The wild-type protein and four active-site mutants of xylanase II from Trichoderma reesei that catalyzes the hydrolysis of glycosidic bonds in xylan have successfully been crystallized. The crystallization of several structures including ligand-free and protein ligand complexes containing the substrate (xylohexaose) or product (xylotriose) are detailed.
Keywords: xylanase II, Trichoderma reesei
Abstract
Xylanase II from Trichoderma reesei catalyzes the hydrolysis of glycosidic bonds in xylan. Crystallographic studies of this commercially important enzyme have been initiated to investigate its reaction mechanism, substrate binding and dependence on basic pH conditions. The wild-type protein was heterologously expressed in an Escherichia coli host using the defined medium and four active-site amino acids were replaced to abolish its activity (E177Q and E86Q) or to change its pH optimum (N44D and N44H). Cation-exchange and size-exclusion chromatography were used to obtain >90% protein purity. The ligand-free proteins and variant complexes containing substrate (xylohexaose) or product (xylotriose) were crystallized in several different space groups and diffracted to high resolutions (from 1.07 to 1.55 Å).
1. Introduction
The microbial enzyme xylanase (endo-1,4-β-xylanase, EC 3.2.1.8) catalyzes the hydrolysis of xylan into oligosaccharides by attacking the glycosidic bonds connecting β-1,4-linked d-xylose subunits (Saha, 2003 ▶; Polizeli et al., 2005 ▶). Xylanase is widely used in the paper (Kluepfel et al., 1993 ▶) and food industries (Li et al., 2000 ▶). When combined with cellulases, it has further potential applications for the conversion of lignocellulosic biomass into monosaccharides, which can be chemically or biologically converted to biofuels and value-added bioproducts (Gupta et al., 2008 ▶; Langan et al., 2011 ▶). Xylanase II (Xyn II) secreted by the filamentous fungus Trichoderma reesei is particularly attractive for industrial use because it can be produced relatively cheaply and in high yields (Cherry & Fidantsef, 2003 ▶). However, its application is limited by the sensitivity of its catalytic activity to alkaline pH conditions found in many pulping and pre-treatment processes.
We have initiated crystallographic studies of the 21 kDa Xyn II over a range of pH conditions, and in complex with various substrates, in order to gain a better understanding of the enzyme’s catalytic mechanism and its dependency on pH. Native wild-type (nWT) Xyn II has already been crystallized and its structure determined by X-ray crystallography (Törrönen et al., 1994 ▶; Havukainen et al., 1996 ▶). Xyn II belongs to the glycoside hydrolase family 11 (GH11) with a partially closed palm-of-a-hand fold. All GH11 enzymes have two Glu residues (E177 and E86 in Xyn II) with carboxylate side chains that are thought to function as the general acid–base and nucleophile during hydrolysis. In this work, we succeeded in crystallizing a complex of the enzyme after the substrate had been hydrolyzed to form xylotriose (X3). In order to bind substrate without turnover, and therefore to provide a mimic of the Michaelis complex (Zolotnitsky et al., 2004 ▶), we replaced E177 with Q177 in one variant and E86 with Q86 in another. The E177Q substitution cannot provide a proton to attack the glycosidic bond and E86Q cannot act as a nucleophile. We also substituted N44 with D44 or H44 with the aim of changing the electrostatic environment of E177 and consequently its pK a. Here, we report the expression, purification, crystallization and X-ray crystallographic characterization of variants E177Q, E86Q, N44H and N44D in the ligand-free form, and E177Q and N44H in complex with substrate xylohexaose (X6) or product xylotriose (X3), respectively.
