Two nanobodies generated against the VAR2CSA DBL6∊-FCR3 domain involved in pregnancy-associated malaria were selected, expressed, purified and crystallized.
Keywords: pregnancy-associated malaria, VAR2CSA, DBL domain, nanobody
Abstract
The VAR2CSA protein has been closely associated with pregnancy-associated malaria and is recognized as the main adhesin exposed on the surface of Plasmodium falciparum-infected erythrocytes. Chondroitin sulfate A was identified as the main host receptor in the placenta. Single-domain heavy-chain camelid antibodies, more commonly called nanobodies, were selected and produced against the DBL6∊-FCR3 domain of VAR2CSA. Crystals of two specific nanobodies, Nb2907 and Nb2919, identified as strong binders to DBL6∊-FCR3 and the full-length VAR2CSA exposed on the surface of FCR3 P. falciparum-infected erythrocytes, were obtained. Crystals of Nb2907 diffract to 2.45 Å resolution and belong to space group C2 with unit-cell parameters a = 136.1, b = 78.5, c = 103.4 Å, β = 118.8°, whereas Nb2919 crystals diffract to 2.15 Å resolution and belong to space group P43212 with unit-cell parameters a = b = 62.7, c = 167.2 Å.
1. Introduction
Pregnancy-associated malaria is correlated to the cyto-adhesion of Plasmodium falciparum-infected erythrocytes (PEs) to the placenta causing up to 200 000 foetal and infant deaths and at least 10 000 maternal deaths per year. The P. falciparum VAR2CSA protein was identified as the key adhesin responsible for the sequestration of PEs into the placenta through its interaction with chondroitin sulfate A (CSA) (Salanti et al., 2004 ▶). VAR2CSA is a 350 kDa protein composed of six cysteine-rich Duffy binding-like (DBL) domains. Small-angle X-ray scattering experiments showed these domains are folded into a compact structure forming a specific and high-affinity binding site for CSA (Srivastava et al., 2010 ▶). VAR2CSA has been shown to become the target of maternal antibodies after subsequent pregnancies (Fried et al., 1998 ▶; Salanti et al., 2004 ▶). An evasion of immunity to placental malaria would imply the masking of VAR2CSA-specific IgG epitopes by IgM antibodies. The C-terminal part of VAR2CSA, including DBL6∊, would be involved in IgM recognition (Rasti et al., 2006 ▶; Barfod et al., 2011 ▶).
Nanobodies are constituted of the single VHH domain of camelid heavy-chain antibodies. They are small and stable monomeric proteins (14 kDa) displaying a high affinity for their antigen and have been shown to be efficient tools for the crystallization of difficult systems (Baranova et al., 2012 ▶; Steyaert & Kobilka, 2011 ▶) and for the characterization of biological mechanisms (Domanska et al., 2011 ▶).
Nanobodies against the DBL6∊ domain of the VAR2CSA protein from the FCR3 P. falciparum strain were produced to help decipher the role of DBL6∊ in VAR2CSA adhesion to the placenta and evasion of the host immune response. With this aim in mind, structural and biophysical studies of nanobodies in complex with the DBL6∊ domain were initiated in order to describe at the atomic level the receptor binding site or potential epitopes for the host immune system. As part of our ongoing studies, we here present the crystallization of two specific nanobodies able to recognize the isolated DBL6∊-FCR3 domain and VAR2CSA-FCR3 exposed on infected erythrocytes. This implies that the epitope of these nanobodies is available in the compact structure formed by the full-length VAR2CSA.
2. Material and methods
2.1. Expression and purification of DBL6∊-FCR3
The DNA sequence coding for DBL6∊-FCR3 was cloned into a pET15b expression vector that introduces a His tag consisting of six histidines at the N-terminus of the protein. The vector was transformed into the Escherichia coli Rosetta gami strain, which is specifically designed for proteins containing multiple cysteines.
