The small terminase subunit from a lactococcal 936 bacteriophage (strain 454) has been expressed, purified, crystallized and X-ray diffraction data collected to 2.4 Å resolution.
Keywords: small terminase subunit, DNA-binding protein, Siphoviridae bacteriophage
Abstract
Terminases are enzymes that are required for the insertion of a single viral genome into the interior of a viral procapsid by a process referred to as ‘encapsulation or packaging’. Many double-stranded DNA viruses such as bacteriophages T3, T4, T7, λ and SPP1, as well as herpes viruses, utilize terminase enzymes for this purpose. All the terminase enzymes described to date require two subunits, a small subunit referred to as TerS and a large subunit referred to as TerL, for in vivo activity. The TerS and TerL subunits interact with each other to form a functional hetero-oligomeric enzyme complex; however the stoichiometry and oligomeric state have not been determined. We have cloned, expressed and purified recombinant small terminase TerS from a 936 lactococcal bacteriophage strain ASCC454, initially isolated from a dairy factory. The terminase was crystallized using a combination of nanolitre sitting drops and vapour diffusion using sodium malonate as the precipitant, and crystallization optimized using standard vapour-diffusion hanging drops set up in the presence of a nitrogen atmosphere. The crystals belong to the P2 space group, with unit-cell parameters a = 73.93, b = 158.48, c = 74.23 Å, and diffract to 2.42 Å resolution using synchrotron radiation. A self-rotation function calculation revealed that the terminase oligomerizes into an octamer in the asymmetric unit, although size-exclusion chromatography suggests that it is possible for it to form an oligomer of up to 13 subunits.
1. Introduction
Virulent lactococcal bacteriophages are ubiquitous in the dairy environment and their lytic cycle leads to bacterial cell death, resulting in disruption to the manufacture of cheese and yoghurts which in turn can lead to considerable economic losses to the dairy industry. Lactococcus lactis is one of the major fermentative organisms used by the dairy industry and the majority of phages that lead to dairy failures are phage species 936, c2 and P335 (Deveau et al., 2006 ▶; Moineau et al., 1996 ▶; Prevots et al., 1990 ▶). All of these common dairy phage species belong to the Siphoviridae family of double-stranded (ds) DNA viruses characterized by a proteinaceous capsid with icosahedral symmetry attached to a non-contractile tail. A recent study of the population genomics of dairy phages (Castro-Nallar et al., 2012 ▶) sequenced the complete genomes of 28 phages from the 936 species with known host range, geographic and temporal relationships. This study provided insights into the likely method of phage transmission based on a phylodynamic examination of the phage genomes. These genomes are linear dsDNA between 30.9 and 32.1 kbp in length terminated by cohesive ends encoding between 55 and 65 genes. Approximately half of the genome encodes proteins involved in the structure or packaging of DNA into the viral capsid while the remainder of the genome encodes regulatory and other genes of which the majority have no assigned function based on bioinformatic analysis.
A protein crucial to phage assembly which exhibits very little diversity within the lactococcal bacteriophages studied to date is the small terminase subunit (TerS). Bacteriophage terminases act as powerful molecular machines to package viral DNA into proviral capsids by a process referred to as encapsulation (Casjens, 2011 ▶; Teschke, 2012 ▶). These terminase complexes consist of a large hetero-oligomeric complex comprising a small regulatory TerS subunit that specifically binds concatemeric viral DNA and transfers it to the large terminase subunit (TerL) that contains both ATPase and endonuclease activity responsible for ATP-driven translocation of viral DNA into the capsid and cleaving concatemeric DNA to generate mature viral DNA, respectively (Feiss & Rao, 2012 ▶). This terminase complex docks onto a portal protein complex located at the vertex of a preformed viral procapsid and actively injects the viral DNA into the procapsid through a macromolecular conduit of viral proteins to near-crystalline density. The packaged DNA is then sealed within the mature capsid by the addition of portal closure proteins and tail proteins (Camacho et al., 2003 ▶; Casjens & Molineux, 2012 ▶). Such organization of this process of encapsulation is well conserved amongst members of the Siphoviridae family of dsDNA bacteriophages, herpes viruses and adenoviruses.
