Abstract
Peroxiredoxin 4 (Prx4) has been implicated in a wide variety of biological processes, including development, progression of cancer, inflammation, and antioxidant function. The purpose of this study was to provide further insight into its multiple roles at the whole-animal level, using Drosophila. Reduced expression of dPrx4 (up to 90%) resulted in greater sensitivity to oxidative stress, an elevated H2O2 flux, and increases in lipid peroxidation, but no effect on longevity. Overexpression at low levels (<2-fold) gave reduced levels of oxidative damage and tended to show an increase in longevity. Flies expressing dPrx4 globally at high levels (>5-fold) had a dramatically reduced life span (by 20–80%) and increased apoptosis. Analysis of these overexpressors revealed an aberrant redistribution of the dPrx4 protein from the endoplasmic reticulum (ER) to cytosol and hemolymph. In addition to the known proapoptotic effects of the cytosolic form of dPrx4, dPrx4 overexpression triggered an NF-κB-mediated proinflammatory response, similar to that observed in cells under ER stress or when microbially challenged. Finally, we provide the first evidence that dPrx4, on secretion into the hemolymph, elicits a JAK/STAT-mediated response. The effects on fly survival and homeostasis appear to represent a combination of differential effects dictated in large part by dPrx4 subcellular and tissue-specific localization.—Radyuk, S. N., Klichko, V. I., Michalak, K., Orr, W. C. The effect of peroxiredoxin 4 on fly physiology is a complex interplay of antioxidant and signaling functions.
Keywords: aging, stress response, signaling pathways, apoptosis
Six distinct peroxiredoxin forms have been identified, and these have been classified into 3 subtypes, based on their structure and mode of resolution of reactive intermediates. The “classical” 2-cys subtype of peroxiredoxins consists of 4 members, peroxiredoxin 1–4, which share high amino acid homology and similar structure and catalyze thioredoxin-dependent degradation of peroxides. Although peroxidase activity is common to all four peroxiredoxins, their biological functions appear quite divergent, in part due to differences in subcellular localizations. Studies of one of these, peroxiredoxin 4 (Prx4), have generated rather conflicting results. On the basis of work carried out in human cells, Prx4 was identified either as a cytoplasmic protein that attenuates NF-κB activation (1), or as a secretable protein, which serves as a proinflammatory cytokine, activating both NF-κB and JNK (2). A similar discord has been obtained in rodent models, in which one group of researchers found that Prx4 is being secreted to the extracellular space (3), while others failed to detect Prx4 in extracellular space but rather localized it to the endoplasmic reticulum (ER) and cytosol (4, 5). In this latter case, it was found to possess both antioxidant function (6, 7) and cell survival function, independent of its peroxidase activity (8, 9). In most studies, manipulations of Prx4 levels in cell culture or whole animals generally demonstrated a protective antioxidant function (4). For instance, in mice, overexpression of Prx4 in pancreatic islets protected them from diabetes progression by suppressing oxidative stress (10). Conversely, Prx4-knockout mice displayed a significant elevation of oxidative stress, particularly gonadal tissue, leading to testes dystrophy (11).
To address the controversies surrounding the location and function of Prx4, we took advantage of the Drosophila model, which is readily amenable to the manipulation of gene expression. Furthermore, it is a well-established model in aging research, permitting us to broaden our area of functional investigation. The Drosophila ortholog of Prx4, initially referred to as Jafrac2, is also reported to FlyBase (http://www.flybase.org) under various synonyms (CG1274, DPx-4156, and Prx4156). For the sake of clarity, we will henceforth refer to this gene as dPrx4 (the Drosophila form of Prx4). Previously, we identified dPrx4 as a secretable protein with antioxidant function, based on expression in S2 cell culture of a V5 epitope-tagged form (12). Interestingly, another study provided evidence that dPrx4 (Jafrac2) was associated with ER, and under certain conditions, it has been detected in cytosol but not secreted (8). In this present study, the focus is on deciphering dPrx4 function in a whole organism. Using the Drosophila UAS-GAL4 system, we generated fly lines underexpressing and overexpressing dPrx4. To complement this, we conducted studies in parallel on a P-element hypomorph (dprx4), in which expression of dPrx4 is 10 to 20% of wild-type levels. Underexpression of dPrx4 had no or minor positive effects on life span under normal conditions but negative effects when subjected to oxidative stress. High-level global overexpression significantly reduced fly life span. Subsequent molecular analysis revealed that overexpression of dPrx4 results in release of dPrx4 into cytosol and fly hemolymph and tissue-specific apoptosis and activation of the NF-κB and JAK/STAT signaling pathways. Our data also showed that the extracellular form of dPrx4 is required for activation of the JAK/STAT-mediated stress response. The effects on fly survival and homeostasis appear to represent a combination of differential effects dictated in large part by intracellular and tissue localizations of dPrx4.
MATERIALS AND METHODS
Fly strains
The yw reference strain has been maintained in this laboratory for >18 yr. The tubulin (Tub), actin (Act), daughterless (Da), and armadillo (Arm) GAL4 driver lines for broad overexpression and Appl GAL4 driver line for neuronal tissue overexpression, as well as mifepristone-inducible Tub-Switch and Act-Switch drivers were kindly supplied by Blanka Rogina (University of Connecticut Health Science Center, Farmington, CT, USA). The dPrx4 P-element mutant and transgenic fly lines carrying UAS-RNAi constructs are described in Table 1. UAS-dPrx4 transgenic lines were generated by cloning the entire dPrx4 coding region derived from a cDNA (CG1274) into the pUAST vector. The recombinant pUAS-dPrx4 plasmid DNA was sent to the TheBestGene Co. (Chino Hills, CA, USA; http://www.thebestgene.com) for P-element transformation. Three different transgenic lines (UAS-dPrx4-A, -B, and -C) carrying UAS-dPrx4 construct were selected. To exclude background effects, all mutants and transgenic and driver fly lines were backcrossed to our yw control strain in order to obtain genetically homogeneous stocks.
Table 1.
Fly strains obtained from Drosophila public stock centers
| Gene | Stock center | Stock no. | Characteristics | Name |
|---|---|---|---|---|
| dPrx4; synonyms Jafrac2, Dpx4156 | Bloomington, IN, USA | 18489 | P-element insertion 467 bp upstream of the start codon | dprx4 |
| Vienna, Austria | v43254 | RNAi construct GD5909 | RNAi-dPrx4-d | |
| v104006 | RNAi construct KK107748 | RNAi-dPrx4-e | ||
| domeless (dome) | Vienna | v36355 | RNAi construct GD14494 | RNAi-dome-a |
| v19717 | RNAi construct GD2612 | RNAi-dome-b | ||
| hopscotch (hop) | Vienna | v40037 | RNAi construct GD9157 | RNAi-hop-a |
| v102830 | RNAi construct KK105136 | RNAi-hop-b | ||
| Signal-transducer and activator of transcription protein at 92E (Stat92E) | Vienna | v43866 | RNAi construct GD4492 | RNAi-Stat-a |
| v106980 | RNAi construct KK100519 | RNAi-Stat-b |
Procedures
For experiments, flies were collected within 1–2 d after hatching and reared on a standard sucrose-cornmeal medium at 25°C. Overexpression of dPrx4 or underexpression of dome with the global high-level drivers Da, Tub, or Act interfered with normal development at 25°C, but not at 18°C. Consequently, flies overexpressing dPrx4 at high levels, as well as their combinations with RNAi-dome fly lines, were developed at 18°C. After hatching, adults were collected and reared at 25°C. Underexpression of hop and Stat with Da, Tub, or Act GAL4 drivers was lethal even at 18°C, necessitating the use of inducible Tub-Switch or Act-Switch drivers to obtain viable adult progeny. After collection, flies were maintained on regular food for 5 d, followed by transfer to food containing mifepristone. The induction of the P-Switch system was elicited by feeding the flies regular food containing ≥100 μg/ml mifepristone for ≥3 d. Control flies were fed food containing ethanol only (mifepristone solvent).
Oxidative stress was elicited by feeding flies with a 1% sucrose solution containing various concentrations of H2O2 or paraquat, as detailed in figure legends. Heat stress, dehydration, and cold stress were elicited following the protocols (13). Septic injury was performed by pricking flies with a needle dipped in an Escherichia coli suspension (14). Survivorship studies were conducted as described previously (15). Recombinant dPrx4 protein was purified from S2 cells overexpressing this protein, and its purity was assessed by gel electrophoresis and Coomassie staining of ∼5 μg protein (12). Purified dPrx4 protein was then injected into flies using a Hamilton 75 RN digital syringe (Hamilton Co., Reno, NV, USA). Cell-free hemolymph was collected by rupturing fly abdomens and bleeding the contents into Ringer's solution. Hemocytes and other cells were removed by centrifugation at 1000 g for 5–10 min. Both hemolymph and conditioned cell culture medium were concentrated using filter cartridges (Pierce, Rockford, IL, USA) with a 3-kDa cutoff.
Subcellular fractionation and immunoblotting
Crude subcellular fractionation was performed using gradient centrifugation as described previously (12). Proteins for immunoblot analysis were extracted with T-PER tissue protein extraction reagent (Pierce) containing protease inhibitors (Roche, Indianapolis, IN, USA). Immunoblot analyses were essentially performed as described previously (12, 16). Protein levels were quantified by densitometric scanning of the films using a digital imaging analysis system with AlphaEase stand-alone software (Alpha Innotech Corp., San Leandro, CA, USA). Anti-dPrx4 antibodies were raised against purified recombinant dPrx4 protein, produced in E. coli expression system, using services of Covance Research Products, Inc. (Denver, PA, USA) using similar protocols as we have employed in the past (15). Anti-actin antibodies (MP Biomedicals, Solon, OH, USA) were used as a control for loading and purity of hemolymph. Antibodies raised against the CoxVb subunit (mitochondrial) were used to assess purity of the subcellular fractions and hemolymph.
