Abstract
Bioassay-guided fractionation of an ethanol extract of the leaves and inflorescence of Mallotus oppositifolius collected in Madagascar led to the isolation of the two new bioactive dimeric phloroglucinols, mallotojaponins B (1) and C (2), together with the known mallotophenone (3). The structures of the new compounds were determined on the basis of spectroscopic evidence, including their 1- and 2D-NMR spectra, mass spectrometry, and an X-ray crystal structure. Compounds 1 and 2 showed potent antimalarial activity against chloroquine-resistant Plasmodium falciparum, with IC50 values of 0.75 ± 0.30 and 0.14 ± 0.04 μM, while 3 was inactive in this assay. Compounds 1–3 also displayed strong antiproliferative activity against the A2780 human ovarian cancer cell line (IC50 1.10 ± 0.05, 1.3 ± 0.1 and 6.3 ± 0.4 μM, respectively).
The tropical genus Mallotus, a member of the family Euphorbiaceae, contains about 150 species of trees and shrubs.2 It shares membership in the tribe Acalypheae with the genus Macaranga,2 a genus that has afforded several promising bioactive compounds.3,4 Mallotus philippinensis is the source of rottlerin, a natural product which first appears to have been isolated in 1855,5 and has been the subject of numerous biological investigations.6-8
Our ongoing screening of extracts from plants collected in Madagascar as part of the Madagascar International Cooperative Biodiversity Group (ICBG) program for antiproliferative activity towards the A2780 ovarian cancer cell line9 has recently been supplemented with screening for antiplasmodial activity against the malaria parasite Plasmodium falciparum. An ethanol extract of Mallotus oppositifolius (Geiseler) Müll. Arg. (Euphorbiaceae) was found to display strong activity against P. falciparum as well as antiproliferative activity against the A2780 cell line, and this extract was thus selected for further investigation to isolate the active metabolite(s) responsible for the observed activities. M. oppositifolius has been used as a chewing stick in Nigeria,10 and its aqueous and ethanol extracts have been reported to have antifungal activity,11 but no previous work on its constituents has been reported.
RESULTS AND DISCUSSION
Isolation of Bioactive Constituents
Initial dereplication studies using size-exclusion chromatography and HPLC on a small amount of an active hexanes-soluble fraction obtained from the liquid-liquid partition of the active extract indicated the presence of unknown antiproliferative phloroglucinols. Scale up of the isolation to 1 g of extract yielded an antiproliferative hexanes fraction (IC50 6.7 μg/mL), which was subjected to further size-exclusion column chromatography (Sephadex LH-20) to furnish two active fractions with IC50 values of 1.6 and 2.3 μg/mL, and the known phloroglucinol mallotophenone (3, IC50 6.3 ± 0.4 μM). The most active fractions were subjected to HPLC and silica gel column chromatography to yield the two new bioactive phloroglucinols 1 and 2.
Structure Elucidation
Mallotophenone (3) was identified by single-crystal X-ray analysis and by comparison of its spectroscopic data with values reported in the literature.12
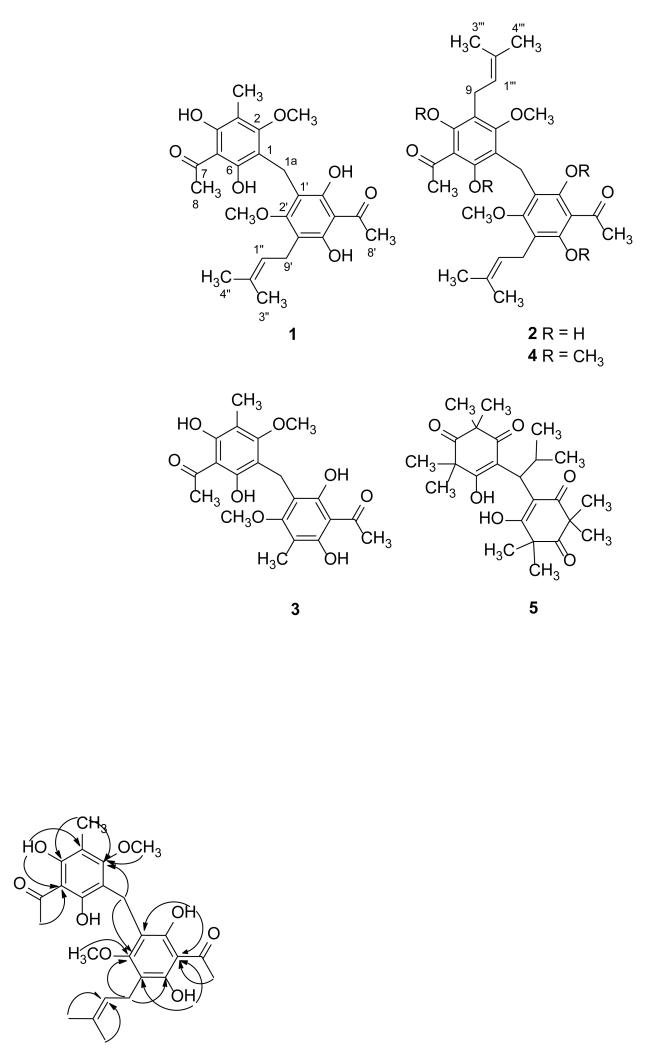