The small terminase proteins from different viruses comprise between 100 and 200 amino acids, are non-homologous in structure and contain a DNA-binding domain, an oligomeric domain and a C-terminal large terminase subunit recognition domain (Casjens & Molineux, 2012 ▶). Biochemical analysis, mass spectrometry, electron microscopy and X-ray crystallography reaveal the small terminases from bacteriophages T4, T7, SPP1, P22 form common high-molecular-weight oligomeric ring-like structures (Bazinet et al., 1988 ▶; Chang et al., 2006 ▶; Lin et al., 1997 ▶; White & Richardson, 1987 ▶). The oligomerization status for these different small terminases whose structures have been determined by X-ray crystallography varies from an octamer for Sf6 gp1 (Zhao et al., 2010 ▶), nonamer for P22 gp3 (Nemecek et al., 2008 ▶; Roy et al., 2011 ▶, 2012 ▶), nonamer for full-length siphovirus SF6 G1P (Büttner et al., 2012 ▶), to 11-mer and 12-mer for truncated versions of T4-like phage terminase subunit (Sun et al., 2012 ▶).
The small terminase subunit of 936 species L. lactis bacteriophage ASCC454 displays no amino-acid sequence similarity to any of the small terminases whose crystal structures have been determined. Its structure will provide important insights into the formation of large macromolecular terminase complexes involving both the TerS and TerL subunits.
2. Materials and methods
2.1. Cloning, expression and purification
The gene sequence encoding the full-length small terminase subunit LLAPH_454_0001 (GenBank accession AFE87597.1; residues 1–174) of L. lactis bacteriophage ASCC454 (GenBank accession JQ740802) was amplified by PCR. DNA coding for the 174 residues of small terminase subunit was cloned into a modified pET43.1a plasmid containing a C-terminal hexa-His tag. The vector-associated sequence introduced MGS at the N-terminus and ASHHHHHH at the C-terminus. Rubidium chloride chemically competent Escherichia coli BL21-AI cells (LifeTechnologies, California, USA) were transformed with the small terminase contruct, and single colonies picked and tested for overexpression and production of the soluble small terminase subunit. The molecular weight of the recombinant small terminase subunit, including the vector-associated sequence, is 21 133.7 Da.
Large-scale expression cultures were performed by inoculating 15 ml of an overnight starter culture into 1.0 l 2YT media containing 100 µg ml−1 ampicillin in a 2.5 l TunAir polypropylene baffled shake flask (Sigma, USA). The culture was incubated at 310 K, 200 rev min−1 until a cell density of 0.6–0.8 (OD600 nm) was attained. Protein expression was induced with the addition of 0.5 mM IPTG and 0.025%(w/v) arabinose at 303 K, 180 rev min−1, for 4 h. Cells were harvested by centrifugation at 6000g for 10 min and the pellets frozen at 193 K.
The cell pellet was resuspended in lysis buffer consisting of 10 mM Tris–HCl pH 8.0, 300 mM NaCl, 10 mM imidazole, 2 mM MgCl2, 5 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), EDTA-free protease inhibitor tablets (1 tablet per 50 ml lysis buffer), 0.5 mg ml−1 lysozyme and 5 units ml−1 benzonase at a ratio of 1.0 g cell pellet to 10 ml lysis buffer. The cells were lysed by three passes through an Emulsiflex C5 homogenizer (Avestin, Canada) operating at 100 MPa and 277 K. The lysate was clarified by centrifugation (20 000 rev min−1 for 20 min at 277 K) and filtered through a 5.0 µm syringe filter.