500 ml of 25 mg ml−1 LB containing 100 µg ml−1 ampicillin and 25 µg ml−1 chloramphenicol was inoculated with 10 ml of preculture and left to grow overnight at 310 K under shaking. When the culture reached an OD600 of 1.0, 0.1 mM IPTG was added and the temperature was decreased to 289 K to allow the overnight expression of soluble protein. The cells were harvested the next day by centrifugation for 15 min at 6490g. The pellet was resuspended in 40 ml of 50 mM phosphate buffer pH 6.3, 50 mM cystamine and the cells were broken by sonicating three times for 5 min at 275 K. The solution obtained was centrifuged for 30 min at 12 096g. The supernatant was loaded onto a 1 ml Ni–NTA column (HisTrap HP, GE Healthcare) equilibrated in 50 mM phosphate buffer pH 6.3, 100 mM NaCl, 30 mM imidazole, 50 mM cystamine. DBL6∊-FCR3 was eluted using 50 mM phosphate buffer pH 6.3, 50 mM NaCl, 400 mM imidazole, 50 mM cystamine. The eluted protein was subsequently loaded onto a Superdex 75 16/60 column (GE Healthcare) equilibrated in 50 mM phosphate buffer pH 7.0, 10 mM cystamine and eluted using the same buffer.
2.2. Generation of nanobodies against DBL6∊
Recombinant DBL6∊-FCR3 was injected subcutaneously every 7 d for 5 weeks in a Lama glama at a concentration of 0.44 mg ml−1 in 50 mM phosphate buffer pH 7.0, 10 mM cystamine. Screening and selection of nanobodies were performed as previously described (Conrath et al., 2005 ▶). Briefly, lymphocytes were purified from the blood sample obtained 1 week after the last immunization. cDNA was synthesized from the acquired RNA using a reverse PCR protocol. A VHH phage display library was constructed using the pMES4 expression and phagemid vector. This vector introduces a pelB signal peptide and a C-terminal His tag consisting of six histidines. Multiple rounds of panning and screening of this library resulted in eight binders to DBL6∊-FCR3. These nanobodies of interest, which are still in the pMES4 vector, were subsequently transformed into the E. coli WK6 expression strain.
2.3. Purification of nanobodies
For nanobody production, a preculture was grown at 310 K with shaking in 25 mg ml−1 LB containing 100 µg ml−1 ampicillin, 2% glucose and 1 mM MgCl2. 1 l of TB (Terrific Broth) supplemented with the same reagents was inoculated with 10 ml preculture. 1 mM IPTG was added once the OD600 reached 0.7. The induced cultures were left overnight for protein expression and secretion to the periplasm at 301 K with shaking. The cells were harvested by centrifugation for 15 min at 6490g. The pellet was resuspended in 15 ml of 200 mM Tris–HCl pH 8.0, 500 mM EDTA and 500 mM sucrose for 1 h under stirring on ice. An osmotic shock was subsequently applied by addition of 30 ml of 50 mM Tris–HCl pH 8.0, 125 mM EDTA and 125 mM sucrose and left for 1 h under stirring on ice. After centrifugation for 30 min at 12 096g the pellet was discarded and the periplasmic extract was loaded onto a 1 ml Ni–NTA column (HisTrap HP, GE Healthcare) equilibrated in 50 mM phosphate buffer pH 7.0, 1 M NaCl and washed with 50 mM phosphate buffer pH 6.0, 1 M NaCl. The nanobodies were eluted using 50 mM phosphate buffer pH 7.0, 1 M NaCl, 0.5 M imidazole and loaded onto a Superdex 75 16/60 column (GE Healthcare) pre-equilibrated in 50 mM Tris–HCl pH 7.5, 150 mM NaCl.
2.4. Crystallization
Crystallization trials were initiated for nanobodies that are able to recognize both the DBL6∊-FCR3 domain and the full-length VAR2CSA-FCR3 exposed on the surface of P. falciparum-infected red blood cells, which implies their epitope is still available in the compact VAR2CSA. The purified nanobodies, which still contain their His tag, were concentrated in 50 mM Tris–HCl pH 7.5, 150 mM NaCl using an ultrafiltration unit (3000 Da cutoff, Centricon). Crystallization conditions for two nanobodies, Nb2907 and Nb2919, were obtained using a Phoenix robot in 96-well Intelli-Plates (Art Robbins Instruments) with commercial screens from Hampton Research (Crystal Screen and Crystal Screen Cryo) and Molecular Dimensions (Proplex, JCSG-plus and Morpheus). The sitting-drop vapour-diffusion method was used with drops consisting of 100 nl nanobody sample mixed with an equal amount of screening solution. 70 µl of the screening solution were placed into the reservoir of the plates and the plates were stored at 292 K.