Compound 1 was obtained as yellowish crystals and gave the molecular formula C25H30O8 as indicated by high-resolution ESIMS analysis, which gave a protonated molecular ion peak at m/z 459.2023 [M+H]+. Its IR spectrum showed an absorption characteristic of a conjugated hydrogen bonded carbonyl group (1620 cm−1). The UV spectrum of 1 was very similar to that of 3, suggesting that the two compounds share the same chromophore. The 1H NMR spectroscopic data of 1 (Table 1) displayed resonances due to an aromatic methyl (δ 2.13, s, 3H), a 3,3-dimethylallyl group (δ 1.68, s, 3H and 1.77, s, 3H; δ 5.21, tq, J = 6.5, 1.4 Hz, 1H and δ 3.31, d, J = 6.5 Hz, 2H), two methoxy groups (δ 3.98, bs, 6H), two acyl methyl groups (δ 2.71, bs, 6H), and one methylene group at δ 3.68 (s, 2H), together with signals for two hydroxy groups, one of which was hydrogen bonded (δ 8.97, s, 1H and 13.64, s, 1H). The 13C NMR data of 1 (Table 2) exhibited 25 carbon signals that were identical with those of 3 except for the replacement of the signal of an aromatic methyl carbon with signals for the carbons of a 3,3-dimethylallyl unit (δ18.0, 22.9, 25.8, 122.7, 132.2). On comparison of the 13C NMR data of 1 with those of 3, the deshielding of the signal for C-3′ (δ114.2 instead of 109.1 in 3) suggested that the 3,3-dimethylallyl group is attached at this position, which is methylated in 3.13 The locations of the methyl, methoxy, methylene, carboxyl, hydroxy and the 3,3-dimethylallyl groups were confirmed by interpretation of the 1D- and 2D-NMR spectroscopic data of 1, including COSY, HSQC, HMBC, and nuclear Overhauser effect spectroscopy experiments. The attachment of the 3,3-dimethylallyl group at C-3′ was confirmed by the observation of the HMBC long-range correlation between the two geminal methyls at δ 1.68 and 1.77 to C-1″ (δ 122.7), and from the methine proton at δ 5.21 to C-3′ (δ 114.2). The methoxy groups were assigned to C-2 and C-2′ due to the long-range correlations (Figure 1) observed between the signals at δ 3.98 and those at δ 157.2 (C-2) and 157.7 (C-2′), between the C-1a methylene proton signal (δ 3.68) and C-2 and C-2′, as well as those observed between the aromatic methyl protons at δ 2.13 and C-2 and between the methylene protons signals at δ 3.31 and C-2′. In the same manner, the acyl group was assigned to C-5 and C-5′ from the HMBC cross peaks between the methyl protons at δ 2.71 (CH3-8 and 8′) and C-5 and C-5′. The two hydroxy groups must be located at C-4, C-4′, and C-6, C-6′, as indicated by the presence of two hydrogen-bonded hydroxy protons and the HMBC long-range correlations between the hydroxy group at δ 8.97 and C-3, C-3′, C-5 and C-5′. Moreover, NOESY correlations were observed between the methoxy protons and H-1″, H-1a, and CH3-9.
Table 1. 1H NMR Data for Compounds 1-3 (500 MHz, CDCl3).
| position | 1 | 2 | 3 |
|---|---|---|---|
| 1a | 3.68 s | 3.68 s | 3.66 s |
| 8 | 2.71 s | 2.70 s | 2.70 s |
| 9 | 2.13 s | 3.31 d (6.3) | 2.11 s |
| 8′ | 2.71 s | 2.70 s | 2.70 s |
| 9′ | 3.31 d (6.5) | 3.31 d (6.3) | 2.11 s |
| OCH3 | 3.98 s 3.98 s |
3.98 s 3.98 s |
3.97 s 3.97 s |
| 1″ | 5.21 (tq, 6.5, 1.4) | 5.21 (br t, 6.0) | |
| 3″ | 1.68 s | 1.68 s | |
| 4″ | 1.77 s | 1.77 s | |
| 1″′ | 5.21 (br t, 6.0) | ||
| 3″′ | 1.68 s | ||
| 4″′ | 1.77 s | ||
| OH | 8.97 s 13.64 s |
9.05s 13.48 s |
8.99 13.66 |
Table 2.
13C NMR Data for Compounds 1-3 (125 MHz, CDCl3)
| carbon | 1 | 2 | 3 |
|---|---|---|---|
| 1 | 108.3 | 108.5 | 108.4 |
| 1a | 18.1 | 17.9 | 18.1 |
| 2 | 157.2 | 157.5 | 157.1 |
| 3 | 109.2 | 114.2 | 109.1 |
| 4 | 162.9 | 162.8 | 163.0 |
| 5 | 110.0 | 109.2 | 110.2 |
| 6 | 159.8 | 159.6 | 159.9 |
| 7 | 205.4 | 205.4 | 205.6 |
| 8 | 33.8 | 33.8 | 34.1 |
| 9 | 8.9 | 22.9 | 9.2 |
| 1′ | 108.5 | 108.5 | 108.4 |
| 2′ | 157.7 | 157.5 | 157.1 |
| 3′ | 114.2 | 114.2 | 109.1 |
| 4′ | 162.9 | 162.8 | 163.0 |
| 5′ | 109.2 | 109.2 | 110.2 |
| 6′ | 159.6 | 159.6 | 159.9 |
| 7′ | 205.4 | 205.4 | 205.6 |
| 8′ | 33.8 | 33.8 | 34.1 |
| 9′ | 22.9 | 22.9 | 9.2 |
| OCH3 | 62.1 63.0 |
63.0 63.0 |
62.2 62.2 |
| 1″ | 122.7 | 122.7 | |
| 2″ | 132.2 | 132.2 | |
| 3″ | 18.0 | 17.9 | |
| 4″ | 25.8 | 25.8 | |
| 1″′ | 122.7 | ||
| 2″′ | 132.2 | ||
| 3″′ | 17.9 | ||
| 4″′ | 25.8 |
Figure 1.