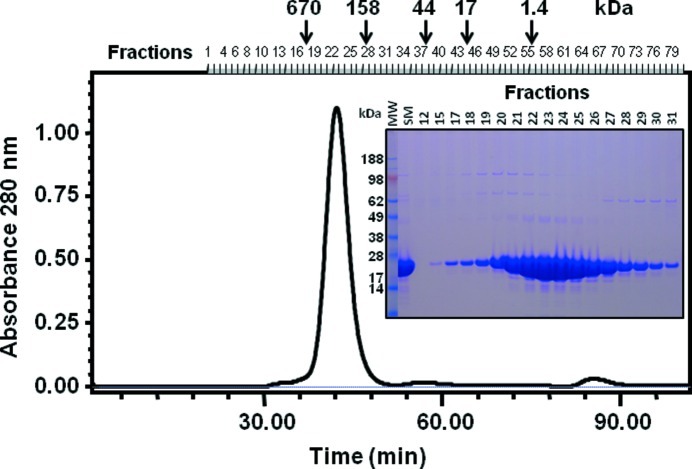
The supernatant containing the small terminase protein was purified using a combination of immobilized Ni affinity chromatography, gel-filtration chromatography and anion-exchange chromatography. Briefly, the clarified bacterial lysate was loaded onto a 5 ml HisTrap column (GE Healthcare) equilibrated in 10 mM Tris–HCl pH 8.0, 300 mM NaCl, 10 mM imidazole, 5 mM DTT at a flow rate of 5.0 ml min−1. The column was subsequently washed with equilibration buffer containing 20 mM imidazole, before eluting the bound small terminase protein with equilibration buffer containing 250 mM imidazole. The small terminase protein was further purified on a HiLoad 16/60 Superdex 200 preparative-grade gel-filtration column (GE Healthcare) pre-equilibrated with 10 mM Tris–HCl pH 8.0, 150 mM NaCl and 5 mM DTT at a flow rate of 1.5 ml min−1 (Fig. 1 ▶). The peak fraction was further purified by anion-exchange chromatography on a Mono-Q 5/5 column (GE Healthcare) equilibrated in 40 mM Tris–HCl buffer pH 8.0 and eluted with a 0–500 mM NaCl gradient in the presence of 10 mM DTT, at a flow rate of 2 ml min−1. Purification of the small terminase protein was monitored by PAGE using 4–12% gradient bis-tris NuPAGE gels and MES electrophoresis buffer systems under reducing conditions (LifeTechnologies, California, USA). The estimation of molecular weight was determined using SeeBlue2 molecular-weight standards (LifeTechnologies, California, USA).
Figure 1.
Size-exclusion chromatography of post-IMAC purified small terminase protein on a Superdex 200 1.6/60 column equilibrated in 10 mM Tris–HCl pH 8.0, 150 mM NaCl and 5 mM DTT. The column had previously been equilibrated with BioRad gel-filtration standards (bovine thyroglobulin, 670 kDa; bovine γ globulin, 158 kDa; chicken ovalbumin, 44 kDa; horse myoglobin, 17 kDa; and vitamin B12, 1.35 kDa). The protein migrated with an apparent molecular weight of ∼290 kDa, representing a higher-order oligomeric form of the small terminase protein. SDS–PAGE analysis of the peak fractions on a 4–12% bis-tris NuPAGE gel under reducing conditions revealed the oligomer dissociated into its monomeric subunits. Abbreviations: MW, molecular-weight standards; SM, starting material.
2.2. Protein crystallization
Nanolitre crystallization trials were performed at the CSIRO Collaborative Crystallization Centre (C3), Parkville, Melbourne (http://www.csiro.au/c3/) using a Phoenix crystallization robot (Art Robbins Instruments, Sunnyvale, California, USA). Screening experiments were performed using a sparse-matrix screen (Newman et al., 2005 ▶) at both 281 and 293 K. The nanolitre crystallization experiments were set up using the sitting-drop method in 96-well Innovadyne SD-2 plates (IDEX Corporation) with a reservoir containing 50 µl precipitant solution. Each drop contained 150 nl protein solution (13–26 mg ml−1) in a buffer of 10 mM Tris–HCl pH 8.0, 150 mM NaCl and 10 mM DTT with an equal volume of the reservoir solution. Initial screening identified conditions that gave rise to numerous small ‘twinned’ plate-like crystals using sodium malonate as the precipitant and either 100 mM HEPES buffer or 10%(v/v) malate–MES–Tris buffer in the pH range 6.5 to 7.5. Randomized screening around these conditions and the addition of additives [1%(v/v) dioxane, 1%(v/v) 2-methyl-2,4-pentanediol and 50 mM guanidine hydrochloride] known to reduce twinning failed to improve the quality of these crystals. We identified that fresh protein preparations in the presence of higher concentrations of the reducing agent 1,4-dithiothreitol gave improved diffraction and this was further optimized by using the hanging-drop vapour-diffusion method performed in a nitrogen glove box at 293 K. A 2 µl droplet of concentrated small terminase (15 mg ml−1) in a buffer of 10 mM Tris–HCl pH 8.0, 150 mM NaCl and 20 mM DTT was mixed in a 1:1 ratio with the reservoir solution and equilibrated against 500 µl reservoir solution using VDX pre-greased 24-well plates (Hampton Research, California, USA). The largest crystals were obtained using a reservoir solution consisting of 100 mM HEPES pH 7.0 containing 1.1 M sodium malonate and 1%(v/v) 2,4-methylpentanediol (Fig. 2 ▶).