2. Materials and methods
2.1. Cloning, expression and purification of heterologous Xyn II
The recombinant wild-type (rWT) gene of Xyn II from T. reesei was synthesized by DNA2.0 (Menlo Park, CA, USA) with codon usage optimized for protein expression in Escherichia coli. Genes encoding the rWT, N44D and N44H proteins were synthesized to express amino acids 2–190 of the native, secreted protein, with the N-terminal glutamate residue replaced by methionine, and eight amino acids containing a hexa-histidine (His6) tag attached to the C-terminus. For untagged protein production, all genes were synthesized to encode amino acids 2–190 of this construct with the N-terminal glutamate residue replaced by methionine. Although the xylanases secreted from T. reesei and T. longibrachiatum have the same protein sequence, they are different species (Kuhls et al., 1997 ▶) and the DNA sequence of the T. reesei gene (Torronen et al., 1992 ▶) is not identical to that in T. longibrachiatum (http://genome.jgi.doe.gov/Trilo1/Trilo1.home.html). Therefore, we refer to this protein as the T. reesei xylanase II enzyme.
E. coli BL21-gold (Agilent Tech, Santa Clara, CA, USA) cells transformed with the pJexpress401 (DNA2.0) vector containing each gene were isolated on lysogeny broth (LB) agar medium containing kanamycin (50 µg ml−1). A single colony was picked and inoculated into 40 ml LB as the starting culture. After growing overnight at 310 K, 4 l Enfors minimal media (Törnkvist et al., 1996 ▶) was inoculated with the above starting culture and was incubated until the OD600 reached 0.6. After adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to 0.5 mM, the culture was grown overnight at 295 K. The cells were centrifuged at 6000g for 30 min at 277 K and stored at 193 K.
Approximately 20 g of cells were resuspended in 100 ml lysis buffer [25 mM MES–NaOH, one tablet of protease inhibitor cocktail from Sigma–Aldrich (St Louis, MO, USA), pH 6.0] and lysed by sonication on ice. The cell lysate was clarified by centrifugation at 15 000g for 1 h at 277 K. The supernatant was filtered through a 0.45 µm filter and loaded onto a 5 ml HiTrap SP cation-exchange column from GE Healthcare (Pittsburgh, PA, USA), which had been pre-equilibrated with 50 mM MES–NaOH, pH 6.0. The protein was eluted using the linear gradient of 0.2–1.0 M NaCl in 50 mM MES–NaOH, pH 6.0. The eluted protein was concentrated using an Amicon Ultra-15 (5000 Da cut-off) concentrator from Millipore (Billerica, MA, USA) to a volume of 1.0 ml and then loaded onto a HiLoad 16/60 Superdex 75 preparative-grade size-exclusion column (GE Healthcare), which was pre-equilibrated with the final buffer [0.1 M Tris–HCl, 0.1 M NaCl, 1 mM dithiothreitol (DTT), pH 8.5]. The purified protein was concentrated using an Amicon Ultra-4 (5000 Da cut-off) concentrator to reach the concentration of 40 mg ml−1 and stored at 193 K. The variants were produced using the same protocol. The protein concentrations were determined by measuring UV absorption at 280 nm using molar absorption coefficients calculated from their amino-acid sequences (∊280 = 58 330 M −1 cm−1). The protein purity was estimated by SDS–PAGE with Coomassie Blue staining.
2.2. Crystallization
Crystallization of the rWT enzyme and its variants (30 mg ml−1) in the ligand-free form was achieved using the following condition: 15–20% PEG 8000, 0.2 M NaI, 0.1 M MES, pH 6.0. In this condition, the pH is more physiological compared to the previously published condition to crystallize the native enzyme (nWT) (Kovalevsky et al., 2011 ▶). We did not observe substrate binding by soaking oligosaccharides in crystallization drops that already contained crystals in the ligand-free form. We were also unsuccessful in obtaining crystals under similar crystallization conditions when the enzyme and substrate were mixed beforehand in co-crystallization studies. We therefore screened crystallization conditions using a number of commercial screens including Crystal Screen HT and Index HT (Hampton Research, Aliso Viejo, CA, USA), and The JCSG+ Suite and The PEGs Suite (Qiagen, Valencia, CA, USA) in co-crystallization trials. The best diffracting crystals were found after replacing NaI with CaCl2 for the N44H–X3 complex: 18% PEG 8000, 0.2 M CaCl2, 0.1 M MES, pH 6.0, 50 mM xylohexaose. For the E177Q–X6 complex, the initial hit for de novo crystallization only produced crystal clusters, which could not be separated (Index No. 88: 0.2 M ammonium citrate tribasic pH 7.0, 20% PEG 3350, 50 mM xylohexaose). To obtain single crystals of this complex, multiple rounds of microseeding were performed: the crystal clusters were ground thoroughly in a 2 µl reservoir solution to obtain tiny seeds. The seed solution was diluted 10 000-fold to make a stock. About 0.2 µl of the diluted seed stock was added into a 2 µl crystallization solution with decreased precipitant concentration, which was pre-equilibrated overnight.