Quantitative RT-PCR
Primers specific for antimicrobial peptide (AMP) genes were as reported previously (14). Primers for Turandot A (TotA) were 5′ ACTGCTCTTATGTGCTTTGC (forward) and 5′ CCTGGGTGCTATTGATTTTG (reverse). Real-time PCR analysis was performed using Rotor-Gene RG-3000 and software (Corbett Research, Sydney, NSW, Australia), as described previously (14). Briefly, each RNA sample was isolated from 20–30 flies, followed by qRT-PCR, as described previously (14). PCR reactions were performed with SYBR Green (Molecular Probes, Grand Island, NY, USA) added to PCR reaction mixtures. Signals obtained for each gene were standardized against signals obtained for a housekeeping gene, ribosomal protein 49 (rp49), which exhibits only minor changes during aging (17), in parallel reactions.
H2O2 and 4-hydroxynonenal (HNE) assays
Steady-state levels of hydrogen peroxide were determined by using the Amplex Red hydrogen peroxide/peroxidase assay kit (Invitrogen, Grand Island, NY, USA) as described previously (18, 19). Levels of lipid peroxidation were determined by measuring levels of HNE by immunoblotting using anti-HNE antibodies (Alpha Diagnostics, Owings Mills, MD, USA), which recognize HNE-protein adducts, essentially as described previously (20, 21).
TUNEL labeling
Assessment of tissue-specific DNA fragmentation was made in cryosections prepared from whole flies using the in situ cell death detection kit, TMR red (Roche), as specified previously (19). Images were acquired by fluorescence microscopy (Nikon, Melville, NY, USA), using MetaMorph software (Molecular Devices, Sunnyvale, CA, USA).
Statistical analysis
Differences in the mRNA, protein, H2O2, and HNE levels were assessed by paired Student's t tests using Microsoft Excel software (Microsoft, Redmond, WA, USA). The mean survivorship time and statistical significance of differences between survival curves were assessed by the log rank test using Prism for Macintosh 4.0a software (GraphPad Software, San Diego, CA, USA). The significance of differences between survival curves was determined by pairwise comparisons of each experimental combination and its corresponding driver-only and responder-only controls.
RESULTS
Expression of dPrx4 during aging and in response to different stresses
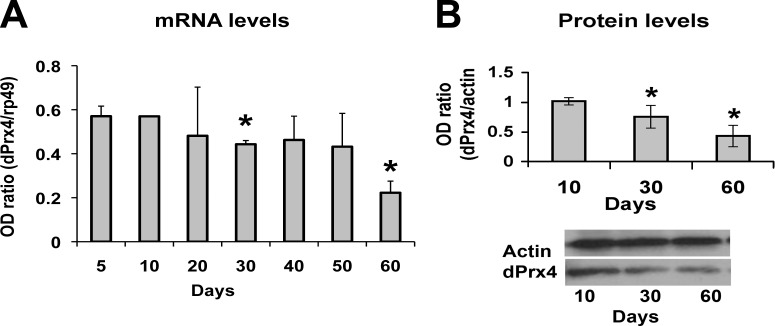
We first examined the effect of aging on dPrx4 expression in whole flies ranging from 5 to 60 d of age (mean life span of these wild-type flies is in the 55- to 60-d range). As observed in Fig. 1, dPrx4 levels exhibited a gradual decline with age at both the RNA and protein levels, reaching levels below 50% that of young flies by d 60 (Fig. 1). Subsequently, we measured dPrx4 protein levels in young flies after exposure to a battery of stressors that included hydrogen peroxide, paraquat, cadmium, heat stress, and bacterial infection. None of these treatments were observed to have an effect on dPrx4 levels (Supplemental Fig. S4 and data not shown), which suggests that the dPrx4 activity is not regulated at the transcriptional level.
Figure 1.
Expression of dPrx4 during aging. A) dPrx4 mRNA expression determined by qRT-PCR. RNA was isolated from flies collected at 10-d intervals. Signals obtained with dPrx4 primers were normalized against signals obtained with rp49 primers. Results are means ± sd (n=3). Statistically significant differences were observed between 5- and 30-d-old flies (P=0.016) and 5- and 60-d-old flies (P=0.02), as indicated by asterisks. B) Immunoblot analysis of protein preparations made from 10-, 30-, and 60-d-old flies. Immunoblots were performed using anti-dPrx4 antibodies, reprobed with anti-actin antibodies to control for loading. The results are mean ± sd (n=3). Statistically significant differences were observed between 10- and 30-d-old flies (P=0.047) and 10- and 60-d-old flies (P=0.014). *P < 0.05.
Effects of reduced expression and ectopic overexpression of dPrx4 on fly survivorship under normal and stress conditions
The observed reduction in dPrx4 expression in older flies led us to postulate that compensation of this relative deficiency might provide positive effects on survivorship. To test this, we investigated the phenotypes of flies displaying different levels of dPrx4. To determine the effects of reduced expression, we used a P-element mutant strain dprx4 (Table 1). There are two differentially spliced mRNA species (1031 and 1138 bp) derived from the dprx4 gene and, as described in the FlyBase database (http://www.flybase.org), the P-element insertion disrupts the first intron of isoform A and the 5′UTR sequence of isoform B, resulting in reduced dPrx4 expression (Fig. 2A and Table 1). Alternatively, the dPrx4 reduction was achieved using UAS-RNAi hairpin fly lines (Fig. 2B and Table 1). The RNAi-dPrx4 fly lines were also tested for down-regulation of the potential off-target genes CG6888 and CG1633 using specific antibodies developed to these genes. No significant silencing of these genes by dPrx4 RNAi constructs was found (Supplemental Fig. S1).
Figure 2.
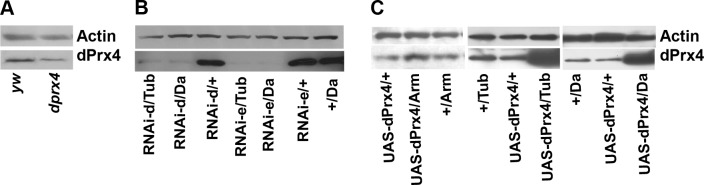
Immunoblot analysis of dPrx4 levels in flies underexpressing and overexpressing dPrx4. All immunoblots were performed with anti-dPrx4 antibodies and reprobed with anti-actin antibodies to control for loading. A) dPrx4 expression in the P-element mutant. Protein was extracted from 10-d-old control (yw) and homozygous mutant (dprx4) flies, and subjected to immunoblot analysis. About 80% reduction in the dPrx4 expression was observed. B) dPrx4 expression in the RNAi mutants. Shown are representative images for two UAS-RNAi transgenic lines (RNAi-d and RNAi-e) crossed to Da- or Tub-GAL4 drivers. dPrx4 protein levels were virtually undetectable in experimental flies (<5%), in contrast to the driver (+/Da) and transgene controls (RNAi-d or RNAi-e/+). C) Immunoblot analysis of dPrx4 overexpression. Shown is a representative image for one (UAS-dPrx4-A) of 3 different transgenic lines. Similar levels of dPrx4 were observed with two other transgenic lines. More detailed presentation of dPrx4 overexpression is shown in Supplemental Fig. S2. Driver (+/Da-GAL4, +/Tub-GAL4, and +/Arm-GAL4) and transgene controls (UAS-dPrx4/+) express ∼10 times less dPrx4 protein compared to the experimental fly lines obtained with high-level global Da- or Tub-GAL4 (UAS-dPrx4/Da, UAS-dPrx4/Tub) drivers, while <2 times overexpression was achieved with low-level Arm-GAL4 driver (UAS-dPrx4/Arm).
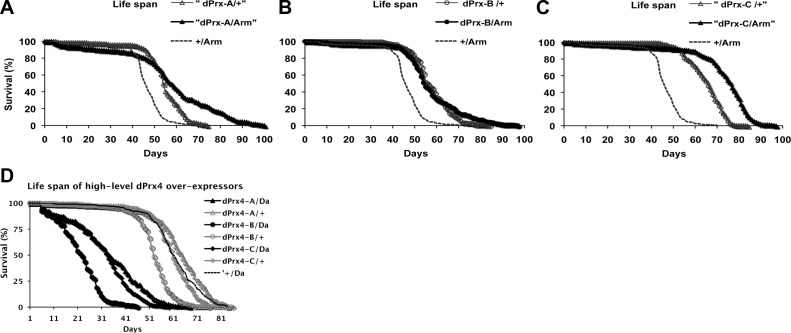
Different levels of overexpression of dPrx4 were achieved by crossing UAS-dPrx4 transgenics to different GAL4 enhancer lines, Da-GAL4 and Tub-GAL4 (high level) and Arm-GAL4 (low level). An increase of ∼6–10-fold in dPrx4 expression was verified in flies crossed to the ubiquitous high-level expression drivers, Da-GAL4 or Tub-GAL4 (Fig. 2C and Supplemental Fig. S2), while overexpression in response to the armadillo driver did not exceed 2-fold.