Important HMBC correlations observed in 1.
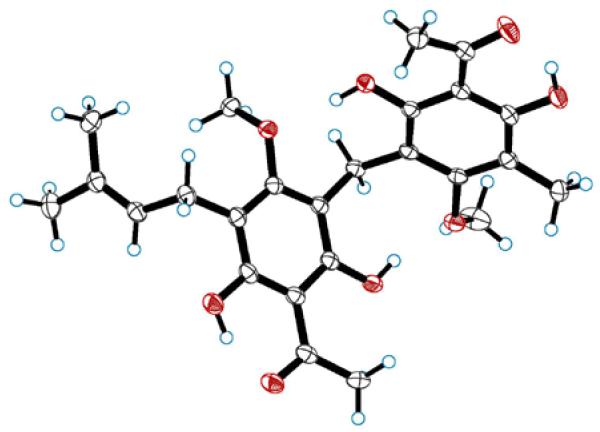
The structure of 1 was confirmed by single-crystal X-ray diffraction (Figure 2). Compound 1 was thus assigned as 3′-(3,3-dimethylallyl)-1′-(5-acetyl-6-hydroxy-3-methyl-2-methoxybenzyl)-2′-methoxyphloracetophenone, and has been named mallotojaponin B based on its relationship to mallotojaponin, seen here as mallotojaponin A.14
Figure 2.
Anisotropic displacement ellipsoid drawing (50%) of 1.
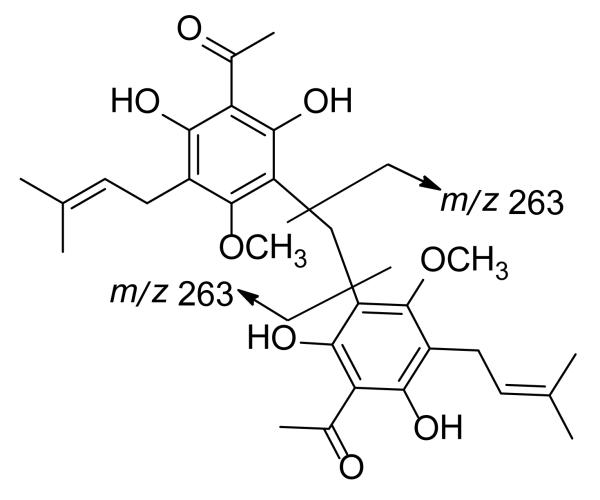
Compound 2, named mallotojaponin C, gave the molecular formula C29H36O8 as determined by positive ion HRESIMS (m/z 513.2499, [M+H]+, requires for C29H37O8, 513.2488). Similarly to 1, the IR and UV spectra of 2 were indicative of the presence of a prenylated phloroglucinol. The 1H and 13C NMR spectroscopic data of 2 were superposable upon those of 1 (Tables 1 and 2), except for the replacement of the signal due to the aromatic methyl group with the signals for a second 3,3-dimethylallyl group. Thus, the signal due to the methine of the 3,3-dimethylallyl group (δ 5.21, brt, J = 6.0 Hz) integrated for two protons (H-1″ and H-1″′) while the signal at δ 3.68 (s, H2-1a) also integrated for two protons. In addition, the broad triplet observed for the 1H NMR signal of the methine protons at δ 5.21 suggested the presence of two overlapping signals (H-1″ and H-1″′). These data coupled with the high-resolution mass spectra allowed the conclusion to be made that compound 2 was a symmetrical dimer with two 3,3-dimethylallyl units, one each at C-3 and C-3′. This was also confirmed by the observation of the base peak at m/z 263 in its mass spectrum (Figure 3). Comparison of the 13C NMR spectroscopic data of 2 with those of 1 demonstrated that the methyl group at C-3 of 1 is replaced by a 3,3-dimethylallyl unit in 2. Also, HMBC long-range correlations were observed from H-1″ and H-1″′ to C-3 and C-3′, respectively, and from H-9 and H-9′ to C-2, C-4 and to C-2′ and C-4′, respectively. The locations of the hydroxy groups at C-4 (C-4′) and C-6 (C-6′), the methoxyl groups at C-2 (C-2′), the acetyl group at C-5 (C-5′), and the methylene at C-1 (C-1′) were elucidated in the same manner as for 1. These data led to the assignment of the structure of 2 as 1-methylene-bis-4-methoxy-6-hydroxy-3-(3,3-dimethylallyl)-2-methoxyacetophenone.
Figure 3.
Mass fragmentation observed for 2.
