Abstract
Light chain amyloidosis is one of the unique examples within amyloid diseases where the amyloidogenic precursor is a protein that escapes the quality control machinery and is secreted from the cells to be circulated in the bloodstream. The immunoglobulin light chains are produced by an abnormally proliferative monoclonal population of plasma cells that under normal conditions produce immunoglobulin molecules such as IgG, IgM or IgA. Once the light chains are in circulation, the proteins misfold and deposit as amyloid fibrils in numerous tissues and organs, causing organ failure and death. While there is a correlation between the thermodynamic stability of the protein and the kinetics of amyloid formation, we have recently found that this correlation applies within a thermodynamic range, and it is only a helpful correlation when comparing mutants from the same protein. Light chain amyloidosis poses unique challenges because each patient has a unique protein sequence as a result of the selection of a germline gene and the incorporation of somatic mutations. The exact location of the misfolding process is unknown as well as the full characterization of all of the toxic species populated during the amyloid formation process in light chain amyloidosis.
Keywords: light chain amyloidosis, immunoglobulin light chain, somatic mutations, amyloid formation, dimer
Introduction
In order to understand the molecular determinants that drive light chain amyloidosis, it is important to understand the structural basis of immunoglobulin light chains. There have been some recent reviews and book chapters that have covered these topics extensively [1, 2]. Some work has been done to understand the nature of the gene selection that may contribute to the amyloidogenic potential of light chains, which has been reviewed as well. From the point of view of medicinal chemistry, it is important to understand the structure and thermodynamic stability of immunoglobulin light chains, in particular those that are amyloidogenic. While this topic has also been extensively surveyed, this review will explore new understanding of the complex interplay between thermodynamic stability and amyloidogenic potential. We will also examine the recent advances made in the perception of the nature of the toxic species and the numerous cellular, tissue, and animal models that have been used to understand this misfolding disease.
Immunoglobulin structure
Immunoglobulin molecules, such as IgG, which is the simplest form of an immunoglobulin, consists of a heterotetramer formed by two heterodimers of an immunoglobulin light chain and an immunoglobulin heavy chain linked together via disulfide bonds. The immunoglobulin heavy chain has a variable domain and three different constant domains in an IgG molecule, while the immunoglobulin light chain has a variable domain and only one constant domain. The source of sequence variability in immunoglobulin molecules comes from combinatorial pairing of the variable (V) genes (discussed below) and the junction (J) genes, making it possible to generate at least 90,000 different heavy chains and 3000 different light chain sequences. In addition, somatic mutations improve the antibody affinity for the antigen, leading to further immunoglobulin sequence variation. An immunoglobulin light chain is composed of an N-terminal variable domain and a C-terminal constant domain. There are two major light chain variable domain families: kappa and lambda. The light chain variable domains are not uniformly variable throughout their lengths. Three small regions, the hypervariable regions or complementarity determining regions (CDR), show much more variability than the rest of the domain. These regions vary both in size and in sequence among different VL germline isotypes; they determine the specificity of the antigen-antibody interactions.
The remaining regions of the light chain variable domain, four framework regions (FRs), have homologous amino acid sequences, particularly within the kappa and lambda families.
The overall structure of the immunoglobulin variable domain is an immunoglobulin fold with 9 β-strands (A, B, C, C’, C”, D, E, F, and G) packed tightly against each other in two antiparallel β-sheets joined together by a disulfide bridge in a form of a Greek key β-barrel. The immunoglobulin constant domain is simpler than the variable domain, lacking β-strands C’ and C” and sharing great structural homology with immunoglobulin fold domains such as β2-microglobulin. The N- and C- termini strands (A and G, respectively) interact in a parallel manner [3]. The CDRs form three unstructured loops between amino acids 24-34, 50-56 and 89-95 that contain the sequence that will recognize the antigen, and as we mentioned above, are highly variable and acquire a high rate of somatic hypermutation. Within immunoglobulin structures such as IgG, the light chain variable domain interacts with the heavy chain variable domain through β-strands C, C’, F and G. Immunoglobulin light chains are secreted from plasma cells and are found in circulation. When these proteins are found in urine, they are sometimes referred to as Bence-Jones proteins. Heavy chains are unable to be secreted alone due to their structural complexity, so they are always present in circulation as part of an intact immunoglobulin molecule complexed with light chains.
Germline sequence bias and somatic mutations
There are 40 kappa and 33 lambda germline genes available to form a light chain variable domain. In addition, there are 5 kappa Junction (JK) and 4 lambda Junction (JL) genes. The Junction genes make up β-strand G within the variable domain structure, so they are essential for the correct folding of the domain and add to the overall variability of the protein. In AL amyloidosis, there is an overrepresentation of specific germline genes: κI, λ1, λII, λIII, and λVI [4]. As we mentioned before, the process of somatic hypermutation adds to the complexity of AL amyloidosis because it means that each patient possesses a unique amyloidogenic protein sequence: a combination of different germline genes and different somatic mutations at different positions of the protein and mutations to different amino acids. These protein sequence variations could result in varying propensities to form toxic species and form amyloid fibrils, and may be involved in the diverse organ involvement found in different AL patients as well as the range of severity of the disease. In AL, λ is overrepresented (λ/κ=3:1) as compared to healthy individuals or multiple myeloma (MM) patients (λ/κ=1:2) [5]. In addition, the light chain variable domain germline donor gene usage in AL is biased even within the overrepresented germline genes [6-10]. The three studies by Comenzo, Abraham and Prokaeva agree that in AL amyloidosis, the light chain variable germline donor gene usage comprises VλII 2a2, VλIII 3r, VλVI 6a, and VλI O18/O8. There are slight differences in the sample size, sample selection, and the frequency of use of each germline donor gene in each of these studies. Comenzo and co-workers demonstrated 30% of AL VL genes used the VλVI 6a germline donor [8]. Abraham and co-workers found that most κ patients selected for their study used the VκI subgroup (77%) [7]. A similar observation has been made by Prokaeva and co-workers [9]. While there are some strong correlations with these small subset of germline donor genes and AL amyloidosis, none of these are absolute and the determinants for AL amyloidosis are most certainly multifactorial.
The mutational diversity among AL proteins has been well documented. Several studies have compared amino acid sequences of AL proteins, searching for common mutations or regions of high mutational frequency. In an analysis of 121 κI light chains (37 of which were amyloidogenic), Stevens found four structural features that render a light protein more likely to be amyloidogenic [11]. All of these structural features involved loss or gain of certain residues, including a mutation that introduces a glycosylation site, mutations of Arg61 or Ile27b, and mutations of Pro residues in β-turns.
More recently, we undertook a sequence study with 141 κ and λ AL light chain sequences to catalogue the non-conservative mutations in these proteins and modeled their positions onto known light chain structures to correlate structural regions (β-strands or loops) with potentially destabilizing mutations [4]. This study confirmed that the total number of non-conservative mutations may be less important than their location as an amyloidogenic determinant for light chain proteins. Additionally, the patients’ free light chain levels, an indicator of disease progression [12], were also assessed in a subset of the analyzed protein sequences. A correlation between non-conservative mutations in certain regions and free light chain levels was revealed, suggesting that patients with initial low free light chain levels have acquired mutations in their light chains that rendered these proteins to be more amyloidogenic than light chains from patients with higher free light chain levels. We propose that analyzing the location of these mutations in large numbers of AL protein sequences could further advance our understanding of the determinants for amyloidogenicity and lead to a prognostic factor for AL amyloidosis disease progression.
Protein thermodynamic stability and amyloidogenesis
Studies using variable domain proteins from AL patients have shown that mutations in the variable domain that reduce thermodynamic stability are more prone to form amyloid fibrils, most likely due to the population of partially folded states [13-15]. In an analysis linking mutations and stability, Hurle et al. analyzed 36 sequences (18 κ and 18 λ) in search of rare amino acid replacements that occurred in structurally significant regions of the proteins [13]. They proceeded to create single-point mutants incorporating the rare residues into a non-amyloidogenic Bence Jones light chain protein to determine whether the amino acids destabilized the protein significantly enough to induce unfolding. Four of the six mutations were destabilizing, leading to the conclusion that some mutations are involved in amyloidogenicity. Further evidence comes from reports that show that AL proteins are less stable than their non-amyloidogenic counterparts, such as multiple myeloma proteins [16, 17]. There are several possible sources of protein destabilization for AL proteins: 1) somatic mutations (particularly mutations that result in non-conservative amino acid changes) that cause the protein to sample partial unfolded states, 2) loss of the interaction with the heavy chain due to mutations, and 3) environmental factors within the cell such as pH and accessory molecules. The general conclusion from these studies is that somatic mutations have a global destabilizing effect on AL proteins and as a consequence these proteins require less energy to unfold [13-15]. The propensity to form amyloid fibrils in vitro for some immunoglobulin variable domains appears to be inversely correlated with their free energy of unfolding, suggesting that both stabilizing and destabilizing interactions within the immunoglobulin variable domain can influence the kinetics of amyloid fibril formation [16-18].
