The high-resolution crystal structure of human Dim2/TXNL4B indicated that a region previously reported to be responsible for its interaction with Prp6 is unlikely to facilitate this interaction.
Keywords: Dim2, TXNL4B, pre-mRNA splicing, human Dim1, U5-15kD
Abstract
TXNL4A (thioredoxin-like 4A) is an essential protein conserved from yeast to humans and is a component of the pre-mRNA splicing machinery. TXNL4B was identified as a TXNL4-family protein that also interacts with Prp6, an integral component of the U4/U6·U5 tri-snRNP complex, and has been shown to function in pre-mRNA splicing. A crystal structure of TXNL4B was determined at 1.33 Å resolution and refined to an R work of 0.13 and an R free of 0.18 with one native dimer in the asymmetric unit. Residues 1–33 of TXNL4B have previously been reported to be responsible for its interaction with Prp6. However, this region extends to the β-sheet core of the thioredoxin-fold structure of TXNL4B. This suggests that the interpretation of the previously reported GST pull-down results without considering the structure and stability of TXNL4B is debatable.
1. Introduction
The removal of introns from pre-messenger RNA is an essential step in the expression of most eukaryotic genes (Toor et al., 2009 ▶). After recognition of the 5′ and 3′ ends of the introns, the actual excision of the introns is catalyzed by the spliceosome, which is one of the largest molecular complexes in the cell. The spliceosome contains four small nuclear ribonucleoprotein (snRNP) particles (U1, U2, U4/U6 and U5) and numerous auxiliary proteins (see, for example, Krämer, 1996 ▶; Zhou et al., 2002 ▶). Besides the seven highly conserved common core proteins (B/B′, D1, D2, D3, E, F and G Sm proteins), snRNPs also contains highly conserved important proteins specific for individual snRNP particles (see, for example, Will & Lührmann, 1997 ▶). Structural information on snRNP proteins is essential in order to understand many aspects of the pre-mRNA splicing mechanisms. To date, much has been learned about the interaction and function of distinct components of the spliceosome by structural investigations. These include structural studies of the complex of the U1A RNA-binding domain and part of the U1 snRNA (Oubridge et al., 1994 ▶; Howe et al., 1994 ▶), structural studies of the U2B′′–U2A′ complex associated with a fragment of U2 snRNA (Price et al., 1998 ▶) and structural studies of the evolutionally highly conserved U5 snRNP-specific protein TXNL4A (also known as U5-15kD and human Dim1; Reuter & Ficner, 1999 ▶; Zhang et al., 1999 ▶, 2003 ▶). However, very little is known about the structures and interactions of other snRNP proteins. While we were studying human Dim1/TXNL4A, we identified a previously undescribed protein Dim2, which is a paralogue of Dim1 (Zhang et al., 1999 ▶); it has now been named TXNL4B by the HUGO Gene Nomenclature Committee. Unlike TXNL4A, TXNL4B could only be identified in human-, mouse- and aspen-expressed sequence tags (ESTs) at the time of its discovery. Previously, we have reported that TXNL4A interacts with multiple proteins that are involved in pre-mRNA splicing as well as alternative splicing (Zhang et al., 2000 ▶). The crystal structure of TXNL4A was reported by Reuter et al. (1999 ▶) and previous studies indicated that a 14-residue C-terminal tail of TXNL4A was important for its function but had little effect on the structure and stability of the protein (Zhang et al., 2003 ▶). A crystal structure of TXNL4B at 2.5 Å resolution has also been reported (Simeoni et al., 2005 ▶). In addition, it has been reported that the first 33 amino acids of TXNL4B are responsible for its interaction with Prp6 (Sun et al., 2004 ▶). To better understand the structural and functional differences between TXNL4A and TXNL4B, we sought to obtain a high-resolution structure of TXNL4B. Here, we report the crystal structure of TXNL4B at 1.33 Å resolution. Regions of TXNL4B that have previously been reported as being responsible for its function and its interaction with Prp6 are also discussed in the light of protein structure and stability.