2.3. Data collection and analysis
Crystals were transferred into a reservoir solution with an additional 25% glycerol as the cryoprotectant for a few seconds before being flash-cooled by direct immersion into liquid nitrogen. Data sets were collected on beamline ID19 at 100 K at the Advanced Photon Source, Argonne, IL. Diffraction data were processed using HKL-2000 (Otwinowski & Minor, 1997 ▶), then transformed from the SCA format to the MTZ format with 5% of the data flagged as the test set using the CCP4 program suite (Winn et al., 2011 ▶). Structures were solved by the molecular-replacement method using Phaser (McCoy et al., 2007 ▶) in the program suite Phenix (Adams et al., 2010 ▶). The initial phasing information and the structure solution came from the Xyn II structure (PDB code 2dfb; Watanabe et al., 2006 ▶).
3. Results and discussion
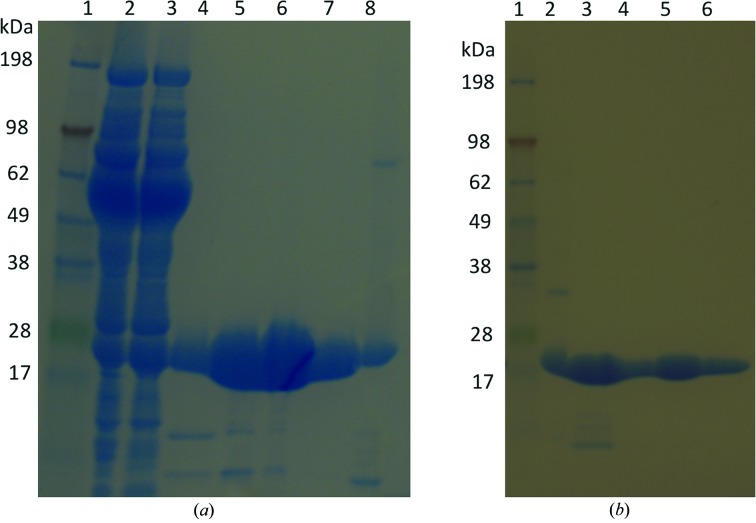
T. reesei cells express Xyn II with a leader peptide that is cleaved during secretion, producing a 21 kDa protein with an N-terminal pyroglutamate residue. Previously, this protein has been expressed in E. coli fused to a 36-amino-acid tag at the N-terminus (He et al., 2009 ▶) or an N-terminal periplasmic signal peptide (Le et al., 2011 ▶). We designed new expression vectors to produce high levels of proteins without N-terminal extensions for crystallography. Following Xyn II expression in E. coli BL21-gold cells grown in LB medium, SDS–PAGE analysis showed that most expressed protein partitioned into the insoluble portion of cell lysate, probably forming inclusion bodies as reported for a previous cytosolic expression system (Le et al., 2011 ▶). However, a substantial portion of the protein remained soluble and active inside cells grown in medium containing 50 mM glycylglycine (in LB) or glycerol [in Terrific Broth or Enfors minimal medium (Törnkvist et al., 1996 ▶)] (Fig. 1 ▶ a). Therefore, protein for crystallography was expressed using Enfors minimal medium at 295 K.
Figure 1.