To determine if the germline sequences, particularly those over-represented in AL amyloidosis, are prone to generating inherently more amyloidogenic AL proteins, two studies tested κ and λ germline proteins. Baden et al. compared AL-09, an amyloidogenic protein that has 7 somatic mutations, to its germline protein κI O18/O8 [19]. The germline protein was more thermodynamically stable than its amyloidogenic counterpart, and although κI O18/O8 was able to form fibrils, its fibril formation kinetics were significantly slower than AL-09. Additionally, fibril formation of AL protein BIF and MM protein GAL (also of the κI O18/O8 germline) was compared at 37°C, but only BIF formed fibrils [16].
Because the λ6a germline is expressed almost exclusively in AL amyloidosis patients (it is one of the last germline genes screened in the process of germline gene selection) and is rarely found in the normal light chain repertoire [7-9], del Pozo Yauner et al. hypothesized that this germline would be as unstable as AL proteins. However, experiments revealed that the λ6a germline protein was more stable than Wil, an AL protein from that germline with 11 somatic mutations [20]. The λ6a germline also had significantly slower fibril formation kinetics than Wil.
Which germline protein is more amyloidogenic? When compared to AL-09 and κI O18/O8, Wil and λ6a demonstrated a comparable increase in thermodynamic stability, but faster amyloid fibril formation kinetics (14 hours for λ6a compared to 216 hours for κI O18/O8 at 37°C). Additionally, λ6a was able to form fibrils in the absence of seeds, while κI O18/O8 required seeds for fibril formation. This may indicate an increase in amyloidogenic potential for the λ6a germline and for any AL proteins derived from this germline gene, supporting the observation that the λ6a germline is over represented in AL amyloidosis patients. More studies are necessary to verify that AL amyloidosis prone germline sequences are more amyloidogenic than normal immunoglobulin repertoire germline sequences.
To determine if a single mutation is enough to render a protein amyloidogenic, Davis et al. studied AL protein SMA and MM protein LEN, belonging to the kIV germline. Only eight residues differ between these two proteins, and each SMA mutation was introduced into LEN to assess the individual effects on fibrillogenesis. Of the mutations tested, only P40L, located in a loop region between β-strand C and C’, was able to form Thioflavine T (ThT) positive fibrils in unseeded reactions [21]. Although stability data were not reported for these mutants, it is likely that the P40L mutant was less stable than wild-type LEN because Pro40 (very favorable for loops and turns) is conserved among 98% of all κ and λ germline sequences.
In vitro fibril formation studies have revealed that AL proteins form fibrils under a variety of solution conditions with varying kinetics and morphology of fibrils. AL-09 is unique because it forms amyloid fibrils with very similar kinetics across a wide variety of solution conditions [22]. Additionally, AL-09's fibril formation kinetics are significantly faster than other AL proteins, particularly those derived from κI O18/O8. We propose that the altered dimer interface populated by AL-09 (discussed at length in the next section) facilitates the initial misfolding events that trigger amyloid fibril formation, while the other proteins require stochastic conformational fluctuations to populate the appropriate misfolded intermediate that leads the amyloid formation reaction. In the case of κIV light chains from both amyloidosis and multiple myeloma patients, it has been shown that amyloid formation is enhanced at low pH while amorphous aggregation occurred around neutral pH; all of these reactions populated different partially folded intermediates [23-27].
Another link between thermodynamic stability and fibril formation is found in the recently analyzed κI O18/O8 and λ6a germline proteins. These proteins were significantly more stable than all AL amyloidogenic proteins that have been studied to date [19, 20]. The Tm values (melting temperatures defined as the temperature in which 50% of the proteins are unfolded) for the germline proteins were increased by 15°C and 11.6°C, respectively, over the corresponding AL proteins analyzed in each study. Both κI O18/O8 and λ6a germline proteins had slower fibril formation kinetics than their amyloidogenic counterparts.
Del Pozo Yauner and colleagues incorporated an R25G mutation into the λ6a germline protein with junction region JL2 (6aJL2-R25G), as this mutation is found in 25% of amyloidogenic λ6 light chains and presumably represents an allotypic variant [20, 28, 29]. This mutation resulted in a 6°C decrease in Tm value for the mutated protein, and 6aJL2-R25G fibril formation had a much shorter lag time and faster growth rate than the λ6a germline protein. The authors explain that the R25G mutation may affect the structure of complementarity determining region 1 (CDR1), resulting in an altered conformation and increased amyloidogenicity [29].
Further research on the κI O18/O8 germline protein and amyloidogenic AL-09 also connected thermodynamic stability to fibril formation. Baden et al. undertook a systematic restorative mutational analysis of the non-conservative mutations of AL-09, which are all located in the dimer interface [18]. Of the three non-conservative restorative mutations (I34N, Q42K and H87Y), restoring the His87 mutation to the Tyr87 residue found in the germline sequence increased the thermodynamic stability and decreased the fibril formation kinetics to the same levels as κI O18/O8. Significant structural alterations were also observed with this restorative mutant (as is discussed in the section below). Restoring the Asn34 residue had intermediate effects on stability and fibril formation propensity, while reintroducing Lys42 did not appear to alter the thermodynamics to any extent.
In complementary experiments introducing the His87 residue from the amyloidogenic protein into κI O18/O8, this protein was only destabilized half as much as AL-09. κI Y87H also had intermediate fibril formation kinetics between those measured for κI O18/O8 and AL-09, indicating that this mutation alone may not have been sufficient for the amyloidogenicity observed in AL-09. However, introducing a second mutation into κI O18/O8 (Ile34, in addition to His87) completely destabilized the protein and exhibited the same fast fibril formation kinetics as amyloidogenic AL-09. These results suggest that rather than a single mutation that causes amyloidogenesis, it is probable that a combination of destabilizing and compensatory mutations leads to fibrillogenicity among AL proteins.
In a recent comparative analysis we conducted with amyloidogenic light chain proteins from AL amyloidosis patients we have observed that the correlation between thermodynamic stability and kinetics of fibril formation is maintained within a certain range. One of the proteins in our analysis, AL-T03 (protein from a patient that received autologous stem cell transplant for treatment) appears to be the least thermodynamically stable of all the AL proteins -to the best of our knowledge- with a Tm of 29.8°C. AL-T03 is incapable of forming amyloid fibrils in vitro at pH 7.4 and is only able to form fibrils marginally at pH 2. Why is the AL-T03 protein incapable of forming fibrils at pH 7.4, 37°C? Under these experimental conditions, the protein is primarily unfolded, so it marginally populates partially-folded states that trigger amyloid fibril formation (Poshusta, Katoh, Gertz, Dispenzieri and Ramirez-Alvarado, unpublished observations). These studies suggest that lack of stability per se does not trigger amyloid fibril formation. The protein is required to populate a critical concentration of partially folded states to sustain amyloid fibril formation.
Other groups have studied fibril formation using different AL and MM proteins. Jto, an MM protein, and Wil, an AL protein, are both light chain proteins from the λ6a germline that differ by 19 amino acids. Fibrils were formed with both Jto and Wil at 37°C, pH 7.5 [17]. Jto displayed slower kinetics than Wil. In addition, Jto fibrils appeared more rigid and shorter than fibrils formed by Wil. Certain ionic interactions may affect fibrillogenesis and be crucial to maintain the structure and stability of light chain proteins. Wall et al. noted an ionic interaction between Asp29 and Arg68 in MM protein Jto, whereas AL protein Wil has neutral amino acids in these positions [30]. To test the importance of this ionic interaction, mutations were made to Jto to introduce the neutral residues (from Wil) at these sites (JtoD29A, JtoR68S). The thermodynamic stability of these mutants was the same, and the rate of fibril formation for JtoD29A was the same as that for Jto. However, fibril formation kinetics were much faster for JtoR68S, and the X-ray crystal structure of this mutant revealed several side-chain differences compared to Jto and JtoD29A. These differences changed the electrostatic potential surface and increased the amount of solvent-exposed hydrophobic surface for the protein. These results highlight critical structural features such as ionic interactions that participate in the stability and fibrillogenicity of AL proteins.