2. Materials and methods
2.1. Cloning, expression, purification, crystallization, data collection and processing
The cloning, expression, purification and crystallization of TXNL4B have been reported previously (Jin et al., 2005 ▶). Since then, using a crystal obtained under the same conditions, we have collected a high-resolution data set for TXNL4B which diffracted to 1.33 Å. The new data set was collected at low temperature using the same cryoprotectant (Jin et al., 2005 ▶). The data were collected using a MAR 300 CCD on the SER-CAT 22ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory. The distance between the crystal and the detector was 89 mm and 180 frames were collected with 1° oscillation and 1 s exposure. The diffraction data were processed using the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶).
2.2. Structure determination and refinement
Shortly after we reported the crystallization and crystal characterization of TXNL4B, Simeoni et al. (2005 ▶) reported a crystal structure of Dim2/TXNL4B at 2.5 Å resolution (PDB entry 1xbs). Using the 2.5 Å resolution monomeric TXNL4B as a template, a structural solution for the high-resolution data was derived by molecular-replacement calculations using the program Phaser (McCoy et al., 2005 ▶; Storoni et al., 2004 ▶). Structure refinement was carried out with REFMAC5 (Murshudov et al., 2011 ▶) as implemented in CCP4 (Winn et al., 2011 ▶). The refinement was alternated with manual model building and model improvement using Coot (Emsley & Cowtan, 2004 ▶).
The final structure was refined to 1.33 Å resolution and the final model was checked using PROCHECK (Laskowski et al., 1993 ▶) and MolProbity (Chen et al., 2010 ▶). The structure model was also checked using a shake-and-omit protocol by introducing random errors of up to 0.3 Å into the coordinates of the final refined structure using the program PDBSET distributed with CCP4 (Winn et al., 2011 ▶). For each region to be checked, the concerned region of the shaken structure was manually omitted and 20 cycles of restrained refinements were carried out using REFMAC5 (Murshudov et al., 2011 ▶). This was followed by inspecting the F o − F c map together with the final refined structure. The coordinates and structure factors of TXNL4B have been deposited in the Protein Data Bank (Berman et al., 2002 ▶) with accession code 4in0.
Structure-based sequence alignment and corresponding structural superpositions were carried out using the program MUSTANG (Konagurthu et al., 2006 ▶), and root-mean-square deviations (r.m.s.d.s) were calculated using FAST (Zhu & Weng, 2005 ▶). Molecular graphics were prepared using the programs RasMol (Sayle & Milner-White, 1995 ▶), MolScript (Kraulis, 1991 ▶) and Raster3D (Merritt & Murphy, 1994 ▶).
3. Results and discussion
3.1. Structure quality
The best TXNL4B crystals diffracted to 1.33 Å resolution and a complete diffraction data set was collected from one crystal (Table 1 ▶). A structural solution was readily obtained by molecular replacement. The structure of the initial solution contained most of the backbone fitted in the electron density and the refinement was carried out without much difficulty. The final refined structure of TXNL4B adopted a typical thioredoxin fold. It gave R and R free values of 0.13 and 0.18, respectively, for all data to 1.33 Å resolution (Table 1 ▶ and Fig. 1 ▶). The r.m.s.d.s from ideal empirical values (Engh & Huber, 1991 ▶) were 0.022 Å for bond lengths and 2.2° for bond angles, with no main-chain bond length or bond angle deviating from the ‘ideal’ small-molecule values by more than six times the standard deviation. All residues were in the allowed regions of the Ramachandran plot calculated with MolProbity (Chen et al., 2010 ▶) and 99.28% of all residues were in favored regions. In the final refined structure there were two TXNL4B molecules in the asymmetric unit. It included 282 protein residues, 382 water molecules and one sucrose molecule. For both monomers, residues Gly1 and Asp143–Ile149 could not be located in the electron-density map and were not included in the refined structure. The last residue is Tyr142 for both chains and the main-chain atoms plus Cβ could be located in the map. All atoms in residues 2–141 were included in the final structure, except for Phe2 of chain B, for which side-chain atoms beyond Cγ could not be located.