Biological Activities
Compounds 1-3 were evaluated for their activity against P. falciparum Dd2 (a chloroquine/mefloquine-resistant strain). Compounds 1 and 2 showed submicromolar activity with half-maximum inhibitory concentration (IC50) values of 0.75 ± 0.30 and 0.14 ± 0.04 μM, respectively, while compound 3 was not active. In addition to their cytostatic activity in inhibiting the growth of P. falciparum, compounds 1 and 2 also showed cytocidal activity vs. P. falciparum. Using a newly developed rapid assay for determination of cytocidal activity,15 compound 1 was found to have median lethal dose (LD50) values of 14.6 ± 0.7 and 6.7 ± 0.2 μM vs. the drug-sensitive HB3 strain and the drug-resistant Dd2 strain, respectively, while compound 2 had LD50 values of 0.81 ± 0.05 and 0.80 ± 0.02 μM vs the same two strains. In these same assays, chloroquine exhibited LD50 values of 0.10 ± 0.01 and 15.3 ± 0.9 μM (HB3 vs. Dd2), so for the drug-resistant Dd2 strain, compound 2, in particular, is significantly more cytocidally potent than chloroquine.
Compounds 1 and 2 were further evaluated for their gametocytocidal activity against late stage gametocytes (the stage responsible for malaria transmission) using the chloroquine-sensitive NF54 strain to generate gametocytes. This strain was used because it forms gametocytes in culture much better than the chloroquine-sensitive HB3 strain. Only compound 2 showed gametocytocidal activity with an IC50 value of 3.6 ± 0.2 μM. This activity is comparable to the current antimalarial drug artesunate (IC50 value of 2.3 μM) and to NPC1161B, an antimalarial drug currently in the development pipeline (IC50 value of 3.8 μM).16,17 The IC50 value determined for compound 2 against asexual stages in the NF54 strain was 0.07 ± 0.01 μM.
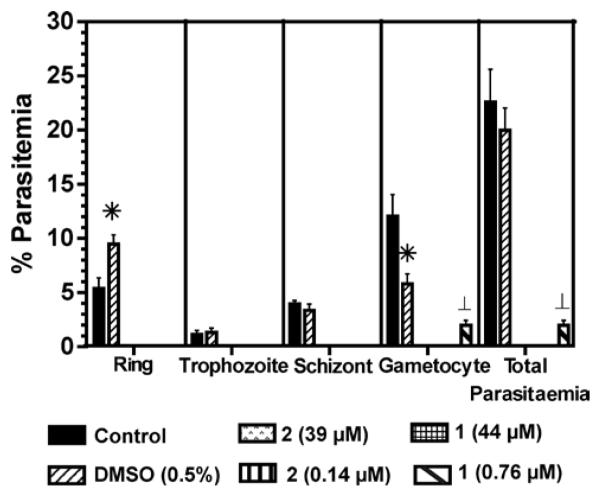
In order to address if compounds 1 and 2 were able also to prevent gametocytogenesis, P. falciparum in vitro cultures were treated with 0.76 μM (~IC50) or 44 μM (~IC100) of compound 1 and 0.14 μM (~IC50) or 39 μM (~IC100) of compound 2 for 13 days, as described in the Experimental Section. On day 13, cultures were recovered and smeared for microscopic examination. Neither asexual intraerythrocytic stages nor gametocytes were observed in cultures treated with 0.14 μM of compound 2 (Figure 4). Compound 1 cleared asexual intraerythrocytic stages in vitro at 0.76 μM, and the presence of mature gametocytes was reduced 80% as compared to untreated parasites (control). This is the first report on mallotojaponin derivatives showing antimalarial activity.
Figure 4.
P. falciparum blood stage distribution after 13 days of treatment at the indicated concentrations to assess antimalarial activity against early stage gametocytes. The control is untreated parasites. DMSO (drug vehicle at 0.5%) was included as a second control since it affects the blood-stages distribution but does not affect the total parasitemia. No parasite growth was observed at doses of 2 of 0.14 and 39 μM and of 1 at 44 μM. Results are presented as means of two independent experiments ± S.E.M., (*) p ≤ 0.02 and (⊥) p ≤ 0.005 compared with the control group.