Figure 2.

Crystals of the lactococcal bacteriophage small terminase, grown at 293 K in 100 mM HEPES buffer pH 7.0 containing 1.1 M sodium malonate and 1%(v/v) 2,4-methylpentanediol. The dimensions of this crystal are 0.40 × 0.40 × 0.02 mm.
2.3. X-ray diffraction data collection and analysis
Prior to data collection, single crystals were transferred into a cryoprotectant solution of 100 mM HEPES pH 7.0 containing 1.1 M sodium malonate and 30%(w/v) sucrose for 5 min. The crystals were then frozen under the stream of nitrogen gas cooled to 100 K and exposed to X-rays. Diffraction data sets were collected on the MX2 micro-focus beamline at the Australian Synchrotron (Melbourne, Australia) and processed with the program XDS (Kabsch, 2010 ▶). The obtained hkl intensities were input into the CCP4 program package (Winn et al., 2011 ▶) for space-group examination, intensity scaling and reduction. A self-rotation function was calculated with GLRF (Tong & Rossmann, 1997 ▶) to check the local symmetry in the crystal using data in the resolution range of 10–3.0 Å with an integration radius of 30 Å. A summary of diffraction data statistics is shown in Table 1 ▶.
Table 1. X-ray diffraction data-collection statistics.
The values in parentheses are for the highest-resolution bin (approximate interval of 0.1 Å).
| Diffraction source | Beamline MX2, Australian Synchrotron |
| X-ray wavelength (Å) | 0.9537 |
| Temperature (K) | 100 |
| Space group | P2 |
| Unit-cell parameters (Å, °) | a = 73.93, b = 158.48, c = 74.23, α = γ = 90, β = 96.38 |
| Maximum resolution (Å) | 2.42 (2.55–2.42) |
| Total observations | 451502 (55498) |
| Unique reflections | 63769 (8922) |
| Multiplicity | 7.1 (6.2) |
| Completeness (%) | 98.9 (94.6) |
| R merge † (%) | 9.4 (64.8) |
| R p.i.m. ‡ (%) | 4.1 (29.2) |
| Mean I/σ(I) | 13.8 (3.1) |
| Average mosaicity | 0.24 |
| B factor estimated from Wilson plot (Å2) | 40.0 |
R
merge = 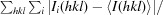
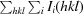 , where I is the observation of reflection intensity and 〈I〉 is the weighted average intensity for multiple and symmetry-related measurements.
, where I is the observation of reflection intensity and 〈I〉 is the weighted average intensity for multiple and symmetry-related measurements.
Multiplicity-weighted R
merge from all reflections. For a definition, see Evans (2006 ▶). R
p.i.m. = 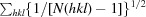
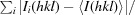
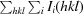 .
.
3. Results and discussion
A number of crystals were screened for data collection with synchrotron radiation. Auto-indexing, Laue-group examination and inspection of the systematic extinction of collected data sets revealed that the crystals belong to space group P2 but with large variations of the cell length along the unique b axis, indicating non-isomorphism among the crystals. The best crystal diffracted beyond 2.4 Å resolution (Fig. 3 ▶) and a complete data set was obtained to a resolution of 2.42 Å by collecting 360 frames with a rotation increment of 1°. The data set has a cumulative merging R factor of 9.4% and completeness of 98.9%. The cell parameters of the crystal are a = 73.93, b = 158.48, c = 74.23 Å, and β = 96.38° (Table 1 ▶).
Figure 3.
A representative 1.0° oscillation image of data collected from a crystal of the lactococcal bacteriophage small terminase. The centre of the edge of the diffraction image corresponds to 2.1 Å resolution. The image was collected using an ADSC Quantum 315 detector from a crystal frozen at 100 K on the MX2 beamline, at the Australian Synchrotron, with an X-ray wavelength of 0.9537 Å and exposure time of 1 s at a crystal-to-detector distance of 330 mm.