Table 1. Summary of data-collection, structure-determination and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Wavelength (Å) | 1.000 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 39.33, b = 63.64, c = 51.15, β = 92.29 |
| Resolution (Å) | 30.57–1.33 (1.38–1.33) |
| No. of unique reflections | 56401 (5177) |
| Completeness (%) | 97.0 (89.6) |
| Average multiplicity | 3.6 (3.2) |
| 〈I〉/〈σ(I)〉 | 20.9 (4.4) |
| Molecules per asymmetric unit | 2 |
| Matthews coefficient (Å3 Da−1) | 1.88 |
| Solvent content (%) | 34.6 |
| R merge † | 0.042 (0.210) |
| Structure refinement | |
| Resolution range (Å) | 30.57–1.33 (1.36–1.33) |
| R work/R free ‡ | 0.13/0.18 |
| No. of residues/protein atoms | 282/2439 |
| No. of water atoms | 382 |
| No. of ligand atoms | 23 |
| Average B factor (Å2) | 20.1 |
| Overall B (from Wilson plot) (Å2) | 12.1 |
| MolProbity scores | |
| All-atom clashscore | 10.1 |
| Bad rotamers (%) | 0.78 |
| Ramachandran outliers (%) | 0.0 |
| Ramachandran favored (%) | 99.3 |
| R.m.s.d. from ideal geometry§ | |
| Bond lengths (Å) | 0.022 |
| Bond angles (°) | 2.2 |
R
merge = 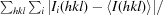
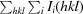 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
4% of the reflections from thin shells in the center of each reflection bin were used to calculate R free.
Empirical ideal geometry parameters are those derived by Engh & Huber (1991 ▶).
Figure 1.
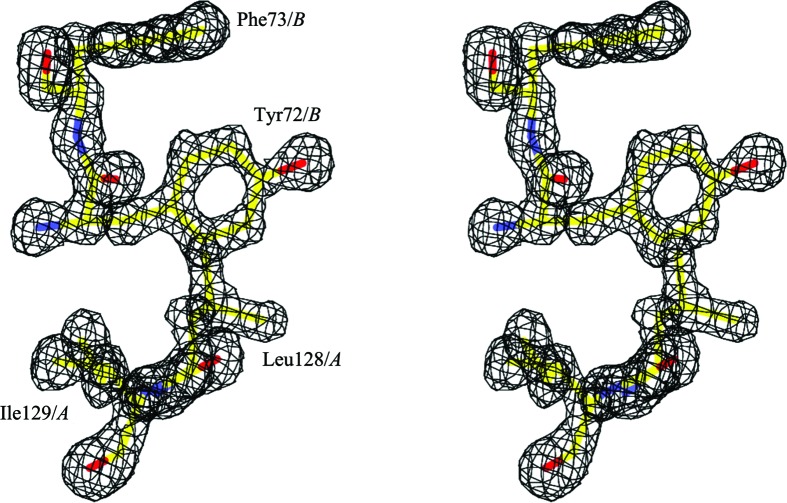
A stereoview of a shake-and-omit F o − F c electron-density map contoured at 0.3 e Å−3. The map was calculated with Leu128 and Ile129 from protomer A and Tyr72 and Phe73 from protomer B omitted. The omitted residues in the final structure are shown in a stick representation with the CPK coloring scheme.