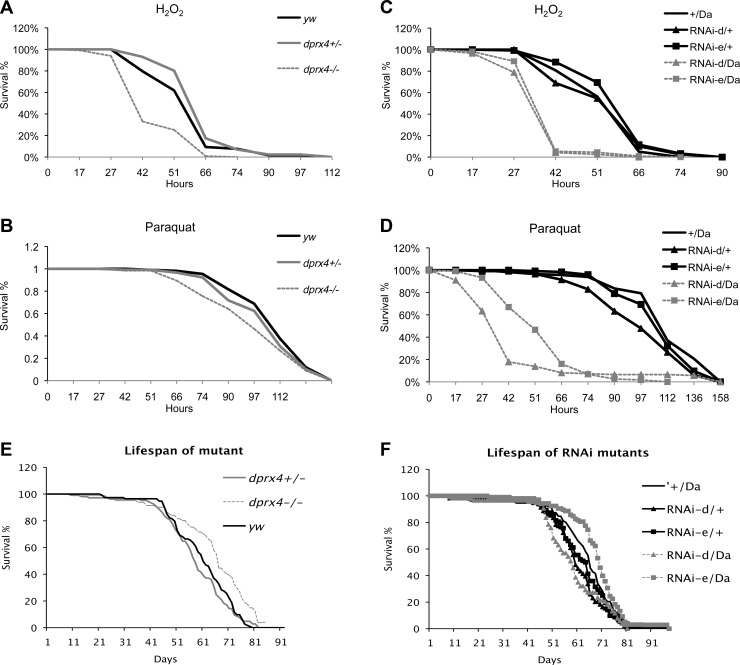
Underexpression of dPrx4 decreased resistance to oxidative stress. The hypomorphic dprx4 mutant was somewhat more sensitive to hydrogen peroxide and paraquat relative to controls (Fig. 3A, B), while more severe effects of dPrx4 underexpression were observed in the RNAi mutants, which precipitously died after exposure to H2O2 or paraquat (Fig. 3C, D). It seems that dramatic effects were observed when dPrx4 was nearly absent (RNAi mutants), while lesser reduction of dPrx4 in the P element dprx4 mutant showed less prominent effects on fly viability under oxidative stress. However, overexpression of dPrx4 at high levels with Da-GAL4 or Tub-GAL4 drivers or at low levels (Arm-GAL4) or with a tissue-specific (Appl-GAL4) driver provided little if any significant effects on viability under stress conditions (representative graphs are shown in Supplemental Fig. S3).
Figure 3.
Effects of underexpression of dPrx4 on survivorship under normal and oxidative stress conditions. A–D) Approximately 100–150 flies of each group were fed 1% sucrose solution containing 0.5 M H2O2 (A, C) or 10 mM paraquat (B, D). Results are representative of 2–3 independent experiments. Control yw, dprx4+/−, and dprx4−/− are heterozygous and homozygous P-element mutants, respectively. RNAi experimental flies (RNAi-d or RNAi-e/Da) are indicated by gray dashed lines. +/Da and RNAi-d or RNAi-e/+ are driver and transgene controls. Statistically significant differences (P<0.05) determined by log rank test were found between dprx4−/−mutant and yw control treated with H2O2 (A) and experimental and control RNAi mutants treated with H2O2 and paraquat (C, D). E) Life span of dprx4 mutant. F) Life spans of RNAi mutants. Shown are representative data of 3 and 2 independent experiments. In each experiment, 200–250 flies for each fly line were used. Statistical data are shown in Tables 2 and 3.
Although flies underexpressing dPrx4 were sensitive to oxidants, their life span under normal conditions did not differ substantially from controls or was even marginally extended (Fig. 3E, F and Tables 2 and 3). Overexpression of dPrx4 globally at low levels with Arm-GAL4 driver had also only minor positive effects on survivorship under normal conditions (Fig. 4 and Table 4). In contrast, global high-level overexpression of dPrx4 using the Da-GAL or Tub-GAL4 drivers had severe negative effects on fly development and adult survivorship under normal conditions. No viable progeny were obtained at 25°C; however, experimental adults were obtained when developed at 18°C. These adult flies were viable at 25°C but their life spans were shortened by 20–80% compared to relative controls (Fig. 4B and Table 4).
Table 2.
Mean age of P-element mutant flies underexpressing dPrx4
| Experiment | Age (d) |
Percentage vs. yw | |
|---|---|---|---|
| yw | dprx4 | ||
| 1 | 59.0 | 69.5 | 117* |
| 2 | 62.5 | 65.8 | 105 |
| 3 | 50.5 | 54.8 | 108 |
| 4 | 52.6 | 56.9 | 108* |
P < 0.05 between control (yw) and mutant (dprx4) flies, determined by log-rank test.
Table 3.
Mean age of RNAi mutant flies underexpressing dPrx4
| Genotype | Age (d) | Percentage |
|
|---|---|---|---|
| Vs. transgene/+ | Vs. driver/+ | ||
| RNAi–dPrx4-d/+ | 52.1 | ||
| RNAi–dPrx4-e/+ | 53.9 | ||
| +/Da | 58.4 | ||
| RNAi–dPrx4-d/Da | 52.1 | 100; 109* | 111;* 103 |
| RNAi–dPrx4-e/Da | 60.3 | 112;* 103 | 103;* 108 |
P < 0.05 vs. control flies, determined by log-rank test.
Figure 4.
Effects of overexpression of dPrx4 on fly life span. Three different UAS-dPrx4 transgenic flies (dPrx-A, dPrx-B, or dPrx-C) were crossed to low-level global (Arm-GAL4) driver (A) or high-level global (Da-GAL4) driver (B). Controls (dPrx4-A, dPrx4-B, or dPrx4-C/+ and dPrx4-C/Arm or dPrx4-C/Da) were obtained by crossing UAS transgenes or driver lines to yw flies. Shown are representative data of 2 independent experiments, summarized in Table 4. In each experiment, 200–250 flies for each fly line were used. Statistical data are shown in Table 4.
Table 4.
Mean age of flies overexpressing dPrx4
| Overexpression level and genotype | Age (d) | Percentage |
|
|---|---|---|---|
| Vs. transgene/+ | Vs. driver/+ | ||
| High level | |||
| UAS-dPrx4-A/+ | 57.6; 58.2 | ||
| UAS-dPrx4-B/+ | 54.1; 53.9 | ||
| UAS-dPrx4-C/+ | 64.5; 55.3 | ||
| +/Arm | 45.8; 53.1 | ||
| UAS-dPrx4-A/Arm | 56.8; 54.3 | 98; 93* | 124;* 102 |
| UAS-dPrx4-B/Arm | 57.4; 57.2 | 106; 107 | 125;* 108* |
| UAS-dPrx4-C/Arm | 72.3; 59.7 | 112;* 108* | 158;* 112* |
| Low level | |||
| UAS-dPrx4-A/+ | 50.1; 59.5 | ||
| UAS-dPrx4-B/+ | 70.2; 51.6 | ||
| UAS-dPrx4-C/+ | 56.8; 62.7 | ||
| +/Da | 49.6; 60.4 | ||
| UAS-dPrx4-A/Da | 10.3; 30.5 | 21;* 51* | 21;* 51* |
| UAS-dPrx4-B/Da | 26.4; 21.1 | 38;* 41* | 53;* 35* |
| UAS-dPrx4-C/Da | 40.1; 32,8 | 71;* 52* | 81;* 54* |
Age column lists values obtained for 2 independent cohorts. Percentage columns list overall percentage of the experimentals vs. the transgene/+ and driver/+ controls.
P < 0.05 vs. control flies, determined by log-rank test.
Subcellular distribution of dPrx4
In previous studies using S2 cells, we detected the presence of a recombinant V5-tagged dPrx4 protein, in both cell lysates and the culture medium (12). On the basis of these results, we postulated that dPrx4 is a secretable protein. Others determined that dPrx4 was not extracellular but was associated with the ER, where, on UV stimulation, it was released into the cytosol and promoted cell death by antagonizing IAP1 activity (8). Having since generated anti-dPrx4 antibodies, we confirmed the presence of high levels of extracellular dPrx4 derived from dPrx4-V5 overexpressing cells. However, in the nonoverexpressing control cells, there was little secretion, and most of the dPrx4 remained associated with the cells (Fig. 5A). To see whether the relative abundance of dPrx4 could affect its subcellular distribution, we then assessed the presence of dPrx4 in cell-free hemolymph derived from our various experimental and control flies. Prx4 was almost undetectable in the hemolymph isolated from control flies but was found in significant amounts in the hemolymph from flies overexpressing dPrx4 globally at high levels (Fig. 5B). On the basis of the absence of signals corresponding to either anti-actin (cytoskeleton) or anti-CoxVb (mitochondria) antibodies in our hemolymph samples, we infer that the presence of dPrx4 in extracellular liquids is not because of cell lysis and is genuinely due to secretion. In cellular fractions derived from whole flies, we determined that endogenous dPrx4 localized primarily to the light mitochondrial fraction (LMF), which normally contains some mitochondria and is enriched for ER (Fig. 5B). Thus, we concluded that dPrx4 normally resides in the ER and can be released into other compartments when overexpressed or in response to various insults, such as UV stimulation (8). Comparing the distribution of dPrx4 protein and its effects on fly survival led us to suggest that its accumulation in the extracellular space or cytosol in flies overexpressing this protein may be responsible for detrimental effects on fly survivorship.
Figure 5.