As we mentioned before, immunoglobulin light chains are composed of a variable domain and a constant domain [31]. For many years, AL amyloidosis researchers have conducted their biochemical and biophysical studies with the variable domain based on a report that stated that amyloid deposits from AL amyloidosis patients were formed primarily by the variable domain and small regions of the constant domain [32]. Lavatelli et al. described in 2008 the proteomics analysis of fat aspirates from 6 lambda and 1 kappa AL amyloidosis patients. In these samples, full length light chains were observed in the amyloid deposits [33]. In 2009, evidence from Vrana et al. demonstrated with a large cohort of AL amyloidosis patient tissue samples (25 lambda and 5 kappa) that full length proteins are present in the amyloid deposits of these patients [34] confirming the Lavatelli results from fat aspirates, and suggesting that while some patients may have amyloid deposits formed of truncated light chains, many patients may have full length light chains in their deposits. The function of the constant domain in the pathophysiology of AL amyloidosis is not well established. Only recently, the role of constant domain in thermal stability and aggregation has been characterized for a λ6a full length AL protein and its corresponding variable and constant domain proteins [35]. This has prompted our laboratory to currently explore the behavior of full length AL proteins and compare it to studies we have previously conducted with their corresponding variable domains.
Structural determinants of amyloidogenicity in light chain proteins
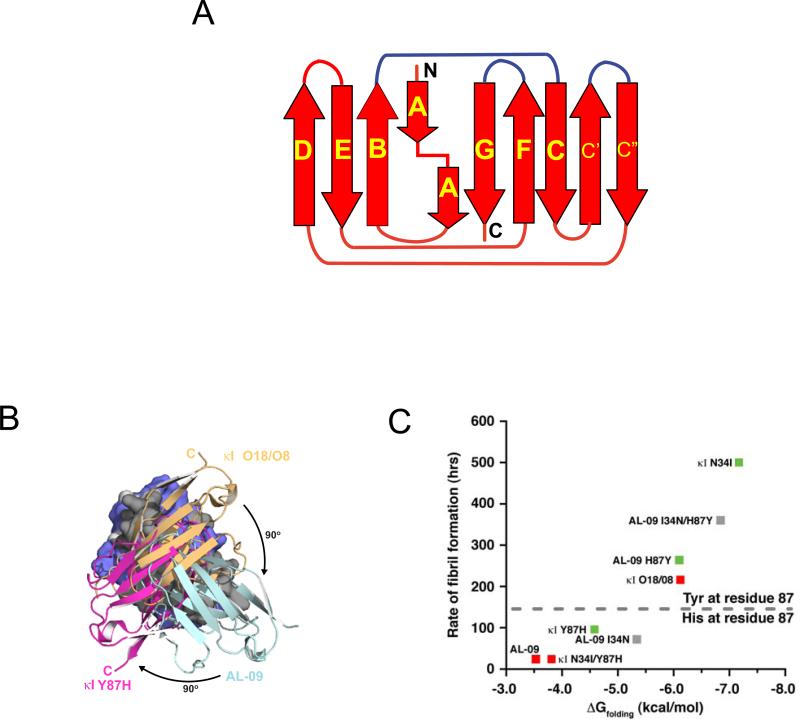
Structural studies have shown that most variable domains from AL amyloidosis patients crystallize as monomers or dimers with the expected antiparallel β-sheet immunoglobulin fold [30, 36-42]. The dimer observed is homologous to the conformation occurring between light and heavy chains in immunoglobulin molecules such as IgG. The germline κI O18/O8 crystallizes as a canonical dimer while the amyloidogenic protein AL-09 adopts an altered dimer with a 90° rotation with respect to the canonical dimer structure [19]. Restorative mutational analysis showed that a single mutation in AL-09 (AL-09 H87Y) stabilized the protein, delayed fibril formation, and changed its conformation from the altered dimer to the canonical dimer interface [18]. We have recently reported that the reciprocal mutant κI Y87H, in which we mutated the germline residue towards the residue found in AL-09, crystallized as a canonical dimer. However, using solution Nuclear Magnetic Resonance spectroscopy, we showed that this protein adopts a different dimer interface rotated 180° from the canonical dimer interface and 90° from the AL-09 altered dimer interface [43]. The different dimer structures could be compared to the hands on a clock moving in intervals of 90° (Figure 1). Sequence alignments of the variable domains of 50 κ and 91 λ AL light chains variable domains revealed that non-conservative mutations on the dimer interface, especially Histidine mutations, are very common in AL proteins, including Y87H as found in AL-09 [4]. Taken together with our structural analysis of AL-09, AL-09 H87Y, and κI Y87H, our results suggest that dynamic dimerization could occur frequently in AL proteins. Our structural studies show that light chains are able to dimerize in different conformations; the residues in the dimer interface determine whether or not a dimer conformation will be favored or if the numerous interfaces will be populated at the same time. This dynamic dimerization has the potential to occur even in the context of immunoglobulin molecules such as IgG depending on the somatic mutations that are acquired in the dimer interface region between the variable domains of both the light and the heavy chains.
Figure 1.
A) Schematic representation of the light chain variable domain. The b-strands are labeled as arrows and with their respective letter names. The regions corresponding to the complementarity determining regions (CDRs) are colored in blue. B) Overlay of three different dimer orientations found in crystal structures of κI O18/O8, AL-09, and NMR structure of κI Y87H. C) Rates of fibril formation are inversely correlated with ΔGfolding. Mutation of Y87 to Histidine stabilizes an altered dimer interface and promotes faster rates of amyloid formation. HSQC analysis was used to categorize each protein as single state (green) or promiscuous states (red); others were not determined (gray). Figures B and C adapted from (Peterson et al., 2010, Fig. 4 and 5, respectively), with permission from Elsevier. Copyright © 2010.
In summary, while it has been identified that certain germline gene products are overrepresented in AL amyloidosis and it has now been reported that some of these germline proteins are capable of forming amyloid fibrils, the determinants of amyloid formation in AL amyloidosis is multifactorial. The mutational analysis conducted by a number of laboratories points to a correlation between thermodynamic stability and amyloid formation; most importantly, it appears that the population of partially folded states appears to be critical in the initiation of the amyloid fibril reaction and certain somatic mutations appear to promote these partially folded states more than others.
Environmental factors affecting amyloid formation in AL amyloidosis
In addition to studying the characteristics that make a soluble light chain protein more amyloidogenic, a tremendous amount of research has and is currently being done with respect to the factors that affect fibril formation both in vitro and in vivo. The in vitro factors include temperature, pH, ionic strength, agitation, protein concentration, and pressure, which all destabilize the protein in order to populate partially-folded states that are prone to aggregation [44]. The rate of fibril formation for amyloidogenic light chain protein SMA was shown to be highly accelerated at pH 2 [24]. Both AL protein SMA and MM protein LEN formed fibrils at pH 2 with agitation, but SMA displayed faster kinetics [45]. Amorphous aggregation of SMA was observed in samples from pH 4 to 7, while fibrils were observed in samples at pH ≤3 implying that SMA formed different partially-folded intermediates depending on the pH of the solution. At pH 4.5, 30 mM NaCl, SMA formed annular aggregates whereas at high ionic strength, fibrils and amorphous deposits were the predominant species [46]. At pH 7, the fibril formation kinetics of MM light chain LEN was faster with lower protein concentrations and increased concentrations of urea (0 to 3 M) [27].
The use of renal solutes has shed light onto the destabilizing and compensatory effects that different reagents can have on amyloid fibril formation. Urea, a known protein denaturant, at a concentration of 1.5 M decreased the thermodynamic stability and reduced the lag time of fibril formation of both SMA and LEN, while betaine and sorbitol (organic osmolytes) had the opposite effect at a concentration of 0.5 M, showing the interplay between stabilizing and denaturing forces that may occur in physiological environments [47].
Other denaturant studies indicate that SMA fibril formation kinetics were dependent on the concentration of guanidine hydrochloride (GuHCl) [48]. The reaction at 2 M GuHCl had the fastest amyloid fibril formation kinetics, whereas amorphous aggregates were formed at lower concentrations of GuHCl. Additionally, GuHCl affected the intermediate structures in fibril formation, determined by circular dichroism spectroscopy. At 1 M GuHCl, amorphous aggregates were formed by native-like intermediate structures, while at 2 M, amyloid fibrils were generated through an unfolded intermediate.
Protein concentration is another factor that influences fibril formation kinetics for SMA and LEN; solutions with low concentrations of LEN showed faster kinetics of amyloid fibril formation than solutions with higher concentrations [26]. This behavior of low protein concentration enhancement for fibril formation was also observed for SMA in the presence of 2 M GuHCl, where low protein concentration is more favorable for amyloid fibril formation [48]. This phenomenon could be explained by several mechanisms: i. The monomer is populated at low concentrations and is more amyloidogenic than the dimeric structure; ii. At high protein concentrations, there are more off-pathway mechanisms (i.e. amorphous aggregation) competing with the amyloid fibril formation pathway [49]. In order to reduce the lag phase of amyloid fibril formation, the addition of a seed or nucleus of preformed fibrils to soluble protein solutions accelerates the kinetics of fibril formation [50]. This has been observed with several AL proteins. The addition of 5% seeds in a SMA fibril formation reaction decreased the lag time by half when compared to an unseeded reaction [21]. In the case of AL-103, the presence of 0.4% seeds significantly accelerates the kinetics of fibril formation under a number of experimental conditions [22, 51].