3.2. Monomer structure
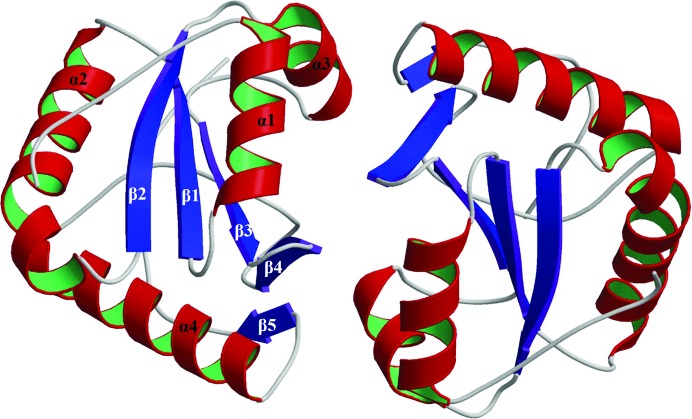
Dim2/TXNL4B has been reported to be a homodimer in solution (PDB entry 1xbs; Simeoni et al., 2005 ▶). In this study, the TXNL4B crystals belonged to space group P21 and the asymmetric unit contained a biological dimer. The two monomers of Dim2 are largely superimposable (with an r.m.s.d. of 1.17 Å calculated using the program FAST; Zhu & Weng, 2005 ▶). Fig. 2 ▶ shows a ribbon diagram of the TXNL4B homodimer in a unit cell. Simeoni and coworkers obtained Dim2/TXNL4B crystals in space group P41212 and reported a crystal structure determined at 2.5 Å resolution with one TXNL4B molecule in the asymmetric unit with residues Asn138–Ile149 missing (PDB entry 1xbs; Simeoni et al., 2005 ▶). That study also reported a biological unit for TXNL4B which contained two monomers and a similar monomer–monomer interface as observed in the dimer in the asymmetric unit in the present study. The r.m.s.d. between monomer A of this study and PDB entry 1xbs is 0.85 Å and the r.m.s.d. between monomer B of this study and PDB entry 1xbs is 0.77 Å.
Figure 2.
A ribbon-diagram presentation of a TXNL4B dimer. The secondary-structure elements in monomer B as assigned in the RasMol output file for MolScript input are labeled α for α-helices and β for β-strands and are numbered from the N-terminus to the C-terminus.
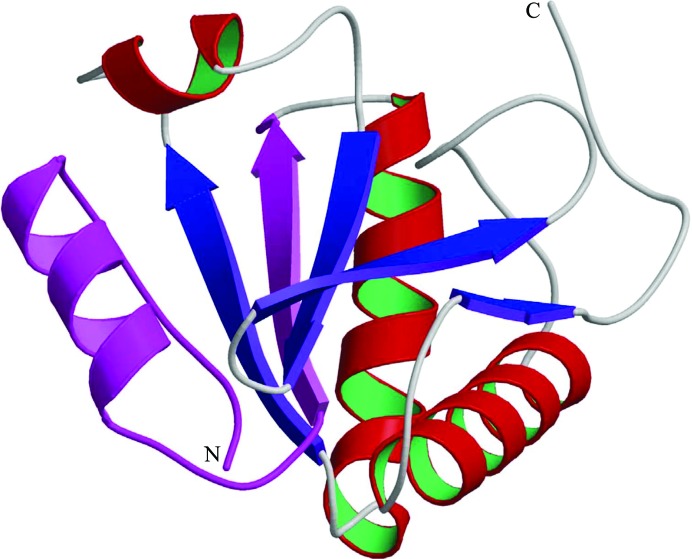
Previously, Sun and coworkers performed GST pull-down experiments using GST fusions of different deletion mutants of DLP/TXNL4B and concluded that the amino-terminal 33 residues of TXNL4B were responsible for its interaction with Prp6 (Sun et al., 2004 ▶). In light of the structure, however, deleting the first 33 residues could have destroyed the native structure of TXNL4B as this involves removing a whole strand at the center of the β-sheet core of the protein (Fig. 3 ▶). Thus, the effect of the deletion on the pull-down experiment was most likely to be because the TXNL4B deletion mutant was unable to fold into a structure with a surface similar to that of the native protein without the deleted region. Mapping the Prp6 interaction to this region based on the GST pull-down results may be misleading.
Figure 3.
Monomer of Dim2/TXNL4B. A ribbon diagram of monomer A of the asymmetric unit is shown with the N- and C-termini of the structure indicated. α-Helices are shown in red and green and β-strands are shown in blue, except for residues 2–33 which have been reported to be responsible for the interaction of Dim2/TXNL4B with Prp6 and are shown in magenta.