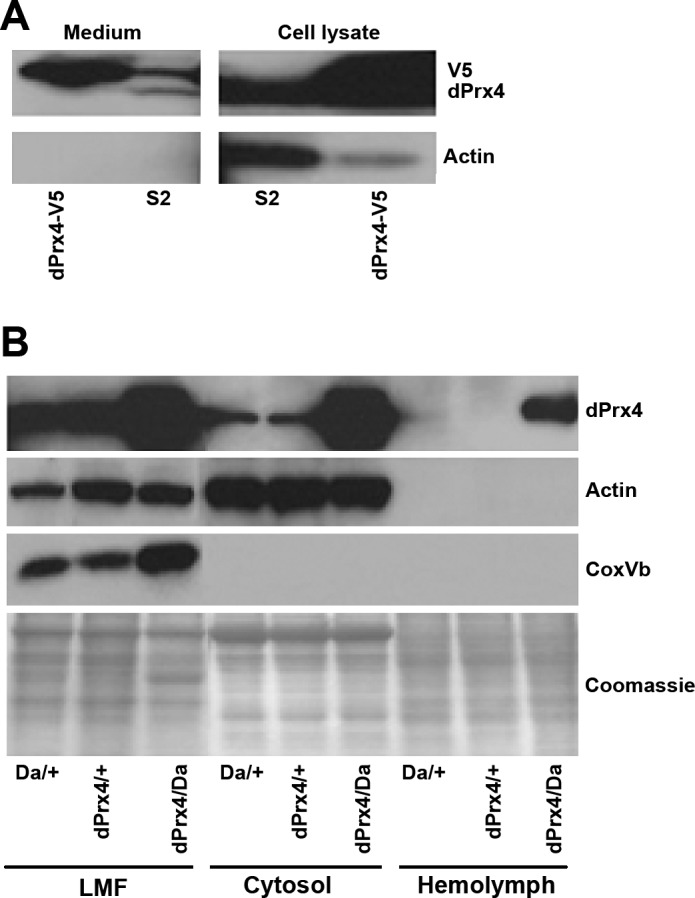
Analysis of localization of Drosophila dPrx4 protein. A) Immunoblot analysis of conditioned medium and cell lysates prepared from S2 cells overexpressing dPrx4-V5 protein (dPrx4-V5) and control cells (S2). Blots were developed with anti-dPrx4 antibodies that recognize both endogenous and V5-tagged dPrx4 protein. To control for an absence of intracellular proteins that potentially could contaminate conditioned medium due to cell lysis, blots were redeveloped with anti-actin antibodies. B) Immunoblot analysis of protein lysates extracted from the light mitochondrial fraction (LMF, contains some mitochondria but is particularly enriched for ER), cytosol, and hemolymph prepared from driver and transgene controls (Da/+, dPrx4/+) and flies overexpressing dPrx4 (dPrx4/Da). Signals were obtained with anti-dPrx4, anti-actin (cytoskeletal) and anti-Cox-Vb (mitochondrial protein) to control for purity of cytosolic fraction and hemolymph. Staining with Coomassie (bottom panel) was performed to control for loading.
dPrx4 protects from oxidative stress but causes a shift in proinflammatory response and tissue-specific apoptosis
Because the endogenous dPrx4 is mostly localized to the ER under normal conditions, we examined markers of oxidative stress in this compartment. Analysis of samples prepared from flies underexpressing and overexpressing dPrx4 revealed that markers of oxidative stress, such as HNE and H2O2 levels in the ER, were clearly elevated in the dprx4 mutant (Fig. 6A). In contrast, overexpression of dPrx4 provided protection from lipid peroxidation, as was determined using anti-HNE antibodies, but did not reduce H2O2 levels (Fig. 6B). The apparent lack of effect on H2O2 steady-state levels in the overexpressor suggests that endogenous dPrx4 activity is relatively effective at controlling oxidative stress, unlike in the mutant, where dPrx4 levels fell below the threshold necessary to withstand a prooxidizing shift.
Figure 6.
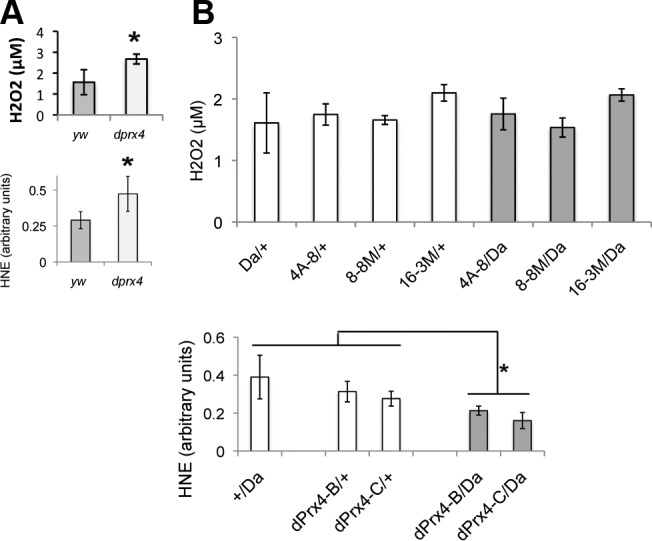
Levels of H2O2 and HNE in subcellular fractions enriched for ER in dprx4 mutant (A) and flies overexpressing dPrx4 (B). LMF organelle fraction, enriched for ER, was pelleted at 17,000 g and analyzed for H2O2 levels by Amplex Red assay. H2O2 concentrations were normalized to the total protein levels and plotted on the y axis. Aliquots of the same lysates were also resolved by PAGE, followed by immunoblot analysis with anti-HNE antibodies. Densitometry signals obtained with anti-HNE antibodies were normalized against densitometry signals obtained with Coomassie staining and plotted on the y axis as arbitrary units of signal intensity. All groups of flies were 9–10 d old. Differences in H2O2 and HNE levels between the dprx4 mutant and yw control were found to be statistically significant (P=0.034 and P=0.046, respectively), as indicated by an asterisk. Also statistically significant were the differences in HNE levels between a driver control and flies overexpressing dPrx4 (P=0.049 and P=0.047), as well as transgene controls and flies overexpressing dPrx4 (P=0.022 and P=0.013). No statistically significant differences in H2O2 levels were observed between controls and experimental flies. Results are means of 3 independent experiments. *P < 0.05.
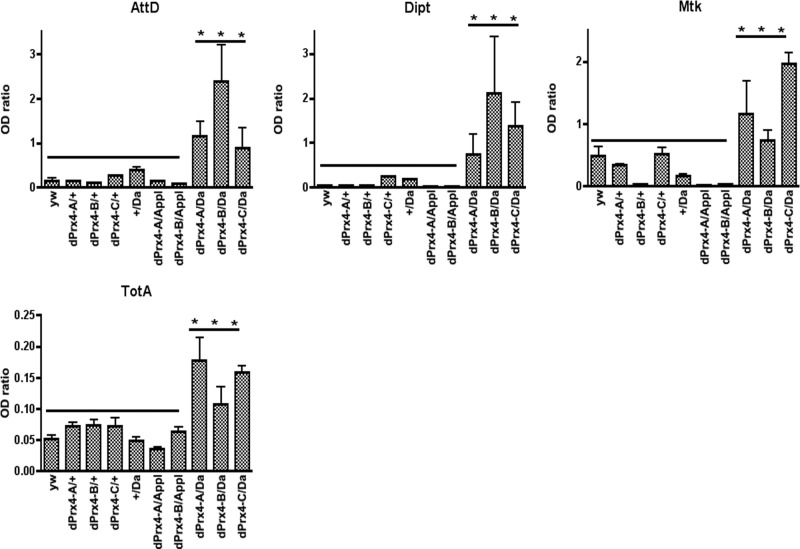
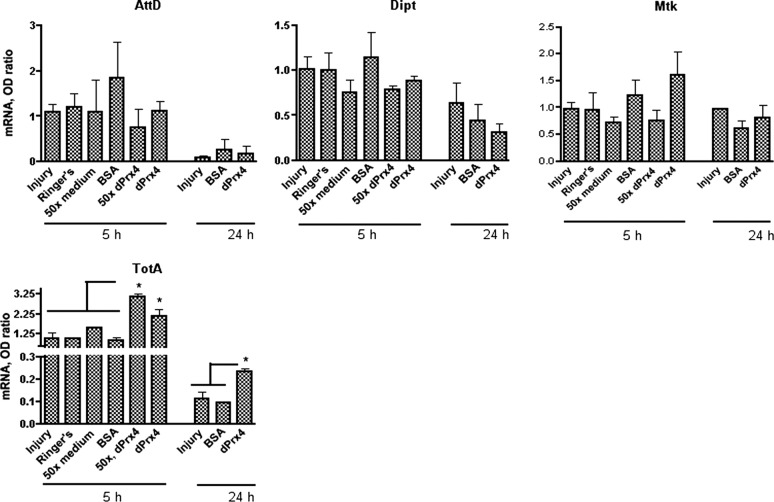
The misexpression of dPrx4 in the cytosol or hemolymph led us to hypothesize that these forms may contribute to the deleterious effects on survivorship observed in the dPrx4 overexpressors. This prompted an assessment of both stress response (JNK and JAK/STAT) and immune response (Toll and Imd) pathways in the dPrx4 overexpressor flies to detect any disruption in homeostasis as revealed by the expression of “readout” genes that are specific for each pathway. As observed in Fig. 7, flies overexpressing dPrx4 at high levels exhibited a severalfold elevation in the expression of AMPs, normally triggered by the Imd and Toll immune pathways, AttD, Dipt, and Mtk (22, 23), while no effects on the AMP expression was observed in flies overexpresssing dPrx4 at low levels or in a tissue-specific manner (Fig. 7 and not shown). The expression of a downstream target of the JAK/STAT pathway, the cytokine-like protein TotA (24), was also induced in flies overexpressing high levels of dPrx4 (Fig. 7). No changes were found in the levels of Puckered, a readout for the JNK pathway (ref. 25 and data not shown).
Figure 7.