The effect of surfaces on amyloid fibril formation is extremely important, although it is not always studied in a systematic way. Zhu et al. studied the effect of mica on SMA fibril formation. They found that SMA has faster fibrillation and requires less protein for the reaction on mica. They also discovered different fibril growth mechanisms; on a mica surface, protofibrils were observed, while in solution, fibrils were present. While it is hard to correlate, these surface experiments may be relevant in vivo since AL amyloid deposits are associated with the extracellular matrix in the basement membrane of tissues [52].
An early study of interactions between a urine-derived light chain from a MM patient and phospholipid vesicles demonstrated that light chains associate with lipids through two distinct events: one presumably represents a collision limited binding event followed by a slower reorganization event. The secondary event is believed to be associated with the maximization of both electrostatic and hydrophobic interactions between the protein and the membranes. The structure of the light chain remains the same in the presence of the phospholipid vesicles as determined by circular dichroism (CD), suggesting that any conformational changes that may occur are not global and therefore not detected by CD [53]. An additional effort to understand the role of components of the plasma membrane reported a role for lipids in amyloid formation for AL amyloidosis. The authors found that a higher protein to lipid vesicles ratio slowed SMA amyloid fibril formation kinetics [54]. SMA fibrillation was inhibited by increasing cholesterol concentrations above 10%. Additionally, the presence of cholesterol-containing lipid vesicles and high Ca2+, Mg2+, or Zn2+concentrations were shown to decrease SMA fibril formation kinetics. While this is the only study describing the role of lipids in AL amyloid fibril formation, it suggests that amyloid deposition is influenced by the combined effects of cations and membrane surfaces.
The effect of salts and glycosaminoglycans
Glycosaminoglycans (GAGs) are a component of the extracellular matrix [55] and have been found extensively in amyloid deposits. They are long, unbranched, negatively-charged, highly polymorphic molecules composed by repeating subunits of N-acetylglucosamine or N-acetylgalactosamine and uronic acid. The type of disaccharide, the number of disaccharide units, and the number of sulfate molecules per disaccharide varies in vivo, increasing the diversity of the GAG molecule as response of its functional status. Ohishi et al. found that GAGs are an integral part of AL amyloid fibrils and that the amount of GAGs increased 10-fold in tissues from AL amyloidosis patients, suggesting that GAGs not only play a role interacting with amyloid fibrils but the presence of the amyloid fibrils affect GAG levels [56]. Early in vitro studies using HPLC chromatography and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) showed an interaction between light chain proteins and various GAGs [57]. Keeling and co-workers incubated renal mesangial cells embedded in matrigel (a gelatinous mixture resembling the extracellular matrix) in the presence of AL amyloidosis light chains. These cells show extracellular deposition of amyloid fibrils formed by AL light chains after 96 hours of incubation [58]. Their results suggest that the presence of the extracellular matrix mixture allowed the observation of the extracellular amyloid, although it is also possible that it may have promoted it. Trinkaus-Randall and co-workers showed that the internalization of light chains in cardiac fibroblasts affects GAG regulation and localization and induces GAG sulfation, suggesting that the presence of amyloidogenic light chains causes an injury-like response in these cells[59]. In another in vitro study, our laboratory showed that dermatan sulfate accelerated AL-09 amyloid fibril formation, whereas chondroitin sulfate A inhibited fibril formation and yielded a spherical intermediate [60]. More recently, we have found that GAGs enhance AL-103 amyloid fibril formation in vitro by a transient electrostatic interaction with the soluble precursor protein [51]. It has been shown that the presence of GAGs play a role in amyloid formation and this process is highly affected by GAG sulfation [51, 59, 61].
Further studies of GAG influence on AL fibrillogenesis reflecting their true heterogeneity in humans both in disaccharide composition and multiple sulfate moieties may reveal important clues about the mechanism of amyloidogenesis and the role that GAGs play in in vivo amyloid deposition in the extracellular deposition.
The presence of salts can greatly affect amyloid fibril formation. The Hofmeister series is a tool to understand salt ionic effects that ranks ions according to their ability to stabilize or destabilize a protein [62, 63]. A proof of principle study was done with the amyloidogenic variable domain protein AL-12 to determine the role of physiologically relevant anions and cations from the Hofmeister series on protein stability and amyloid fibril formation. The presence of various salts with AL-12 did not affect the secondary structure but increased the thermodynamic stability of the protein [64], and all salts enhanced amyloid fibril formation. Reactions with SO42- and Mg2+ showed the largest enhancement of amyloid fibril formation. We recently performed a systematic analysis of the effect of different concentrations of NaCl on amyloid fibril formation using two similar amyloidogenic light chains. AL-09 readily formed fibrils across a wide range of salt concentrations; however, the amyloidogenic light chain AL-103 (90% sequence identity to AL-09) showed a roughly inverse dependence of the fibril formation rate on salt concentration [22]. Future studies with various AL proteins and salts will help determine how sulfate ions enhance amyloid fibril formation and will shed light onto the role of glycosaminoglycan sulfation on fibril formation in vivo. The interplay between electrostatic interactions and possible crowding effects from glycosaminoglycans should be dissected to determine their contributions to the process.
Cellular toxicity studies
One of the most important questions in amyloidosis research is to determine the most toxic species of the amyloid fibril formation reaction. For years, researchers assumed that amyloid fibril deposits were highly toxic to the cells near them by blocking the exchange of nutrients, creating a mechanical barrier around the cells, and by attracting macrophages that ultimately caused tissue damage. Later experiments conducted with soluble fractions from preparations of amyloid affected tissue showed that soluble species were as toxic as or more toxic than insoluble amyloid fibrils. Recent work done by our laboratory and others has shown that the presence of soluble AL proteins in cell culture induces apoptosis [65, 66]. In particular, we were able to demonstrate that the light chain species present in cell culture at the time of maximum apoptotic activity are primarily light chain monomer and dimers. Future experiments are required to compare the toxicity of these soluble species with the toxic effect that fibrils may have to cells in culture.
Teng and co-workers have reported that light chains that cause AL amyloidosis or light chain deposition disease (LCDD) interact with small invaginations of the plasma membrane in human renal mesangial cells with ultra structural features of caveolae [67]. Both AL light chains and LCDD light chains compete for the same receptor, co-localize in the plasma membrane of human renal mesangial cells, and crosslink with filamin and talin, both components of caveolin-1. Receptor-mediated internalization is proposed by these authors, although the receptor was not identified. Monis et al. performed internalization experiments that suggested that the amyloidogenic light chains are not internalized using a membrane receptor but an active mechanism such as constitutive pinocytosis into an endosomal/lysosomal pathway mediated by microtubules [68]. Interestingly, there was a significant difference between the internalization of truncated, monomeric, and dimeric light chains. Truncated and monomeric light chains are easily internalized, while dimeric light chains are rarely internalized under the conditions used to conduct the experiments by these authors. It is worth mentioning that the nomenclature of monomeric and dimeric is derived from mass spectrometry data where they are assessing the presence of covalently bound light chains. No association experiments were reported for these proteins, although the concentrations used for the experiments (1.39 μM) are close to the concentrations reported by Qin et al. for a monomer SMA variable domain (5 μM) [48]. Monis et al. suggest that different cell types such as rat cardiac fibroblasts and renal mesangial cells may have different modes of internalization for AL proteins [68].
Internalization studies comparing the internalization rates of several full length light chains have shown that AL-09 internalizes into cardiomyocytes within 24 h, migrating into lysosomal compartments. Full length κI O18/O8 is delayed in this process. Different restorative AL-09 mutants internalize with rates that correlate with their corresponding variable domain amyloid fibril formation rates (Levinson, Olatoye, Randles, Howell, DiCostanzo, and Ramirez-Alvarado, under review). These comparative studies are allowing us to fully characterize the biophysical, biochemical, and cellular properties of amyloidogenic light chains to get the entire picture of the role of somatic mutations in the disease process.
Given the different modes of internalization reported for human mesangial cells and rat cardiac fibroblasts, more experiments using different amyloidogenic proteins are needed. Moreover, it would be important to couple these comparison studies with the accurate determination of the oligomerization state of the starting protein used in the experiments as the oligomerization state of the protein may determine the mode of internalization. This appears quite clear from tissue culture studies of the toxicity induced by TTR or Aβ peptides [69-71].