Compounds 1 and 2 also displayed strong antiproliferative activity against the A2780 human ovarian cancer cell line with IC50 values of 1.10 ± 0.05 and 1.3 ± 0.1 μM, respectively. Since mallotophenone (3) showed weaker activity (IC50 6.3 ± 0.4 μM), it appears that the presence of the 3,3-dimethylallyl group in the molecule enhances the activity; however, this effect was more pronounced in the malaria parasite. Phloroglucinols have been reported to have a wide range of biological activities.14,18-22 Previous investigation showed that 3′-(3,3-dimethylallyl)-1′-(5-acetyl-6-hydroxy-3-methyl-2-methoxybenzyl)-2′-hydroxyphloracetophenone, a 2′-hydroxylated derivative of mallotojaponin B (1), displayed cytotoxic activities against KB and mouse leukemia L-5178Y (ED50 0.58 and 0.74 μg/mL, respectively).12
The discovery of the potent antimalarial and gametocytocidal activities of compound 2 raised the question of its possible mechanism of action. It has been proposed that acylphloroglucinols can function as antimalarial agents by acting as radical-generating species or by inhibiting hemozoin formation.23 The latter may occur through binding to pre-crystalline forms of heme via π-π interactions (given their electron rich structures) and/or coordination with the Fe3+ center of hematin (such as an Fe—O interaction with the phenolic moiety of 2).23 As a test of the importance of the phenolic hydroxy groups of 2, the compound was converted to its tetramethyl ether 4 by treatment with potassium carbonate and excess methyl iodide. Compound 4 was active but was fifteen times less potent than its parent compound 2 against P. falciparum, with IC50 values of 2.2 ± 0.5 μM and 2.5 ± 0.5 μM to the drug-sensitive NF54 cell line and the drug-resistant Dd2 cell line, respectively. It thus appears that the phenolic hydroxy groups of 2 are important for its antimalarial activity, supporting the idea that coordination to Fe3+ and/or radical generation plays an important role in the antimalarial activity of the phloroglucinols in general and of compound 2 in particular. This conclusion is also supported by the recent report of the modest antiplasmodial activity of watsonianone A (5), a related compound lacking phenolic hydroxyl groups isolated from the Australian tree Corymbia watsoniana.24
Bioactive phloroglucinols have been detected in the Aspidiaceae, Cannabinaceae, Clusiaceae, Compositae, Crassulaceae, Euphorbiaceae, Fagaceae, Guttiferae, Lauraceae, Myrtaceae, Rosaceae and Rutaceae families.18 From the present study, it may be concluded that Mallotus japonicus and M. oppositifolius are two Euphorbiaceous species that produce dimeric phloroglucinols such as mallotophenone (3) and related compounds.12,25,26 It is also noteworthy that an extract of Mallotus japonicus inhibited production of pro-inflammatory cytokines during macrophage activation, and mallotojaponin was one of the bioactive phloroglucinol derivatives isolated from this extract that showed this effect.27 Mallotojaponin was also shown to inhibit HIV-reverse transcriptase activity.28
Experimental Section
General Experimental Procedures
Optical rotations were recorded on a JASCO P-2000 polarimeter. IR and UV spectra were measured on MIDAC M-series FTIR and Shimadzu UV-1201 spectrophotometers, respectively. 1H and 13C NMR spectra were recorded on a JEOL Eclipse 500 spectrometer in CDCl3 with TMS as internal standard. Mass spectra were obtained on JEOL JMS-HX-110, Agilent 6220 LC-TOF-MS. Preparative HPLC was performed using Shimadzu LC-10AT pumps coupled with a semipreparative Varian Dynamax C18 column (5 μm, 250×10 mm), a Shimadzu SPD M10A diode array detector (DAD), and a SCL-10A system controller.
Plant Material
Leaves and inflorescences of Mallotus oppositifolius (Geiseler) Müll. Arg. (collection: Richard Randrianaivo et al. 1425) were collected at an elevation of 137 m in December 2006 near the village of Befarafara in the dry forest of Solanampilana, 35 km north of Daraina, Antsiranana, Sava region, 13°05′42″S 049°34′57″E, northern Madagascar. The sample collected was from a shrub 3 m tall, with white flowers. The plant taxonomy was determined by Dr. Gordon McPherson (Missouri Botanical Garden). Duplicate voucher specimens were deposited at the Centre National d’Application des Recherches Pharmaceutiques (CNARP), the Herbarium of the Parc Botanique et Zoologique de Tsimbazaza, Antananarivo, Madagascar (TAN), the Missouri Botanical Garden, St. Louis, Missouri (MO), and the Museum National d’Histoire Naturelle in Paris, France (P).
Antiproliferative Bioassay
The A2780 ovarian cancer cell line assay was performed at Virginia Tech as previously reported.29 The A2780 cell line is a drug-sensitive ovarian cancer cell line.30
Intraerythrocytic Stages Antimalarial Bioassay
The effect of each fraction and pure compound on parasite growth of Dd2 strain was measured in a 72 h growth assay in the presence of drug as described previously with minor modifications.31,32 Briefly, ring stage parasite cultures (200 μL per well, with 1% hematocrit and 1% parasitemia) were then grown for 72 h in the presence of increasing concentrations of the drug in a 5.05% CO2, 4.93% O2 and 90.2% N2 gas mixture at 37 °C. After 72 h in culture, parasite viability was determined by DNA quantitation using SYBR Green I (50 μL of SYBR Green I in lysis buffer at 0.4 μL of SYBR Green I/mL of lysis buffer).32 The half-maximum inhibitory concentration (IC50) calculation was performed with GraFit software using a nonlinear regression curve fitting. IC50 values are the average of three independent determinations with each determination in duplicate, and are expressed ± S.E.M.
Intraerythrocytic Stages Cytocidal Antimalarial Bioassay
The effectiveness of compounds 1 and 2 at killing intraerythrocytic stages of the chloroquine-sensitive HB3 and chloroquine-resistant Dd2 strains of P. falciparum was performed as previously reported.15 LD50 values are the average of three replicate determinations and are expressed ± S.E.M.