2.5. Data collection and analysis
Crystals of the two nanobodies were directly mounted from the drops in a cryoloop and flash-cooled in the nitrogen-gas cryostream without additional cryoprotectant. A complete diffraction data set was collected from one crystal of Nb2907 on PROXIMA-1 of the SOLEIL synchrotron (Gif-sur-Yvette, France) using a PILATUS 6M detector (see Table 1 ▶). 1000 images were collected with an oscillation step of 0.2° and 0.4 s exposure time. The data were integrated, scaled and merged using the XDS package (Kabsch, 2010 ▶). The merged intensities were converted to structure-factor amplitudes using the program TRUNCATE from the CCP4 package (Winn et al., 2011 ▶).
Table 1. Data-collection statistics.
Values for the outer shell are given in parentheses.
| Nb2907 | Nb2919 | |
|---|---|---|
| Diffraction source | PROXIMA-1 (SOLEIL, France) | X06DA (SLS, Switzerland) |
| Wavelength (Å) | 0.98 | 1.00 |
| Detector | PILATUS 6M | MAR Mosaic CCD |
| Crystal–detector distance (mm) | 509.1 | 199.8 |
| Space group | C2 | P43212 |
| Unit-cell dimensions (Å, °) | a = 136.1, b = 78.5, c = 103.4, α = 90, β = 118.8, γ = 90 | a = 62.7, b = 62.7, c = 167.2, α = β = γ = 90 |
| Mosaicity (°) | 0.31 | 0.75 |
| Resolution range (Å) | 47.46–2.45 (2.58–2.45) | 34.78–2.15 (2.27–2.15) |
| R merge † | 0.097 (0.50) | 0.078 (0.37) |
| R meas ‡ | 0.10 (0.60) | 0.09 (0.44) |
| Total No. of reflections | 125228 (16670) | 59781 (8464) |
| No. of unique reflections | 34820 (4969) | 18702 (2667) |
| Completeness (%) | 98.9 (98.1) | 98.9 (98.8) |
| Redundancy | 3.6 (3.4) | 3.2 (3.2) |
| 〈I/σ(I)〉 | 11.0 (2.3) | 8.1 (2.4) |
| Overall B factor from Wilson plot (Å2) | 54.1 | 35.8 |
R
merge = 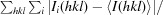
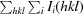 .
.
R
meas = 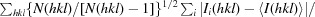
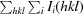 .
.
For nanobody Nb2919 data were recorded for several crystals of Nb2919 on beamline X06DA (SLS, Villigen, Switzerland) using a MAR Mosaic CCD detector (see Table 1 ▶). The data set considered to be the best consists of 120 images measured using an oscillation step of 1° and 1.0 s exposure time. Data were integrated using iMOSFLM (Battye et al., 2011 ▶) and subsequently scaled and merged using SCALA from the CCP4 package (Winn et al., 2011 ▶). Reduction to structure-factor amplitudes was performed with TRUNCATE. Matthews coefficients were calculated with the program MATTHEWS_COEF (Winn et al., 2011 ▶; Matthews, 1968 ▶). Molecular-replacement calculations were carried out with Phaser (McCoy et al., 2007 ▶) using PDB entry 1yc7 (83% sequence identity, Conrath et al., 2005 ▶) from which the CDR (complementary determining region) loops were removed. Self-rotation functions were calculated with MOLREP (Vagin & Teplyakov, 1997 ▶) using a Patterson integration radius of 20 Å and a resolution cutoff of 10.0–3.0 Å.
3. Results and discussion
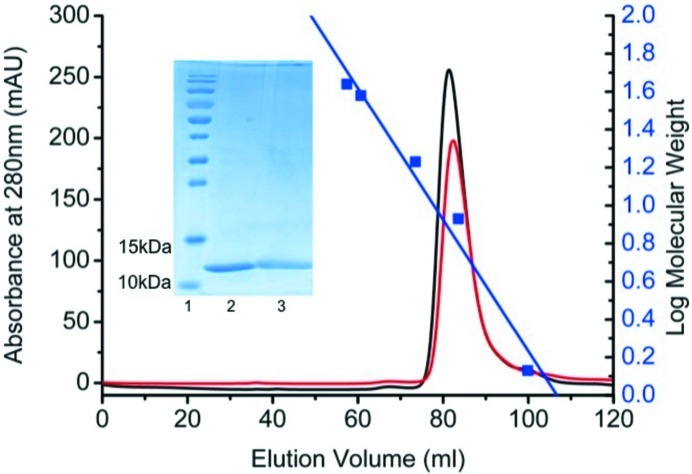
Nanobodies were generated against the DBL6∊-FCR3 domain of the P. falciparum VAR2CSA protein and strong binders were identified by ELISA. These latter have been found to be equally active against the full-length VAR2CSA exposed on infected erythrocytes (results not shown). Two nanobodies against DBL6∊-FCR3, Nb2907 and Nb2919, were crystallized. The amino-acid sequences of nanobodies Nb2907 and Nb2919 are 98% identical (Fig. 1 ▶) and both nanobodies are thus expected to bind the same epitope. Preliminary isothermal titration calorimetry (ITC) experiments (using Nb2907 and Nb2919) indicate affinities of 30 and 20 nM, respectively, for DBL6∊-FCR3. Both nanobodies were purified to homogeneity and behave as monomers in size-exclusion chromatography (Fig. 2 ▶).