While these cell culture studies have been helpful (mainly due to the lack of an animal model for AL amyloidosis), it is important to emphasize that cells in culture are isolated from working organ systems and do not truly reflect the pathophysiology of AL amyloidosis.
Human tissue toxicity studies
The first tissues affected by AL protein deposition are the blood vessels. It was previously shown that AL amyloidosis patients present with early endothelial microcirculatory dysfunction [72], and that light chain amyloid infiltration in epicardial coronary arteries occurs in almost all of the AL amyloidosis patients analyzed [73].
Another report showed that the presence of light chain is associated with histological evidence of myocardial ischemia (decrease in the blood supply) in the majority of AL patients studied [74]. These findings suggest that microvascular dysfunction is central to AL pathophysiology, yet its underlying mechanism and its role in the general pathophysiology of the disease are unknown. Migrino et al. recently reported an increase in protein oxidation in AL amyloidosis patients. When arterioles were exposed to amyloidogenic light chains, they observed higher levels of superoxide and impaired dilation to sodium nitroprusside [75] and increased apoptosis that could be reverted by treatment with superoxide dismutase [76]. Human arterioles are physiologically relevant to early AL pathophysiology and offer an important tissue system to study tissue dysfunction caused by AL light chains.
Models of secretion and tissue damage
Arendt and co-workers established the first amyloidogenic human cell line system, ALMC-1 and ALMC-2 [77]. They used plasma cells from an AL patient isolated both pre- (ALMC-1) and post- (ALMC-2) stem cell transplant. These cell lines secrete a full length λ6a light chain protein called ALMC. While there is some genetic variation between ALMC-1 and ALMC-2, the protein sequences from both cell lines are 100% identical. The protein secreted from these cell lines is fully folded with a β-sheet structure and is as stable as other full length proteins characterized to date [78]. ALMC has the ability to form amyloid fibrils in vitro. To date, ALMC-1 and ALMC-2 are the only human-derived systems that secrete a significant amount of protein for biophysical studies. We expect that future studies using these cell lines will advance our understanding of the cellular microenvironment and its possible role in the misfolding of light chain proteins.
Currently, there is no reported animal model for AL amyloidosis that displays the full pathophysiology of the disease. The only reported animal models are described below. Hrnic and co-workers created a highly artificial model which had some success. This model involves injecting large amounts of solubilized human amyloid deposits into the animal. A mass forms consisting largely of fibrillar material surrounded by macrophages and neutrophils which are ultimately resorbed [79]. Shi and co-workers reported an animal model in which wild type and dominant negative p38α transgenic mice were initially injected with amyloidogenic light chains through the tail vein followed by systemic intravenous infusion via the use of an osmotic minipump for 7 days. Wild type animals with fully active p38α presented an increase in the Bax/Bcl2 ratio and a very modest increase in cellular apoptosis as determined by TUNEL staining [65]. More recently, Ward and colleagues developed a new transgenic mouse model expressing an amyloidogenic λ6 light chain using the cytomegalovirus (CMV) promoter; by 24-30 months, 83% of the transgenic mice developed amyloid deposits in the stomach [80]. Interestingly, immunohistochemistry detected human λ6 light chain in other epithelial tissues, but not the liver or bone marrow. There was also no cardiac or kidney deposition or dysfunction as is generally observed in AL amyloidosis patients. Treatment with doxycycline decreased amyloid fibril formation in these transgenic mice. The authors conclude that this transgenic model could be useful for testing modulators of fibril inhibition, although it does not seem to recapitulate the phenotype of AL amyloidosis.
Buxbaum in 2009 analyzed the possible causes for the lack of success of amyloidosis animal models. He speculated that one possible reason for the lack of success is that the amyloidogenic potential of a particular light chain structure can only be realized when the protein is synthesized and secreted to a level equal to or greater than the critical concentration required for its aggregation under physiologic conditions. This may require a degree of clonal expansion in vivo that has not yet been achieved experimentally. Since AL amyloidosis is systemic, deposition frequently takes place at locations distant from the site of synthesis, so an organismal rather than a tissue culture approach should be favored. Ideal models should allow the development of more precise notions of pathogenesis and the roles of proteins other than the actual precursor in facilitating or inhibiting amyloid generation and deposition. One would also like the model to display both clinical and pathologic phenotypes that resemble those seen in human patients, including its temporal pattern of development, i.e. tissue distribution of deposits.
The use of worms (Caenorhabditis elegans) and flies (Drosophila melanogaster) to study amyloidoses has allowed the study of some disease processes, but Buxbaum argues that the relationship between the cellular and molecular phenotype and human disease may be problematic, although it may facilitate the in vivo screen for inhibitors of amyloidosis [81].
Strategies to reduce AL light chain levels and toxicity
Current treatments for AL amyloidosis target the malignant plasma cell population in the bone marrow. These treatments include chemotherapy and autologous stem cell transplant. These treatments are somewhat successful, whilst they are poorly tolerated by some AL amyloidosis patients due to their organ dysfunction. New therapeutic strategies targeting the amyloidogenic light chains and the AL amyloid fibrils are currently in development, and their efficacy is being studied.
Small molecules have been tested in search of fibril formation inhibitors. Congo red is a histological dye that binds to amyloid fibrils and presents a green birefringence under polarized light [82]. AL-09 fibril formation was inhibited by Congo red at a 1:1 molar ratio [60]. In contrast, Congo red did not inhibit fibril formation of SMA suggesting some specificity in the role of Congo red as an inhibitor [83]. More research is needed to find effective fibril inhibitors for a variety of AL proteins both in vitro and using cell culture systems.
A murine monoclonal antibody (mAb 11-1F4) that binds to light chain fibrils but not soluble proteins was generated and characterized by Solomon and co-workers [84, 85]. Immunohistochemical analysis revealed that mAb 11-1F4 recognized light chain fibrils regardless of their variable domain subgroup. The specificity of this antibody for AL light chain fibrils (κI, κII, κIV, λ1, λ3, λ6, λ8) was shown by Europium-Linked Immunosorbant Assay (EuLISA) where an EC50 value (concentration of antibody at half maximum binding) for binding was ~130 ± 39 nM. Peptide mapping determined that the cryptic epitope is located in the first 18 amino acids of the variable light chain domain; a prolyl residue at position 8 is necessary. A competition EuLISA was set up with mAb 11-1F4, and recombinant Wil (λ6 AL protein) fibrils were inhibited by a 50-fold molar excess of soluble LEN (1-22) peptide.
Small interference RNA (siRNA) can be used to reduce the amount of messenger RNA for amyloidogenic light chains. Phipps and co-workers transfected SP2/O mouse myeloma cells with a construct encoding the λ6 AL light chain Wil under control of the cytomegalovirus promoter, using the λ2-producing myeloma cell line RPMI 8226 as a control. The siRNAs were designed specifically to the V, J, or C portions of the molecules. Forty eight hrs after exposure to the siRNAs, the authors observed a 40% reduction in messenger RNA and light chain production with a greater effect observed in the 8226 cells [86]. A more recent report by Hovey and co-workers utilized siRNA molecules targeting the CDR2, CDR3, and a portion of the constant domain of the AL protein AL-009 κI. The siRNAs were transfected into SP2/O mouse myeloma cells for their studies. The siRNA treatment yielded a reduction of 80% of messenger RNA and protein in serum [87].
Outlook
During the past 10 years, our knowledge of the molecular mechanisms of AL amyloidosis has greatly increased as a result of recent research about the role of mutations, dimerization structures, different species populated in AL amyloid fibril formation, the cellular microenvironment, and light chain-associated cell and tissue toxicity. However, much remains to be understood for this complex disease. Each new discovery prompts additional questions, and reinforces the need for innovative multidisciplinary research. In particular, the development of effective transgenic organismal models and misfolding-related therapeutic strategies, especially targeting light chain toxic species, is necessary. It is clear that a multidisciplinary research effort is required to analyze all aspects of the disease and provide a deeper understanding of its pathogenesis, ultimately leading to a successful therapeutic intervention.
Acknowledgements
I thank Kristi Simmons for her critical reading of the manuscript. The Ramirez-Alvarado team has been supported by the National Institutes of Health grants R01 GM071514, R01 CA111345, the American Heart Association grant AHA 06-30077N, the Mayo Foundation and the generous support of amyloidosis patients and their families.
Footnotes
This article is published as part of a themed issue on Protein Misfolding in Conformational Disorders, Guest Edited by Cláudio M. Gomes.