SDS–PAGE analysis of Xyn II purification. (a) SP cation-exchange chromatography: lane 1, protein ladder; lane 2, cell lysate; lane 3, supernatant; lanes 4–8, eluted peaks. (b) Size-exclusion chromatography: lane 1, protein ladder; lanes 2–6, different eluted peaks.
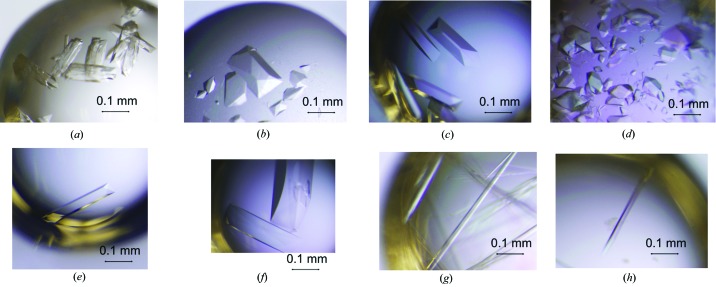
Our initial expression vectors encoded proteins with C-terminal His6 tags for affinity purification. However, using conditions that successfully produced crystals of native Xyn II protein (Kovalevsky et al., 2011 ▶), the His6-tagged rWT protein could only be crystallized in the ligand-free form as clusters that could not be separated without damaging the crystal quality (Fig. 2 ▶ a). Therefore, constructs without the His6 tag were used for protein expression and we designed a new purification protocol for the untagged proteins.
Figure 2.
Crystal photos of rWT and mutants. (a) rWT with N-terminal His tag. (b) rWT without His tag. (c) N44D in the ligand-free state. (d) N44H in the ligand-free state. (e) E177Q in the ligand-free state. (f) E86Q in the ligand-free state. (g) N44H–X3 complex. (h) E177Q–X6 complex crystals obtained after multiple rounds of microseeding.
Although most E. coli proteins have low pI values (Han & Lee, 2006 ▶), we calculated a value of 8.7 for Xyn II from its amino-acid sequence. We exploited this significant difference in pI between Xyn II and bacterial proteins to use sulfopropyl (SP) cation-exchange chromatography as the first purification step, achieving purities above 80% (Fig. 1 ▶ a). We then used size-exclusion chromatography for final purification. After this two-step purification process, SDS–PAGE analysis showed that the final purity was above 90% (Fig. 1 ▶ b).
The ligand-free form of rWT Xyn II and its variants (N44D, N44H, E177Q and E86Q) were crystallized in the space group P212121 with one molecule in the asymmetric unit and similar asymmetric unit volumes (Fig. 2 ▶). Because N44H is still catalytically active, when N44H was co-crystallized with xylohexaose, we only found xylotriose bound in the active site. The asymmetric unit volume for the N44H–X3 crystal was significantly smaller, because of the reduction in a and c cell dimensions (Table 1 ▶) compared to its ligand-free form, indicating a more compact, closed conformation for the enzyme’s fold and reduced water content. In contrast, the E177Q–X6 complex was crystallized with different cell dimensions in space group P21221 and the asymmetric unit contained one molecule. This crystal therefore lost a screw axis. After molecular replacement with the structure 2dfb and structure refinement, we found clear electron density showing the presence of six xylose units of the intact X6. Symmetry analysis shows that the reducing terminus of X6 would have a steric clash with one of its symmetry-related neighbors if the screw axis were still present (Fig. 3 ▶) forcing adoption of a simple twofold rotation axis.
Table 1. Statistics of data processing.