Effects of dPrx4 overexpression on activation of the immune and stress response genes. All flies were 6–7 d old. Total RNA was isolated from yw, driver (+/Da), and transgene (dPrx4-A, dPrx4-B, or dPrx4-C) controls and flies overexpressing dPrx4 with global Da-GAL4 driver (dPrx4-A, dPrx4-B, or dPrx4-C/Da) or tissue-specific Appl-GAL4 driver (dPrx4-A, dPrx4-B, or dPrx4-C/Appl). RT-PCR was performed with primers specific for AttD, Dipt, and Mtk genes that are activated via the Imd (AttD and Dipt) and the Toll (Mtk) immune pathways, as well as with primers specific for the TotA gene, which is activated via the JAK/STAT pathway. Signals obtained for each gene were standardized against that obtained for the housekeeping gene rp49. Differences in the levels of AttD, Dipt, Mtk, and TotA in samples isolated from flies overexpressing dPrx4 at elevated levels with Da-GAL4 driver were statistically significant, as indicated by asterisks. Results are means ± sd of 4 independent experiments. *P < 0.05.
To investigate further the role of dPrx4 in apoptosis in adult flies, we determined apoptotic patterns using TUNEL analysis. No significant differences were observed in the incidence of apoptosis between dprx4 mutant and control flies, but a dramatic increase in the number of cells undergoing DNA degradation in fat body tissue and to lesser extent in thoracic muscles was noted in flies overexpressing dPrx4 with global high-level driver Da-GAL4 (Fig. 8;).
Figure 8.
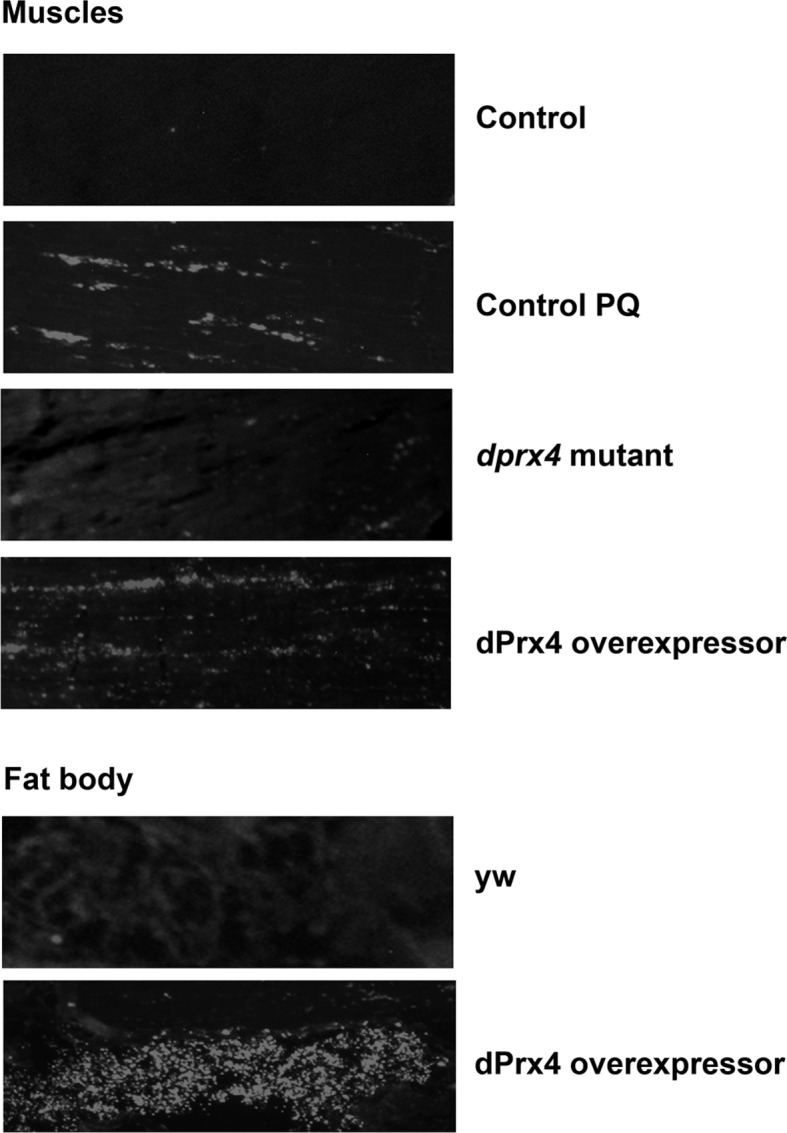
TUNEL analysis of cell death in yw control and flies underexpressing or overexpressing dPrx4. Representative images show DNA fragmentation in cryosections made from 10-d-old control flies (yw strain), dprx4 mutant (dprx4 mutant), and high-level overexpressor UAS-dPrx4/Da-GAL4, designated as dPrx4. Control flies were reared under normal conditions or fed 10 mM paraquat for 6 h (control PQ). Shown are images of the abdomen and thoracic regions, where differences in DNA fragmentation between dprx4 mutant, overexpressor, and control were detected.
Extracellular form of dPrx4 activates the JAK/STAT pathway
To determine whether the observed effects are due to the presence of dPrx4 in the hemolymph, purified dPrx4 protein was injected directly into the body cavity. Flies injected with dPrx4 were characterized by induction of totA gene expression at 5 and 24 h postinjection (Fig. 9). We did not find significant differences in AMP levels in the injected flies (Fig. 9), suggesting that the extracellular form of dPrx4 is not involved in activation of the immune pathways.
Figure 9.
Effects of recombinant dPrx4 protein injected into flies on activation of the immune and stress response genes. One-week-old yw flies were injured with a sterile needle (injury) or injected with 100 nl Ringer's solution, 50× concentrated conditioned medium from S2 cell culture (50× medium), or conditioned medium from S2 cells overexpressing dPrx4-V5 protein (50×, dPrx4), Ringer's solution with 50 μg/ml BSA (BSA), or 50 μg/ml purified dPrx4-V5 protein in Ringer's solution (dPrx4). Flies were incubated for 5 and 24 h, followed by RNA isolation, and qRT-PCR was performed. RT-PCR was performed with primers specific for AMP genes, AttD, Dipt, and Mtk, readouts of the Imd and the Toll immune pathways, as well as with primers specific for the TotA gene, which is primarily activated via the JAK/STAT pathway. Differences in the mRNA levels of TotA between samples isolated from flies injected with solutions containing dPrx4 recombinant protein [conditioned medium containing dPrx4 (50×, dPrx4) and purified dPrx4 in Ringer's solution (dPrx4)] and samples isolated from injured flies and flies injected with various solutions that did not contain dPrx4 protein [Ringer's, conditioned medium from regular cell culture (50× medium), BSA] were statistically significant, as indicated by asterisks. Results are means ± sd of 3 or 4 independent experiments. *P < 0.05.
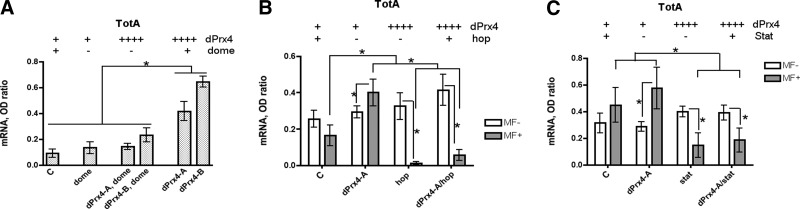
The up-regulation of totA gene in response to dPrx4 overexpression or injection of this protein in hemolymph suggested that the extracellular form of dPrx4 is involved in activating JAK/STAT signaling. The JAK/STAT signaling transmits the signals from the extracellular milieu that are recognized by the receptor Dome, leading to activation of the kinase Hop and the transcription factor Stat92E, which then translocates to the nucleus and triggers the expression of the cytokine-like protein Tot. Epistatic analysis using RNAi mutants for the genes representing the JAK/STAT pathway, dome, hop, and Stat92E showed that the activity of these genes is required for activation of TotA, elicited by overexpression of dPrx4 protein (Fig. 10). As demonstrated in Fig. 10, up-regulation of TotA, normally observed in flies overexpressing dPrx4, was abolished in flies underexpressing Dome, Hop, or Stat92E. The data suggest that dPrx4 acts upstream of the transmembrane receptor Dome to activate JAK/STAT signaling, consistent with a role of the extracellular form of dPrx4 in triggering JAK/STAT-mediated signaling (Fig. 9).
Figure 10.
Effects of dPrx4 on induction of the stress response gene TotA in the JAK/STAT pathway mutants. All flies were 1 wk old. Two different RNAi lines (see Table 1) were used for each gene, giving comparable results; hence, average values are presented. Real-time RT-PCR analysis was performed with primers specific for the totA gene, and signals were normalized against those obtained for rp49. A) Flies underexpressing receptor domeless and overexpressing dPrx4 (UAS-dPrx4-A or B/RNAi-dome; Da-GAL4/+) are indicated here as dPrx4-A, dome and dPrx4-B, dome. Also indicated are values for fly lines underexpressing domeless with normal dPrx4 levels (dome), fly lines overexpressing dPrx4 with normal domeless levels (dPrx4-A and dPrx4-B), and a control (C) containing the Da-Gal4 driver only. Flies were reared at 18°C during development, and upon eclosion, adults were collected and maintained at 25°C. B, C) Flies underexpressing either hopscotch or Stat92E in the presence or absence of Prx4 overexpression were generated using the pSwitch system. Over or underexpression of genes was achieved by feeding 5- to 7-d-old flies food containing mifepristone dissolved in ethanol (MF+) or food containing only ethanol as a control (MF–). After 3 d of feeding, RNA was isolated, and the totA gene expression was analyzed by qRT-PCR. B) Flies overexpressing dPrx4 and underexpressing hopscotch (UAS-dPrx4-A/RNAi-hop; Act-Switch/+) are designated here as dPrx4-A/hop. Also indicated are values for flies overexpressing dPrx4 with normal hopscotch levels (dPrx4-A), flies underexpressing hopscotch with normal dPrx4 levels (hop) and a control (C) containing the Act-Switch driver. C) Flies overexpressing dPrx4 and underexpressing Stat92E (UAS-dPrx4-A/RNAi-Stat; Tub-Switch/+) are designated here as dPrx4-A/stat. Values are also provided for flies underexpressing Stat92E with normal levels of Prx4 (stat), flies overexpressing dPrx4 with normal levels of Stat92E (dPrx4-A), and control flies containing the Tub-Switch driver by itself (C). Symbols above graphs represent the relative level of each gene: –for underexpression; ++++ for overexpression; + for wild type levels. All analyses were done in triplicate with 2 independent cohorts of flies. Results are means ± sd. *P < 0.05.