Late Gametocyte Stage Antimalarial Bioassay
To test compounds for their effectiveness in killing late stage (stage V) gametocytes, late stage gametocytes were generated using a combination of established methods.16,33 Initial gametocyte cultures were developed from the P. falciparum NF54 strain (chloroquine-sensitive strain). All cultures were maintained in 75 cm2 culture flasks in a reduced oxygen environment (5% O2, 5% CO2, 95% N2) at 37 °C. Parasitaemia was calculated by counting the percentage of infected RBCs by Giemsa staining of thin smears and light microscopy. Asexual stages were synchronized by sorbitol treatment at least two days before setting gametocyte cultures.34 Thin blood smears were made and stained with Giemsa to check parasite development on days 4, 8, 12, and 13 after the initial subculture. On day nine of the gametocyte cultures, parasites were treated with 5% sorbitol for 10 min at 37 °C to start removing asexual stages. Sorbitol treatment was performed for four consecutive days, which effectively removes >99% of asexual parasites. Gametocyte recovery and concentration was achieved on day 13 using a NycoPrep™ 1.077 cushion16 and the number of gametocytes was calculated using a Neubauer chamber and 30,000 to 50,000 gametocytes per well were added to the black flat-bottom half-area 96-well plates containing drug candidates in a 100 μL final volume. The plate was incubated in a humidified chamber at 37 °C and low oxygen conditions (5% O2, 5% CO2, 95% N2) for 72 h. AlamarBlue® was added on day 16 post-induction at 10% of the well volume.33 The plate was returned to the chamber for an additional 24 h and then was read in a microplate reader at 585 nm after excitation at 540 nm. IC50 values were calculated using a dose-response curve fitting with GraFit. IC50 values are the average of two independent determinations, each determination in duplicate and are expressed ± S.E.M.
Early Gametocyte Stage Antimalarial Bioassay
To test efficacy in preventing gametocytogenesis, 24-well plates were set at 0.75% parasitemia (NF54 strain) and 1% hematocrit and cultured for 13 days with or without the presence of compound 1 and 2. The plate was incubated in a humidified chamber at 37°C and low oxygen conditions for the duration of the experiment. Medium or medium supplemented with drug was replaced on days 4, 6, 8, and 9-12. On day 13, each well was recovered and the parasitaemia (both asexual and sexual) was calculated from Giemsa stained smears.
Extraction and Isolation
A ground sample of M. oppositifolius leaves and inflorescences (137 g) was extracted with ethanol at room temperature to yield 6.0 g of crude ethanol extract, designated MG 4129. A total of 1.8 g of this extract was made available to Virginia Polytechnic Institute and State University. In order to locate the biological activity and to have an idea about the types of metabolites responsible for the activity of the active fraction, 100 mg of the crude ethanol extract of M. oppositifolius were subjected to a liquid-liquid partition using hexanes, EtOAc and H2O to afford 42.5 mg of an active hexanes fraction (IC50 6.7 μg/mL). Size-exclusion chromatography on Sephadex LH-20 of the hexanes fraction eluted with MeOH-CH2Cl2 gave mallotophenone (3, IC50 6.3 ± 0.4μM) and two active fractions (Fr. 3, 13 mg; IC50 2.3 μg/mL and Fr. 4, 8.4 mg; IC50 1.6 μg/mL). High-performance liquid chromatography (HPLC) on a C18 column with a solvent gradient from water-MeOH (system I): 40:60 to 30:70 for 10 min, to 20:80 from 10 to 15 min, to 15:85 from 15 to 20 min, keep 15:85 for 5 min, to 10:90 from 25 to 30 min, and to 0:100 from 30 min to 35 min, ending with 100% MeOH to 50 min of fractions 3 and 4 showed the presence of two major and active phloroglucinols (tR: 39.79 min; IC50 0.5 μg/mL and tR: 44.15 min; IC50 0.61 μg/mL). To isolate more material for structure elucidation and for bioctivity evaluations, the isolation was scaled up by starting with 1 g of ethanol extract. Liquid-liquid partion (hexanes, 3 × 200 mL) followed by Sephadex LH-20 of the hexanes fraction (407 mg) afforded two active fractions (Fr.3, 125.2 mg; IC50 2.4 μg/mL and Fr.4, 137.1 mg; IC50 2.2 μg/mL). Mallotophenone (3) was obtained from fractions 3 and 4 by precipitation. The mother liquid of fraction 3 was subjected to silica gel column chromatography to give compounds 1 (8.3 mg), and 2 (6 mg). Also, HPLC of the mother liquid of fraction 4 on a C18 column using isocratic 100% MeOH gave two active peaks (tR: 39.79 min and 44.15 min), which were purified by silica gel CC to yield compounds 1 (3.1 mg) and 2 (5 mg).
Mallotojaponin B (1)
Colorless prisms (EtOAc-hexane), mp 175 ± 1 °C; UV (MeOH) λmax (log ε): 283 (4.06) nm; IR (film) νmax 3450, 3223, 1620, 1596, 1405, 1283, 1128 cm−1; 1H NMR and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 459.2023 [M+H]+ (calcd for C25H31O8, 459.2019).
X-ray Crystallography of 1
A colorless prism of 1 was centered on the goniometer of an Oxford Diffraction SuperNova A diffractometer operating with CuKα radiation. The data collection routine, unit cell refinement, and data processing were carried out with the program CrysAlisPro.35 The Laue symmetry and systematic absences were consistent with the monoclinic space group P21/c. The structure was solved using SHELXS-9736 and refined using SHELXL-9736 via OLEX2.37 The final refinement model involved anisotropic displacement parameters for non-hydrogen atoms. A riding model was used for the aromatic and alkyl hydrogens. The –OH hydrogen positions were located from the residual electron density map and refined independently.