Figure 1.

Sequence alignment of Nb2907 and Nb2919. The complementary determining regions (CDR) are shown in red boxes.
Figure 2.
Nanobody production. Size-exclusion chromatography elution profile of the recombinant Nb2907 (red) and Nb2919 (black) on a Superdex HR 16/60 column equilibrated with a buffer consisting of 50 mM Tris–HCl pH 7.5, 150 mM NaCl. The blue squares represent the standard used to determine the relationship between elution volume and molecular weight and consists of vitamin B12 (1.35 kDa), ubiquitin (8.6 kDa), myoglobin (17 kDa), DBL6∊ (38 kDa) and ovalbumin (44 kDa). Inset: SDS–PAGE of PageRuler Prestained Protein Ladder (lane 1, Thermo Scientific), purified Nb2907 (lane 2) and Nb2919 (lane 3) on 15% SDS–PAGE.
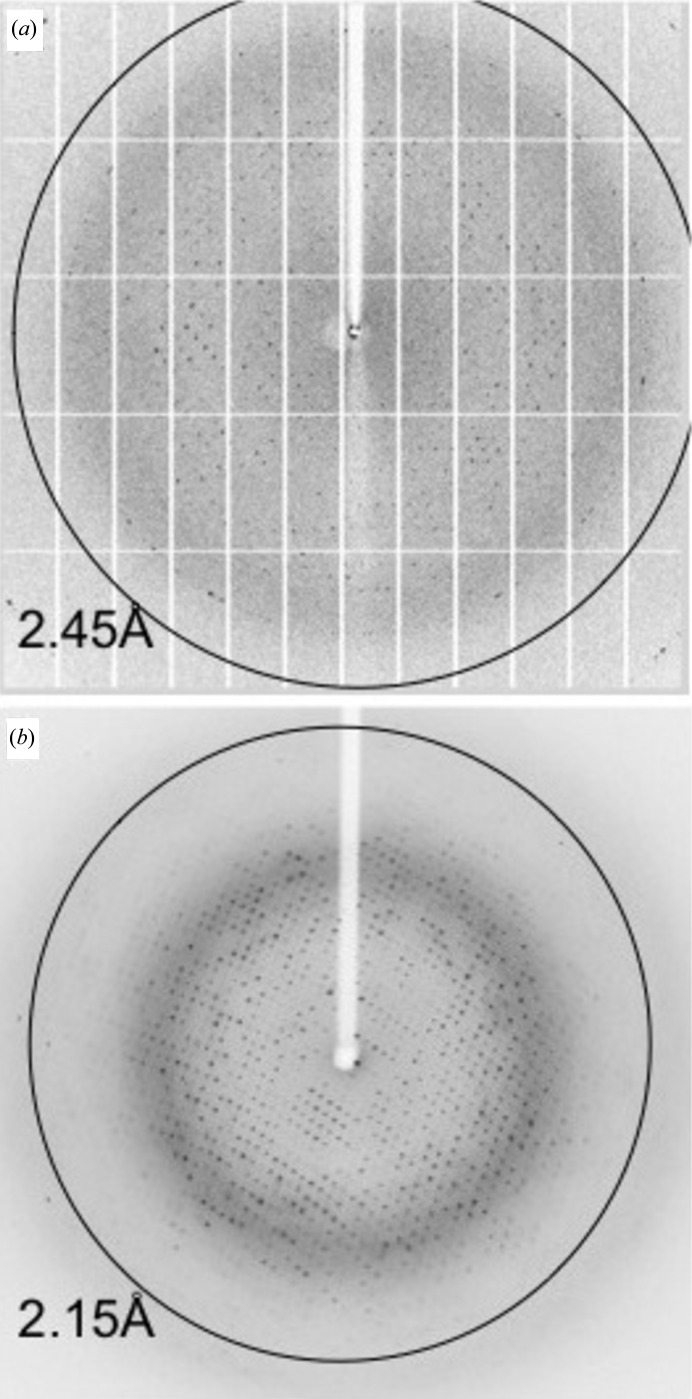
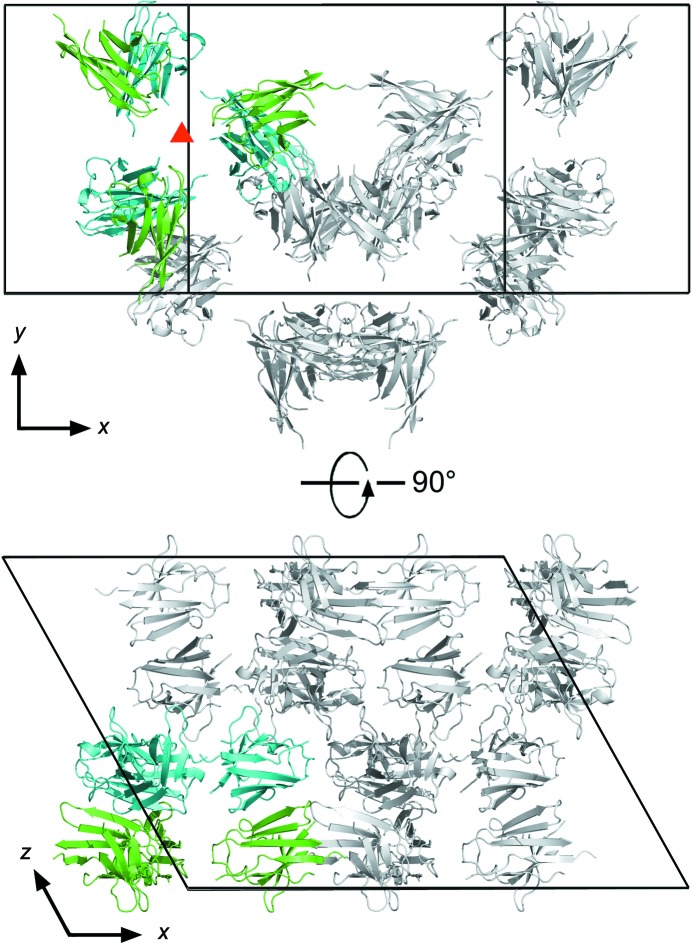
Crystals of Nb2907 (Fig. 3 ▶ a) grew after 1 month from a protein solution of 25 mg ml−1 (50 mM Tris–HCl pH 7.5, 150 mM NaCl) and a precipitant solution consisting of 0.1 M HEPES pH 7.5, 4.3 M NaCl. The size of the crystals is 0.08 × 0.06 × 0.04 mm. The crystals diffract to 2.45 Å resolution (Fig. 4 ▶ a) and belong to space group C2 with unit-cell dimensions a = 136.1, b = 78.5, c = 103.4 Å, β = 118.8°. Matthews coefficient analysis indicates that the number of nanobody molecules present in the asymmetric unit most likely lies between six and eight, corresponding to Matthews coefficients of 2.88 and 2.16 Å3 Da−1 and solvent contents between 57.3 and 43.1%, respectively (combined probability 0.84), although there is a small probability for five or nine molecules as well. Molecular replacement using PDB entry 1yc7 as a search model identified six molecules in the asymmetric unit [log likelihood gain (LLG) of 784 for the six-molecule solution compared to 644 and 640 for searches for five and seven molecules, respectively, and a translation function Z-score of 21.5 compared to 5.1 for a seventh molecule]. The molecules are packed such that a hexamer is formed consisting of three pairs of Nb2907 molecules that pack with their β-sheets upon each other. These pairs then pack with their CDR loops facing each other around a non-crystallographic threefold axis oriented perpendicular to the a–b plane of the unit cell (Fig. 5 ▶). This local threefold axis is observed in the self-rotation function as a peak with height 80% of the crystallographic twofold axis and located at θ = 0°, ϕ = 0°, χ = 120°.
Figure 3.