3.3. Comparison of TXNL4B with TXNL4A
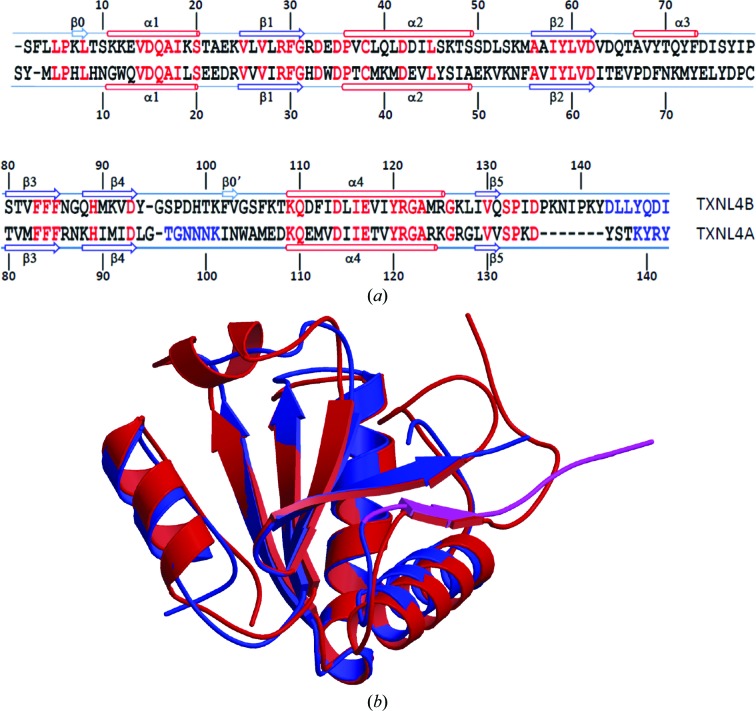
The r.m.s.d. between monomer A of TXNL4B in this study and the reported structure of TXNL4A (PDB entry 1qgv; Reuter & Ficner, 1999 ▶) is 1.34 Å and the r.m.s.d. between monomer B and TXNL4A is 1.35 Å. Although the percentage sequence identity is only 38%, the overall structures of TXNL4A and TXNL4B are conserved (Fig. 4 ▶). However, the region connecting strands β2 and β3 forms a more stable α-helix (α3) in TXNL4B, while this region is generally disordered in TXNL4A (Zhang et al., 2003 ▶). The C-terminal tail of the TXNL4 proteins after the last α-helix contains a short β-strand. However, deleting this tail did not change the structure, stability or cellular localization of TXNL4A (Zhang et al., 1999 ▶, 2003 ▶), although this region is functionally important and its removal resulted in a dominant negative form of the protein (Zhang et al., 1999 ▶). Compared with TXNL4A, TXNL4B extends this tail by seven more amino acids. Sun and coworkers showed that removing this region does not abrogate interaction between TXNL4B and Prp6. Based on structural alignment, this region in TXNL4B is not likely to affect the structure or the stability of the protein. Whether it is functionally important remains to be investigated.
Figure 4.
Comparison between TXNL4A and TXNL4B. (a) Sequence alignment based on structure superposition by MUSTANG. Single-letter amino-acid codes are used. Red indicates identical residues in the two proteins. Rather than structural alignment, blue indicates amino acids that were not located in the electron-density maps. Secondary-structure elements as assigned by STRIDE (Heinig & Frishman, 2004 ▶) are shown above the sequence for TXNL4B and below the sequence for TXNL4A. β-Strands are shown as arrows and α-helices are shown as cylinders. They are numbered from the N-terminus of the proteins, except for two strands in TXNL4B that consist of only two residues, which are numbered separately. These two short strands are not shown as strands in (b). (b) Structure comparison between TXNL4A and TXNL4B. Each structure is represented as a ribbon diagram, with monomer A of TXNL4B shown in red and TXNL4A shown in blue, except for residues Leu129–Thr138 (shown in magenta) which are within the functionally important 14-residue tail of TXNL4A, the removal of which is known to result in a dominant negative form of the protein.
