Abstract
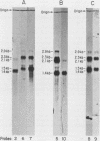
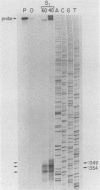
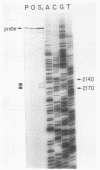
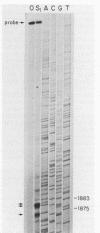
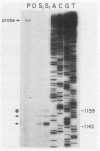
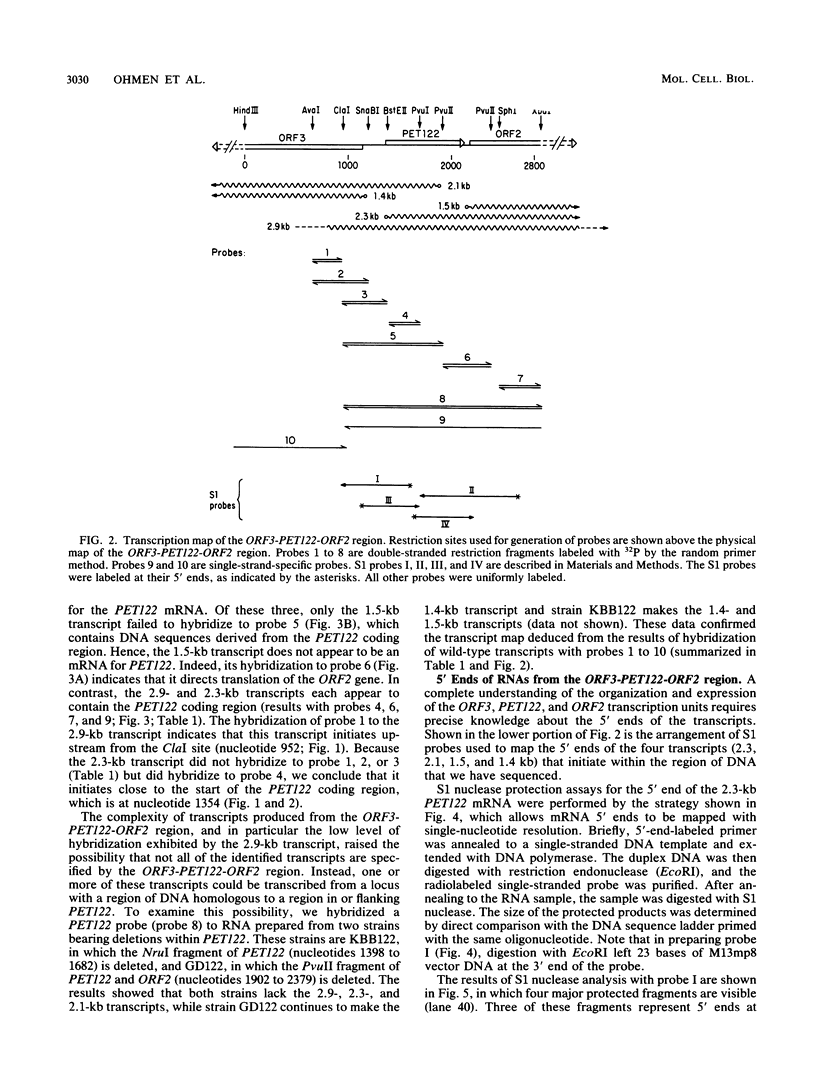
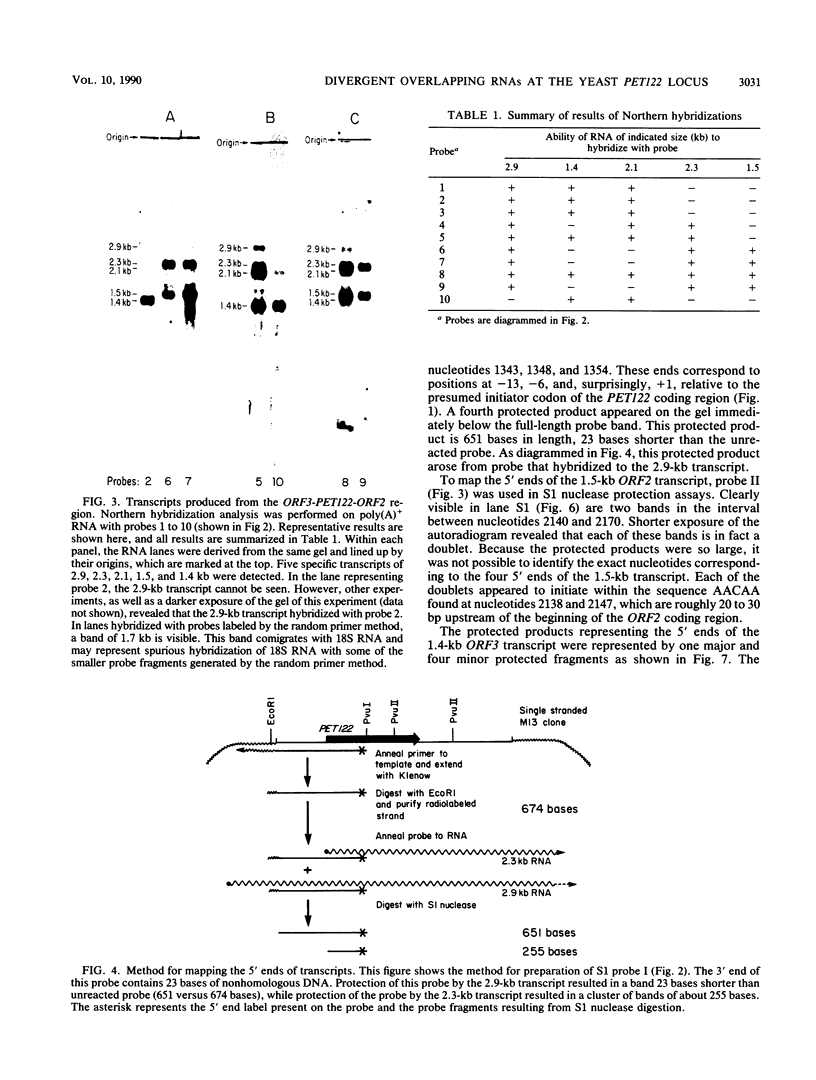
PET122 is one of three nuclear genes specifically required for translation of the mitochondrial mRNA for cytochrome c oxidase subunit III in Saccharomyces cerevisiae. The nucleotide sequence of 2,862 base pairs (bp) of yeast genomic DNA encompassing the PET122 locus shows very close spacing between the PET122 gene (254 codons) and two unidentified open reading frames, termed ORF2 and ORF3. ORF2 is encoded by the same strand of DNA as PET122 and is located 53 bp downstream of PET122, while ORF3 is encoded on the opposite strand and is located 215 bp upstream of PET122. Five transcripts, with sizes of 2.9, 2.3, 2.1, 1.5, and 1.4 kilobases (kb), are produced from this locus. The 2.1- and 1.4-kb transcripts encode ORF3, the 1.5-kb transcript encodes ORF2, and the 2.9- and 2.3-kb transcripts encode PET122. A particularly interesting feature of the ORF3-PET122-ORF2 transcription unit is a 535-base overlap between the 2.3-kb PET122 transcript produced from one strand and a 2.1-kb ORF3 transcript produced from the opposite strand. Similarly, the 2.9-kb PET122 transcript overlaps the 2.1-kb ORF3 transcript by more than 900 bases and the 1.5-kb ORF3 transcript by at least 200 bases. Hence, these pairs of transcripts are antisense to one another and have the potential to regulate, in an interdependent fashion, the posttranscriptional expression of ORF3 and PET122.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Beck C. F., Warren R. A. Divergent promoters, a common form of gene organization. Microbiol Rev. 1988 Sep;52(3):318–326. doi: 10.1128/mr.52.3.318-326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Donahue T. F. Sequence and structural features associated with translational initiator regions in yeast--a review. Gene. 1987;59(1):1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- Clements J. M., Laz T. M., Sherman F. Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Oct;8(10):4533–4536. doi: 10.1128/mcb.8.10.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Specific translational activation by nuclear gene products occurs in the 5' untranslated leader of a yeast mitochondrial mRNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2677–2681. doi: 10.1073/pnas.85.8.