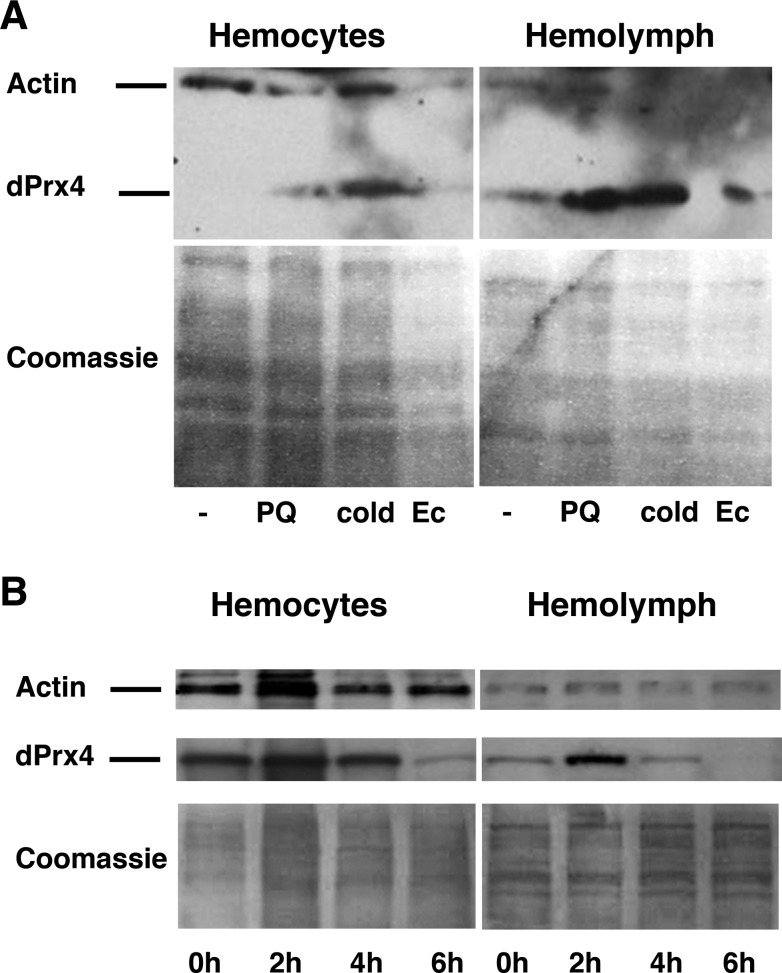
It is known that the cytokine-like protein Tot is up-regulated in response to a variety of stressors (13). Therefore, we investigated whether other stimuli might cause the release of dPrx4 into the hemolymph and, indeed, found a prominent increase in the dPrx levels in the hemolymph of flies subjected to septic injury, cold, or paraquat treatment (Fig. 11A). The increase of dPrx4 in hemolymph was not due to up-regulation of dPrx4 expression, as the total dPrx4 levels in the whole body lysates did not change (Supplemental Fig. S4A–C). However, we did observe the induction of dPrx4 in hemocytes (Fig. 11A). Furthermore, the induction of dPrx4 in hemocytes and release of this protein into hemolymph dramatically increased peaking at 2 h postinfection, followed by gradual reduction of the dPrx4 levels by 4 and 6 h (Fig. 11B). No cellular proteins (actin) were found in hemolymph, suggesting that the presence of dPrx4 in samples taken from stressed flies is indeed due to release of dPrx4 into extracellular space. Furthermore, RT-PCR analysis of totA gene expression showed that dPrx4 activity is required for activation of JAK/STAT signaling in response to either septic injury or paraquat, as strong activation of TotA, observed in the control, was abrogated in dPrx4 RNAi mutants (Fig. 12). No up-regulation of TotA or release of dPrx4 into hemolymph was observed in response to agents that cause ER stress, such as tunicamycin (Supplemental Fig. S4D, E); no release of dPrx4 protein into hemolymph was observed in response to heat (Supplemental Fig. S4D).
Figure 11.
dPrx4 is released into fly hemolymph under stress conditions. A) Ten-day-old yw flies were exposed to cold, paraquat (PQ) or septically injured (Ec) as specified in Materials and Methods. Untreated (−) animals served as a control. Protein samples were prepared from hemolymph and hemocytes. B) Ten-day-old yw flies were septically injured with E. coli, followed by hemolymph and hemocyte collection at different time intervals. Proteins were resolved by PAGE. Anti-actin antibodies were used to control for purity of the hemolymph fraction. Staining with Coomassie and/or anti-actin antibodies (for hemocytes) was used to control for loading.
Figure 12.
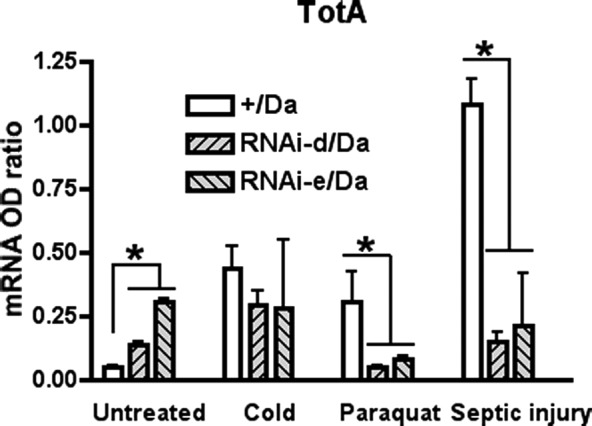
Activation of stress response gene TotA is abolished in flies underexpressing dPrx4. Ten-day-old driver control (+/Da) and RNAi mutants underexpressing dPrx4 (RNAi-dPrx4-d/Da or RNAi-dPrx4-e/Da) were exposed to paraquat, cold, or septically injured with E. coli suspension, as specified in Materials and Methods. Untreated animals served as a control. Total RNA was isolated, followed by qRT-PCR analysis of totA. Small but significant differences in the basal levels of TotA observed in untreated mutants and control can be accounted, perhaps, for partial activity of the JAK/STAT signaling due to mild stress caused by the reduction of dPrx4 activity in RNAi mutants. *P < 0.05.
DISCUSSION
The major finding of the study is that Prx4 may confer both negative and positive effects on fly survival, and these effects depend not only on the levels of dPrx4 expression but also on its spatial distribution. Similar to the situation in mammals (8, 9, 26–29), reports on the subcellular localization of Prx4 in Drosophila are conflicting, described either as primarily secretable (12), or as primarily associated with the ER (8). On the basis of results obtained in the present study with S2 cells and flies, these disparities are likely due to the differences in the expression levels of dPrx4 and the redistribution of this protein under different stress conditions. Thus, in mutant and wild-type flies, dPrx4 expression is largely restricted to the ER under normal conditions, and there is no detectable expression in the extracellular milieu. On the other hand, overexpression of dPrx4 in flies or in cell culture leads to a significant level of secretion (Fig. 5). It would appear then that Prx4 in Drosophila is normally present in the ER, from where it can be released to the cytosol on stresses, such as UV exposure (8) or when overexpressed, or secreted into the extracellular milieu when expressed at higher levels (Fig. 5) or in response to stimuli, such as infection or paraquat toxicity (Fig. 11). Infectious stimuli can also cause other types of subcellular redistribution of dPrx4, such as its occurrence in phagosomes, vesicles within Drosophila hemocyte-like phagocytic S2 cells from which immune processes are initiated (30).
Analysis of subcellular fractions enriched for ER revealed a clear antioxidant function for dPrx4 in this organelle. Thus, lipid peroxidation was reduced in samples prepared from dPrx4 overexpressing flies, while levels of both H2O2 and other peroxidation products were elevated in the dprx4 mutant flies (Fig. 6). Moreover, the dprx4 mutant flies exhibited reduced survivorship on exposure to oxidants relative to controls (Fig. 3). Similarly, knockdown of Prx4 in human HT1080 cells resulted in reduced cell viability when challenged with H2O2, although in this case, no effects on H2O2 turnover or ER redox were noted (9). The peroxidatic activity of Prx4 in ER was shown to be important for removing H2O2 produced by Ero1 during disulfide formation (6). Antioxidant Prx4 function was also demonstrated in the whole organism, where the Prx4-knockout mice did exhibit a clear increase in oxidative stress markers (8-oxo-dG and HNE), which is accompanied by testicular cell abnormalities (11). Otherwise, like the dprx4 mutant flies, the adult phenotype of the mouse knockout appears to be quite mild, at least under nonstress conditions. While it is presumed that Prx4 plays an important role in dampening H2O2 levels in the ER, its absence does not have detrimental effects in most tissues under normoxic conditions. Indeed in dprx4 mutant flies 10–20% Prx4 activity (Fig. 2) appears to be sufficient to support normal longevity in the absence of stress (Fig. 3E, F).