References
- 1.Baden EM, Sikkink LA, Ramirez-Alvarado M. Light chain amyloidosis - current findings and future prospects. Curr Protein Pept Sci. 2009;10(5):500–8. doi: 10.2174/138920309789351949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiCostanzo AC, Ramirez-Alvarado M. Current and New Perspectives on the Molecular and Cellular Mechanisms of Amyloid Formation and Toxicity in Light Chain Amyloidosis. InTech; 2011. pp. 1–22. [Google Scholar]
- 3.Branden C, Tooze J. Introduction to protein structure. 2nd ed. Garland Publishing, Inc; New York: 1999. p. 410. [Google Scholar]
- 4.Poshusta TL, Sikkink LA, Leung N, Clark RJ, Dispenzieri A, Ramirez-Alvarado M. Mutations in specific structural regions of immunoglobulin light chains are associated with free light chain levels in patients with AL amyloidosis. PLoS One. 2009;4(4):e5169. doi: 10.1371/journal.pone.0005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyle RA, Gertz MA. Primary systemic amyloidosis: clinical and laboratory features in 474 cases. Semin Hematol. 1995;32(1):45–59. [PubMed] [Google Scholar]
- 6.Perfetti V, Casarini S, Palladini G, Vignarelli MC, Klersy C, Diegoli M, Ascari E, Merlini G. Analysis of V(lambda)-J(lambda) expression in plasma cells from primary (AL) amyloidosis and normal bone marrow identifies 3r (lambdaIII) as a new amyloid-associated germline gene segment. Blood. 2002;100(3):948–53. doi: 10.1182/blood-2002-01-0114. [DOI] [PubMed] [Google Scholar]
- 7.Abraham RS, Geyer SM, Price-Troska TL, Allmer C, Kyle RA, Gertz MA, Fonseca R. Immunoglobulin light chain variable (V) region genes influence clinical presentation and outcome in light chain-associated amyloidosis (AL). Blood. 2003;101(10):3801–08. doi: 10.1182/blood-2002-09-2707. [DOI] [PubMed] [Google Scholar]
- 8.Comenzo RL, Zhang Y, Martinez C, Osman K, Herrera GA. The tropism of organ involvement in primary systemic amyloidosis: contributions of Ig V(L) germ line gene use and clonal plasma cell burden. Blood. 2001;98(3):714–20. doi: 10.1182/blood.v98.3.714. [DOI] [PubMed] [Google Scholar]
- 9.Prokaeva T, Spencer B, Kaut M, Ozonoff A, Doros G, Connors LH, Skinner M, Seldin DC. Soft tissue, joint, and bone manifestations of AL amyloidosis: clinical presentation, molecular features, and survival. Arthritis Rheum. 2007;56(11):3858–68. doi: 10.1002/art.22959. [DOI] [PubMed] [Google Scholar]
- 10.Perfetti V, Palladini G, Casarini S, Navazza V, Rognoni P, Obici L, Invernizzi R, Perlini S, Klersy C, Merlini G. The repertoire of lambda light chains causing predominant amyloid heart involvement and identification of a preferentially involved germline gene, IGLV1-44. Blood. 2012;119(1):144–50. doi: 10.1182/blood-2011-05-355784. [DOI] [PubMed] [Google Scholar]
- 11.Stevens FJ. Four structural risk factors identify most fibril-forming kappa light chains. Amyloid. 2000;7(3):200–211. doi: 10.3109/13506120009146835. [DOI] [PubMed] [Google Scholar]
- 12.Dispenzieri A, Zhang L, Katzmann JA, Snyder M, Blood E, Degoey R, Henderson K, Kyle RA, Oken MM, Bradwell AR, Greipp PR. Appraisal of immunoglobulin free light chain as a marker of response. Blood. 2008;111(10):4908–15. doi: 10.1182/blood-2008-02-138602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurle MR, Helms LR, Li L, Chan W, Wetzel R. A role for destabilizing amino acid replacements in light-chain amyloidosis. Proc Natl Acad Sci U S A. 1994;91(12):5446–50. doi: 10.1073/pnas.91.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens PW, Raffen R, Hanson DK, Deng Y-L, Berrios-Hammond M, Westholm FA, Murphy C, Eulitz M, Wetzel R, Solomon A, Schiffer M, Stevens FJ. Recombinant immunoglobulin variable domains generated from synthetic genes provide a system for in vitro characterization of light-chain amyloid proteins. Protein Sci. 1995;4:421–32. doi: 10.1002/pro.5560040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wetzel R. Domain stability in immunoglobulin light chain deposition disorders. Adv Protein Chem. 1997;50:183–242. doi: 10.1016/s0065-3233(08)60322-8. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y, Wall JS, Meyer J, Murphy C, Randolph TW, Manning MC, Solomon A, Carpenter JF. Thermodynamic modulation of light chain amyloid fibril formation. J Biol Chem. 2000;275(3):1570–4. doi: 10.1074/jbc.275.3.1570. [DOI] [PubMed] [Google Scholar]
- 17.Wall J, Schell M, Murphy C, Hrncic R, Stevens FJ, Solomon A. Thermodynamic instability of human lambda 6 light chains: correlation with fibrillogenicity. Biochemistry. 1999;38(42):14101–8. doi: 10.1021/bi991131j. [DOI] [PubMed] [Google Scholar]
- 18.Baden EM, Randles EG, Aboagye AK, Thompson JR, Ramirez-Alvarado M. Structural insights into the role of mutations in amyloidogenesis. J Biol Chem. 2008;283(45):30950–6. doi: 10.1074/jbc.M804822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baden EM, Owen BA, Peterson FC, Volkman BF, Ramirez-Alvarado M, Thompson JR. Altered dimer interface decreases stability in an amyloidogenic protein. J Biol Chem. 2008;283(23):15853–60. doi: 10.1074/jbc.M705347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Pozo Yauner L, Ortiz E, Sanchez R, Sanchez-Lopez R, Guereca L, Murphy CL, Allen A, Wall JS, Fernandez-Velasco DA, Solomon A, Becerril B. Influence of the germline sequence on the thermodynamic stability and fibrillogenicity of human lambda 6 light chains. Proteins. 2008;72(2):684–692. doi: 10.1002/prot.21934. [DOI] [PubMed] [Google Scholar]
- 21.Davis PD, Raffen R, Dul LJ, Vogen MS, Williamson KE, Stevens JF, Argon Y. Inhibition of amyloid fiber assembly by both BiP and its target peptide. Immunity. 2000;13(4):433–42. doi: 10.1016/s1074-7613(00)00043-1. [DOI] [PubMed] [Google Scholar]
- 22.Martin DJ, Ramirez-Alvarado M. Comparison of amyloid fibril formation by two closely related immunoglobulin light chain variable domains. Amyloid. 2010;17(3-4):129–36. doi: 10.3109/13506129.2010.530081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ionescu-Zanetti C, Khurana R, Gillespie JR, Petrick JS, Trabachino LC, Minert LJ, Carter SA, Fink AL. Monitoring the assembly of Ig light-chain amyloid fibrils by atomic force microscopy. Proc Natl Acad Sci U S A. 1999;96(23):13175–9. doi: 10.1073/pnas.96.23.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khurana R, Gillespie JR, Talapatra A, Minert LJ, Ionescu-Zanetti C, Millett I, Fink AL. Partially folded intermediates as critical precursors of light chain amyloid fibrils and amorphous aggregates. Biochemistry. 2001;40(12):3525–35. doi: 10.1021/bi001782b. [DOI] [PubMed] [Google Scholar]
- 25.Souillac PO, Uversky VN, Fink AL. Structural transformations of oligomeric intermediates in the fibrillation of immunoglobulin light chain LEN. Biochemistry. 2003;42(26):8094–104. doi: 10.1021/bi034652m. [DOI] [PubMed] [Google Scholar]
- 26.Souillac PO, Uversky VN, Millett IS, Khurana R, Doniach S, Fink AL. Elucidation of the molecular mechanism during the early events in immunoglobulin light chain amyloid fibrillation. Evidence for an off- pathway oligomer at acidic pH. J Biol Chem. 2002;277(15):12666–79. doi: 10.1074/jbc.M109229200. [DOI] [PubMed] [Google Scholar]
- 27.Souillac PO, Uversky VN, Millett IS, Khurana R, Doniach S, Fink AL. Effect of association state and conformational stability on the kinetics of immunoglobulin light chain amyloid fibril formation at physiological pH. J Biol Chem. 2002;277(15):12657–65. doi: 10.1074/jbc.M109230200. [DOI] [PubMed] [Google Scholar]
- 28.Ch'ang LY, Yen CP, Besl L, Schell M, Solomon A. Identification and characterization of a functional human Ig V lambda VI germline gene. Mol Immunol. 1994;31(7):531–6. doi: 10.1016/0161-5890(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 29.del Pozo Yauner L, Ortiz E, Becerril B. The CDR1 of the human lambdaVI light chains adopts a new canonical structure. Proteins. 2006;62(1):122–9. doi: 10.1002/prot.20779. [DOI] [PubMed] [Google Scholar]