Crystal Data
Colorless prism; C25H30O8, Mr = 458.49, monoclinic, P21/c a = 12.88614(12) Å, b = 12.34409(7) Å, c = 14.84644(13) Å, β = 110.4761(10)°, V = 2212.38(3) Å3, 37837 reflections, 317 parameters; crystal size: 0.4149 × 0.1458 × 0.1287 mm3. The final indices were R1 = 0.0367, wR2 = 0.0973 [I > 2σ(I)]. Crystallographic data for compound 1 have been deposited as supplementary material at the Cambridge Crystallographic Data Centre (Deposition No. CCDC 874720).38
Mallotojaponin C (2)
Amorphous powder; UV (MeOH) λmax (log ε) 283 (4.12) nm; IR (film) νmax 3440, 3220, 1620, 1595, 1434, 1405, 1280, 1121 cm−1; 1H NMR and 13C NMR data, see Tables 1 and 2; positive HRESIMS m/z 513.2499 [M+H]+ (calcd for C29H37O8, 513.2488).
Mallotophenone (3)
The structure of 3 was identified by single crystal X-ray analysis and by comparison of its spectroscopic data with values reported in the literature.12 Crystallographic data for compound 3 has been deposited as supplementary material at the Cambridge Crystallographic Data Centre (Deposition No. CCDC 874719).38
Methylation of Mallotojaponin B (2)
Compound 2 (0.9 mg) was dissolved in acetone (1.5 mL) and treated with K2CO3 (120 mg) and methyl iodide (100 μL). The mixture was stirred at room temperature for 17 hours. The reaction mixture was evaporated, dissolved in water and extracted with EtOAc to give compound 4 (1 mg). The structure of 4 was confirmed by interpretation of its 1H NMR spectrum and by HRESIMS (see Supporting Information).
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the Fogarty International Center, the National Cancer Institute, the National Science Foundation, the National Heart, Lung and Blood Institute, the National Institute of Mental Health, the Office of Dietary Supplements, and the Office of the Director of NIH, under Cooperative Agreement U01 TW000313 with the International Cooperative Biodiversity Groups. This project was also supported by the National Research Initiative of the Cooperative State Research, Education and Extension Service, USDA, Grant #2008-35621-04732, and by the National Center for Complementary and Alternative Medicine under Cooperative Agreement U01 TW000313-19S1. This work was also supported by the National Science Foundation under Grant no. CHE-0619382 for purchase of the Bruker Avance 600 NMR spectrometer and Grant no. CHE-0722638 for the purchase of the Agilent 6220 mass spectrometer. We thank Mr. B. Bebout for obtaining the mass spectra and Dr. Hugo Azurmendi for assistance with the NMR spectra. Field work essential for this project was conducted under a collaborative agreement between the Missouri Botanical Garden and the Parc Botanique et Zoologique de Tsimbazaza and a multilateral agreement between the ICBG partners, including the Centre National d’Applications des Recherches Pharmaceutiques. We gratefully acknowledge courtesies extended by the Government of Madagascar (Ministère des Eaux et Forêts). All support is gratefully acknowledged.
Footnotes
Dedicated to Dr. Lester A. Mitscher, of the University of Kansas, for his pioneering work on the discovery of bioactive natural products and their derivatives.
ASSOCIATED CONTENT
1H and 13C NMR spectra of compounds 1 and 2 and 1H NMR spectrum of 4, and an ORTEP drawing of 3. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- (1).Biodiversity Conservation and Drug Discovery in Madagascar, Part 53. For Part 52, see Dai Y, Harinantenaina L, Brodie PJ, Birkinshaw C, Randrianasolo S, Ratsimbason M, Rasamison VE, Shen Y, TenDyke K, Kingston DGI. Chem. Biodiversity. doi: 10.1002/cbdv.201200156. accepted for publication.
- (2).Radcliffe-Smith A. Genera Euphorbiacearum. Royal Botanic Gardens; Kew, UK: 2001. p. 464. [Google Scholar]
- (3).Yoder BJ, Cao S, Norris A, Miller JS, Ratovoson F, Razafitsalama J, Andriantsiferana R, Rasamison VE, Kingston DGI. J. Nat. Prod. 2007;70:342–346. doi: 10.1021/np060484y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Beutler JA, Shoemaker RH, Johnson T, Boyd MR. J. Nat. Prod. 1998;61:1509–1512. doi: 10.1021/np980208m. [DOI] [PubMed] [Google Scholar]
- (5).McGookin A, Reed FP, Robertson A. J. Chem. Soc. 1937:748–755. [Google Scholar]
- (6).Liao Y-F, Hung Y-C, Chang W-H, Tsay GJ, Hour T-C, Hung H-C, Liu G-Y. Life Sci. 2005;77:707–719. doi: 10.1016/j.lfs.2005.01.010. [DOI] [PubMed] [Google Scholar]
- (7).Sharma V. J. Plant Biochem. Biotechnol. 2011;20:190–195. [Google Scholar]
- (8).Valacchi G, Pecorelli A, Sticozzi C, Torricelli C, Muscettola M, Aldinucci C, Maioli E. Chem. Biol. Drug Des. 2011;77:460–470. doi: 10.1111/j.1747-0285.2011.01121.x. [DOI] [PubMed] [Google Scholar]
- (9).Kingston DGI. J. Nat. Prod. 2011;74:496–511. doi: 10.1021/np100550t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Adekunle AA, Ikumapayi AM. West Indian Med. J. 2006;55:219–223. doi: 10.1590/s0043-31442006000400003. [DOI] [PubMed] [Google Scholar]
- (11).Okwu DE, Ekei O. Global J. Pure Appl. Sci. 2003;9:235–238. [Google Scholar]
- (12).Arisawa M, Fujita A, Hayashi T, Morita N, Kawano N, Koshimura S. J. Nat. Prod. 1985;48:455–459. doi: 10.1021/np50039a014. [DOI] [PubMed] [Google Scholar]
- (13).Owing to symmetry considerations, the assignment of the 3,3-dimethylallyl group to the C-3′ position as opposed to the C-3 position is arbitrary.