Numbers in parentheses represent values in the highest resolution.
| Crystals | E177Q ligand-free | E177Q–X6 | N44H ligand-free | N44H–X3 | rWT ligand-free | N44D ligand-free | E86Q ligand-free |
|---|---|---|---|---|---|---|---|
| Space group | P212121 | P21221 | P212121 | P212121 | P212121 | P212121 | |
| Unit-cell dimensions (Å, °) | a = 48.2, b = 59.2, c = 69.7 | a = 38.5, b = 45.5, c = 111.4 | a = 48.3, b = 59.0, c = 69.7 | a = 42.4, b = 59.6, c = 62.1 | a = 47.8, b = 59.7, c = 69.7 | a = 48.3, b = 59.6, c = 69.9 | a = 48.5, b = 59.3, c = 69.9 |
| α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 | |
| Molecules per asymmetric unit | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Wavelength (Å) | 1.5417 | 0.9793 | 0.9793 | 0.9793 | 1.54 | 0.9793 | 0.9793 |
| Resolution (Å) | 1.5 | 1.15 | 1.10 | 1.55 | 1.65 | 1.07 | 1.07 |
| Unique reflections | 32599 | 63877 | 81313 | 23195 | 24538 | 88792 | 86222 |
| Redundancy | 3.7 (3.4) | 6.5 (6.9) | 4.3(3.7) | 3.8 (3.6) | 4.0 (3.8) | 3.8 (3.2) | 5.1 (3.2) |
| Completeness (%) | 99.6 (97.5) | 90.8 (84.2) | 99.8 (98.6) | 98.5(99.7) | 99.4 (98.8) | 99.4 (98.3) | 96.1(96.6) |
| R sym † | 0.069 (0.408) | 0.070 (0.622) | 0.051(0.629) | 0.099(0.521) | 0.093 (0.651) | 0.051 (0.260) | 0.127(0.398) |
| 〈I/σ(I)〉 | 15.2 (2.8) | 27.6 (3.4) | 26.7(1.9) | 13.8(2.7) | 19.2 (2.1) | 19.6 (4.2) | 12.4(2.4) |
| Mosaicity (°) | 0.57 | 0.47 | 0.44 | 0.72 | 1.87 | 0.12 | 1.30 |
R
sym = 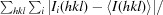
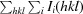 , where Ii is the measured intensities and 〈I〉 is the mean intensity of all measured observations equivalent to reflection Ii.
, where Ii is the measured intensities and 〈I〉 is the mean intensity of all measured observations equivalent to reflection Ii.
Figure 3.
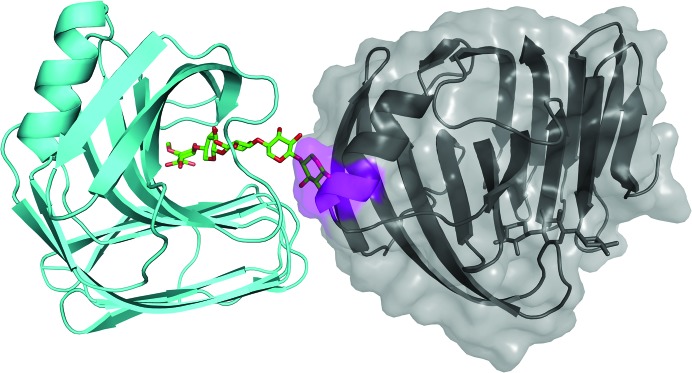
In the crystal structure of the E177Q–X6 complex, a xylose at one end of the active site would protrude into a neighboring molecule according to the original space group P212121. Therefore, one screw axis will change to a rotational axis to prevent such packing collision and the new space group is P21221.
Our results present the first crystallographic study of Xyn II with active-site amino acids replaced in order to obtain complexes with substrate and product. The X-ray diffraction data are of very high resolution for GH11 family xylanases and will undoubtedly provide insight into the mechanism of retaining glycosidases. We also expect that the new expression, purification and crystallization protocols developed in this study will provide a detailed understanding of why activity and substrate affinity drop sharply at high pH values. Refinement of the above structures is in process and the detailed structural analysis will be reported in a separate publication.