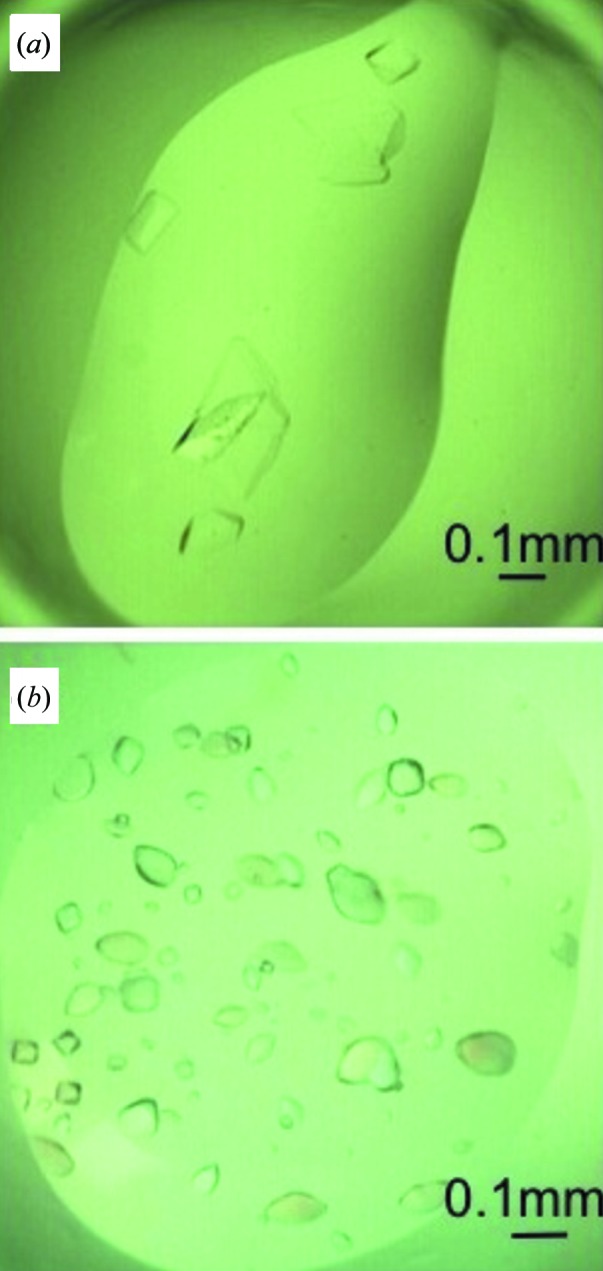
Nanobody crystals. (a) Crystals of Nb2907 grown in 0.1 M HEPES pH 7.5, 4.3 M NaCl. (b) Crystals of Nb2919 grown in 0.1 M HEPES pH 7.5, 0.1 M NaCl, 1.6 M ammonium sulfate. All crystals were grown in 96-well Intelli-Plates (Art Robbins Instruments) (see §2 for further details).
Figure 4.
Diffraction pattern collected at 100 K from the single crystals of (a) Nb2907 and (b) Nb2919 that were used for data collection. The maximum resolution is in each case indicated by a circle. See §2 for further details.
Figure 5.
Crystal packing of Nb2907. Shown are two orientations of the unit cell of Nb2907 rotated 90°. The six nanobodies present in one asymmetric unit are coloured while the symmetry mates that complete the unit cell are shown in grey. The local threefold axis perpendicular to the x–y plane is indicated by a red triangle. One set of nanobodies related by this local threefold axis is shown in green, the other set in cyan. CDR loops (not present in the model) point towards the local threefold axis.
Crystals of Nb2919 (Fig. 3 ▶ b) observable after 4 d from a protein solution of 23 mg ml−1 (50 mM Tris–HCl pH 7.5, 150 mM NaCl) and a precipitant solution consisting of 0.1 M HEPES pH 7.5, 0.1 M NaCl, 1.6 M ammonium sulfate. The size of the crystals is 0.09 × 0.06 × 0.06 mm. The crystals belong to space group P43212 with unit-cell dimensions a = b = 62.7, c = 167.2 Å and diffract up to 2.15 Å resolution (Fig. 4 ▶ b). Calculation of Matthews coefficients suggest that either two or three Nb2919 molecules are present in the asymmetric unit with a Matthews coefficient of 3.16 or 2.11 Å3 Da−1 and a solvent content of 61.1 or 41.6%, respectively. Molecular-replacement calculations yielded a solution consisting of two molecules in the asymmetric unit (LLG 619 and translation function Z-score 26.3). Again, solutions containing only a single or three molecules in the asymmetric unit scored significantly worse (LLG 170 and 356, and translation function Z-score 15.7 and 5.8, respectively).