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman J. C., van Heeswijk W. C., Grivell L. A. Identification of two factors which bind to the upstream sequences of a number of nuclear genes coding for mitochondrial proteins and to genetic elements important for cell division in yeast. Nucleic Acids Res. 1988 Aug 11;16(15):7287–7301. doi: 10.1093/nar/16.15.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone C., Agostinelli M., Frontali L. Mitochondrial translation products during release from glucose repression in Saccharomyces cerevisiae. J Bacteriol. 1983 Mar;153(3):1125–1132. doi: 10.1128/jb.153.3.1125-1132.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989 Aug;3(8):1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Hahn S., Pinkham J., Wei R., Miller R., Guarente L. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol Cell Biol. 1988 Feb;8(2):655–663. doi: 10.1128/mcb.8.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrin D. L., Schmidt G. W. Rapid, reversible staining of northern blots prior to hybridization. Biotechniques. 1988 Mar;6(3):196-7, 199-200. [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloeckener-Gruissem B., McEwen J. E., Poyton R. O. Identification of a third nuclear protein-coding gene required specifically for posttranscriptional expression of the mitochondrial COX3 gene is Saccharomyces cerevisiae. J Bacteriol. 1988 Mar;170(3):1399–1402. doi: 10.1128/jb.170.3.1399-1402.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloeckener-Gruissem B., McEwen J. E., Poyton R. O. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae: multiple trans-acting nuclear genes exert specific effects on expression of each of the cytochrome c oxidase subunits encoded on mitochondrial DNA. Curr Genet. 1987;12(5):311–322. doi: 10.1007/BF00405753. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig A., Padmanaban G., Rabinowitz M. Regulation of the nuclear-coded peptides of yeast cytochrome c oxidase. Biochemistry. 1982 Jan 19;21(2):309–316. doi: 10.1021/bi00531a017. [DOI] [PubMed] [Google Scholar]
- McEwen J. E., Ko C., Kloeckner-Gruissem B., Poyton R. O. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants in 34 complementation groups. J Biol Chem. 1986 Sep 5;261(25):11872–11879. [PubMed] [Google Scholar]