It has been known that the small terminase subunit oligomerizes in solution. Oligomers with a subunit number of 8, 9, 10, 11 or 12 have been found in the crystallographic structures of other small terminases (Zhao et al., 2010 ▶; Nemecek et al., 2008 ▶; Roy et al., 2011 ▶, 2012 ▶; Büttner et al., 2012 ▶; Sun et al., 2012 ▶). Biochemical evidence using size-exclusion chromatography suggests that the L. lactis bacteriophage ASCC454 small terminase subunit exists as an oligomer in solution, with a molecular weight of ∼290 kDa. Calculation of the Matthews coefficient (Matthews, 1968 ▶) showed that the most probable oligomeric state of the small terminase subunit in the crystal is an octamer or nonamer, corresponding to 2.57 or 2.29 Å3 Da−1, with a solvent content of 52 or 46%, respectively.
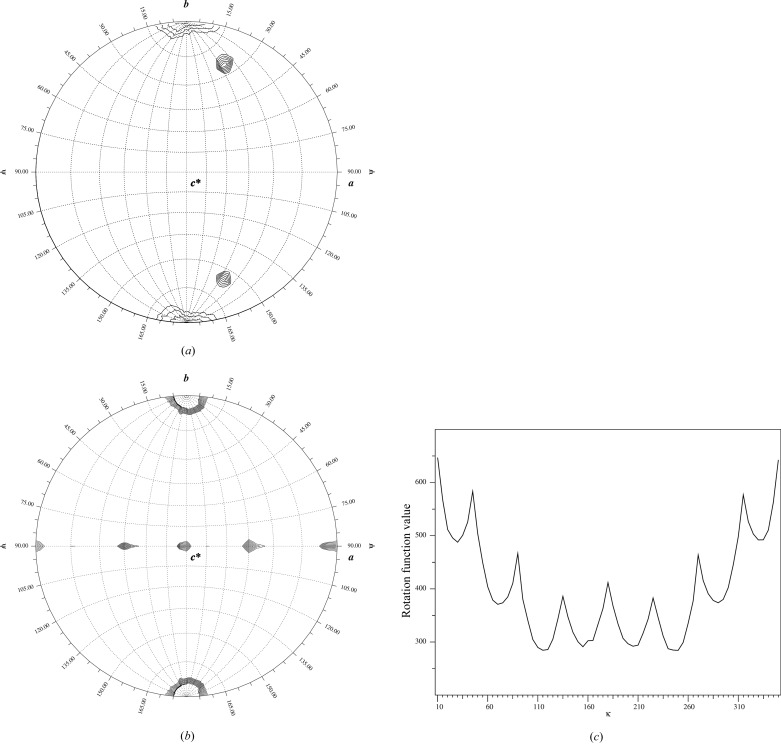
To check the non-crystallographic symmetry (NCS) in the crystal, the self-rotation function was calculated using the polar angle system (Rossmann & Blow, 1962 ▶). Because of the monoclinic space group of the crystal, ψ is defined as the angle from the Cartesian y axis in the calculation, while κ is the self-rotation angle and ϕ is the angle between the x axis and the projection of the self-rotation axis on the xz plane. Calculation with κ = 45° revealed clear peaks at (ϕ = 41°, ψ = 25°) and (ϕ = 41°, ψ = 155°) suggesting the existence of eightfold symmetry in the crystal (Fig. 4 ▶ a). Peaks corresponding to the twofold rotation axes of the octamer were revealed in the self-rotation function calculated with κ =180°, in which neighbouring NCS peaks are distant from each other by 45° (Fig. 4 ▶ b). The eightfold symmetry was further manifested by a one-dimensional search for κ in the range of 0 to 360° with the orientation of (ϕ = 41°, ψ = 25°), which resulted in a one-dimensional κ plot showing clearly a periodic maximum at the interval of every 45° (Fig. 4 ▶ c). It was therefore concluded that the terminase exists as an octamer in the crystal.
Figure 4.
NCS analysis of the diffraction data. (a) The stereographic plot of the self-rotation function calculated with κ = 45°, using the program GLRF (Tong & Rossmann, 1997 ▶). Apart from the peak at the origin, the two peaks at (ϕ, ψ) = (41°, 25°) and (41°, 155°) are symmetry-related to each other, which corresponds to a local eightfold axis. (b) Self-rotation function calculated with κ = 180°, showing the local twofold axes, which are ψ = 90° with an interval of 45°. The maps are contoured at 4.04σ (a) and 2.0σ (b) in steps of 0.1σ. (c) The one-dimensional κ plot showing the intensity of the self-rotation function with (ϕ = 41°,ψ = 25°) for κ between 10 and 350°. The origin peak is truncated for clarity.