- (14).Arisawa M, Fujita A, Morita N, Okuyama T, Nishino H. J. Nat. Prod. 1991;54:1409–1412. doi: 10.1021/np50077a029. [DOI] [PubMed] [Google Scholar]
- (15).Paguio MF, Bogle KL, Roepe PD. Mol. Biochem. Parasitol. 2011;178:1–6. doi: 10.1016/j.molbiopara.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lelievre J, Almela MJ, Lozano S, Miguel C, Franco V, Leroy D, Herreros E. PLoS One. 2012;7:e35019. doi: 10.1371/journal.pone.0035019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Peatey CL, Leroy D, Gardiner DL, Trenholme KR. Malar. J. 2012;11:34–37. doi: 10.1186/1475-2875-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Singh IP, Bharate SB. Nat. Prod. Rep. 2006;23:558–591. doi: 10.1039/b600518g. [DOI] [PubMed] [Google Scholar]
- (19).Costa ES, Hiruma-Lima CA, Lima EO, Sucupira GC, Bertolin AO, Lolis SF, Andrade FDP, Vilegas W, Souza-Brito ARM. Phytother. Res. 2008;22:705–707. doi: 10.1002/ptr.2397. [DOI] [PubMed] [Google Scholar]
- (20).Cottiglia F, Casu L, Leonti M, Caboni P, Floris C, Busonera B, Farci P, Ouhtit A, Sanna G. J. Nat. Prod. 2012;75:225–229. doi: 10.1021/np2009219. [DOI] [PubMed] [Google Scholar]
- (21).Farombi EO, Ogundipe OO, Uhunwangho ES, Adeyanju MA, Moody JO. Phytother. Res. 2003;17:713–716. doi: 10.1002/ptr.1050. [DOI] [PubMed] [Google Scholar]
- (22).Mavar-Manga H, Haddad M, Pieters L, Baccelli C, Penge A, Quetin-Leclercq J. J. Ethnopharmacol. 2008;115:25–29. doi: 10.1016/j.jep.2007.08.043. [DOI] [PubMed] [Google Scholar]
- (23).Verotta L. Phytochem. Rev. 2002;1:389–407. [Google Scholar]
- (24).Carroll AR, Avery VM, Duffy S, Forster PI, Guymer GP. Org. Biomol. Chem. 2013;11:453–458. doi: 10.1039/c2ob26931g. [DOI] [PubMed] [Google Scholar]
- (25).Arisawa M, Fujita A, Hayashi T, Hayashi K, Ochiai H, Morita N. Chem. Pharm. Bull. 1990;38:1624–1628. doi: 10.1248/cpb.38.1624. [DOI] [PubMed] [Google Scholar]
- (26).Arisawa M, Fujita A, Morita N. J. Nat. Prod. 1990;53:638–643. [Google Scholar]
- (27).Ishii R, Horie M, Saito K, Arisawa M, Kitanaka S. Biochim. Biophys. Acta. 2003;1620:108–118. doi: 10.1016/s0304-4165(02)00514-7. [DOI] [PubMed] [Google Scholar]
- (28).Nakane H, Arisawa M, Fujita A, Koshimura S, Ono K. FEBS Lett. 1991;286:83–85. doi: 10.1016/0014-5793(91)80946-z. [DOI] [PubMed] [Google Scholar]
- (29).Cao S, Brodie PJ, Miller JS, Randrianaivo R, Ratovoson F, Birkinshaw C, Andriantsiferana R, Rasamison VE, Kingston DGI. J. Nat. Prod. 2007;70:679–681. doi: 10.1021/np060627g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Louie KG, Behrens BC, Kinsella TJ, Hamilton TC, Grotzinger KR, McKoy WM, Winker MA, Ozols RF. Cancer Res. 1985;45:2110–2115. [PubMed] [Google Scholar]
- (31).Bennett TN, Paguio M, Gligorijevic B, Seudieu C, Kosar AD, Davidson E, Roepe PD. Antimicrob. Agents Chemother. 2004;48:1807–1810. doi: 10.1128/AAC.48.5.1807-1810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. Antimicrob. Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Tanaka TQ, Williamson KC. Mol. Biochem. Parasitol. 2011;177:160–163. doi: 10.1016/j.molbiopara.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Lambros C, Vanderberg JP. J. Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- (35).CrysAlisPro Software System, v1.171.35.11. Agilent Technologies UK Ltd; Oxford, UK: 2011. [Google Scholar]
- (36).Sheldrick GM. Acta Cryst. 2008;A64:112–122. [Google Scholar]
- (37).Dolomanov OV, Bourhis LJ, Gildea RJ, Howard JAK, Puschmann H. J. Appl. Cryst. 2009;42:339–341. doi: 10.1107/S0021889811041161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Crystallographic data for the structures reported in this paper have been deposited with the Cambridge Crystallographic Data Centre. Copies of the data can be obtained, free of charge, on application to the Director, CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (fax: +44-(0)1223-336033 or deposit@ccdc.cam.ac.uk).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.