In agreement with antioxidant capacity of dPrx4 to dampen oxidative stress in ER, low-level overexpression of this protein had slight life span extension effects under normal conditions (Fig. 4A). In a similar vein, it has been observed in mice that Prx4 overexpression in pancreatic islets suppressed oxidative stress and alleviated symptoms of diabetes (10). In sharp contrast, when dPrx4 was overexpressed globally at high levels in flies, life span was significantly reduced (Fig. 4B). We reasoned that such negative effects could be due to the overreduced status of the ER, affecting protein-folding capacity. Although Prx4 has recently been shown to compensate for ER oxidoreductin 1 (Ero1) function in reoxidation of PDI during oxidative protein folding formation in a recycling reaction coupled with H2O2 (5, 31), it is also considered to play a key role in the maintenance of H2O2 levels. Thus, while low-level overexpression of Prx4 may have slight beneficial effects, high levels may compromise oxidative protein folding by virtue of its potential effect on H2O2 fluxes. Alternatively, the observed negative effects on fly life span could arise due to the deleterious action of dPrx4 when released to other compartments (cytosolic or extracellular), affecting cell viability or other critical functions.
As described by Tenev et al. (8), the presence of dPrx4 in the cytosol appears to antagonize the activity of the inhibitor of apoptosis protein (dIAP1) and thus liberate proapoptotic caspases, resulting in cell death. In our studies involving overexpression of dPrx4 in S2 cells, the viability of S2 cells overexpressing dPrx4 was not adversely affected, nor were there any observable proapoptotic changes under either normal or oxidative stress conditions (ref. 12 and unpublished results). Moreover, S2 cells overexpressing dPrx4 were more resistant to H2O2 or paraquat (ref. 12 and unpublished results). In contrast, global overexpression of dPrx4 at high levels in flies led to a significant increase in the incidence of apoptosis in muscle and particularly in fat body tissues (Fig. 8). While the increase in the number of apoptotic cells in thoracic muscles was documented previously in old flies and flies exposed to oxidants (32) and in flies underexpressing mitochondrial peroxiredoxins (19), the induction of apoptosis in fat tissue seems to be a unique characteristic of flies overexpressing Prx4. Proapoptotic changes in flies expressing high levels of dPrx4 correlated with high amounts of this protein in the cytosol (Fig. 5B), which is in agreement with the data obtained by Tenev et al. (8), who showed a similar proapoptotic effect on release of dPrx4 from ER in response to UV or ER stress-inducing agents. ER stress can also be caused by protein overload or unfolding (33). It is plausible then that the large amounts of dPrx4 in high-level overexpressors could elicit ER stress, and the consequent release of this protein into cytosol would promote apoptosis and accelerated death. This apparently did not happen in flies underexpressing dPrx4 or overexpressing this protein at low levels. ER stress is also known to activate NF-κB pathways (34). In Drosophila, ER stress causes induction of immunity-related genes that are regulated through activation of the NF-κB transcription factors Dif and Relish (35). The observed increase in the levels of Dif- and Relish-dependent immunity response genes, AMPs, in flies overexpressing dPrx4 (Fig. 9) supports the notion that ER stress, caused by dPrx4 overload and perhaps unfolding, underlies the detrimental effects on fly survivorship. It also indicates that constitutive overproduction of AMPs is harmful for fly physiology. In Drosophila, high-level constitutive production of these immunity effectors was observed in old flies (36, 37), as well as in flies with overactivated NF-κB pathway, where it resulted in premature aging (38). Surprisingly, we did not obtain similar effects with flies fed tunicamycin, an ER stress-causing agent, although issues of bioavailability will need to be addressed before any conclusions may be drawn.
ER stress and its downstream sequelae do not appear sufficient to explain the effects of dPrx4 high-level overexpression. For instance, apoptosis and cell death were not observed in S2 cells overexpressing dPrx4. One plausible explanation is that the threshold protein levels have not been reached in S2 dPrx4-overexpressing cells. On the other hand, the role of extracellular dPrx4 form in the observed effects cannot be ruled out. Because the detrimental effects of dPrx4 in S2 cells overexpressing this protein were not observed, then the simplest interpretation is that it is due to the dilution of secreted dPrx4 by large volumes of cell culture medium.
On the basis of cell culture studies of the human homologue, Prx4 (TRANK) was identified as a potential proinflammatory cytokine, activating NF-κB and JNK, and this activation occurred when Prx4 was added to cell culture medium and interacted with an as yet to be identified receptor (2). Using the Drosophila targets of NF-κB pathways (Toll and Imd) and stress response pathways (JNK and JAK/STAT) as readouts, we determined that dPrx4 injected into fly hemolymph directs the induction of the cytokine-like protein TotA, but not the immune effectors, AMPs, or Puckered, a target of the JNK pathway (Fig. 8 and not shown). Tot is induced in response to a variety of stressors, including heat, mechanical stress, ROS, and bacteria (13, 24). If activation of stress response pathways by extracellular dPrx4 occurs, as demonstrated in the present study, dPrx4 secretion into extracellular space should be physiologically relevant. Indeed, we found that at least some stresses cause secretion of dPrx4 into hemolymph, where it is required for JAK/STAT-mediated response to stress and induction of TotA (Fig. 12). TotA protein is produced in the fat body, and its expression there is regulated by the transcription factors Stat92E and Relish (39). Presumably, that is why we did not observe activation of TotA by S2 cells, which have a macrophage-like phenotype (data not shown). Thus, the data obtained in this study indicate that extracellular dPrx4 elicits up-regulation of the stress response protein TotA via activation of the JAK/STAT pathway. dPrx4 mutants that failed to mount such a response (Fig. 12) died rapidly under oxidative stress (Fig. 3C, D), although it is not yet possible to rule out a deficiency in antioxidant function (Fig. 6). Epistatic analysis using mutants for the JAK/STAT pathway placed dPrx4 upstream of the transmembrane receptor Domeless. It is still unclear whether dPrx4 directly interacts with Domeless or functions via interactions with other factors in the fly hemolymph, such as the hemocyte-produced cytokine-like protein Unpaired (Upd3), which has been previously shown to be required for JAK/STAT-mediated activation of Turandot A in response to infection (39). It is also not yet determined whether these putative interactions are dependent on peroxidatic, chaperone, or other yet unknown functions of dPrx4. It is also unclear whether overproduction of Tot observed in the dPrx4 overexpressor is a marker of stress response and/or has additional effects on fly physiology. Initial attempts to address the TotA effects on fly physiology by overexpressing this gene have not been conclusive (13).
It is notable that fat body-specific apoptosis is observed in flies with high levels of dPrx4. Indeed, since the fat body is a major producer of AMPs and Tot, it is tempting to postulate that dPrx4 acts as a core regulator of responses to stress in this tissue. The data obtained in this study together with working hypotheses are summarized in the following scheme (Fig. 13). In summary, the net effect of dPrx4 on the fly entails a combination of differential effects that are specified in part through expression levels and localization.
Figure 13.
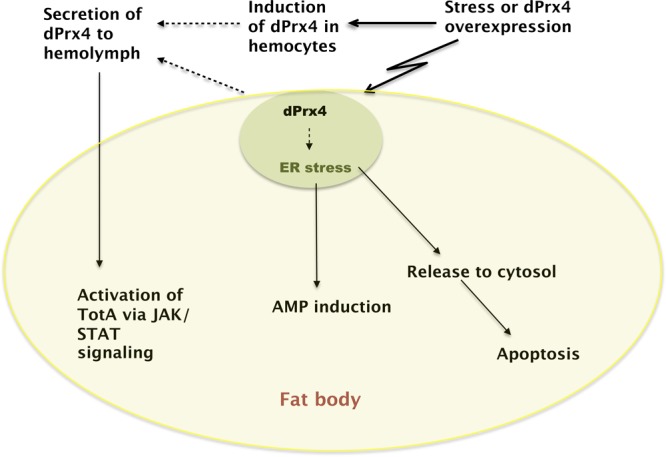
Proposed model of dPrx4 functions. Normally, dPrx4 resides in ER, where it is presumed to serve an important antioxidant function. Various stresses can elicit the release of dPrx4 to cytosol, causing induction of apoptosis. Overexpression of dPrx4 at high levels causes activation of NF-κB-mediated responses, resulting in production of AMPs, primary immune effectors. Various stresses can also elicit induction of dPrx4 in hemocytes and its secretion into extracellular space, where dPrx4 interacts with a yet to be identified factor, triggering activation of the JAK/STAT signaling pathways and inducing TotA, its downstream cytokine-like effector. In the absence of infection, overexpressed dPrx4 protein elicits responses that are similar to those observed after exposure to microbes, plausibly mediated through ER stress engendered by dPrx4 overload. All downstream events, specifically NF-κB and JAK/STAT-mediated responses, as well as tissue-specific apoptosis, are integrated in the fat body tissue.
