assay lies in that our strategy allows for site-specific determination of relative binding affinities of proteins towards different nucleotides (e.g. ATP and GTP). Thus, nucleotide-protein interaction studies can be extended to quantitative surveys of specific binding regions in proteins of interest. Furthermore, it can be envisaged that similar nucleotide-affinity probes can be generally applied for the capture and characterization of proteins that can bind to other nucleotides, and such experiments are currently being pursued in our laboratory.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (R01 ES019873 to Y.W.) and California Tobacco-related Disease Research Program (20DT-0040 to Y.X.). The authors also would like to thank Dr. Xiaoli Dong for help with the setup of LC-MS/MS systems.
Footnotes
Supporting Information Available: Detailed experimental procedures, protein identification data, and heatmap. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]; (b) Takai Y, Sasaki T, Matozaki T. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 2.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 3.Dong X, Xiao Y, Jiang X, Wang Y. J. Proteome Res. 2011;10:5463–5471. doi: 10.1021/pr200718p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor P, Nielsen PA, Trelle MB, Andersen MB, Vorm O, Moran MF, Kislinger T. J. Proteome Res. 2009;8:1610–1616. doi: 10.1021/pr800986c. [DOI] [PubMed] [Google Scholar]
- 5.Ito J, Heazlewood JL, Millar AH. J. Proteome Res. 2006;5:3459–3469. doi: 10.1021/pr060403j. [DOI] [PubMed] [Google Scholar]
- 6.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Stemmann O, Mann M. Mol. Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Adam GC, Sorensen EJ, Cravatt BF. Mol. Cell. Proteomics. 2002;1:828–835. doi: 10.1074/mcp.t200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 8.Ohta T, Nagano K, Yoshida M. Proc. Natl. Acad. Sci. U. S. A. 1986;83:2071–2075. doi: 10.1073/pnas.83.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaneda M, Masuda S, Tomohiro T, Hatanaka Y. Chem Bio Chem. 2007;8:595–598. doi: 10.1002/cbic.200600527. [DOI] [PubMed] [Google Scholar]
- 10.(a) Qiu H, Wang Y. Anal. Chem. 2007;79:5547–5556. doi: 10.1021/ac0622375. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Biochemistry (Mosc.) 2006;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 11.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 12.Saraste M, Sibbald PR, Wittinghofer A. Trends Biochem. Sci. 1990;15:430–444. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 13.Deyrup AT, Krishnan S, Cockburn BN, Schwartz NB. J. Biol. Chem. 1998;273:9450–9456. doi: 10.1074/jbc.273.16.9450. [DOI] [PubMed] [Google Scholar]
- 14.(a) Borisov OV, Goshe MB, Conrads TP, Rakov VS, Veenstra TD, Smith RD. Anal. Chem. 2002;74:2284–2292. doi: 10.1021/ac010974p. [DOI] [PubMed] [Google Scholar]; (b) Sioud S, Genestie B, Jahouh F, Martin P, Banoub J. Rapid Commun. Mass Spectrom. 2009;23:1941–1956. doi: 10.1002/rcm.4091. [DOI] [PubMed] [Google Scholar]
- 15.Rybak JN, Scheurer SB, Neri D, Elia G. Proteomics. 2004;4:2296–2299. doi: 10.1002/pmic.200300780. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch JD, Eslamizar L, Filanoski BJ, Malekzadeh N, Haugland RP, Beechem JM, Haugland RP. Anal. Biochem. 2002;308:343–357. doi: 10.1016/s0003-2697(02)00201-4. [DOI] [PubMed] [Google Scholar]
- 17.Jakobi R, Traugh JA. Physiol. Chem. Phys. Med. NMR. 1995;27:293–301. [PubMed] [Google Scholar]
- 18.Soti C, Vermes A, Haystead TAJ, Csermely P. Eur. J. Biochem. 2003;270:2421–2428. doi: 10.1046/j.1432-1033.2003.03610.x. [DOI] [PubMed] [Google Scholar]
- 19.Cox J, Mann M. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 20.Zhang T, Li S, Zhang Y, Zhong C, Lai Z, Ding J. Structure (London, England : 1993) 2009;17:602–610. doi: 10.1016/j.str.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Pillwein K, Chiba P, Knoflach A, Czermak B, Schuchter K, Gersdorf E, Ausserer B, Murr C, Goebl R, Stockhammer G, Maier H, Kostron H. Cancer Res. 1990;50:1576–1579. [PubMed] [Google Scholar]
- 22.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Mann M, Daub H. Mol. Cell. Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Nat. Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]; (b) Duncan James S, Whittle Martin C, Nakamura K, Abell Amy N, Midland Alicia A, Zawistowski Jon S, Johnson Nancy L, Granger Deborah A, Jordan Nicole V, Darr David B, Usary J, Kuan P-F, Smalley David M, Major B, He X, Hoadley Katherine A, Zhou B, Sharpless Norman E, Perou Charles M, Kim William Y, Gomez Shawn M, Chen X, Jin J, Frye Stephen V, Earp HS, Graves Lee M, Johnson Gary L. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Regan L, Blot J, Fry A. Cell Div. 2007;2:25. doi: 10.1186/1747-1028-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta A. Biochim. Biophys. Acta. 1970;220:51–60. doi: 10.1016/0005-2744(70)90228-7. [DOI] [PubMed] [Google Scholar]
- 26.Srinivasan N, Antonelli M, Jacob G, Korn I, Romero F, Jedlicki A, Dhanaraj V, Sayed MF-R, Blundell TL, Allende CC, Allende JE. Protein Eng. 1999;12:119–127. doi: 10.1093/protein/12.2.119. [DOI] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Lempicki RA. Nat. Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Coleman C, Stoller R, Drake J, Chabner B. Blood. 1975;46:791–803. [PubMed] [Google Scholar]
- 29.Takahashi-Iniguez T, Garcia-Arellano H, Trujillo-Roldan MA, Flores M. Biochem. Biophys. Res. Commun. 2011;404:443–447. doi: 10.1016/j.bbrc.2010.11.141. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz D, Gygi SP. Nat. Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 31.Hanks SK, Hunter T. The FASEB Journal. 1995;9:576–596. [PubMed] [Google Scholar]
- 32.(a) Johnson LN, Noble MEM, Owen DJ. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]; (b) Nolen B, Taylor S, Ghosh G. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 33.Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 34.Schricker R, Magdolen V, Strobel G, Bogengruber E, Breitenbach M, Bandlow W. J. Biol. Chem. 1995;270:31103–31110. doi: 10.1074/jbc.270.52.31103. [DOI] [PubMed] [Google Scholar]
- 35.(a) Panayiotou C, Solaroli N, Johansson M, Karlsson A. Int. J. Biochem. Cell Biol. 2010;42:62–69. doi: 10.1016/j.biocel.2009.09.007. [DOI] [PubMed] [Google Scholar]; (b) Liu R, Strom A-L, Zhai J, Gal J, Bao S, Gong W, Zhu H. Int. J. Biochem. Cell Biol. 2009;41:1371–1380. doi: 10.1016/j.biocel.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koller-Eichhorn R, Marquardt T, Gail R, Wittinghofer A, Kostrewa D, Kutay U, Kambach C. J. Biol. Chem. 2007;282:19928–19937. doi: 10.1074/jbc.M700541200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.