- Ohmen J. D., Kloeckener-Gruissem B., McEwen J. E. Molecular cloning and nucleotide sequence of the nuclear PET122 gene required for expression of the mitochondrial COX3 gene in S. cerevisiae. Nucleic Acids Res. 1988 Nov 25;16(22):10783–10802. doi: 10.1093/nar/16.22.10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M. A., Gould J., Kim S., Kane M. Y., Hereford L. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986 May 23;45(4):537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984 Jul;37(3):969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- Simons R. W. Naturally occurring antisense RNA control--a brief review. Gene. 1988 Dec 10;72(1-2):35–44. doi: 10.1016/0378-1119(88)90125-4. [DOI] [PubMed] [Google Scholar]
- Smith M. M., Andrésson O. S. DNA sequences of yeast H3 and H4 histone genes from two non-allelic gene sets encode identical H3 and H4 proteins. J Mol Biol. 1983 Sep 25;169(3):663–690. doi: 10.1016/s0022-2836(83)80164-8. [DOI] [PubMed] [Google Scholar]
- Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu Rev Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M., Manzara T., Pichersky E., Cashmore A., Gruissem W. Genomic organization, sequence analysis and expression of all five genes encoding the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase from tomato. Mol Gen Genet. 1987 Sep;209(2):247–256. doi: 10.1007/BF00329650. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Myers A. M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Tzamarias D., Roussou I., Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989 Jun 16;57(6):947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zennaro E., Grimaldi L., Baldacci G., Frontali L. Mitochondrial transcription and processing of transcripts during release from glucose repression in 'resting cells' of Saccharomyces cerevisiae. Eur J Biochem. 1985 Feb 15;147(1):191–196. doi: 10.1111/j.1432-1033.1985.tb08736.x. [DOI] [PubMed] [Google Scholar]
- Zitomer R. S., Walthall D. A., Rymond B. C., Hollenberg C. P. Saccharomyces cerevisiae ribosomes recognize non-AUG initiation codons. Mol Cell Biol. 1984 Jul;4(7):1191–1197. doi: 10.1128/mcb.4.7.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol A. R., Mol J. N., Stuitje A. R. Modulation of eukaryotic gene expression by complementary RNA or DNA sequences. Biotechniques. 1988 Nov-Dec;6(10):958–976. [PubMed] [Google Scholar]