Despite a 98% sequence identity between Nb2907 and Nb2919, corresponding to a single amino-acid substitution in CDR1 and CDR2 and a single substitution in the framework region (Fig. 1 ▶), their crystals grow in different conditions and belong to different space groups. It is tempting to speculate that these amino acids help to dictate the crystal packing, in agreement with our molecular-replacement results. The resulting structures are contributing to the growing repertoire of CDR loop conformations of camelid VHH domains and will be used in later stages to obtain more insight into VAR2CSA adhesion to the placenta, including structure determination of DBL6∊–nanobody complexes.
Once the influence of the nanobodies on the interaction between DBL6∊ and CSA/IgM has been determined, the structure of the DBL6∊–nanobody complexes will help clarify how VAR2CSA binds to the placenta and elucidate the role of DBL6∊ in the compact structure of VAR2CSA. These results will eventually open perspectives for further structural and functional studies on the VAR2CSA protein.
Acknowledgments
We thank N. Buys and K. Willibal for their technical support for nanobody screening and selection, and Dr A. Wohlkonig and Dr S. Soror for their assistance with data collection for Nb2919. This work was also supported by the Vlaams Interuniversitair Instituut voor Biotechnology (VIB), the Onderzoeksfonds of the Vrije Universiteit Brussel (OZR-VUB), the Fund for Scientific Research of Flanders (FWO-Vlaanderen), the Institute for the Encouragement of Scientific Research and Innovation of Brussels (ISRIB) FWO-Vlaanderen and the Hercules Foundation. AV is a recipient of a PhD fellowship from FWO-Vlaanderen. The Labex GR-Ex is funded by the program ‘Investissements d’avenir’. The authors acknowledge the use of the PROXIMA-1 beamline at SOLEIL (Gif-Sur-Yvette, France) and the X06DA beamline at SLS (Villigen, Switzerland).
References
- Baranova, E., Fronzes, R., Garcia-Pino, A., Van Gerven, N., Papapostolou, D., Péhau-Arnaudet, G., Pardon, E., Steyaert, J., Howorka, S. & Remaut, H. (2012). Nature (London), 487, 119–122. [DOI] [PubMed]
- Barfod, L., Dalgaard, M., Pleman, S., Ofori, M., Pleass, R. & Hviid, L. (2011). Proc. Natl Acad. Sci. USA, 108, 12485–12490. [DOI] [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Conrath, K., Vincke, C., Stijlemans, B., Schymkowitz, J., Decanniere, K., Wyns, L., Muyldermans, S. & Loris, R. (2005). J. Mol. Biol. 350, 112–125. [DOI] [PubMed]
- Domanska, K., Vanderhaegen, S., Srinivasan, V., Pardon, E., Dupeux, F., Marquez, J., Giorgetti, S., Stoppini, M., Wyns, L., Bellotti, V. & Steyaert, J. (2011). Proc. Natl Acad. Sci. USA, 108, 1314–1319. [DOI] [PMC free article] [PubMed]
- Fried, M., Nosten, F., Brockman, A., Brabin, B. J. & Duffy, P. E. (1998). Nature (London), 395, 851–852. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Rasti, N., Namusoke, F., Che, A., Chen, Q., Staalsoe, T., Klinkert, M., Mirembe, F., Kironde, F. & Wahlgren, M. (2006). Proc. Natl Acad. Sci. USA, 103, 13795–13800. [DOI] [PMC free article] [PubMed]
- Salanti, A., Dahlbäck, M., Turner, L., Nielsen, M. A., Barfod, L., Magistrado, P., Jensen, A. T., Lavstsen, T., Ofori, M. F., Marsh, K., Hviid, L. & Theander, T. G. (2004). J. Exp. Med. 200, 1197–1203. [DOI] [PMC free article] [PubMed]
- Srivastava, A. et al. (2010). Proc. Natl Acad. Sci. USA, 107, 4884–4889.
- Steyaert, J. & Kobilka, B. K. (2011). Curr. Opin. Struct. Biol. 21, 567–572. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst. 30, 1022–1025.
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.