Acknowledgments
We thank the staff at the ID19 beamline at the Advanced Photon Source at Argonne National Laboratory for assistance with data collection. Argonne is operated by the University of Chicago Argonne, LLC, for the US Department of Energy’s Office of Science under contract No. DE-AC02-06CH11357. This research was supported by the Laboratory Directed Research and Development Program (LDRD) at Oak Ridge National Laboratory, which is managed by UT-Battelle, LLC, for the US Department of Energy’s Office of Science under contract No. DE-AC05-00OR22725. This research used facilities provided by Oak Ridge National Laboratory’s Center for Structural Molecular Biology (CSMB), which is supported by the Office of Biological and Environmental Research in the DOE Office of Science.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Cherry, J. R. & Fidantsef, A. L. (2003). Curr. Opin. Biotechnol. 14, 438–443. [DOI] [PubMed]
- Gupta, R., Kim, T. H. & Lee, Y. Y. (2008). Appl. Biochem. Biotechnol. 148, 59–70. [DOI] [PubMed]
- Han, M. & Lee, S. Y. (2006). Microbiol. Mol. Biol. Rev. 70, 362–439. [DOI] [PMC free article] [PubMed]
- Havukainen, R., Torronen, A., Laitinen, T. & Rouvinen, J. (1996). Biochemistry, 35, 9617–9624. [DOI] [PubMed]
- He, J., Yu, B., Zhang, K. Y., Ding, X. M. & Chen, D. W. (2009). Protein Expr. Purif. 67, 1–6.
- Kluepfel, D., Shareck, F., Senior, D. J., Bernier, R. L. & Morosoli, R. (1993). Industrial Microorganisms: Basic and Applied Molecular Genetics, pp. 137–142, edited by R. H. Batlz, G. Hegeman & P. L. Skatrud. Washington DC: ASM Press.
- Kovalevsky, A. Y., Hanson, B. L., Seaver, S., Fisher, S. Z., Mustyakimov, M. & Langan, P. (2011). Acta Cryst. F67, 283–286. [DOI] [PMC free article] [PubMed]
- Kuhls, K., Lieckfeldt, E., Samuels, G. J., Meyer, W., Kubicek, C. P. & Borner, T. (1997). Mycologia, 89, 442–460.
- Langan, P., Gnanakaran, S., Rector, K. D., Pawley, N., Fox, D. T., Cho, D. W. & Hammel, K. E. (2011). Energ. Environ. Sci. 4, 3820–3833.
- Le, Y. L., Peng, J. J., Wu, H. W., Sun, J. Z. & Shao, W. L. (2011). PLos ONE, 6, e18489. [DOI] [PMC free article] [PubMed]
- Li, K. C., Azadi, P., Collins, R., Tolan, J., Kim, J. S. & Eriksson, K. L. (2000). Enzyme Microb. Technol. 27, 89–94. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Method Enzymol, 276, 307–326. [DOI] [PubMed]
- Polizeli, M. L. T. M., Rizzatti, A. C. S., Monti, R., Terenzi, H. F., Jorge, J. A. & Amorim, D. S. (2005). Appl. Microbiol. Biotechnol. 67, 577–591. [DOI] [PubMed]
- Saha, B. C. (2003). J. Ind. Microbiol. Biotechnol. 30, 279–291. [DOI] [PubMed]
- Törnkvist, M., Larsson, G. & Enfors, S. O. (1996). Bioprocess Eng. 15, 231–237.
- Törrönen, A., Harkki, A. & Rouvinen, J. (1994). EMBO J. 13, 2493–2501. [DOI] [PMC free article] [PubMed]
- Torronen, A., Mach, R. L., Messner, R., Gonzalez, R., Kalkkinen, N., Harkki, A. & Kubicek, C. P. (1992). Bio-Technol, 10, 1461–1465. [DOI] [PubMed]
- Watanabe, N., Akiba, T., Kanai, R. & Harata, K. (2006). Acta Cryst. D62, 784–792. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zolotnitsky, G., Cogan, U., Adir, N., Solomon, V., Shoham, G. & Shoham, Y. (2004). Proc. Natl Acad. Sci. USA, 101, 11275–11280. [DOI] [PMC free article] [PubMed]