- 30.Wall JS, Gupta V, Wilkerson M, Schell M, Loris R, Adams P, Solomon A, Stevens F, Dealwis C. Structural basis of light chain amyloidogenicity: comparison of the thermodynamic properties, fibrillogenic potential and tertiary structural features of four Vlambda6 proteins. J Mol Recognit. 2004;17(4):323–31. doi: 10.1002/jmr.681. [DOI] [PubMed] [Google Scholar]
- 31.Schroeder HW, Jr., Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol. 2010;125(2 Suppl 2):S41–52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen KE, Sletten K, Westermark P. Fragments of the constant region of immunoglobulin light chains are constituents of AL-amyloid proteins. Biochem Biophys Res Commun. 1998;251(2):642–7. doi: 10.1006/bbrc.1998.9508. [DOI] [PubMed] [Google Scholar]
- 33.Lavatelli F, Perlman DH, Spencer B, Prokaeva T, McComb ME, Theberge R, Connors LH, Bellotti V, Seldin DC, Merlini G, Skinner M, Costello CE. Amyloidogenic and associated proteins in systemic amyloidosis proteome of adipose tissue. Mol Cell Proteomics. 2008;7(8):1570–83. doi: 10.1074/mcp.M700545-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vrana JA, Gamez JD, Madden BJ, Theis JD, Bergen HR, 3rd, Dogan A. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood. 2009;114(24):4957–9. doi: 10.1182/blood-2009-07-230722. [DOI] [PubMed] [Google Scholar]
- 35.Klimtchuk ES, Gursky O, Patel RS, Laporte KL, Connors LH, Skinner M, Seldin DC. The critical role of the constant region in thermal stability and aggregation of amyloidogenic immunoglobulin light chain. Biochemistry. 2010;49(45):9848–57. doi: 10.1021/bi101351c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alim MA, Yamaki S, Hossain MS, Takeda K, Kozima M, Izumi T, Takashi I, Shinoda T. Structural relationship of kappa-type light chains with AL amyloidosis: multiple deletions found in a VkappaIV protein. Clin Exp Immunol. 1999;118(3):344–8. doi: 10.1046/j.1365-2249.1999.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourne PC, Ramsland PA, Shan L, Fan ZC, DeWitt CR, Shultz BB, Terzyan SS, Moomaw CR, Slaughter CA, Guddat LW, Edmundson AB. Three-dimensional structure of an immunoglobulin light-chain dimer with amyloidogenic properties. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 5):815–23. doi: 10.1107/s0907444902004183. [DOI] [PubMed] [Google Scholar]
- 38.Epp O, Lattman EE, Schiffer M, Huber R, Palm W. The molecular structure of a dimer composed of the variable portions of the Bence-Jones protein REI refined at 2.0-A resolution. Biochemistry. 1975;14(22):4943–52. doi: 10.1021/bi00693a025. [DOI] [PubMed] [Google Scholar]
- 39.Huang DB, Chang CH, Ainsworth C, Brunger AT, Eulitz M, Solomon A, Stevens FJ, Schiffer M. Comparison of crystal structures of two homologous proteins: structural origin of altered domain interactions in immunoglobulin light-chain dimers. Biochemistry. 1994;33(49):14848–57. doi: 10.1021/bi00253a024. [DOI] [PubMed] [Google Scholar]
- 40.Pokkuluri PR, Solomon A, Weiss DT, Stevens FJ, Schiffer M. Tertiary structure of human lambda 6 light chains. Amyloid. 1999;6(3):165–71. doi: 10.3109/13506129909007322. [DOI] [PubMed] [Google Scholar]
- 41.Schormann N, Murrell JR, Liepnieks JJ, Benson MD. Tertiary structure of an amyloid immunoglobulin light chain protein: a proposed model for amyloid fibril formation. Proc Natl Acad Sci U S A. 1995;92(21):9490–4. doi: 10.1073/pnas.92.21.9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernandez-Santoyo A, del Pozo Yauner L, Fuentes-Silva D, Ortiz E, Rudino-Pinera E, Sanchez-Lopez R, Horjales E, Becerril B, Rodriguez-Romero A. A single mutation at the sheet switch region results in conformational changes favoring lambda6 light-chain fibrillogenesis. J Mol Biol. 2010;396(2):280–92. doi: 10.1016/j.jmb.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 43.Peterson FC, Baden EM, Owen BA, Volkman BF, Ramirez-Alvarado M. A single mutation promotes amyloidogenicity through a highly promiscuous dimer interface. Structure. 2010;18(5):563–70. doi: 10.1016/j.str.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson CM. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc Natl Acad Sci U S A. 1999;96(7):3590–4. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khurana R, Souillac PO, Coats AC, Minert LJ, Ionescu-Zanetti C, Carter SA, Solomon A, Fink AL. A model for amyloid formation in immunoglobulin light chains based on comparison of amyloidogenic and benign proteins and specific antibody binding. Amyloid. 2003;10:97–109. doi: 10.3109/13506120309041731. [DOI] [PubMed] [Google Scholar]
- 46.Zhu M, Han S, Zhou F, Carter SA, Fink AL. Annular oligomeric amyloid intermediates observed by in situ atomic force microscopy. J Biol Chem. 2004;279(23):24452–9. doi: 10.1074/jbc.M400004200. [DOI] [PubMed] [Google Scholar]
- 47.Kim YS, Cape SP, Chi E, Raffen R, Wilkins-Stevens P, Stevens FJ, Manning MC, Randolph TW, Solomon A, Carpenter JF. Counteracting effects of renal solutes on amyloid fibril formation by immunoglobulin light chains. J Biol Chem. 2001;276(2):1626–33. doi: 10.1074/jbc.M007766200. [DOI] [PubMed] [Google Scholar]
- 48.Qin Z, Hu D, Zhu M, Fink AL. Structural characterization of the partially folded intermediates of an immunoglobulin light chain leading to amyloid fibrillation and amorphous aggregation. Biochemistry. 2007;46(11):3521–31. doi: 10.1021/bi061716v. [DOI] [PubMed] [Google Scholar]
- 49.Powers ET, Powers DL. Mechanisms of protein fibril formation: nucleated polymerization with competing off-pathway aggregation. Biophys J. 2008;94(2):379–91. doi: 10.1529/biophysj.107.117168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harper JD, Lansbury PT., Jr. Models of amyloid seeding in Alzheimer's disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 51.Martin DJ, Ramirez-Alvarado M. Glycosaminoglycans promote fibril formation by amyloidogenic immunoglobulin light chains through a transient interaction. Biophys Chem. 2011;158(1):81–9. doi: 10.1016/j.bpc.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu M, Souillac PO, Ionescu-Zanetti C, Carter SA, Fink AL. Surface-catalyzed amyloid fibril formation. J Biol Chem. 2002;277(52):50914–22. doi: 10.1074/jbc.M207225200. [DOI] [PubMed] [Google Scholar]
- 53.Wall JS, Ayoub FM, O'Shea PS. A study of the interactions of an immunoglobulin light chain with artificial and B-lymphocyte membranes. Front Biosci. 1996;1:a46–58. doi: 10.2741/a105. [DOI] [PubMed] [Google Scholar]
- 54.Meng X, Fink AL, Uversky VN. The effect of membranes on the in vitro fibrillation of an amyloidogenic light-chain variable-domain SMA. J Mol Biol. 2008;381(4):989–99. doi: 10.1016/j.jmb.2008.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bosman FT, Stamenkovic I. Functional structure and composition of the extracellular matrix. J Pathol. 2003;200(4):423–8. doi: 10.1002/path.1437. [DOI] [PubMed] [Google Scholar]
- 56.Ohishi H, Skinner M, Sato-Araki N, Okuyama T, Gejyo F, Kimura A, Cohen AS, Schmid K. Glycosaminoglycans of the hemodialysis-associated carpal synovial amyloid and of amyloid-rich tissues and fibrils of heart, liver, and spleen. Clin Chem. 1990;36(1):88–91. [PubMed] [Google Scholar]
- 57.Jiang X, Myatt E, Lykos P, Stevens FJ. Interaction between glycosaminoglycans and immunoglobulin light chains. Biochemistry. 1997;36(43):13187–94. doi: 10.1021/bi970408h. [DOI] [PubMed] [Google Scholar]
- 58.Keeling J, Teng J, Herrera GA. AL-amyloidosis and light-chain deposition disease light chains induce divergent phenotypic transformations of human mesangial cells. Lab Invest. 2004;84(10):1322–38. doi: 10.1038/labinvest.3700161. [DOI] [PubMed] [Google Scholar]