Supplementary Material
PDB reference: TXNL4B, 4in0
Acknowledgments
X-ray diffraction data were collected on the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory. Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract W-31-109-Eng-38. The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
References
- Berman, H. M. et al. (2002). Acta Cryst. D58, 899–907. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Engh, R. A. & Huber, R. (1991). Acta Cryst. A47, 392–400.
- Heinig, M. & Frishman, D. (2004). Nucleic Acids Res. 32, W500–W502. [DOI] [PMC free article] [PubMed]
- Howe, P. W., Nagai, K., Neuhaus, D. & Varani, G. (1994). EMBO J. 13, 3873–3881. [DOI] [PMC free article] [PubMed]
- Jin, T., Howard, A. J., Golemis, E. A., Wang, Y. & Zhang, Y.-Z. (2005). Acta Cryst. F61, 282–284. [DOI] [PMC free article] [PubMed]
- Konagurthu, A. S., Whisstock, J. C., Stuckey, P. J. & Lesk, A. M. (2006). Proteins, 64, 559–574. [DOI] [PubMed]
- Krämer, A. (1996). Annu. Rev. Biochem. 65, 367–409. [DOI] [PubMed]
- Kraulis, P. J. (1991). J. Appl. Cryst. 24, 946–950.
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Merritt, E. A. & Murphy, M. E. P. (1994). Acta Cryst. D50, 869–873. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Oubridge, C., Ito, N., Evans, P. R., Teo, C.-H. & Nagai, K. (1994). Nature (London), 372, 432–438. [DOI] [PubMed]
- Price, S. R., Evans, P. R. & Nagai, K. (1998). Nature (London), 394, 645–650. [DOI] [PubMed]
- Reuter, K. & Ficner, R. (1999). Acta Cryst. D55, 888–890. [DOI] [PubMed]
- Reuter, K., Nottrott, S., Fabrizio, P., Lührmann, R. & Ficner, R. (1999). J. Mol. Biol. 294, 515–525. [DOI] [PubMed]
- Sayle, R. A. & Milner-White, E. J. (1995). Trends Biochem. Sci. 20, 374–376. [DOI] [PubMed]
- Simeoni, F., Arvai, A., Bello, P., Gondeau, C., Hopfner, K., Neyroz, P., Heitz, F., Tainer, J. & Divita, G. (2005). Biochemistry, 44, 11997–12008. [DOI] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Sun, X., Zhang, H., Wang, D., Ma, D., Shen, Y. & Shang, Y. (2004). J. Biol. Chem. 279, 32839–32847. [DOI] [PubMed]
- Toor, N., Keating, K. S. & Pyle, A. M. (2009). Curr. Opin. Struct. Biol. 19, 260–266. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Will, C. L. & Lührmann, R. (1997). Curr. Opin. Cell Biol. 9, 320–328. [DOI] [PubMed]
- Zhang, Y., Cheng, H., Gould, K. L., Golemis, E. A. & Roder, H. (2003). Biochemistry, 42, 9609–9618. [DOI] [PubMed]
- Zhang, Y., Gould, K. L., Dunbrack, R. L. Jr, Cheng, H., Roder, H. & Golemis, E. A. (1999). Physiol. Genomics, 1, 109–118. [DOI] [PubMed]
- Zhang, Y., Lindblom, T., Chang, A., Sudol, M., Sluder, A. E. & Golemis, E. A. (2000). Gene, 257, 33–43. [DOI] [PubMed]
- Zhou, Z., Licklider, L. J., Gygi, S. P. & Reed, R. (2002). Nature (London), 419, 182–185. [DOI] [PubMed]
- Zhu, J. & Weng, Z. (2005). Proteins, 58, 618–627. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: TXNL4B, 4in0