Acknowledgments
This work was supported by the grants RO1 AG15122 (W.C.O.) and R01 AG032342 (S.N.R.) from the National Institute on Aging, U.S. National Institutes of Health.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- Act
- actin
- AMP
- antimicrobial peptide
- Arm
- armadillo
- Da
- daughterless
- ER
- endoplasmic reticulum
- HNE
- 4-hydroxynonenal
- LMF
- light mitochondrial fraction
- Prx4
- peroxiredoxin 4
- rp49
- ribosomal protein 49
- TotA
- Turandot A
- Tub
- tubulin
REFERENCES
- 1. Jin D. Y., Chae H. Z., Rhee S. G., Jeang K. T. (1997) Regulatory role for a novel human thioredoxin peroxidase in NF-κB activation. J. Biol. Chem. 272, 30952–30961 [DOI] [PubMed] [Google Scholar]
- 2. Haridas V., Ni J., Meager A., Su J., Yu G. L., Zhai Y., Kyaw H., Akama K. T., Hu J., Van Eldik L. J., Aggarwal B. B. (1998) TRANK, a novel cytokine that activates NF-κB and c-Jun N-terminal kinase. J. Immunol. 161, 1–6 [PubMed] [Google Scholar]
- 3. Matsumoto A., Okado A., Fujii T., Fujii J., Egashira M., Niikawa N., Taniguchi N. (1999) Cloning of the peroxiredoxin gene family in rats and characterization of the fourth member. FEBS Lett. 443, 246–250 [DOI] [PubMed] [Google Scholar]
- 4. Wong C. M., Chun A. C., Kok K. H., Zhou Y., Fung P. C., Kung H. F., Jeang K. T., Jin D. Y. (2000) Characterization of human and mouse peroxiredoxin IV: evidence for inhibition by Prx-IV of epidermal growth factor- and p53-induced reactive oxygen species. Antioxidants Redox Signal. 2, 507–518 [DOI] [PubMed] [Google Scholar]
- 5. Tavender T. J., Springate J. J., Bulleid N. J. (2010) Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 29, 4185–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tavender T. J., Bulleid N. J. (2010) Peroxiredoxin IV protects cells from oxidative stress by removing H2O2 produced during disulphide formation. J. Cell Sci. 123, 2672–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giguere P., Turcotte M. E., Hamelin E., Parent A., Brisson J., Laroche G., Labrecque P., Dupuis G., Parent J. L. (2007) Peroxiredoxin-4 interacts with and regulates the thromboxane A2 receptor. FEBS Lett. 581, 3863–3868 [DOI] [PubMed] [Google Scholar]
- 8. Tenev T., Zachariou A., Wilson R., Paul A., Meier P. (2002) Jafrac2 is an IAP antagonist that promotes cell death by liberating Dronc from DIAP1. EMBO J. 21, 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tavender T. J., Sheppard A. M., Bulleid N. J. (2008) Peroxiredoxin IV is an endoplasmic reticulum-localized enzyme forming oligomeric complexes in human cells. Biochem. J. 411, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding Y., Yamada S., Wang K. Y., Shimajiri S., Guo X., Tanimoto A., Murata Y., Kitajima S., Watanabe T., Izumi H., Kohno K., Sasaguri Y. (2010) Overexpression of peroxiredoxin 4 protects against high-dose streptozotocin-induced diabetes by suppressing oxidative stress and cytokines in transgenic mice. Antioxid. Redox Signal. 13, 1477–1490 [DOI] [PubMed] [Google Scholar]
- 11. Iuchi Y., Okada F., Tsunoda S., Kibe N., Shirasawa N., Ikawa M., Okabe M., Ikeda Y., Fujii J. (2009) Peroxiredoxin 4 knockout results in elevated spermatogenic cell death via oxidative stress. Biochem. J. 419, 149–158 [DOI] [PubMed] [Google Scholar]
- 12. Radyuk S. N., Klichko V. I., Spinola B., Sohal R. S., Orr W. C. (2001) The peroxiredoxin gene family in Drosophila melanogaster. Free Radic. Biol. Med. 31, 1090–1100 [DOI] [PubMed] [Google Scholar]
- 13. Ekengren S., Tryselius Y., Dushay M. S., Liu G., Steiner H., Hultmark D. (2001) A humoral stress response in Drosophila. Curr. Biol. 11, 1479. [DOI] [PubMed] [Google Scholar]
- 14. Radyuk S. N., Michalak K., Klichko V. I., Benes J., Orr W. C. (2010) Peroxiredoxin 5 modulates immune response in Drosophila. Biochim. Biophys. Acta 1800, 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Orr W. C., Radyuk S. N., Prabhudesai L., Toroser D., Benes J. J., Luchak J. M., Mockett R. J., Rebrin I., Hubbard J. G., Sohal R. S. (2005) Overexpression of glutamate-cysteine ligase extends life span in Drosophila melanogaster. J. Biol. Chem. 280, 37331–37338 [DOI] [PubMed] [Google Scholar]
- 16. Michalak K., Orr W. C., Radyuk S. N. (2008) Drosophila peroxiredoxin 5 is the second gene in a dicistronic operon. Biochem. Biophys. Res. Commun. 368, 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shikama N., Brack C. (1996) Changes in the expression of genes involved in protein synthesis during Drosophila aging. Gerontology 42, 123–136 [DOI] [PubMed] [Google Scholar]
- 18. Berndt A., Kottke T., Breitkreuz H., Dvorsky R., Hennig S., Alexander M., Wolf E. (2007) A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J. Biol. Chem. 282, 13011–13021 [DOI] [PubMed] [Google Scholar]
- 19. Radyuk S. N., Rebrin I., Klichko V. I., Sohal B. H., Michalak K., Benes J., Sohal R. S., Orr W. C. (2010) Mitochondrial peroxiredoxins are critical for the maintenance of redox state and the survival of adult Drosophila. Free Radic. Biol. Med. 49, 1892–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toroser D., Orr W. C., Sohal R. S. (2007) Carbonylation of mitochondrial proteins in Drosophila melanogaster during aging. Biochem. Biophys. Res. Commun. 363, 418–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zoltowski B. D., Vaidya A. T., Top D., Widom J., Young M. W., Crane B. R. (2011) Structure of full-length Drosophila cryptochrome. Nature 480, 396–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lemaitre B., Nicolas E., Michaut L., Reichhart J. M., Hoffmann J. A. (1996) The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 [DOI] [PubMed] [Google Scholar]
- 23. Lemaitre B., Reichhart J. M., Hoffmann J. A. (1997) Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. U. S. A. 94, 14614–14619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ekengren S., Hultmark D. (2001) A family of Turandot-related genes in the humoral stress response of Drosophila. Biochem. Biophys. Res. Commun. 284, 998–1003 [DOI] [PubMed] [Google Scholar]
- 25. Martin-Blanco E., Gampel A., Ring J., Virdee K., Kirov N., Tolkovsky A. M., Martinez-Arias A. (1998) puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12, 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuki S., Sasagawa I., Iuchi Y., Fujii J. (2002) Impaired expression of peroxiredoxin 4 in damaged testes by artificial cryptorchidism. Redox. Rep. 7, 276–278 [DOI] [PubMed] [Google Scholar]
- 27. Sasagawa I., Matsuki S., Suzuki Y., Iuchi Y., Tohya K., Kimura M., Nakada T., Fujii J. (2001) Possible involvement of the membrane-bound form of peroxiredoxin 4 in acrosome formation during spermiogenesis of rats. Eur. J. Biochem. 268, 3053–3061 [DOI] [PubMed] [Google Scholar]
- 28. Okado-Matsumoto A., Matsumoto A., Fujii J., Taniguchi N. (2000) Peroxiredoxin IV is a secretable protein with heparin-binding properties under reduced conditions. J. Biochem. 127, 493–501 [DOI] [PubMed] [Google Scholar]
- 29. Palande K., Roovers O., Gits J., Verwijmeren C., Iuchi Y., Fujii J., Neel B. G., Karisch R., Tavernier J., Touw I. P. (2011) Peroxiredoxin-controlled G-CSF signalling at the endoplasmic reticulum-early endosome interface. J. Cell Sci. 124, 3695–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stuart L. M., Boulais J., Charriere G. M., Hennessy E. J., Brunet S., Jutras I., Goyette G., Rondeau C., Letarte S., Huang H., Ye P., Morales F., Kocks C., Bader J. S., Desjardins M., Ezekowitz R. A. (2007) A systems biology analysis of the Drosophila phagosome. Nature 445, 95–101 [DOI] [PubMed] [Google Scholar]
- 31. Zito E., Melo E. P., Yang Y., Wahlander A., Neubert T. A., Ron D. (2010) Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol. Cell 40, 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng J., Edelman S. W., Tharmarajah G., Walker D. W., Pletcher S. D., Seroude L. (2005) Differential patterns of apoptosis in response to aging in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 102, 12083–12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuang E., Wan Q., Li X., Xu H., Liu Q., Qi Y. (2005) ER Ca2+ depletion triggers apoptotic signals for endoplasmic reticulum (ER) overload response induced by overexpressed reticulon 3 (RTN3/HAP). J. Cell. Physiol. 204, 549–559 [DOI] [PubMed] [Google Scholar]
- 34. Pahl H. L., Baeuerle P. A. (1997) The ER-overload response: activation of NF-κB. Trends Biochem. Sci. 22, 63–67 [DOI] [PubMed] [Google Scholar]
- 35. Boltz K. A., Carney G. E. (2008) Loss of p24 function in Drosophila melanogaster causes a stress response and increased levels of NF-κB-regulated gene products. BMC Genomics 9, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Landis G. N., Abdueva D., Skvortsov D., Yang J., Rabin B. E., Carrick J., Tavare S., Tower J. (2004) Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 101, 7663–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sowell R. A., Hersberger K. E., Kaufman T. C., Clemmer D. E. (2007) Examining the proteome of Drosophila across organism lifespan. J. Proteome Res. 6, 3637–3647 [DOI] [PubMed] [Google Scholar]
- 38. Libert S., Chao Y., Chu X., Pletcher S. D. (2006) Trade-offs between longevity and pathogen resistance in Drosophila melanogaster are mediated by NF-κB signaling. Aging Cell 5, 533–543 [DOI] [PubMed] [Google Scholar]
- 39. Agaisse H., Petersen U. M., Boutros M., Mathey-Prevot B., Perrimon N. (2003) Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 5, 441–450 [DOI] [PubMed] [Google Scholar]