- 59.Trinkaus-Randall V, Walsh MT, Steeves S, Monis G, Connors LH, Skinner M. Cellular response of cardiac fibroblasts to amyloidogenic light chains. Am J Pathol. 2005;166(1):197–208. doi: 10.1016/S0002-9440(10)62244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLaughlin RW, De Stigter JK, Sikkink LA, Baden EM, Ramirez-Alvarado M. The effects of sodium sulfate, glycosaminoglycans, and Congo red on the structure, stability, and amyloid formation of an immunoglobulin light-chain protein. Protein Sci. 2006;15(7):1710–22. doi: 10.1110/ps.051997606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren R, Hong Z, Gong H, Laporte K, Skinner M, Seldin DC, Costello CE, Connors LH, Trinkaus-Randall V. Role of glycosaminoglycan sulfation in the formation of immunoglobulin light chain amyloid oligomers and fibrils. J Biol Chem. 2011;285(48):37672–82. doi: 10.1074/jbc.M110.149575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cacace MG, Landau EM, Ramsden JJ. The Hofmeister series: salt and solvent effects on interfacial phenomena. Q Rev Biophys. 1997;30(3):241–77. doi: 10.1017/s0033583597003363. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Furyk S, Bergbreiter DE, Cremer PS. Specific ion effects on the water solubility of macromolecules: PNIPAM and the Hofmeister series. J Am Chem Soc. 2005;127(41):14505–10. doi: 10.1021/ja0546424. [DOI] [PubMed] [Google Scholar]
- 64.Sikkink LA, Ramirez-Alvarado M. Salts enhance both protein stability and amyloid formation of an immunoglobulin light chain. Biophys Chem. 2008;135(1-3):25–31. doi: 10.1016/j.bpc.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi J, Guan J, Jiang B, Brenner DA, Del Monte F, Ward JE, Connors LH, Sawyer DB, Semigran MJ, Macgillivray TE, Seldin DC, Falk R, Liao R. Amyloidogenic light chains induce cardiomyocyte contractile dysfunction and apoptosis via a non-canonical p38alpha MAPK pathway. Proc Natl Acad Sci U S A. 2010;107(9):4188–93. doi: 10.1073/pnas.0912263107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sikkink LA, Ramirez-Alvarado M. Cytotoxicity of amyloidogenic immunoglobulin light chains in cell culture. Cell Death and Disease. 2010;1:e98. doi: 10.1038/cddis.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teng J, Russell WJ, Gu X, Cardelli J, Jones ML, Herrera GA. Different types of glomerulopathic light chains interact with mesangial cells using a common receptor but exhibit different intracellular trafficking patterns. Lab Invest. 2004;84(4):440–51. doi: 10.1038/labinvest.3700069. [DOI] [PubMed] [Google Scholar]
- 68.Monis GF, Schultz C, Ren R, Eberhard J, Costello C, Connors L, Skinner M, Trinkaus-Randall V. Role of endocytic inhibitory drugs on internalization of amyloidogenic light chains by cardiac fibroblasts. Am J Pathol. 2006;169(6):1939–52. doi: 10.2353/ajpath.2006.060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bateman DA, Chakrabartty A. Cell surface binding and internalization of abeta modulated by degree of aggregation. Int J Alzheimers Dis. 2011;2011:962352. doi: 10.4061/2011/962352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartley DM, Walsh DM, Ye CP, Diehl T, Vasquez S, Vassilev PM, Teplow DB, Selkoe DJ. Protofibrillar intermediates of amyloid beta-protein induce acute electrophysiological changes and progressive neurotoxicity in cortical neurons. J Neurosci. 1999;19(20):8876–84. doi: 10.1523/JNEUROSCI.19-20-08876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reixach N, Deechongkit S, Jiang X, Kelly JW, Buxbaum JN. Tissue damage in the amyloidoses: Transthyretin monomers and nonnative oligomers are the major cytotoxic species in tissue culture. Proc Natl Acad Sci U S A. 2004;101(9):2817–22. doi: 10.1073/pnas.0400062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berghoff M, Kathpal M, Khan F, Skinner M, Falk R, Freeman R. Endothelial dysfunction precedes C-fiber abnormalities in primary (AL) amyloidosis. Ann Neurol. 2003;53(6):725–30. doi: 10.1002/ana.10552. [DOI] [PubMed] [Google Scholar]
- 73.Wittich CM, Neben-Wittich MA, Mueller PS, Gertz MA, Edwards WD. Deposition of amyloid proteins in the epicardial coronary arteries of 58 patients with primary systemic amyloidosis. Cardiovasc Pathol. 2007;16(2):75–8. doi: 10.1016/j.carpath.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 74.Neben-Wittich MA, Wittich CM, Mueller PS, Larson DR, Gertz MA, Edwards WD. Obstructive intramural coronary amyloidosis and myocardial ischemia are common in primary amyloidosis. Am J Med. 2005;118(11):1287. doi: 10.1016/j.amjmed.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 75.Migrino RQ, Hari P, Gutterman DD, Bright M, Truran S, Schlundt B, Phillips SA. Systemic and microvascular oxidative stress induced by light chain amyloidosis. Int J Cardiol. 2010;145(1):67–8. doi: 10.1016/j.ijcard.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Migrino RQ, Truran S, Gutterman DD, Franco DA, Bright M, Schlundt B, Timmons M, Motta A, Phillips SA, Hari P. Human microvascular dysfunction and apoptotic injury induced by AL amyloidosis light chain proteins. Am J Physiol Heart Circ Physiol. 2011;301(6):H2305–12. doi: 10.1152/ajpheart.00503.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arendt BK, Ramirez-Alvarado M, Sikkink LA, Keats JJ, Ahmann GJ, Dispenzieri A, Fonseca R, Ketterling RP, Knudson RA, Mulvihill EM, Tschumper RC, Wu X, Zeldenrust SR, Jelinek DF. Biologic and genetic characterization of the novel amyloidogenic lambda light chain-secreting human cell lines, ALMC-1 and ALMC-2. Blood. 2008;112(5):1931–41. doi: 10.1182/blood-2008-03-143040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sikkink LA, Ramirez-Alvarado M. Biochemical and aggregation analysis of Bence Jones proteins from different light chain diseases. Amyloid. 2008;15(1):29–39. doi: 10.1080/13506120701815324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hrncic R, Wall J, Wolfenbarger DA, Murphy CL, Schell M, Weiss DT, Solomon A. Antibody-mediated resolution of light chain-associated amyloid deposits. Am J Pathol. 2000;157(4):1239–46. doi: 10.1016/S0002-9440(10)64639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ward JE, Ren R, Toraldo G, Soohoo P, Guan J, O'Hara C, Jasuja R, Trinkaus-Randall V, Liao R, Connors LH, Seldin DC. Doxycycline reduces fibril formation in a transgenic mouse model of AL amyloidosis. Blood. 2011;118(25):6610–7. doi: 10.1182/blood-2011-04-351643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buxbaum JN. Animal models of human amyloidoses: are transgenic mice worth the time and trouble? FEBS Lett. 2009;583(16):2663–73. doi: 10.1016/j.febslet.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sipe JD, Cohen AS. Review: history of the amyloid fibril. J Struct Biol. 2000;130(2-3):88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 83.Kim YS, Randolph TW, Manning MC, Stevens FJ, Carpenter JF. Congo red populates partially unfolded states of an amyloidogenic protein to enhance aggregation and amyloid fibril formation. J Biol Chem. 2003;278(12):10842–50. doi: 10.1074/jbc.M212540200. [DOI] [PubMed] [Google Scholar]
- 84.O'Nuallain B, Allen A, Kennel SJ, Weiss DT, Solomon A, Wall JS. Localization of a conformational epitope common to non-native and fibrillar immunoglobulin light chains. Biochemistry. 2007;46(5):1240–7. doi: 10.1021/bi0616605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Solomon A, Weiss DT, Wall JS. Immunotherapy in systemic primary (AL) amyloidosis using amyloid-reactive monoclonal antibodies. Cancer Biother Radiopharm. 2003;18(6):853–60. doi: 10.1089/108497803322702824. [DOI] [PubMed] [Google Scholar]
- 86.Phipps JE, Kestler DP, Foster JS, Kennel SJ, Donnell R, Weiss DT, Solomon A, Wall JS. Inhibition of pathologic immunoglobulin-free light chain production by small interfering RNA molecules. Exp Hematol. 2010;38(11):1006–13. doi: 10.1016/j.exphem.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hovey BM, Ward JE, Soo Hoo P, O'Hara CJ, Connors LH, Seldin DC. Preclinical development of siRNA therapeutics for AL amyloidosis. Gene Ther. 2011;18(12):1150–6. doi: 10.1038/gt.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]