Acknowledgments
This work was supported by grants from CSIRO’s Materials Science and Engineering Capability Development Fund and CSIRO’s Transformational Biology Capability Platform. The authors acknowledge use of the CSIRO Collaborative Crystallization Centre and the technical assistance provided by Dr Janet Newman, Dr Shane Seabrook and Ms Sarina Meuseberger. We thank the staff at the MX2 beamline of the Australian Synchrotron for their help with data collection.
References
- Bazinet, C., Benbasat, J., King, J., Carazo, J. M. & Carrascosa, J. L. (1988). Biochemistry, 27, 1849–1856. [DOI] [PubMed]
- Büttner, C. R., Chechik, M., Ortiz-Lombardía, M., Smits, C., Ebong, I. O., Chechik, V., Jeschke, G., Dykeman, E., Benini, S., Robinson, C. V., Alonso, J. C. & Antson, A. A. (2012). Proc. Natl Acad. Sci. USA, 109, 811–816. [DOI] [PMC free article] [PubMed]
- Camacho, A. G., Gual, A., Lurz, R., Tavares, P. & Alonso, J. C. (2003). J. Biol. Chem. 278, 23251–23259. [DOI] [PubMed]
- Casjens, S. R. (2011). Nature Rev. Microbiol. 9, 647–657. [DOI] [PubMed]
- Casjens, S. R. & Molineux, I. J. (2012). Adv. Exp. Med. Biol. 726, 143–179. [DOI] [PubMed]
- Castro-Nallar, E., Chen, H., Gladman, S., Moore, S. C., Seeman, T., Powell, I. B., Hillier, A., Crandall, K. A. & Chandry, P. S. (2012). Genome Biol. Evol. doi: 10.1093/gbe/evs017. [DOI] [PMC free article] [PubMed]
- Chang, J., Weigele, P., King, J., Chiu, W. & Jiang, W. (2006). Structure, 14, 1073–1082. [DOI] [PubMed]
- Deveau, H., Labrie, S. J., Chopin, M. C. & Moineau, S. (2006). Appl. Environ. Microbiol. 72, 4338–4346. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Feiss, M. & Rao, V. B. (2012). Adv. Exp. Med. Biol. 726, 489–509. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Lin, H., Simon, M. N. & Black, L. W. (1997). J. Biol. Chem. 272, 3495–3501. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Moineau, S., Borkaev, M., Holler, B. J., Walker, S. A., Kondo, J. K., Vedamuthu, E. R. & Vandenbergh, P. A. (1996). J. Dairy Sci. 79, 2104–2111.
- Nemecek, D., Lander, G. C., Johnson, J. E., Casjens, S. R. & Thomas, G. J. (2008). J. Mol. Biol. 383, 494–501. [DOI] [PMC free article] [PubMed]
- Newman, J., Egan, D., Walter, T. S., Meged, R., Berry, I., Ben Jelloul, M., Sussman, J. L., Stuart, D. I. & Perrakis, A. (2005). Acta Cryst. D61, 1426–1431. [DOI] [PubMed]
- Prevots, F., Mata, M. & Ritzenthaler, P. (1990). Appl. Environ. Microbiol. 56, 2180–2185. [DOI] [PMC free article] [PubMed]
- Rossmann, M. G. & Blow, D. M. (1962). Acta Cryst. 15, 24–31.
- Roy, A., Bhardwaj, A. & Cingolani, G. (2011). Acta Cryst. F67, 104–110. [DOI] [PMC free article] [PubMed]
- Roy, A., Bhardwaj, A., Datta, P., Lander, G. C. & Cingolani, G. (2012). Structure, 20, 1403–1413. [DOI] [PMC free article] [PubMed]
- Sun, S., Gao, S., Kondabagil, K., Xiang, Y., Rossmann, M. G. & Rao, V. B. (2012). Proc. Natl Acad. Sci. USA, 109, 817–822. [DOI] [PMC free article] [PubMed]
- Teschke, C. M. (2012). Structure, 20, 1291–1292. [DOI] [PMC free article] [PubMed]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol. 276, 594–611. [PubMed]
- White, J. H. & Richardson, C. C. (1987). J. Biol. Chem. 262, 8845–8850. [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zhao, H., Finch, C. J., Sequeira, R. D., Johnson, B. A., Johnson, J. E., Casjens, S. R. & Tang, L. (2010). Proc. Natl Acad. Sci. USA, 107, 1971–1976. [DOI] [PMC free article] [PubMed]